Abstract
A large range of microorganisms can associate with plants, resulting in neutral, friendly or hostile interactions. The ability of plants to recognize compatible and incompatible microorganisms and to limit or promote their colonization is therefore crucial for their survival. Elaborated communication networks determine the degree of association between the host plant and the invading microorganism. Central to these regulations of plant microbe interactions, phytohormones modulate microorganism plant associations and coordinate cellular and metabolic responses associated to the progression of microorganisms across different plant tissues. We review here hormonal regulations, focusing on auxin and cytokinin phytohormones, involved in the interactions between plant roots and soil microorganisms, including bacterial and fungi associations, either beneficial (symbiotic) or detrimental (pathogenic). The aim is to highlight similarities and differences in cytokinin/auxin functions amongst various compatible versus incompatible associations.
Keywords: auxin, cytokinin, pathogens, endomycorrhiza, ectomycorrhiza, Rhizobium, root nitrogen fixing symbiosis, legume nodulation
Introduction
Plant–microorganism interactions have received more and more attention due to the benefits they confer to crop productivity by improving nutrient uptake, increasing plant growth and conferring biotic and abiotic stress tolerance (Yang et al., 2009; de Zelicourt et al., 2013; Grover et al., 2013). Identifying communication systems and signals that determine the beneficial or detrimental outcomes of plant–microorganism interactions is a key to improve defense responses without decreasing beneficial (e.g., symbiotic) associations.
Different symbiotic associations with plant roots exist, either with fungi or bacteria (Figure 1). These symbioses are mutualistic, leading to reciprocal exchanges between fungi or bacterial microorganisms and host plants: soil nutrients or fixed atmospheric nitrogen versus carbon skeletons generated through photosynthesis (Udvardi and Poole, 2013; Schweiger and Müller, 2015). These interactions can be established between ectomycorrhizal (ECM) fungi from the Basidiomycota and Ascomycota phyla and many forest trees (Anderson and Cairney, 2007; Diagne et al., 2013; Raudaskoski and Kothe, 2015), arbuscular endomycorrhizal (AM) fungi from the Glomeromycota phylum with most of land plants (Schüβler et al., 2001; Smith and Read, 2010; Foo et al., 2013; Gutjahr and Parniske, 2013), and nitrogen-fixing bacteria such as Rhizobium sp. and Frankia sp. with specific species belonging to the Rosid family, leading to the formation of new root lateral organs called nodules (Dénarié et al., 1992; Franssen et al., 1992; Soltis et al., 1995; Perret et al., 2000; Santi et al., 2013; Svistoonoff et al., 2014) (Figure 1). Unlike AM fungi and N2-fixing bacteria, ECM fungi do not enter inside plant host root tissues and cells. A mycelial mantle is formed by the fungi around short lateral roots and develops between root epidermal and cortical cells, to form a highly branched structure, called the Hartig net (Peterson and Massicotte, 2004; Anderson and Cairney, 2007; Raudaskoski and Kothe, 2015) (Figure 1).
FIGURE 1.
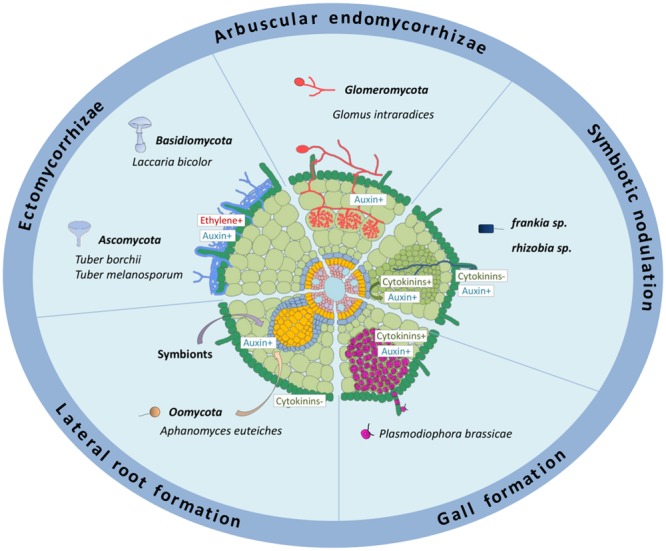
Roles of auxins and cytokinins in different biotic interactions affecting the root system. The schematized root represents different interactions occurring on root systems from different plant species: ectomycorrhizae, formed mainly on tree roots; arbuscular endomycorrhyzae, occurring in most land plants; symbiotic nodulation in legumes (≪rhizobia≫) or actinorhizal plants (≪Frankia≫); galls formed in roots of some Brassicaceae plants. The effect of symbionts and oomycete pathogens on lateral root development is also depicted. Auxin metabolism and/or response is positively associated to ECM, AM, rhizobia and Frankia symbiotic interactions (either in relation to infections and/or to nodule organogenesis), to the interaction with several root pathogens (acting either positively or negatively on pathogenic susceptibility), and positively associated to root gall formation in response to P. brassicae. Auxins produced by different rhizospheric microorganisms stimulate lateral root formation. Cytokinin metabolism and/or response is associated to rhizobia nodulation, either positively in the cortex or negatively in the epidermis, to the interaction with several root pathogens (acting mostly positively on pathogenic susceptibility), and positively associated to root gall formation in response to P. brassicae. +, indicates a positive effect; -, a negative effect. Dark green: root hairs and epidermis; pale green, cortex; middle green, nodule primordium; blue, endodermis; yellow, pericycle; pink, stele tissues including vascular bundles.
In all symbiotic interactions, the recognition of microorg-anisms and plant roots as symbiotic partners is the first critical step to allow a tight beneficial metabolic association (Bonfante and Requena, 2011; Gough and Cullimore, 2011; Geurts et al., 2012; Singh and Parniske, 2012; Genre et al., 2013; Raudaskoski and Kothe, 2015). In the case of nitrogen fixing symbioses, the formation of a new organ, the root nodule, is additionally essential to generate microaerobic conditions allowing differentiated bacteria to fix atmospheric nitrogen (e.g., production of leghemoglobin proteins that chelate oxygen, preventing inhibition of the bacterial nitrogenase enzyme ensuring nitrogen fixation; Ferguson et al., 2010; Desbrosses and Stougaard, 2011; Oldroyd, 2013) (Figure 1). Exchanges of molecular signals, including on the one hand flavonoids and phytohormones produced by legume plants, and on the other hand Myccorhization Factors or Nodulation Factors (NFs) respectively secreted by AM fungi or rhizobia, are required for the recognition of symbiotic partners. In addition, these signaling pathways participate in decreasing root defense responses, preparing root tissues for infection by the symbiont, and in the case of nitrogen fixing bacteria, initiating root cortical cell divisions leading to nodule organogenesis (Oldroyd et al., 2011; Oldroyd, 2013; Gourion et al., 2015) (Figure 1).
Foiling plant defenses is therefore critical for a successful host infection, either for symbiotic partners or pathogens (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012; De Vleesschauwer et al., 2014). To penetrate, colonize and hijack nutrients from host plants, pathogenic bacteria and fungi that infect roots have developed different strategies, including the modification of phytohormonal responses to their advantage. Some pathogens are able to directly synthesize phytohormones affecting plant growth and development, forming new sinks in which nutrients are easily accessible (e.g., Agrobacterium tumefaciens galls, or Plasmodiophora brassicae clubroot galls; Figure 1). Therefore, keeping the control over hormonal pools and signaling pathways is crucial for host plants to both establish beneficial microorganism interactions and prevent pathogenic invasions.
Auxin and Cytokinin Regulations in Plant – Fungus Symbioses
The two types of plant-fungal symbioses, AM and ECM, display different hormonal regulation requirements, notably regarding auxins and cytokinins. A production of cytokinins (isopentenyl and cis-zeatin) by ECM fungi was identified (Morrison et al., 2015). In addition, ECM fungi, such as Laccaria bicolor, Tuber borchii and T. melanosporum, produce measurable amounts of auxin (indole-3-acetic acid; IAA), resulting in morphological changes of host roots (Cistus incanus) either depending on a direct (contact) or indirect (diffusible signal) interaction with the fungus (Karabaghli et al., 1998; Felten et al., 2009; Splivallo et al., 2009). Accordingly, the presence of fungal mycelia reduces root growth and increases root branching of the host plant, as observed in C. incanus and Populus trichocarpa, and similarly as an exogenous auxin treatment does (Felten et al., 2009; Splivallo et al., 2009). Interestingly, in Arabidopsis thaliana, the ectomycorrhizal truffle mycelium also promotes root hair growth as well as lateral root formation, indicating that these may not be symbiosis-specific traits (Splivallo et al., 2009). In addition to the production of auxins, L. bicolor is able to release ethylene which activates the plant auxin synthesis pathway (Splivallo et al., 2009). The production of these two hormones is required to promote root hair growth to an equivalent level as the truffle mycelium does. Altogether, these observations suggest that ethylene production by the ECM fungi may induce auxin production in the host plant, therefore reinforcing the effect of direct auxin production on root development and notably on the promotion of lateral root formation which will then be infected by new mycelia (Figure 1). The local activation of auxin responses in the first tier of root tip columella cells of poplar and Arabidopsis in response to an indirect contact with L. bicolor has been documented using the auxin-response reporter construct DR5::GFP, and this activation was inhibited by a Polar Auxin Transport inhibitor (PAT; Splivallo et al., 2009). In addition, transcriptomic analyses in poplar roots inoculated with the ECM fungus L. bicolor revealed an increased expression of auxin-related genes such as members of the GH3 (Gretchen Hagen3) gene family involved in auxin conjugation, as well as of P. trichocarpa PtaPIN4 and PtaPIN9 auxin efflux carriers essential for PAT (Felten et al., 2009). Interestingly, L. bicolor inoculation induces lateral root formation in wild-type A. thaliana but not in the pin2 mutant (AtPIN2 is the closest Arabidopsis relative of PtaPIN9 in poplar). This result is consistent with the essential role of PAT in controlling lateral root development induced by the presence of the symbiotic ECM fungus (Felten et al., 2009).
In contrast to the ECM symbioses, no comprehensive change was observed in auxin levels in Tropaeolum majus upon inoculation with different AM strains (Jentschel et al., 2007). Nevertheless, a role of auxins in the AM symbiosis was proposed notably in relation to the stimulating effect of the AM inoculation on lateral root formation (Fusconi, 2014). Indeed, several mutants affected in auxin-related developmental responses, such as the Pisum sativum bushy mutant that displays a lower IAA concentration in shoots and roots and the tomato (Solanum lycopersicum) diageotropica auxin-resistant and polycotyledon hyperactive PAT mutants show a reduced mycorrhizal colonization (Hanlon and Coenen, 2011; Foo, 2013) (Figure 1). However, the expression of the strigolactone (SL) biosynthetic PsCCD8 (Carotenoid Cleavage Dioxygenases 8) gene is decreased in the bushy mutant (Foo et al., 2013), suggesting that the auxin effect on AM symbiosis may be at least partly due to a decrease in the SL biosynthesis. Several auxin-responsive genes were identified as induced in AM roots, such as a specific GH3 tomato gene expressed in cells colonized by fungi (Liao et al., 2015). As the symbiotic expression of this marker could be disconnected from its auxin-induction, this suggests that an AM signaling GH3-related response may have evolved at least partially independently of auxin signaling. The expression of the DR5 auxin response reporter was additionally detected in S. lycopersicum, Medicago truncatula, and Oryza sativa root cells containing arbuscules (Etemadi et al., 2014). Finally, although no analysis of the AM colonization capacity of mutants directly affected in auxin perception or polar transport is yet available, overexpression of a microRNA (miR393) that indirectly downregulates the expression of auxin receptor genes (i.e., Transport Inhibitor Response1 and Auxin-related F-Box genes) led to the formation of underdeveloped arbuscules in S. lycopersicum, M. truncatula, and O. sativa roots, suggesting that hampering auxin perception in arbuscule-containing cells negatively affects their formation (Etemadi et al., 2014).
Cytokinins were also proposed to be involved in the AM symbiosis since an increase of cytokinin levels in leaves and roots was detected in AM infected plants (Allen et al., 1980). However, it remains unclear if the cytokinins were produced by the host plant or by the AM fungus (Allen et al., 1980; Barker and Tagu, 2000; Shaul-Keinan et al., 2002). No AM phenotype was detected in the M. truncatula cre1 (cytokinin response 1) mutant defective in a cytokinin receptor essential for symbiotic nodulation (Plet et al., 2011; Laffont et al., 2015), suggesting that at least the CRE1-dependent cytokinin signaling is not essential for the AM symbiotic interaction.
Auxin and Cytokinin Regulations of Nitrogen-Fixing Root Nodule Symbioses
Several studies have highlighted the involvement of auxin and cytokinin phytohormones in the regulation of the Rhizobium nitrogen-fixing symbiotic interaction. Allen et al. (1953) showed that an exogenous application of a PAT inhibitor could induce the formation of nodule-like structures on alfalfa roots, in the absence of Rhizobium. However, the structure of these organs can be considered as more similar to roots than to legume nodules. The inhibition of PAT was found to also induce pseudonodule formation in M. truncatula roots (Rightmyer and Long, 2011), further indicating that nodule organogenesis involved a local auxin accumulation. It was later shown that combined auxin and cytokinin exogenous treatment on pea roots led to cortical cell divisions, which occur at the onset of nodule organogenesis (Libbenga et al., 1973) (Figure 1). The positive effect of cytokinins in the initiation of nodule organogenesis was additionally documented in different legumes where exogenous applications of cytokinins induce cortical cell divisions, amyloplast accumulation, and the expression of early nodulation markers (early nodulins; Torrey, 1961; Dehio and de Bruijn, 1992; Bauer et al., 1996; Jiménez-Zurdo et al., 2000; Mathesius et al., 2000; Murray et al., 2007; Tirichine et al., 2007). In addition, the ectopic expression of a CytoKinin oXidase/deshydrogenase gene from A. thaliana (AtCKX3), involved in the degradation of the cytokinin bioactive pool, or the downregulation of either a cytokinin activation gene from M. truncatula (MtLOG1, standing for LOnely Guy 1) or an Iso-PentenylTransferase biosynthetic gene from L. japonicus (LjIPT3), lead to a reduced nodule organogenesis (Lohar et al., 2004; Chen Y. et al., 2014; Mortier et al., 2014), suggesting that endogenous cytokinins act positively on nodulation. However, a reduced nodulation is also observed when the MtLOG1 gene is overexpressed (Mortier et al., 2014) and in a L. japonicus mutant affecting the NF-induced CKX3 gene (Reid et al., 2016), indicating that a tight regulation of cytokinin levels is required and/or that a negative symbiotic function of cytokinins exists. As rhizobia can secrete bioactive auxins (Camerini et al., 2008; Bianco and Defez, 2010) and cytokinins (Phillips and Torrey, 1972; Sturtevant and Taller, 1989), it was proposed that these two hormones could contribute to the induction of nodule formation, in addition to other bacterial symbiotic signals such as NFs. Indeed, a Rhizobium nod- strain, unable to synthesize NFs and form nodules but genetically modified to secrete the trans-zeatin cytokinin, is able to induce the formation of nodule-like structures expressing nodulation markers (Cooper and Long, 1994). The secretion of cytokinins by wild-type rhizobia does not seem, however, essential for nodulation, even though it might have a minor contribution (Kisiala et al., 2013; Podlešáková et al., 2013). In agreement, van Zeijl et al. (2015) showed that cytokinins are accumulated in wild-type roots in the absence of Rhizobium following a 3 h NF treatment. This suggests that the cytokinin accumulation required for nodulation mainly originates from the host plant. Concerning auxins, the over-production of this phytohormone in Rhizobium positively regulates nodulation and nodule meristem size (Camerini et al., 2008), and auxin-response reporter DR5 and/or GH3 fusions revealed that rhizobia or NFs can locally inhibit PAT and induce a local auxin accumulation in dividing cortical cells and nodule primordia in M. truncatula, L. japonicus, T. repens, and Vicia sativa (Mathesius et al., 1998; Boot et al., 1999; Pacios-Bras et al., 2003; Breakspear et al., 2014; Ng et al., 2015) (Figure 2). Interestingly, the accumulation of auxin (Indole-3-Acetic Acid, IAA) in Rhizobium inoculated plants was found to be dependent on cytokinin signaling pathways (Ng et al., 2015).
FIGURE 2.
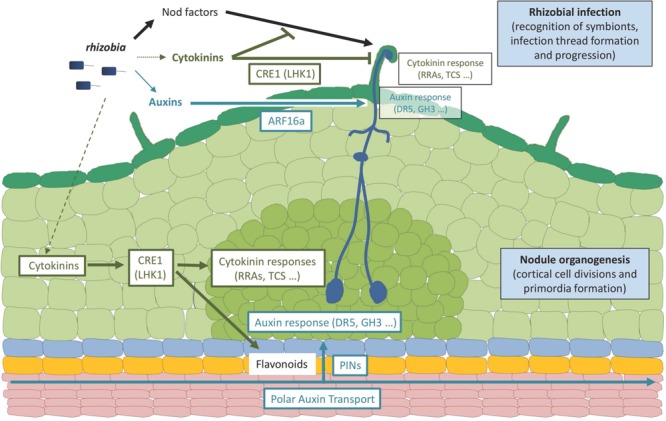
Roles of auxins and cytokinins in legume-rhizobia symbiotic interactions. The scheme represents an early rhizobial infection event in a legume (Medicago truncatula) root leading to the formation of an indeterminate nodule. Rhizobia produce specific Nod factor signaling molecules as well as auxin and cytokinin hormones. A major contribution of Nod factor signals in establishing rhizobial infection events (i.e., symbiont recognition, infection thread formation and progression) in root hairs has been demonstrated, whereas cytokinins produced by rhizobia may only have a minor contribution, and a role for a WT-level bacterial production of auxins remains to be established. Plant and/or bacterial hormones lead to the activation of cytokinin and auxin responses in infected epidermal cells, respectively, monitored by Type A RRs (RRA) genes or the TCS (Two Component output Sensor) cytokinin-response marker and the GH3 (Gretchen Hagen3) auxin responsive gene or the DR5 auxin-response marker. The activation of cytokinin responses in the root epidermis may lead to an inhibition of Nod factor signaling and/or rhizobial infections whereas the activation of auxin responses may promote rhizobial infections. The activation of these root epidermal cytokinin responses depends on the CRE1 (Cytokinin Response 1) receptor (LHK1, standing for Lotus Histidine Kinase1, being its functional homolog in Lotus japonicus) whereas the activation of the auxin epidermal responses depends on ARF16a (Auxin Response Factor 16a, a transcription factor). A cytokinin response is additionally activated in dividing cortical cells and nodule primordia, also depending on the CRE1 (LHK1) receptor, promoting nodule organogenesis even in the absence of symbiotic signals (rhizobia or Nod factors). An activation of auxin responses is observed in the cortex downstream of the CRE1-cytokinin pathway, involving local changes in polar auxin transport associated to the activation of specific flavonoid metabolite production and changes in PIN (PIN-formed) auxin-efflux carriers expression and accumulation. Ultimately, the activation of these auxin and cytokinin responses is linked to modulations of the cell cycle leading either to infection thread formation (epidermis) or cell divisions (cortex). It remains open if the auxin and/or cytokinin hormonal pools may, in addition to originate from plants and/or bacteria, accumulate upon rhizobial/Nod factors activation specifically in the infected epidermis, in the activated cortex, or in both root tissues. In addition, divergences in these hormonal responses are observed between determinate and indeterminate nodule types (e.g., L. japonicus versus M. truncatula model legumes), notably regarding the role of CRE1 versus LHK1 cytokinin receptors in regulating infections in the epidermis, the involvement of polar auxin transport in root inner tissues, and the contribution of a local auxin biosynthesis in these different root tissues. Dark green: root hairs and epidermis; pale green, cortex; middle green, nodule primordium; blue, endodermis; yellow, pericycle; pink, stele tissues including vascular bundles.
The involvement of cytokinin signaling pathways in the regulation of nodulation was first highlighted using a RNAi approach specifically targeting different putative cytokinin receptors in M. truncatula roots, revealing that only the silencing of MtCRE1 led to cytokinin insensitive roots which developed a reduced number of nodules (Gonzalez-Rizzo et al., 2006) (Figure 2). Similarly, the lhk1/hit1 (lotus histidine kinase 1/hyperinfected 1) mutant of L. japonicus, affecting the closest homolog of MtCRE1, showed a strongly reduced nodulation capacity associated with a hyperinfection phenotype (Murray et al., 2007). Strikingly, the L. japonicus snf2 (spontaneous nodule formed 2) mutant carrying a gain of function mutation in the LHK1 cytokinin receptor led to the formation of “spontaneous nodules” in the absence of Rhizobium (Tirichine et al., 2007) (Figure 2). Altogether, these results unambiguously indicate that cytokinins and the LHK1/CRE1 pathway play a positive role in nodulation by inducing cortical cell divisions and nodule organogenesis (Figures 1 and 2). The fact that the lhk1 and cre1 mutants still form some nodules strongly suggest that a functional redundancy exists within the cytokinin receptor CHK family. Accordingly, mutants affecting other cytokinin receptors than cre1/lhk1 also have reduced nodulation efficiencies, even though to a lesser extent (Held et al., 2014; Boivin et al., 2016). In addition, these reduced nodulation phenotypes are stronger when LHK1 or CRE1 is affected, further indicating a predominant role for CRE1/LHK1 in nodule initiation.
As previously mentioned, compatible rhizobia can locally inhibit auxin accumulation and/or PAT. Accordingly, in L. japonicus, the induction of an auxin response is observed both in Rhizobium-induced nodules and in snf2 spontaneous nodules (Suzaki et al., 2012, 2013), indicating that this response is associated at least with early nodule organogenesis, downstream of the LHK1 cytokinin signaling pathway (Figure 2). The inhibitory effect of a Rhizobium application on PAT in M. truncatula roots is not observed in the cre1 mutant, who displays an increased PAT capacity and accumulate excessively polarly localized MtPINs auxin efflux carriers (Plet et al., 2011) (Figure 2). Recently, Ng et al. (2015) showed that the nodulation defect of the cre1 mutant could be partially complemented by an auxin transport inhibitor, as well as by specific flavonoids able to inhibit PAT (such as naringenin, isoliquiritigenin, and kaempferol; Figure 2). Overall, these data highlight that the activation of auxin responses and flavonoid accumulation downstream of cytokinins and LHK1/CRE1 is tightly associated with dividing cortical cells and nodule primordia formation.
Type B Response Regulators (RRBs; Heyl et al., 2013) are transcription factors directly regulating cytokinin primary response genes such as the Nodulation Signaling Pathway 2 (NSP2) gene that encodes a GRAS-type transcription factor essential for early nodulation (Kaló et al., 2005), directly linking NF and cytokinin signaling pathways (Ariel et al., 2012). Other direct cytokinin signaling targets are associated to negative feedback regulations, such as the Cytokinin Oxidase/deshydrogenase 1 (CKX1) gene involved in cytokinin degradation and type A Response Regulators (RRAs) thought to act as negative regulators of cytokinin signaling. Several RRA genes, used as markers of the activation of cytokinin responses, are associated with symbiotic nodulation in M. truncatula, such as MtRRA4 that is expressed in dividing root cortical cells, nodule primordia and meristems, (Plet et al., 2011) (Figure 2) and MtRRA9 and MtRRA11 that are upregulated both in the root epidermis and in the root cortex in response to rhizobia or NFs (Op den Camp et al., 2011) (Figure 2). An RNAi construct targeting MtRRA9 as well as MtRRA4, MtRRA5, and MtRRA11 significantly reduces nodule formation in M. truncatula, suggesting a positive role of RRAs in nodulation (Op den Camp et al., 2011). As observed when affecting the cytokinin pool, the downregulation of the type A RR MtRRA9 reduces nodulation, suggesting a negative function of cytokinins in nodulation (Op den Camp et al., 2011). Different versions of the “Two Component output Sensor” (TCS) reporter, used as a proxy to monitor the activation of the cytokinin primary response, revealed either an expression only in nodule primordia (TCS reporter, van Zeijl et al., 2015), in contrast to MtRRA9 and MtRRA11 (Op den Camp et al., 2011), or first in the epidermis and then in the cortex (TCSn reporter, Jardineau et al., 2016) (Figure 2). In L. japonicus, the TCS reporter expression was detected both in the infected root epidermis and in cortical cells during nodule organogenesis (Held et al., 2014), consistent with the induction of LjRRA4, LjRRA6, and LjRRA8 genes in response to a NF treatment (Op den Camp et al., 2011), but following a different kinetic, being first expressed in the root cortex and then in the epidermis. The expression of several cytokinin biosynthesis and signaling genes, including CRE1 and MtRRA2, MtRRA8, MtRRA9 and MtRRA10, is rapidly upregulated by NFs or Rhizobium in the M. truncatula root epidermis and/or in isolated root hairs (Breakspear et al., 2014; Liu et al., 2015; Jardineau et al., 2016) (Figure 2). Accordingly, ProLHK1:GUS and ProCRE1: GUS fusions were detected in response to Rhizobium both in the root epidermis and in dividing cortical cells (Held et al., 2014; Boivin et al., 2016; Jardineau et al., 2016). This epidermal expression correlates with the Rhizobium hyperinfected phenotype observed in the hit1/lhk1 mutant, suggesting that in addition to a positive role in nodule organogenesis, a negative function of cytokinins regulating epidermal infections may exist (Murray et al., 2007; Held et al., 2014) (Figures 1 and 2). Interestingly, in M. truncatula, the cytokinin/CRE1 pathway negatively regulates the NF-induction of the ENOD11 epidermal infection marker (Figure 2), and reducing the cytokinin pool specifically in the epidermis positively impacts nodulation (Jardineau et al., 2016) (Figure 1).
Several auxin-related genes, such as members of the Auxin Response Factor (ARF16a), GH3 (GH3.1), Indole-3-Acetic Acid (IAA9) and Small Auxin Up RNA (SAUR1) families, are also induced in the root epidermis and/or root hairs by Rhizobium and/or NFs (Breakspear et al., 2014; Jardineau et al., 2016) (Figure 2). In agreement, the auxin-response reporter DR5 fusion was detected in Rhizobium infected root hairs as well as the auxin-responsive GH3.1 gene, suggesting a role for auxins in epidermal infections (Breakspear et al., 2014; Laplaze et al., 2015) (Figures 1 and 2). Accordingly, an arf16a mutant shows a reduced rhizobial infection efficiency even though the number of nodule primordia and differentiated nodules remains unchanged, suggesting a positive role of auxins in the earliest stages of the rhizobial infection. A local accumulation of auxins depending on the AUX1 influx carrier was linked to Frankia nitrogen-fixing bacterial infections in the Casuarina glauca actinorhizal host plant forming symbiotic nodules evolutionary related to the legume nodulation (Péret et al., 2007; Perrine-Walker et al., 2010). Using a dominant-negative version of an auxin signaling repressor gene expressed in actinorhizal nodules, Indole-3-Acetic Acid7 (IAA7), to specifically inhibit auxin signaling in Frankia-infected cells, an increased actinorhizal nodulation was observed (Champion et al., 2015). This suggests a model where auxins, notably produced by Frankia symbiotic bacteria, induce the degradation of IAA7 and thus activate auxin-signaling, ultimately leading to an inhibitory negative feedback on nodulation. Finally, an activation of flavonoid pathways was also detected in the root epidermis and in root hairs in response to rhizobia and/or NFs (Breakspear et al., 2014; Chen et al., 2015; Jardineau et al., 2016), pointing to potential connections with cytokinin and auxin responses as recently described in early nodule organogenesis (Ng et al., 2015). Overall, these data indicate that in addition to their roles in nodule organogenesis, auxins, cytokinins and flavonoids may also regulate symbiotic bacterial infection (Figures 1 and 2).
Cytokinins and potentially auxins additionally likely regulate later symbiotic nodulation stages such as the nitrogen fixation metabolism, as suggested by transcriptomic analyses of laser-dissected differentiated nodule zones (Roux et al., 2014). Concerning auxins, it was recently reported that an auxin-overexpressing S. meliloti free-living strain showed transcriptome changes reminiscent of a differentiated nitrogen-fixing bacteroid, suggesting that auxins may affect bacteria differentiation depending on an unknown signaling pathway (Defez et al., 2016). Changes in auxin metabolism and response occurring in a wild-type Rhizobium strain during its differentiation within nodules however remain to be demonstrated. Concerning cytokinins, the downregulation of the Iso-PentenylTransferase 3 (LjIPT3) gene in L. japonicus differentiated nodules, decreases nodule nitrogenase activity (Chen Y. et al., 2014). Recent studies, respectively using a ckx3 mutant or cre1 and related chk cytokinin receptor mutants, indicate that cytokinins affect nitrogen fixation efficiency both in L. japonicus and in M. truncatula (Boivin et al., 2016; Reid et al., 2016). Interestingly, a genomic clone corresponding to the closest CHK cytokinin receptor from the Arabidopsis non–symbiotic plant, AHK4/CRE1, is able to rescue early cre1 nodulation defects (i.e., the number of nodules per plants) but not to complement the nitrogen fixation deficiency (Boivin et al., 2016). Noteworthy, three L. japonicus hemoglobin encoding genes (LjGLB161, LjGLB2 and LjGLB3-1) are transcriptionally regulated by cytokinins (Bustos-Sanmamed et al., 2011), and in O. sativa, the expression of some hemoglobin genes may be directly regulated by RRB cytokinin signaling transcription factors (Ross et al., 2004). This may imply that a potential direct connection between cytokinins and the nitrogen fixation metabolism exists. Finally, it was recently proposed that cytokinins could be involved in a systemic shoot-to-root negative regulation of nodulation (Sasaki et al., 2014). More detailed analyses are therefore required to understand and integrate the various roles of auxins and cytokinins at these different symbiotic nodulation stages.
Auxin and Cytokinin Regulations in Plant–Pathogen Interactions
In the past decades, most of the studies on plant pathogen interactions have focused on how the pathogens infect plant aerial organs (e.g., leaves). Auxins and cytokinins have been recently highlighted to act in defense responses either depending on other defense-related hormones such as Salicylic Acid and Jasmonic Acid, or independently (reviewed in Naseem and Dandekar, 2012). It should be noted that the function of auxins and cytokinins in defense and immunity responses largely depends on the plant and organ involved, probably because of their divergent developmental roles notably in shoots versus roots. It is only recently that data have been gained using root pathosystems (Chen Y.C. et al., 2014). Some root pathogens are able to synthesize auxin- and cytokinin-like molecules (Estruch et al., 1991; Argueso et al., 2009), indicating that the production of these two hormones is not restricted to either beneficial (symbiotic) or detrimental (pathogenic) microorganisms. Amongst the best described examples, Agrobacterium tumefaciens and A. rhizogenes are soil pathogenic bacteria targeting dicot plants (Smith and Townsend, 1907; Costantino et al., 1980). These bacteria carry a plasmid containing a Transfer-DNA (T-DNA) region that can be integrated into the plant genome (Liu and Kado, 1979; Kutáček and Rovenská, 1991; Lee et al., 2009). The A. tumefaciens T-DNA encodes two transcripts, named iaaH and iaaM, encoding auxin biosynthetic enzymes (Wood et al., 2001), and the trans-zeatin synthesizing (tzs) gene involved in cytokinin biosynthesis (Akiyoshi et al., 1984, 1987; Hwang et al., 2010). The integration of genes encoding these phytohormonal biosynthetic enzymes into the host plant genome leads to cell proliferation and a gall formation in the case of A. tumefaciens, or to root organogenesis in the case of A. rhizogenes “hairy roots”. Another well studied root pathogen is P. brassicae, which causes the clubroot disease in cruciferous plants such as Brassica napus (rapeseed) and A. thaliana (Hwang et al., 2012). The clubroot disease is characterized by the formation of galls on infected roots (Figure 1), leading to plant premature senescence. A microarray transcriptomic analysis performed on infected A. thaliana plants versus non-infected plants identified amongst differentially expressed genes, phytohormone-associated genes such as members of the auxin-related GH3 gene family, or genes involved in the cytokinin biosynthesis (AtIPT3 and AtIPT8), cytokinin degradation (AtCKX1 and AtCKX6), cytokinin perception (AHK4/CRE1) and cytokinin signaling (ARR5 and ARR10; Siemens et al., 2006). Cytokinins and auxins were additionally functionally associated with early steps of the P. brassicae – Arabidopsis interaction in relation to the re-initiation of cortical cell divisions to form root galls, since an accumulation of isopentenyladenine and an enhanced auxin and cytokinin-related gene expression were identified (Ando et al., 2006; Devos et al., 2006; Schuller et al., 2014) (Figure 1). Interestingly, a link with flavonoid metabolic pathways was additionally highlighted since an accumulation of three types of flavonoids (naringenin, kaempferol and quercetin) was detected during clubroot gall formation (Päsold et al., 2010). Strikingly, an endophytic fungus, Heteroconium chaetospira, was described as a competitor for root cortical cell colonization, suppressing clubroot disease in rapeseed, and upregulating an auxin biosynthesis gene (BnAAO1 for Ascorbic Acid Oxidase; Lahlali et al., 2014).
Amongst well-described plant pathogens infecting the root system, Aphanomyces euteiches is an oomycete causing strong damage to legume crops (Gaulin et al., 2007). Infected roots become brown and necrotic, leading to a reduction in water and nutrient uptake, and later, to leaf chlorosis and plant death (Gaulin et al., 2007). Interestingly, the resistance against this root pathogen is correlated with an increased capacity of the host plant to form lateral roots (Djébali et al., 2009) (Figure 1). The M. truncatula mutant affecting the MtCRE1 cytokinin receptor show an increased rate of survival in response to the pathogen, and this tolerance is correlated with the higher ability of this mutant to form lateral roots (Laffont et al., 2015), a developmental phenotype also observed in Arabidopsis cytokinin receptor mutants (Chang et al., 2013). In addition, a high-density Genome Wide Association Study (GWAS) revealed that a locus linked to Aphanomyces tolerance was potentially encoding an IPT cytokinin biosynthetic enzyme (Bonhomme et al., 2014), further suggesting the involvement of cytokinins in plant-pathogenic interactions (Figure 1).
Fusarium oxysporum is an ascomycete fungus belonging to a broad group containing non-pathogenic as well as pathogenic species. Root pathogenic strains are able to infect a wide range of plants including cotton, tomato, banana and Arabidopsis (Chen Y.C. et al., 2014). A microarray transcriptomic analysis performed in cotton infected roots (Dowd et al., 2004) and a RNAseq analysis performed on F. oxysporum infected Arabidopsis root tissues revealed changes in auxin-related gene expression, such as members of the GH3, PIN, IAA and ARF gene families (Lyons et al., 2015). Both in vitro and in vivo exogenous applications of auxins improve tomato root growth but also prevent F. oxysporum spore germination, suggesting a positive role of auxins in the plant resistance to F. oxysporum (Sharaf and Farrag, 2004).
Finally, one of the most famous and destructive soil-borne bacteria is Ralstonia solanacearum, causing a rapid vascular wilt disease to more than 200 species, including legumes, tomato, potato, tobacco, banana, and Arabidopsis (Genin and Denny, 2012; Peeters et al., 2013; Huet, 2014; Yuliar et al., 2015). A putative plant resistance gene to this pathogen is WAT1 (for Walls Are Thin1), required for secondary cell-wall deposition in M. truncatula (Ranocha et al., 2013). WAT1 is involved in auxin homeostasis in relation to vacuolar auxin transport, and the inactivation of WAT1 confers a broad spectrum resistance to several vascular pathogenic bacteria including R. solanacearum and Xanthomonas campestris (Denancé et al., 2013). Transcriptomic and metabolomic analyses demonstrated a repression of several genes linked to auxin metabolism in wat1 mutant roots, correlated with a decrease of a major form of auxin (indole glucosinolate) and to a reduction in the amount of the auxin precursor tryptophan. Interestingly, crossing of the wat1 mutant with a trp5 mutant carrying a mutation of an anthranilate synthase (ASA1) provokes an over-accumulation of tryptophan, and restores wat1 susceptibility to R. solanacearum. Altogether, these results suggest a positive role of auxins in secondary wall formation, as well as in the susceptibility to pathogenic R. solanacearum infections. In addition to auxins, an upregulation of cytokinin response genes, such as CKX and a few RRAs, was identified in M. truncatula by a transcriptomic approach in response to R. solanacearum (Moreau et al., 2014). This notably includes the MtRRA4 Response Regulator, which is transcriptionally upregulated by both cytokinins and Ralstonia, depending on the MtCRE1 cytokinin receptor. Accordingly, the cre1 mutant shows an increased resistance to R. solanacearum, indicating a role of cytokinins in promoting root susceptibility to the pathogen.
Concluding Remarks
Rhizospheric beneficial and detrimental microbes penetrate into root systems and tissues and trigger major modifications at organ, cellular and molecular levels, notably through modifications of developmental phytohormonal balances. The Table 1 summarizes roles of auxins and cytokinins in different root–microbe interactions. A main feature is that as auxins and cytokinins are critical to regulate cell division and differentiation, these hormones are therefore tightly associated with the formation of new organs such as lateral roots, nodules on legume roots in response to rhizobia, as well as galls for example in response to A. tumefaciens or to P. brassicae infection (Figure 1; Table 1). Most of the rhizospheric microbes, either symbiotic or pathogenic, affect the root system architecture, generally by altering lateral root formation and/or root hair growth (e.g., Laccaria bicolor and rhizobia; Table 1). In agreement, NFs and Myc Factors, and the associated N2-fixing and AM symbionts, induce lateral root development as part of the symbiotic response (Oláh et al., 2005; Maillet et al., 2011) (Figure 1). However, links likely existing with hormones controlling lateral root development, and notably auxins, remain to be identified. Auxins, cytokinins, and their associated signaling pathways are also required for inducing root cortical cell divisions, either in legume plants to generate nodule primordia in response to rhizobia, or in Brassicaceae plants to form galls in response to the P. brassicae pathogen (Figure 1; Table 1). Interestingly, in these distantly related host plants, cortical cell divisions are similarly associated with the accumulation of naringenin and kaempferol flavonoids. Results reported in this review highlight a positive role of auxins and cytokinins in plant root susceptibility to pathogens, except for F. oxysporum, as well as to rhizobia symbiotic bacteria (Table 1). Using competitors of root cortical colonization such as endophytic fungi and/or rhizobia may then be a strategy to prevent root pathogen colonization. Understanding the different pathways used by beneficial and detrimental microbes to alter root system development, invade the root cortex, and sometimes to generate new organs, is a crucial challenge to develop integrated strategies to promote crop protection without altering symbiotic capacities, in the frame of sustainable agriculture and agro-ecology practices.
Table 1.
Summary of known functions of auxins and cytokinins in various root–microbe interactions.
| Root symbionts |
|||||
|---|---|---|---|---|---|
| Fungal symbionts |
Bacterial symbionts |
Root pathogens | |||
| ECM fungi | AM fungi | Rhizobia | Frankia | ||
| Auxins |
|
|
|
|
|
| Cytokinins |
|
|
|
|
|
The columns indicate the different symbiotic and pathogenic interactions and the rows indicate the phytohormones auxins and cytokinins. ECM, EctoMycorrhiza; AM, Arbuscular Mycorrhiza; GH3, Gretchen Hagen3; DR5, Auxin-response reporter; ARF, Auxin Response Factor; Aux/IAA, Indole-3-Acetic Acid Auxin-response protein; SAUR, Small Auxin Up RNA; PIN, Pin-formed protein; PAT, Polar Auxin Transport; RR, Response Regulator cytokinin response gene; CRE1, Cytokinin Response 1 receptor; LHK1, Lotus Histidine Kinase cytokinin receptor; TCS, Two Component output Sensor cytokinin-response reporter; WAT1, Walls Are Thin1; gof, gain of function.
Author Contributions
SB and FF wrote the manuscript with inputs from CFF, and CFF produced the figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SB was the recipient of a fellowship from the ASTREA “Région Ile de France” program. CF-F was the recipient of a Paris-Sud University (France) Ph.D. fellowship.
References
- Akiyoshi D. E., Klee H., Amasino R. M., Nester E. W., Gordon M. P. (1984). T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 81 5994–5998. 10.1073/pnas.81.19.5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi D. E., Regier D. A., Gordon M. P. (1987). Cytokinin production by Agrobacterium and Pseudomonas spp. J. Bacteriol. 169 4242–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E. K., Allen O. N., Newman A. S. (1953). Pseudonodulation of leguminous plants induced by 2-bromo-3,5-dichlorobenzoic acid. Am. J. Bot. 40 429–435. [Google Scholar]
- Allen M. F., Moore T. S., Jr., Christensen M. (1980). Phytohormone changes in Bouteloua gracilis infected by vesicular–arbuscular mycorrhizae: I. Cytokinin increases in the host plant. Can. J. Bot. 58 371–374. 10.1139/b80-038 [DOI] [Google Scholar]
- Anderson I. C., Cairney J. W. G. (2007). Ectomycorrhizal fungi: exploring the mycelial frontier. FEMS Microbiol. Rev. 31 388–406. 10.1111/j.1574-6976.2007.00073.x [DOI] [PubMed] [Google Scholar]
- Ando S., Tsushima S., Tagiri A., Kamachi S., Konagaya K.-I., Hagio T., et al. (2006). Increase in BrAO1 gene expression and aldehyde oxidase activity during clubroot development in Chinese cabbage (Brassica rapa L.). Mol. Plant Pathol. 7 223–234. 10.1111/j.1364-3703.2006.00333.x [DOI] [PubMed] [Google Scholar]
- Argueso C. T., Ferreira F. J., Kieber J. J. (2009). Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ. 32 1147–1160. 10.1111/j.1365-3040.2009.01940.x [DOI] [PubMed] [Google Scholar]
- Ariel F., Brault-Hernandez M., Laffont C., Huault E., Brault M., Plet J., et al. (2012). Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula. Plant Cell 24 3838–3852. 10.1105/tpc.112.103267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker S. J., Tagu D. (2000). The roles of auxins and cytokinins in mycorrhizal symbioses. J. Plant Growth Regul. 19 144–154. [DOI] [PubMed] [Google Scholar]
- Bauer P., Ratet P., Crespi M. D., Schultze M., Kondorosi A. (1996). Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and MsEnod12A expression patterns in alfalfa roots. Plant J. 10 91–105. 10.1046/j.1365-313X.1996.10010091.x [DOI] [Google Scholar]
- Bianco C., Defez R. (2010). Improvement of phosphate solubilization and Medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appl. Environ. Microbiol. 76 4626–4632. 10.1128/AEM.02756-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin S., Kazmierczak T., Brault M., Wen J., Gamas P., Mysore K. S., et al. (2016). Different cytokinin CHK receptors regulate nodule initiation as well as later nodule developmental stages in Medicago truncatula. Plant Cell Environ. 10.1111/pce.12779 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Bonfante P., Requena N. (2011). Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 14 451–457. 10.1016/j.pbi.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Bonhomme M., André O., Badis Y., Ronfort J., Burgarella C., Chantret N., et al. (2014). High-density genome-wide association mapping implicates an F-box encoding gene in Medicago truncatula resistance to Aphanomyces euteiches. New Phytol. 201 1328–1342. 10.1111/nph.12611 [DOI] [PubMed] [Google Scholar]
- Boot K. J. M., van Brussel A. A. N., Tak T., Spaink H. P., Kijne J. W. (1999). Lipochitin oligosaccharides from Rhizobium leguminosarum bv. viciae reduce auxin transport capacity in Vicia sativa subsp. nigra roots. Mol. Plant Microbe Interact. 12 839–844. 10.1094/MPMI.1999.12.10.839 [DOI] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26 4680–4701. 10.1105/tpc.114.133496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos-Sanmamed P., Tovar-Méndez A., Crespi M., Sato S., Tabata S., Becana M. (2011). Regulation of nonsymbiotic and truncated hemoglobin genes of Lotus japonicus in plant organs and in response to nitric oxide and hormones. New Phytol. 189 765–776. 10.1111/j.1469-8137.2010.03527.x [DOI] [PubMed] [Google Scholar]
- Camerini S., Senatore B., Lonardo E., Imperlini E., Bianco C., Moschetti G., et al. (2008). Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch. Microbiol. 190 67–77. 10.1007/s00203-008-0365-7 [DOI] [PubMed] [Google Scholar]
- Champion A., Lucas M., Tromas A., Vaissayre V., Crabos A., Diédhiou I., et al. (2015). Inhibition of auxin signaling in Frankia species-infected cells in Casuarina glauca nodules leads to increased nodulation. Plant Physiol. 167 1149–1157. 10.1104/pp.114.255307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Ramireddy E., Schmülling T. (2013). Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. J. Exp. Bot. 64 5021–5032. 10.1093/jxb/ert291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. S., Liu C. W., Roy S., Cousins D., Stacey N., Murray J. D. (2015). Identification of a core set of rhizobial infection genes using data from single cell-types. Front. Plant Sci. 6:575 10.3389/fpls.2015.00575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen W., Li X., Jiang H., Wu P., Xia K., et al. (2014). Knockdown of LjIPT3 influences nodule development in Lotus japonicus. Plant Cell Physiol. 55 183–193. 10.1093/pcp/pct171 [DOI] [PubMed] [Google Scholar]
- Chen Y. C., Kidd B. N., Carvalhais L. C., Schenk P. M. (2014). Molecular defense responses in roots and the rhizosphere against Fusarium oxysporum. Plant Signal. Behav. 9:e977710 10.4161/15592324.2014.977710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. B., Long S. R. (1994). Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell 6 215–225. 10.1105/tpc.6.2.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino P., Hooykaas P. J., den Dulk-Ras H., Schilperoort R. A. (1980). Tumor formation and rhizogenicity of Agrobacterium rhizogenes carrying Ti plasmids. Gene 11 79–87. 10.1016/0378-1119(80)90088-8 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Xu J., Höfte M. (2014). Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front. Plant Sci. 5:611 10.3389/fpls.2014.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zelicourt A., Al-Yousif M., Hirt H. (2013). Rhizosphere microbes as essential partners for plant stress tolerance. Mol. Plant 6 242–245. 10.1093/mp/sst028 [DOI] [PubMed] [Google Scholar]
- Defez R., Esposito R., Angelini C., Bianco C. (2016). Overproduction of indole-3-acetic acid in free-living rhizobia induces transcriptional changes resembling those occurring in nodule bacteroids. Mol. Plant Microbe Interact. 29 484–495. 10.1094/MPMI-01-16-0010-R [DOI] [PubMed] [Google Scholar]
- Dehio C., de Bruijn F. J. (1992). The early nodulin gene SrEnod2 from Sesbania rostrata is inducible by cytokinin. Plant J. 2 117–128. 10.1046/j.1365-313X.1992.t01-51-00999.x [DOI] [PubMed] [Google Scholar]
- Denancé N., Ranocha P., Oria N., Barlet X., Rivière M.-P., Yadeta K. A., et al. (2013). Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 73 225–239. 10.1111/tpj.12027 [DOI] [PubMed] [Google Scholar]
- Dénarié J., Debellé F., Rosenberg C. (1992). Signaling and host range variation in nodulation. Annu. Rev. Microbiol. 46 497–531. 10.1146/annurev.mi.46.100192.002433 [DOI] [PubMed] [Google Scholar]
- Desbrosses G. J., Stougaard J. (2011). Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10 348–358. 10.1016/j.chom.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Devos S., Laukens K., Deckers P., Van Der Straeten D., Beeckman T., Inzé D., et al. (2006). A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis. Mol. Plant Microbe Interact. 19 1431–1443. 10.1094/MPMI-19-1431 [DOI] [PubMed] [Google Scholar]
- Diagne N., Diouf D., Svistoonoff S., Kane A., Noba K., Franche C., et al. (2013). Casuarina in Africa: distribution, role and importance of arbuscular mycorrhizal, ectomycorrhizal fungi and Frankia on plant development. J. Environ. Manage. 128 204–209. 10.1016/j.jenvman.2013.05.009 [DOI] [PubMed] [Google Scholar]
- Djébali N., Jauneau A., Ameline-Torregrosa C., Chardon F., Jaulneau V., Mathé C., et al. (2009). Partial resistance of Medicago truncatula to Aphanomyces euteiches is associated with protection of the root stele and is controlled by a major QTL rich in proteasome-related genes. Mol. Plant Microbe Interact. 22 1043–1055. 10.1094/MPMI-22-9-1043 [DOI] [PubMed] [Google Scholar]
- Dowd C., Wilson I. W., McFadden H. (2004). Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f. sp. vasinfectum. Mol. Plant Microbe Interact. 17 654–667. 10.1094/MPMI.2004.17.6.654 [DOI] [PubMed] [Google Scholar]
- Estruch J. J., Schell J., Spena A. (1991). The protein encoded by the rolB plant oncogene hydrolyses indole glucosides. EMBO J. 10 3125–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemadi M., Gutjahr C., Couzigou J. M., Zouine M., Lauressergues D., Timmers A., et al. (2014). Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 166 281–292. 10.1104/pp.114.246595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten J., Kohler A., Morin E., Bhalerao R. P., Palme K., Martin F., et al. (2009). The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol. 151 1991–2005. 10.1104/pp.109.147231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B. J., Indrasumunar A., Hayashi S., Lin M.-H., Lin Y.-H., Reid D. E., et al. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52 61–76. 10.1111/j.1744-7909.2010.00899.x [DOI] [PubMed] [Google Scholar]
- Foo E. (2013). Auxin influences strigolactones in pea mycorrhizal symbiosis. J. Plant Physiol. 170 523–528. 10.1016/j.jplph.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Foo E., Ross J. J., Jones W. T., Reid J. B. (2013). Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann. Bot. 111 769–779. 10.1093/aob/mct041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen H. J., Vijn I., Yang W. C., Bisseling T. (1992). Developmental aspects of the Rhizobium-legume symbiosis. Plant Mol. Biol. 19 89–107. 10.1007/BF00015608 [DOI] [PubMed] [Google Scholar]
- Fusconi A. (2014). Regulation of root morphogenesis in arbuscular mycorrhizae: what role do fungal exudates, phosphate, sugars and hormones play in lateral root formation? Ann. Bot. 113 19–33. 10.1093/aob/mct258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin E., Jacquet C., Bottin A., Dumas B. (2007). Root rot disease of legumes caused by Aphanomyces euteiches. Mol. Plant Pathol. 8 539–548. 10.1111/j.1364-3703.2007.00413.x [DOI] [PubMed] [Google Scholar]
- Genin S., Denny T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50 67–89. 10.1146/annurev-phyto-081211-173000 [DOI] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Balzergue C., Puech-Pagès V., Novero M., Rey T., et al. (2013). Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198 190–202. 10.1111/nph.12146 [DOI] [PubMed] [Google Scholar]
- Geurts R., Lillo A., Bisseling T. (2012). Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Curr. Opin. Plant Biol. 15 438–443. 10.1016/j.pbi.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693. 10.1105/tpc.106.043778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough C., Cullimore J. (2011). Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol. Plant Microbe Interact. 24 867–878. 10.1094/MPMI-01-11-0019 [DOI] [PubMed] [Google Scholar]
- Gourion B., Berrabah F., Ratet P., Stacey G. (2015). Rhizobium-legume symbioses: the crucial role of plant immunity. Trends Plant Sci. 20 186–194. 10.1016/j.tplants.2014.11.008 [DOI] [PubMed] [Google Scholar]
- Grover A., Mittal D., Negi M., Lavania D. (2013). Generating high temperature tolerant transgenic plants: achievements and challenges. Plant Sci. 20 38–47. 10.1016/j.plantsci.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Gutjahr C., Parniske M. (2013). Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 29 593–617. 10.1146/annurev-cellbio-101512-122413 [DOI] [PubMed] [Google Scholar]
- Hanlon M. T., Coenen C. (2011). Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytol. 189 701–709. 10.1111/j.1469-8137.2010.03567.x [DOI] [PubMed] [Google Scholar]
- Held M., Hou H., Miri M., Huynh C., Ross L., Hossain M. S., et al. (2014). Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 26 678–694. 10.1105/tpc.113.119362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A., Brault M., Frugier F., Kuderova A., Lindner A.-C., Motyka V., et al. (2013). Nomenclature for members of the two-component signaling pathway of plants. Plant Physiol. 161 1063–1065. 10.1104/pp.112.213207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet G. (2014). Breeding for resistances to Ralstonia solanacearum. Front. Plant Sci. 5:715 10.3389/fpls.2014.00715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.-H., Wang M.-H., Lee Y.-L., Tsai Y.-L., Li Y.-H., Yang F.-J., et al. (2010). Agrobacterium-produced and exogenous cytokinin-modulated Agrobacterium-mediated plant transformation. Mol. Plant Pathol. 11 677–690. 10.1111/j.1364-3703.2010.00637.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.-F., Strelkov S. E., Feng J., Gossen B. D., Howard R. J. (2012). Plasmodiophora brassicae: a review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol. 13 105–113. 10.1111/j.1364-3703.2011.00729.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardineau M. F., Boivin S., Rodde N., Catrice O., Kisiala A., Lepage A., et al. (2016). A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by Nod factors in the Medicago truncatula root epidermis. Plant Physiol. 171 2256–2276. 10.1104/pp.16.00711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentschel K., Thiel D., Rehn F., Ludwig-Müller J. (2007). Arbuscular mycorrhiza enhances auxin levels and alters auxin biosynthesis in Tropaeolum majus during early stages of colonization. Physiol. Plant. 129 320–333. 10.1111/j.1399-3054.2006.00812.x [DOI] [Google Scholar]
- Jiménez-Zurdo J. I., Frugier F., Crespi M. D., Kondorosi A. (2000). Expression profiles of 22 novel molecular markers for organogenetic pathways acting in alfalfa nodule development. Mol. Plant Microbe Interact. 13 96–106. 10.1094/MPMI.2000.13.1.96 [DOI] [PubMed] [Google Scholar]
- Kaló P., Gleason C., Edwards A., Marsh J., Mitra R. M., Hirsch S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. 10.1126/science.1110951 [DOI] [PubMed] [Google Scholar]
- Karabaghli C., Frey-Klett P., Sotta B., Bonnet M., Le Tacon F. (1998). In vitro effects of Laccaria bicolor S238 N and Pseudomonas fluorescens strain BBc6 on rooting of de-rooted shoot hypocotyls of Norway spruce. Tree Physiol. 18 103–111. 10.1093/treephys/18.2.103 [DOI] [PubMed] [Google Scholar]
- Kisiala A., Laffont C., Emery R. J. N., Frugier F. (2013). Bioactive cytokinins are selectively secreted by Sinorhizobium meliloti nodulating and nonnodulating strains. Mol. Plant Microbe Interact. 26 1225–1231. 10.1094/MPMI-02-13-0054-R [DOI] [PubMed] [Google Scholar]
- Kutáček M., Rovenská J. (1991). Auxin synthesis in Agrobacterium tumefaciens and A. tumefaciens-transformed plant tissue. Plant Growth Regul. 10 313–327. 10.1007/BF00024591 [DOI] [Google Scholar]
- Laffont C., Rey T., André O., Novero M., Kazmierczak T., Debellé F., et al. (2015). The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS ONE 10:e0116819 10.1371/journal.pone.0116819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlali R., McGregor L., Song T., Gossen B. D., Narisawa K., Peng G. (2014). Heteroconium chaetospira induces resistance to clubroot via upregulation of host genes involved in jasmonic acid, ethylene, and auxin biosynthesis. PLoS ONE 9:e94144 10.1371/journal.pone.0094144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L., Lucas M., Champion A. (2015). Rhizobial root hair infection requires auxin signaling. Trends Plant Sci. 20 332–334. 10.1016/j.tplants.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Lee C.-W., Efetova M., Engelmann J. C., Kramell R., Wasternack C., Ludwig-Müller J., et al. (2009). Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21 2948–2962. 10.1105/tpc.108.064576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D., Chen X., Chen A., Wang H., Liu J., Liu J., et al. (2015). The characterization of six auxin-induced tomato GH3 genes uncovers a member, SlGH3.4, strongly responsive to arbuscular mycorrhizal symbiosis. Plant Cell Physiol. 56 674–687. 10.1093/pcp/pcu212 [DOI] [PubMed] [Google Scholar]
- Libbenga K. R., van Iren F., Bogers R. J., Schraag-Lamers M. F. (1973). The role of hormones and gradients in the initiation of cortex proliferation and nodule formation in Pisum sativum L. Planta 114 29–39. 10.1007/BF00390282 [DOI] [PubMed] [Google Scholar]
- Liu C.-W., Breakspear A., Roy S., Murray J. D. (2015). Cytokinin responses counterpoint auxin signaling during rhizobial infection. Plant Signal. Behav. 10:e1019982 10.1080/15592324.2015.1019982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. T., Kado C. I. (1979). Indoleacetic acid production: a plasmid function of Agrobacterium tumefaciens C58. Biochem. Biophys. Res. Commun. 90 171–178. 10.1016/0006-291X(79)91605-X [DOI] [PubMed] [Google Scholar]
- Lohar D. P., Schaff J. E., Laskey J. G., Kieber J. J., Bilyeu K. D., Bird D. M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 38 203–214. 10.1111/j.1365-313X.2004.02038.x [DOI] [PubMed] [Google Scholar]
- Lyons R., Stiller J., Powell J., Rusu A., Manners J. M., Kazan K. (2015). Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS ONE 10:e0121902 10.1371/journal.pone.0121902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F., Poinsot V., André O., Puech-Pagès V., Haouy A., Gueunier M., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469 58–63. 10.1038/nature09622 [DOI] [PubMed] [Google Scholar]
- Mathesius U., Charon C., Rolfe B. G., Kondorosi A., Crespi M. (2000). Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol. Plant Microbe Interact. 13 617–628. 10.1094/MPMI.2000.13.6.617 [DOI] [PubMed] [Google Scholar]
- Mathesius U., Schlaman H. R., Spaink H. P., Of Sautter C., Rolfe B. G., Djordjevic M. A. (1998). Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 14 23–34. 10.1046/j.1365-313X.1998.00090.x [DOI] [PubMed] [Google Scholar]
- Moreau S., Fromentin J., Vailleau F., Vernié T., Huguet S., Balzergue S., et al. (2014). The symbiotic transcription factor MtEFD and cytokinins are positively acting in the Medicago truncatula and Ralstonia solanacearum pathogenic interaction. New Phytol. 201 1343–1357. 10.1111/nph.12636 [DOI] [PubMed] [Google Scholar]
- Morrison E. N., Knowles S., Hayward A., Thorn R. G., Saville B. J., Emery R. J. N. (2015). Detection of phytohormones in temperate forest fungi predicts consistent abscisic acid production and a common pathway for cytokinin biosynthesis. Mycologia 107 245–257. 10.3852/14-157 [DOI] [PubMed] [Google Scholar]
- Mortier V., Wasson A., Jaworek P., De Keyser A., Decroos M., Holsters M., et al. (2014). Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytol. 202 582–593. 10.1111/nph.12681 [DOI] [PubMed] [Google Scholar]
- Murray J. D., Karas B. J., Sato S., Tabata S., Amyot L., Szczyglowski K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315 101–104. 10.1126/science.1132514 [DOI] [PubMed] [Google Scholar]
- Naseem M., Dandekar T. (2012). The role of auxin-cytokinin antagonism in plant-pathogen interactions. PLoS Pathog. 8:e1003026 10.1371/journal.ppat.1003026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J. L. P., Hassan S., Truong T. T., Hocart C. H., Laffont C., Frugier F., et al. (2015). Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. Plant Cell 27 2210–2226. 10.1105/tpc.15.00231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oláh B., Brière C., Bécard G., Dénarié J., Gough C. (2005). Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J. 44 195–207. 10.1111/j.1365-313X.2005.02522.x [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11 252–263. 10.1038/nrmicro2990 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Murray J. D., Poole P. S., Downie J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45 119–144. 10.1146/annurev-genet-110410-132549 [DOI] [PubMed] [Google Scholar]
- Op den Camp R. H. M., De Mita S., Lillo A., Cao Q., Limpens E., Bisseling T., et al. (2011). A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-A response regulators1. Plant Physiol. 157 2013–2022. 10.1104/pp.111.187526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios-Bras C., Schlaman H. R. M., Boot K., Admiraal P., Langerak J. M., Stougaard J., et al. (2003). Auxin distribution in Lotus japonicus during root nodule development. Plant Mol. Biol. 52 1169–1180. 10.1023/B:PLAN.0000004308.78057.f5 [DOI] [PubMed] [Google Scholar]
- Päsold S., Siegel I., Seidel C., Ludwig-Müller J. (2010). Flavonoid accumulation in Arabidopsis thaliana root galls caused by the obligate biotrophic pathogen Plasmodiophora brassicae. Mol. Plant Pathol. 11 545–562. 10.1111/j.1364-3703.2010.00628.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N., Guidot A., Vailleau F., Valls M. (2013). Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14 651–662. 10.1111/mpp.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B., Swarup R., Jansen L., Devos G., Auguy F., Collin M., et al. (2007). Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol. 144 1852–1862. 10.1104/pp.107.101337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret X., Staehelin C., Broughton W. J. (2000). Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64 180–201. 10.1128/MMBR.64.1.180-201.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine-Walker F., Doumas P., Lucas M., Vaissayre V., Beauchemin N. J., Band L. R., et al. (2010). Auxin carriers localization drives auxin accumulation in plant cells infected by Frankia in Casuarina glauca actinorhizal nodules. Plant Physiol. 154 1372–1380. 10.1104/pp.110.163394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R. L., Massicotte H. B. (2004). Exploring structural definitions of mycorrhizas, with emphasis on nutrient-exchange interfaces. Can. J. Bot. 82 1074–1088. 10.1139/b04-071 [DOI] [Google Scholar]
- Phillips D. A., Torrey J. G. (1972). Studies on cytokinin production by Rhizobium. Plant Physiol. 49 11–15. 10.1104/pp.49.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28 489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., et al. (2011). MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65 622–633. 10.1111/j.1365-313X.2010.04447.x [DOI] [PubMed] [Google Scholar]
- Podlešáková K., Fardoux J., Patrel D., Bonaldi K., Novák O., Strnad M., et al. (2013). Rhizobial synthesized cytokinins contribute to but are not essential for the symbiotic interaction between photosynthetic Bradyrhizobia and Aeschynomene legumes. Mol. Plant Microbe Interact. 26 1232–1238. 10.1094/MPMI-03-13-0076-R [DOI] [PubMed] [Google Scholar]
- Ranocha P., Dima O., Nagy R., Felten J., Corratgé-Faillie C., Novák O., et al. (2013). Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 4 2625 10.1038/ncomms3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudaskoski M., Kothe E. (2015). Novel findings on the role of signal exchange in arbuscular and ectomycorrhizal symbioses. Mycorrhiza 25 243–252. 10.1007/s00572-014-0607-2 [DOI] [PubMed] [Google Scholar]
- Reid D. E., Heckmann A. B., Novak O., Kelly S., Stougaard J. (2016). CYTOKININ OXIDASE/DESHYDROGENASE3 maintains cytokinin homeostasis during root and nodule development in Lotus japonicus. Plant Physiol. 170 1060–1074. 10.1104/pp.15.00650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rightmyer A. P., Long S. R. (2011). Pseudonodule formation by wild-type and symbiotic mutant Medicago truncatula in response to auxin transport inhibitors. Mol. Plant Microbe Interact. 24 1372–1384. 10.1094/MPMI-04-11-0103 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J. D. G. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49 317–343. 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- Ross E. J. H., Stone J. M., Elowsky C. G., Arredondo-Peter R., Klucas R. V., Sarath G. (2004). Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 55 1721–1731. 10.1093/jxb/erh211 [DOI] [PubMed] [Google Scholar]
- Roux B., Rodde N., Jardinaud M.-F., Timmers T., Sauviac L., Cottret L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77 817–837. 10.1111/tpj.12442 [DOI] [PubMed] [Google Scholar]
- Santi C., Bogusz D., Franche C. (2013). Biological nitrogen fixation in non-legume plants. Ann. Bot. 111 743–767. 10.1093/aob/mct048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5 4983 10.1038/ncomms5983 [DOI] [PubMed] [Google Scholar]
- Schuller A., Kehr J., Ludwig-Müller J. (2014). Laser microdissection coupled to transcriptional profiling of Arabidopsis roots inoculated by Plasmodiophora brassicae indicates a role for brassinosteroids in clubroot formation. Plant Cell Physiol. 55 392–411. 10.1093/pcp/pct174 [DOI] [PubMed] [Google Scholar]
- Schüβler A., Schwarzott D., Walker C. (2001). A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105 1413–1421. 10.1017/S0953756201005196 [DOI] [Google Scholar]
- Schweiger R., Müller C. (2015). Leaf metabolome in arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 26 120–126. 10.1016/j.pbi.2015.06.009 [DOI] [PubMed] [Google Scholar]
- Sharaf E. F., Farrag A. A. (2004). Induced resistance in tomato plants by IAA against Fusarium oxysporum lycopersici. Pol. J. Microbiol. 53 111–116. [PubMed] [Google Scholar]
- Shaul-Keinan O., Gadkar V., Ginzberg I., Grünzweig J. M., Chet I., Elad Y., et al. (2002). Hormone concentrations in tobacco roots change during arbuscular mycorrhizal colonization with Glomus intraradices. New Phytol. 154 501–507. 10.1046/j.1469-8137.2002.00388.x [DOI] [PubMed] [Google Scholar]
- Siemens J., Keller I., Sarx J., Kunz S., Schuller A., Nagel W., et al. (2006). Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant Microbe Interact. 19 480–494. 10.1094/MPMI-19-0480 [DOI] [PubMed] [Google Scholar]
- Singh S., Parniske M. (2012). Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr. Opin. Plant Biol. 15 444–453. 10.1016/j.pbi.2012.04.002 [DOI] [PubMed] [Google Scholar]
- Smith E. F., Townsend C. O. (1907). A plant-tumor of bacterial origin. Science 25 671–673. 10.1126/science.25.643.671 [DOI] [PubMed] [Google Scholar]
- Smith S. E., Read D. J. (2010). Mycorrhizal Symbiosis. Cambridge, MA: Academic Press. [Google Scholar]
- Soltis D. E., Soltis P. S., Morgan D. R., Swensen S. M., Mullin B. C., Dowd J. M., et al. (1995). Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc. Natl. Acad. Sci. U.S.A. 92 2647–2651. 10.1073/pnas.92.7.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splivallo R., Fischer U., Göbel C., Feussner I., Karlovsky P. (2009). Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol. 150 2018–2029. 10.1104/pp.109.141325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant D. B., Taller B. J. (1989). Cytokinin production by Bradyrhizobium japonicum. Plant Physiol. 89 1247–1252. 10.1104/pp.89.4.1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Ito M., Kawaguchi M. (2013). Induction of localized auxin response during spontaneous nodule development in Lotus japonicus. Plant Signal. Behav. 8:e23359 10.4161/psb.23359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Yano K., Ito M., Umehara Y., Suganuma N., Kawaguchi M. (2012). Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139 3997–4006. 10.1242/dev.084079 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S., Hocher V., Gherbi H. (2014). Actinorhizal root nodule symbioses: what is signalling telling on the origins of nodulation? Curr. Opin. Plant Biol. 20 11–18. 10.1016/j.pbi.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L. H., Radutoiu S., Albrektsen A. S., Sato S., et al. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 104–107. 10.1126/science.1132397 [DOI] [PubMed] [Google Scholar]
- Torrey J. G. (1961). Kinetin as trigger for mitosis in mature endomitotic plant cells. Exp. Cell Res. 23 281–299. 10.1016/0014-4827(61)90038-6 [DOI] [PubMed] [Google Scholar]
- Udvardi M., Poole P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64 781–805. 10.1146/annurev-arplant-050312-120235 [DOI] [PubMed] [Google Scholar]
- van Zeijl A., Op den Camp R. H. M., Deinum E. E., Charnikhova T., Franssen H. Op et al. (2015). Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant 8 1213–1226. 10.1016/j.molp.2015.03.010 [DOI] [PubMed] [Google Scholar]
- Wood D. W., Setubal J. C., Kaul R., Monks D. E., Kitajima J. P., Okura V. K., et al. (2001). The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294 2317–2323. 10.1126/science.1066804 [DOI] [PubMed] [Google Scholar]
- Yang J., Kloepper J. W., Ryu C.-M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14 1–4. 10.1016/j.tplants.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Yuliar, Nion Y. A., Toyota K. (2015). Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ. 30 1–11. 10.1264/jsme2.ME14144 [DOI] [PMC free article] [PubMed] [Google Scholar]


