Abstract
The objective of the study is to investigate potential citrullinated autoantigens as targets of anti-citrullinated protein antibodies (ACPAs) response in synovial fluids (SFs) of patients with rheumatoid arthritis (RA). SFs from six RA patients and six osteoarthritis (OA) patients as controls were collected. The citrullinated proteins in SFs were extracted by immunoprecipitation with rabbit anti-citrulline antibodies. Matrix-assisted laser desorption/ionization time of flight mass spectrometry/time of flight mass spectrometry (MALDI-TOF/TOF) mass spectrometry was subsequently performed to discover a characteristic neutral loss to finally determine citrullinated autoantigens. A total of 182 citrullinated peptides and 200 citrullinated sites were identified in RA SFs, while 3 citrullinated peptides and 4 citrullinated sites were identified in OA SFs. The 182 citrullinated peptides from RA SFs and the 3 citrullinated peptides from OA SFs were derived from 83 and 3 autoantigens, respectively. Eighty-three autoantigens except protein-arginine deiminase type-2 (PADI2) and protein-arginine deiminase type-2 (PADI4) were over-citrullinated compared with controls, and the citrullinated sites of PADI2 and PADI4 were different in two groups. Interestingly, citrullinated histone H3.3 (H3F3A) was found in OA controls, but not in RA groups. The differential citrullinated proteins identified in RA SFs suggested potential autoantigens were targeted for ACPAs response and might contribute to the induction and perpetuation of complement activation and joint inflammation in RA.
Electronic supplementary material
The online version of this article (doi:10.1007/s10067-016-3247-4) contains supplementary material, which is available to authorized users.
Keywords: Citrullinated protein, LC-MALDI-TOF/TOF, Rheumatoid arthritis, Synovial fluid
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease characterized by the formation of inflammatory, invasive tissue, and rheumatoid pannus in synovial membranes, subsequently resulting in joint destruction and systemic complications. The related autoimmunity is often associated with certain major histocompatibility complex (MHC) types and the presence of anti-citrullinated protein antibodies (ACPAs) [1]. ACPAs are important biomarkers of RA and can be detected even before the clinical onset of the disease; consequently, they are recognized as a predictive and diagnostic marker. Furthermore, ACPAs in the inflammatory synovium bind to citrullinated autoantigens to form immune complexes (ICs), which lead to the development of inflammation [2–7]. Thus, a simple and effective method is needed to detect citrullinated proteins in the joint fluid from RA patients.
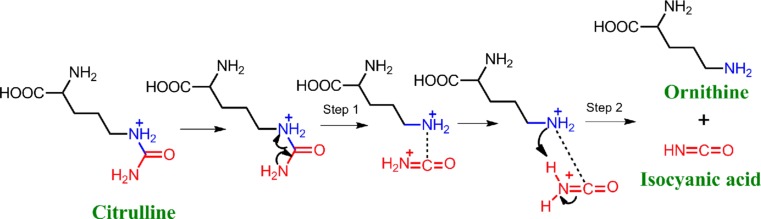
Citrullination is a post-translational modification (PTM) involving the conversion of an arginine residue to a non-coded citrulline residue, catalyzed by peptidylarginine deiminases (PADIs). This PTM leads to the loss of a positive charge and a reduction in hydrogen-bonding ability [8]. The traditional method to detect citrullinated proteins in biological fluids is two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) followed by immunoblotting and Fourier transform ion cyclotron resonance (ICR) mass spectrometry (MS) analysis, which is labor-intensive and time-consuming [9–12]. Moreover, the mass shift of citrullination is very small (+1 Da), which can result in false positives [4,13]. Hao et al. [14] found that one specific signature of citrullination is that the neutral loss of 43 Da from the peptidyl-citrulline can be observed after collision-induced dissociation (CID) during triple quadruple/linear ion trap (Q-Trap) mass spectrometry, which indicates the elimination of isocyanic acid from the citrulline ureido group as shown in (Fig. 1). However, this technology has not been applied to human fluid because the complexity of the protein mixture made detection of low-abundance proteins very difficult. In addition, the low mass resolution of Q-trap MS prevented the reliable peptide and PTM characterization as in the high mass resolution of time-of-flight (TOF) MS. Thus, we first applied liquid chromatography-matrix-assisted laser desorption/ionization (LC-MALDI)-TOF/TOF to detect citrullinated proteins in human RA synovial fluid (SF).
Fig. 1.
Schematic of the citrullinated peptide detection method. Step 1: citrullination of the protein. Step 2: the hydrogen bond is cleaved during CID of the citrullinated peptides, resulting in a signature 43-Da neutral loss from peptidyl-citrulline
In this study, citrullinated proteins were extracted by immunoprecipitation using agarose-conjugated rabbit anti-citrulline antibodies, followed by LC-MALDI-TOF/TOF MS analysis. The peptide sequences and citrullinated sites in RA SFs or osteoarthritis (OA) SFs were identified. The high-energy CID mode of MALDI-TOF/TOF (20 keV collision energy) was adopted to improve fragmentation efficiency for reliable peptide and PTM characterization, thereby enabling the identification of potential autoantigens for ACPAs.
Methods
Human sample collection
Samples of SFs were consecutively obtained from knee joints of patients with RA or patients with OA, as a control, during knee therapeutic arthrocentesis at the Department Osteology, Jinling Hospital, School of Medicine, Nanjing University from January 2011 to January 2012. In order to improve the possibility of the existence of citrullinated antigens, patients of RA fulfilling the criteria (serum ACPA >200 U/ml and synovial fluids ACPA >200 U/ml) were selected and SFs of patients who were diagnosed as OA were collected. Patients who had incomplete data were excluded. The diagnosis of RA was made according to the American College of Rheumatology/European League Against Rheumatism Collaborative Initiative 2010 criteria [15] and the diagnosis of OA was based on the 1986 clinical and radiological criteria for the diagnosis of knee OA developed by the American College of Rheumatology [16]. Finally, six patients with RA were obtained and six patients with OA were selected by random. The patients’ basic information and serologic profile are shown in (Table 1). Informed consent was obtained from all subjects and the study was approved by the local ethics committee (Nanjing, China).
Table 1.
Clinical and laboratory characteristics of the RA and OA patients
| Diagnosis | Sex | Age (years) | ACPA, (U/ml) | RF (IU/ml) | ESR (mm/h) | CRP (mg/dl) | Disease duration (years) | DAS28 |
|---|---|---|---|---|---|---|---|---|
| RA | Female | 61 | >200 | 86.3 | 64 | 25.1 | 3 | 4.28 |
| RA | Female | 79 | >200 | 397 | 109 | 42.7 | 4 | 4.93 |
| RA | Female | 73 | >200 | 40.6 | 44 | 37.5 | 2 | 3.84 |
| RA | Male | 69 | >200 | 24.5 | 29 | 27.3 | 3 | 3.43 |
| RA | Male | 63 | >200 | 142 | 44 | 77.8 | 5 | 3.72 |
| RA | Male | 70 | >200 | 102 | 82 | 31.4 | 2 | 4.16 |
| OA | Female | 68 | <0.5 | <20 | 8 | 8.9 | 2 | _ |
| OA | Female | 53 | <0.5 | <20 | 7 | 7.8 | 3 | _ |
| OA | Female | 63 | <0.5 | <20 | 5 | 2.5 | 2 | _ |
| OA | Male | 69 | <0.5 | <20 | 3 | 0.6 | 5 | _ |
| OA | Male | 54 | <0.5 | <20 | 7 | 4.3 | 4 | _ |
| OA | Male | 58 | <0.5 | <20 | 14 | 9.9 | 3 | _ |
ACPA anticitrullinated protein antibody, RF rheumatoid factor, ESR erythrocyte sedimentation rate, CRP C-reactive protein, DAS28 disease activity score at 28 joints
Immunoprecipitation
All SF samples were centrifuged at 25,000×g for 10 min at 4 °C and the supernatants were stored separately in sterile conditions at −80 °C. EDTA was added at a final concentration of 50 mM, followed by centrifugation at 14,000×g for 10 min at 4 °C, and the supernatant or sample was transferred to a new vial. Protein concentrations were determined by BioSpec-nano (Shimadzu Biotech, Kyoto, Japan), and 1 mg was then subjected to immunoprecipitation [17,18]. Anti-citrulline polyclonal antibody (pAb; Abcam, Cambridge, USA) was cross-linked to protein G-Agarose (Sigma Aldrich, St Louis, MO, USA) with stable amide linkages according to the manufacturer’s instructions. Citrullinated proteins were immunoprecipitated by incubating the supernatant/sample with agarose-conjugated rabbit anti-citrulline antibodies overnight. The samples were washed three times with 50 mM ammonium acetate pH 7.4 and once with H2O, and the bound peptides were then eluted with acetonitrile/H2O (7:3 v/v) containing 5 mM HCl. The eluted peptides were subjected to desalting by C18 tip (SciLifeLab, Shanghai, China). The efficiency of immunoprecipitation was determined on equal amounts of protein/sample (OA, RA, washes of RA, and eluents of RA) by immunoblotting using anti-citrulline pAb.
Protein reduction, alkylation, and enzyme digestion
An additional 937 μL of 50 mM NH4HCO3 was added to the eluted proteins. The proteins were reduced by adding 10 μL of 0.5 M dithiothreitol (DTT) in 50 mM NH4HCO3 to a final concentration of 4 mM and incubated for 20 min at 56 °C. For alkylation, 27 μL of 0.55 M iodoacetamide was added, and the samples were incubated for 15 min at room temperature in the dark. The final concentration of the extracted proteins was approximately 2.4 mg/mL (total volume approximately 1250 μL) according to the optical density at 280 nm. ProteaseMax solution (10 μL of 1 %) was added together with 50 μL of 1 μg/μL LysC before incubation in a hydrated chamber at 37 °C overnight. The reaction was quenched by adding formic acid (FA; Fluka, Sigma) to a final concentration of 0.5 %. The digested peptides were then subjected to desalting by C18 tip (Shimadzu Biotech, Kyoto, Japan). Finally, the desalted peptides were immediately applied to a prominence nano2D-HPLC and Accuspot™ system (Shimadzu Biotech, Kyoto, Japan).
2D-NanoLC fractionation
A 5-μL aliquot of the desalted peptide/sample was loaded directly onto a PolySulfoethyl A Column (1.0 mm × 50 mm, 5 μm) for the first dimensional strong cation exchange separation (A = 10 mM FA, B = 600 mM FA). The samples were then subjected to a second dimensional RP separation using a Capillary EX-Nano MonoCap C-18 column (0.16 mm × 150 mm, 5 μm): A = 5 % acetonitrile + 0.1 % FA, B = 95 % acetonitrile + 0.1 % FA. The flow rate of the system was set at 4 μL/min post-split. The eluent passed through a UV/Vis detector (220 nm) and was mixed with α-cyano 4-hydroxycinnamic acid matrix (CHCA; 5 mg/ml in 50/50 acetonitrile/0.1 % trifluoroacetic acid) and deposited onto a stainless steel MALDI target using the Accuspot™ LC-MALDI deposition robot [19].
LC-MALDI-TOF/TOF MS analysis
Samples were deposited at a volume of ∼1 μL/spot. The signal-to-noise ratio (S/N) was determined using Launchpad version 2.9.1 software (Shimadzu Biotech, Kyoto, Japan). The limit of detection (LOD) was established with a S/N of 3:1. The m/z value was calibrated with 50 fmol each of human angiotensin II and human adrenocorticotropin fragment 18–39 and 250 fmol each of bovine insulin oxidized beta chain and bovine insulin as external standards. The m/z reported in MALDI-TOF/TOF (MALDI-7090, Shimadzu Kratos, Manchester, UK) was set in positive ion mode and a mass range of 1000–4000 Da. The peptide ions with high S/N (S/N > 10:1) were subjected to CID for subsequent MS/MS analysis.
Bioinformatics analysis
The raw MS/MS data were searched using the Mascot engine and then processed with PTM Finder™ Software (Shimadzu Kratos) using the following criteria: database, Swiss-Prot, human; enzyme, LysC; miscleavages, 2; static modifications, carbamidomethylation of cysteine (+57.02 Da); variable modifications, oxidation of methionine (+16.00 Da); neutral loss of isocyanic acid from peptidyl-citrulline (−43.02 Da); precursor ion tolerance, 0.3 Da; fragment ion tolerance, 0.8 Da. At this point, the citrullinated sites were identified by MS/MS analysis of the AA(8)AA ion because an ornithine residue (Orn) was expected to be the product after loss of a carbamyl group. Thus, the neutral loss of isocyanic acid from peptidyl-citrulline could be differentiated from the deamidation of peptidyl-asparagine or peptidyl-glutamine. All entries were filtered using a false positive rate of 1 % at the peptide levels, and false positives were removed. The citrullinated proteins from the RA SFs were further analyzed with DAVID Bioinformatics Resources (David 6.7 software, Bethesda, Maryland, USA) to understand their biological functions.
Results
A total of 182 citrullinated peptides and 200 citrullinated sites were identified in the RA SFs, while only three citrullinated peptides and four citrullinated sites were identified in the OA SFs (Tables 2 and 3). The 182 citrullinated peptides from the RA SFs were derived from 83 autoantigens, and the three citrullinated peptides from the OA SFs were derived from three autoantigens. The autoantigens in the RA SFs were over-citrullinated compared with the controls. Among these, 26 citrullinated proteins identified here have also been validated in previous studies (Table 4), which suggests that this strategy for identifying citrullinated peptides is highly effective.
Table 2.
Citrullinated peptides and their deaminized sites identified by MALDI-TOF-MS in the RA SFs
| Gene name | Protein ID | Peptide sequence | Citrullinated sites |
|---|---|---|---|
| A2M | A2MG_HUMAN | DNGCFRSSGSLLNNAIK | R1081 |
| GNRIAQWQSFQLEGGLK | R174 | ||
| EQAPHCICANGRQTVSWAVTPK | R853 | ||
| FQVDNNNRLLLQQVSLPELPGEYSMK | R1297 | ||
| ACTG1 | ACTG_HUMAN | DLYANTVLSGGTTMYPGIADRMQK | R312 |
| AGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQK | R28 | ||
| ALB | ALBU_HUMAN | AWAVARLSQRFPK | R246 |
| LCTVATLRETYGEMADCCAK | R105 | ||
| VHTECCHGDLLECADDRADLAK | R281 | ||
| RMPCAEDYLSVVLNQLCVLHEK | R469 | ||
| YLYEIARRHPYFYAPELLFFAK | R169 | ||
| CCTESLVNRRPCFSALEVDETYVPK | R509 | ||
| ANXA1 | ANXA1_HUMAN | DITSDTSGDFRNALLSLAK | R177 |
| GTDVNVFNTILTTRSYPQLRRVFQK | R228 | ||
| APOA1 | APOA1_HUMAN | ENGGARLAEYHAK | R212 |
| VEPLRAELQEGARQK | R155 | ||
| DSGRDYVSQFEGSALGK | R51 | ||
| PALEDLRQGLLPVLESFK | R239 | ||
| APOB | APOB_HUMAN | LEGTTRLTRK | R3386 |
| LTTNGRFREHNAK | R1689 | ||
| AEFTGRHDAHLNGK | R3020 | ||
| GNVATEISTERDLGQCDRFK | R207 | ||
| IREVTQRLNGEIQALELPQK | R2449 | ||
| RLIDLSIQNYHTFLIYITELLK | R4519 | ||
| YTYNYEAESSSGVPGTADSRSATRINCK | R75 | ||
| ARHGAP4 | F5GZW3_HUMAN | EEQEVSWTQYTQRK | R486 |
| AERFSSRGGRLGSSREHQSFRK | R73, R77 | ||
| ELLGKTSVRQGLGPASTTSPSPGPRSPK | R889 | ||
| LREAERQEEKRAGRSVPTTTAGATEAGPLRK | R198 | ||
| ARPC1B | ARC1B_HUMAN | QSSQRGLTARERFQNLDK | R294, R299 |
| PTLVILRINRAARCVRWAPNENK | R100 | ||
| C1R | C1R_HUMAN | GFLAYYQAVDLDECASRSK | R149 |
| MQTRAGSRESEQGVYTCTAQGIWK | R420 | ||
| DCGQPRNLPNGDFRYTTTMGVNTYK | R388 | ||
| C1S | C1S_HUMAN | AARLPVAPLRK | R586 |
| C2 | CO2_HUMAN | SSGQWQTPGATRSLSK | R77 |
| C3 | CO3_HUMAN | RRHQQTVTIPPK | R880 |
| VLLDGVQNPRAEDLVGK | R315 | ||
| TVAVRTLDPERLGREGVQK | R945 | ||
| GYTQQLAFRQPSSAFAAFVK | R1060 | ||
| ITHRIHWESASLLRSEETK | R1310 | ||
| PDGVFQEDAPVIHQEMIGGLRNNNEK | R1134 | ||
| C4B | CO4B_HUMAN | ISARFSDGLESNSSTQFEVK | R218 |
| VDFTLSSERDFALLSLQVPLK | R80 | ||
| AAANQMRNFLVRASCRLRLEPGK | R1675 | ||
| SHALQLNNRQIRGLEEELQFSLGSK | R1349 | ||
| C4BPA | C4BPA_HUMAN | PELVNGRLSVDK | R493 |
| NLRWTPYQGCEALCCPEPK | R353 | ||
| C6 | CO6_HUMAN | FRCDSGRCIARK | R150 |
| RSENINHNSAFK | R289 | ||
| SSRTSNPYRVPANLENVGFEVQTAEDDLK | R225 | ||
| C9 | CO9_HUMAN | NFRTEHYEEQIEAFK | R213 |
| CAT | CATA_HUMAN | NAIHTFVQSGSHLAAREK | R522 |
| CD44 | CD44_HUMAN | NGRYSISRTEAADLCK | R41 |
| EQWFGNRWHEGYRQTPK | R407, R413 | ||
| CFH | CFAH_HUMAN | RITCRNGQWSEPPK | R1149, R1153 |
| HGGLYHENMRRPYFPVAVGK | R340 | ||
| RGYRLSSRSHTLRTTCWDGK | R1210, R1215 | ||
| AQTTVTCMENGWSPTPRCIRVK | R441 | ||
| IPCSQPPQIEHGTINSSRSSQESYAHGTK | R885 | ||
| CNMGYEYSERGDAVCTESGWRPLPSCEEK | R246 | ||
| CFHR2 | FHR2_HUMAN | SHSFRAMCQNGK | R254 |
| CFI | CFAI_HUMAN | DNERVFSLQWGEVK | R480 |
| THRYQIWTTVVDWIHPDLK | R389 | ||
| CHMP2A | CHM2A_HUMAN | MDLLFGRRK | R8 |
| DLVRTRRYVRK | R71 | ||
| CLC | LEG10_HUMAN | YQVMVNGQSSYTFDHRIK | R115 |
| CP | CERU_HUMAN | ALYLQYTDETFRTTIEK | R81 |
| NLASRPYTFHSHGITYYK | R115 | ||
| ENLTAPGSDSAVFFEQGTTRIGGSYK | R415 | ||
| DNEDFQESNRMYSVNGYTFGSLPGLSMCAEDRVK | R258 | ||
| NMATRPYSIHAHGVQTESSTVTPTLPGETLTYVWK | R830 | ||
| CPB2 | CBPB2_HUMAN | DHEELSLVASEAVRAIEK | R342 |
| CTLA4 | CTLA4_HUMAN | AMHVAQPAVVLASSRGIASFVCEYASPGK | R51 |
| AQLNLATRTWPCTLLFFLLFIPVFCK | R18 | ||
| ENO1 | ENOA_HUMAN | TGAPCRSERLAK | R403 |
| LAQANGWGVMVSHRSGETEDTFIADLVVGLCTGQIK | R372 | ||
| F2 | THRB_HUMAN | YTACETARTPRDK | R94, R97 |
| DSTRIRITDNMFCAGYK | R541 | ||
| FF3 | AFF3_HUMAN | DFLTDRSNQSHLVGVPK | R111 |
| EAAANGGSGPRAPVGSINARTTSDIAK | R745 | ||
| YTSEDLTSSSRPNGNSLFTSASSSK | R926 | ||
| SPPAAVAVAVSAAAPPPAVPCAPAENAPAPARRSAGK | R606 | ||
| FGA | FIBA_HUMAN | NVRAQLVDMK | R160 |
| GLIDEVNQDFTNRINK | R84 | ||
| FGB | FIBB_HUMAN | REEAPSLRPAPPPISGGGYRARPAK | R60, R72 |
| EDGGGWWYNRCHAANPNGRYYWGGQYTWDMAK | R445 | ||
| FGG | FIBG_HUMAN | YEASILTHDSSIRYLQEIYNSNNQK | R134 |
| YTGNTYRVGDTYERPK | R106 | ||
| WLPSSSPVTGYRVTTTPK | R1573 | ||
| DNRGNLLQCICTGNGRGEWK | R265 | ||
| GC | VTDB_HUMAN | HLSLLTTLSNRVCSQYAAYGEK | R218 |
| HQPQEFPTYVEPTNDEICEAFRK | R149 | ||
| RSDFASNCCSINSPPLYCDSEIDAELK | R445 | ||
| H1FX | H1X_HUMAN | VPWFDQQNGRTYLK | R86 |
| YSQLVVETIRRLGERNGSSLAK | R57, R62 | ||
| H2AFY | H2AY_HUMAN | SIAFPSIGSGRNGFPK | R318 |
| HABP2 | HABP2_HUMAN | EEFHEQSFRVEK | R391 |
| FCEIGSDDCYVGDGYSYRGK | R203 | ||
| LIANTLCNSRQLYDHMIDDSMICAGNLQK | R480 | ||
| HIST2H2AC | H2A2C_HUMAN | TRIIPRHLQLAIRNDEELNK | R89 |
| GNYAERVGAGAPVYMAAVLEYLTAEILELAGNAARDNK | R43, R72 | ||
| HMGB2 | HMGB2_HUMAN | MSSYAFFVQTCREEHK | R24 |
| HNRNPA1L2 | RA1L2_HUMAN | GGNFGGRSSGPYGGGGQYFAK | R284 |
| HP | HPT_HUMAN | VSVNERVMPICLPSK | R261 |
| YVMLPVADQDQCIRHYEGSTVPEK | R311 | ||
| HPR | HPTR_HUMAN | VLVNERVMPICLPSK | R203 |
| HPX | HEMO_HUMAN | NFPSPVDAAFRQGHNSVFLIK | R102 |
| HSP90AA1 | Q8TBA7_HUMAN | AQALRDNSTMGYMAAK | R620 |
| HNDDEQYAWESSAGGSFTVRTDTGEPMGRGTK | R173 | ||
| HSPA1A | HSP71_HUMAN | LLQDFFNGRDLNK | R357 |
| EIAEAYLGYPVTNAVITVPAYFNDSQRQATK | R155 | ||
| HSPA5 | GRP78_HUMAN | SDIDEIVLVGGSTRIPK | R368 |
| RLIGRTWNDPSVQQDIK | R98 | ||
| Ig kappa chain V-II region RPMI 6410 | KV206_HUMAN | VSNRDSGVPDRFSGSGSGTDFTLK | R79 |
| Ig lambda chain V-II region NEI | LV202_HUMAN | RPSGVSNRFSGSK | R56, R63 |
| Ig lambda chain V-II region NIG-84 | LV211_HUMAN | LLIYDVNSRPSGISNRFSGSK | R56, R63 |
| IGHA1 | IGHA1_HUMAN | YLTWASRQEPSQGTTTFAVTSILRVAAEDWK | R282 |
| IGHG3 | IGHG3_HUMAN | SCDTPPPCPRCPEPK | R128 |
| TPLGDTTHTCPRCPEPK | R113 | ||
| ING4 | ING4_HUMAN | WFCPRCSQERK | R241 |
| ITIH2 | ITIH2_HUMAN | RLSNENHGIAQRIYGNQDTSSQLK | R475 |
| TILDDLRAEDHFSVIDFNQNIRTWRNDLISATK | R356, R359 | ||
| KNG1 | KNG1_HUMAN | ICVGCPRDIPTNSPELEETLTHTITK | R268 |
| KRT33B | KT33B_HUMAN | ETMQFLNDRLASYLEK | R66 |
| LBR | LBR_HUMAN | EARREVEVK | R111 |
| PLTSFRQRK | R61 | ||
| SARRSASASHQADIK | R96 | ||
| ELAVRTFEVTPIRAK | R195 | ||
| APRNDLSPASSGNAVYDFFIGRELNPRIGTFDLK | R353 | ||
| LCP1 | PLSL_HUMAN | GDEEGVPAVVIDMSGLREK | R316 |
| ALENDPDCRHVIPMNPNTNDLFNAVGDGIVLCK | R141 | ||
| LGALS3BP | LG3BP_HUMAN | SGGSDRTIAYENK | R514 |
| SQLVYQSRRGPLVK | R436 | ||
| LRG1 | A2GL_HUMAN | ALGHLDLSGNRLRK | R175 |
| LQVLGKDLLLPQPDLRYLFLNGNK | R239 | ||
| MAPRE1 | MARE1_HUMAN | PLTSSSAAPQRPISTQRTAAAPK | R168 |
| MMP8 | MMP8_HUMAN | FYQLPSNQYQSTRK | R52 |
| MNDA | MNDA_HUMAN | INQEEVGLAAPAPTARNK | R119 |
| NCF1 | NCF1_HUMAN | SGQDVSQAQRQIK | R292 |
| STATDITGPIILQTYRAIANYEK | R162 | ||
| ORM1 | A1AG1_HUMAN | EQLGEFYEALDCLRIPK | R167 |
| PABPC1 | PABP1_HUMAN | AVTEMNGRIVATK | R356 |
| PVRIMWSQRDPSLRK | R89, R94 | ||
| ITGMLLEIDNSELLHMLESPESLRSK | R604 | ||
| PADI2 | PADI2_HUMAN | VGVFYVENPFFGQRYIHILGRRK | R225, R233 |
| PADI4 | PADI4_HUMAN | GFRLLLASPRSCYK | R495 |
| TLREHNSFVERCIDWNRELLK | R536 | ||
| PFGPVINGRCCLEEK | R609 | ||
| POFUT2 | OFUT2_HUMAN | VFVATDAVRK | R337 |
| DFIWGHRQDVPSLEGAVRK | R315 | ||
| PPIA | PPIA_HUMAN | EGMNIVEAMERFGSRNGK | R148 |
| PRG4 | PRG4_HUMAN | DQYYNIDVPSRTARAITTRSGQTLSK | R1386, R1391 |
| PRKCD | KPCD_HUMAN | QSMRSEDEAK | R132 |
| SPRDYSNFDQEFLNEK | R628 | ||
| IIGRCTGTAANSRDTIFQK | R216 | ||
| PTPN22 | PTN22_HUMAN | GPRNPPPTWNI | R499 |
| PAESVQSNNSSSFLNFGFANRFSK | R491 | ||
| SAA2 | SAA2_HUMAN | RGPGGAWAAEVISNARENIQRLTGRGAEDSLADQAANK | R80, R89 |
| SERPINA3 | AACT_HUMAN | ADLSGITGARNLAVSQVVHK | R350 |
| SERPINC1 | ANT3_HUMAN | LVSANRLFGDK | R177 |
| IPEATNRRVWELSK | R78 | ||
| ANSRFATTFYQHLADSK | R89 | ||
| SLC22A4 | S22A4_HUMAN | VPLTTSLFFVGVLLGSFVSGQLSDRFGRK | R166 |
| STAT4 | STAT4_HUMAN | NVSTLSNRRFVLCGTNVK | R173 |
| SLQSSSVSERQRNVEHKVAAIK | R139 | ||
| FHGNPMHVAVVISNCLREERRILAAANMPVQGPLEK | R110 | ||
| TAGLN2 | TAGL2_HUMAN | MANRGPAYGLSREVQQK | R4 |
| TF | TRFE_HUMAN | CSTSSLLEACTFRRP | R696 |
| EGYYGYTGAFRCLVEK | R541 | ||
| ADRDQYELLCLDNTRK | R239, R251 | ||
| TNC | TENA_HUMAN | NGRENFYQNWK | R2033 |
| RVTTTRLDAPSQIEVK | R802 | ||
| VEAARNLTVPGSLRAVDIPGLK | R1127 | ||
| PDTEYEVSLISRRGDMSSNPAK | R878 | ||
| ETFTTGLDAPRNLRRVSQTDNSITLEWRNGK | R897 | ||
| TNC | TENA_HUMAN | VPEITRTVSGNTVEYALTDLEPATEYTLRIFAEK | R1866 |
| TNFAIP6 | TSG6_HUMAN | NFLAGRFSHL | R273 |
| VIM | VIME_HUMAN | MALDIEIATYRK | R401 |
| PDLTAALRDVRQQYESVAAK | R270, R273 | ||
| VPRPB | VPRBP_HUMAN | FISGTPRRK | R707 |
| SPFGSSFRTFNATDYK | R1334 |
Citrullinated residues are indicated with a bold R
Table 3.
Citrullinated peptides and their deaminized sites identified by MALDI-TOF-MS in the OA SFs
| Gene name | Protein ID | Peptide sequence | Citrullinated sites |
|---|---|---|---|
| H3F3A | H33_HUMAN | DIQLARRIRGERA | R130 |
| PADI2 | PADI2_HUMAN | GFPVVLDSPRDGNLK | R373 |
| PADI4 | PADI4_HUMAN | TLPVVFDSPRNRGLK | R372, R374 |
Citrullinated residues are indicated with a bold R
Table 4.
26 citrullinated proteins in our study were validated in previous studies
| Protein ID | References |
|---|---|
| Arginine deiminase type-4 | [19] |
| Alpha-1-acid glycoprotein 1 | [12] |
| Alpha-2-macroglobulin | [12] |
| Annexin A1 | [12] |
| Apolipoprotein A-I | [12] |
| Apolipoprotein B-100 | [12] |
| Ceruloplasmin | [12] |
| C4b-binding protein alpha chain | [12] |
| Complement C2 | [12] |
| Complement C4-B | [12] |
| Complement factor H | [12] |
| Enolase | [12,20] |
| Fibrinogen | [20] |
| Fibronectin | [12] |
| Hemopexin | [12] |
| HSP90 | [12] |
| Histone | [8,21] |
| Inter-alpha-trypsin inhibitor heavy chain H2 | [12] |
| Myeloid cell nuclear differentiation antigen | [12] |
| Plastin-2 | [12,22] |
| Proteoglycan 4 | [12] |
| Serotransferrin | [12] |
| Serum albumin | [12] |
| Tenascin | [12] |
| Vitamin D-binding protein | [12] |
| Vimentin | [11] |
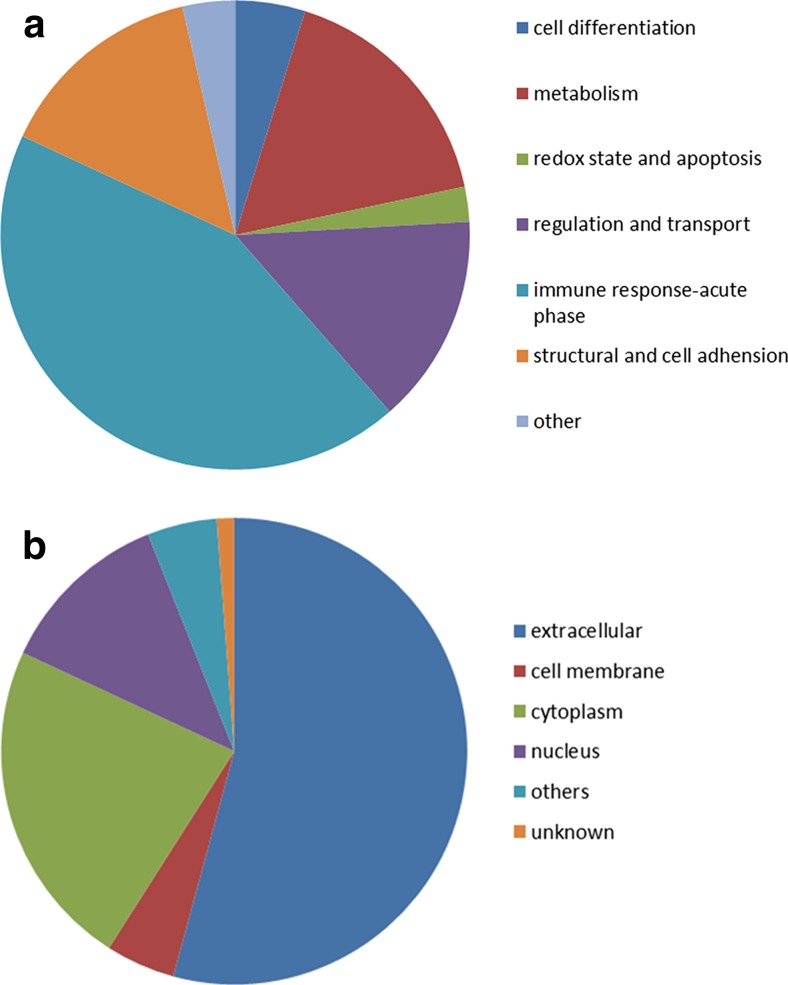
Functional analysis of the identified citrullinated proteins in the RA group was performed with David 6.7 software. The categories of “disease” and “gene ontology” reported a significant enrichment of RA and acute inflammatory response-associated genes, which corresponded to the physiological status of the patients in the present study (Supplement Fig. 1). Furthermore, genes involved in the enriched pathways in the list were associated with complement and coagulation cascades (Supplement Fig. 2). Additionally, proteins that were involved in cell differentiation, metabolism, redox state and apoptosis, regulation and transport, immune response and acute phase, structural and cell adhesion, and other groups based on the NCBI and UniProt database information are shown in (Fig. 2a). Moreover, the proteins were also classified by their subcellular location, as described in (Fig. 2b). The results of our analysis demonstrated that the citrullinated proteins obtained with our protocol provide reliable data on the state of citrullination in RA SF.
Fig. 2.
Classification by a function and b subcellular location of the proteins identified in the RA SFs
Discussion
The major methods used currently to identify citrullinated proteins employ 2D-PAGE followed by immunoblotting and Fourier transform ion cyclotron resonance mass spectrometry analysis. For example, J.B.C. van Beers et al. found 192 proteins including 53 citrullinated proteins with their citrullinated residues in RA SFs [12]. One problem with this method is the small mass shift (+1 Da) from the conversion of peptidyl arginine to Cit, which is challenging for mass detection to distinguish. In the present study, citrullinated proteins were effectively enriched following immunoprecipitation (Supplement Fig. 1). NanoLC was then used to fractionate the tryptic digests of citrullinated proteins to improve the sensitivity and dynamic range of protein identification. With this method, not only are peptides of the same nominal mass isolated by temporal separation, but signal suppression is also reduced because of the separation of low- and high-abundance peptides. Importantly, the unique LC-MALDI peak picking algorithm promotes the MS/MS of selected ions at the apex of the eluting chromatographic peak to allow the most efficient data acquisition. This is not often the case with ES LC-MS/MS, where MS/MS acquisition is often taken on the rising edge of the eluting chromatographic peak. In addition, the high-energy CID mode of MALDI-TOF/TOF (20 keV collision energy, MALDI-7090) allowed us to determine the citrullinated sites more easily, according to the characteristic neutral loss of an isocyanic acid group from peptidyl-citrulline.
A number of chaperone molecules were identified within the SFs, particularly heat shock 70 kDa protein 1A/1B (HSPA1A), glucose-regulated protein 78 kDa (GRP78 or HSPA5), and HSP90AA1, members of the stress-inducible heat-shock protein 70 family. Also, we previously found GRP75 (HSP70) and binding immunoglobulin protein (BiP or GRP78) in RA synovial fibroblast-like synoviocytes (FLSs) [20]. Citrullinated BiP induces anti-CCP and anti-citrullinated fibrinogen antibodies and exacerbates collagen-induced arthritis in mice, and deaminated HSP90 was identified as a diagnostic autoantigen for a potentially serious manifestation of RA [10,21]. Recently, HSPs have been reported that not only act as chaperones during protein folding but also play a role between ubiquitin E3 ligase and the proteasome to inhibit proinflammatory NF-ΚB signaling [22]. In addition, both canonical and non-canonical NF-ΚBs are overexpressed in RA and are associated with the persistence of inflammation in RA [23]. Thus, citrullination of HSP may contribute to the chronic inflammation in the synovium or dysregulation of RA synovial fibroblasts, suggesting that citrullination may correlate with complement activation and the perpetuation of RA.
In a previous study from our group, we also reported that the elevated Annexin A11 in FLSs may be associated with the extensive synovial fibroblast-like synoviocytes hyperplasia. Additionally, in the extracellular environment defined as synovial fluid, we found citrullinated AnnexinA1, another member of the annexin superfamily of structurally related Ca2+-dependent phospholipid-binding proteins. Several other studies have demonstrated that AnnexinA1 is a glucocorticoid-induced molecule that can be transferred into cartilage and can modulate T cell function and the adaptive immune responses relevant to RA [24,25]. Consistent with this, treatment of mice with dexamethasone promotes potent antiarthritic effects that are dynamically attenuated in AnxA1−/− mice [26]. Our observations on citrullinated Annexin A1 reflect the possibility that citrullinated or non-citrullinated Annexin may be a target to minimize glucocorticoid use in RA.
The different citrullinated sites of PADI2 and PADI4 in the two groups suggest new potential biomarkers for RA. PADI2 and PADI4 are the only PAD isotypes expressed in the synovial tissue of patients with RA, and they were reported to induce differentiation and apoptosis [27]. PADI4, found in the cell nucleus, mediated gene transcription by regulating arginine citrullination and methylation in histones H1, H3, and H4 and was autocitrullinated during cell activation [8,28,29]. Interestingly, citrullinated H3F3A was found in the OA controls, but not in the RA group. In addition, histones H1x and H2A were only citrullinated in the RA group. These results suggest that PADI2 and PADI4 represent a heterogeneous subtype with different citrullinated sites targeting multiple structural domains, where the specific citrullinated site may predict a specific disease. The exact mechanism underlying this phenomenon remains to be elucidated.
Although we identified the potential antigens for ACPA, some limitations remain, including the amount of patients was small, thus we pooled all samples per group to gain more sensitivity and to find more citrullinated antigens; results merely compared with previous studies; the validated process used only one method of mass spectrometry and was only on the basis of mass-spectrometry-based proteomics, so we performed DAVID Bioinformatics Resources to classify these genes corresponding to citrullinated proteins, at the same time, estimate and verify the reliability. Further studies will employ western blot to identify some selected potential autoantigens. At the same time, we will collect samples of synovial fluid or serum of RA patients as more as possible and then test antibodies corresponding to autoantigens in synovial fluid or serum of RA patients in order to obtain reliable results from clinical data. These limitations indicate the need for larger validation studies and prospective SFs studies in groups where larger samples are available.
Overall, we demonstrated a simple and efficient strategy for detecting citrullinated proteins and citrullinated sites in human RA SFs. In addition to the previously detected citrullinated proteins in RA SF, the novel citrullinated proteins identified by the data here may represent new antigens for ACPAs, as well as new markers for diagnosis. More importantly, this data will contribute to the search for the etiopathogenesis of, and new therapeutic targets for RA.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 1136 kb)
Compliance with ethical standards
Informed consent was obtained from all subjects and the study was approved by the local ethics committee (Nanjing, China).
Disclosures
None.
Funding
This research was supported by the National Natural Science foundation of China (No. 81470071).
Footnotes
Fei Wang and Fang-Fang Chen contributed equally to this work.
References
- 1.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller S, Radic M (2014) Citrullinated autoantigens: from diagnostic markers to pathogenetic mechanisms. Clin Rev Allergy Immunol 1–8 [DOI] [PubMed]
- 3.Sakkas LI, Bogdanos DP, Katsiari C, Platsoucas CD. Anti-citrullinated peptides as autoantigens in rheumatoid arthritis-relevance to treatment. Autoimmun Rev. 2014;13:1114–1120. doi: 10.1016/j.autrev.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Gudmann NS, Hansen NUB, Jensen ACB, Karsdal MA, Siebuhr AS. Biological relevance of citrullinations: diagnostic, prognostic and therapeutic options. Autoimmunity. 2015;48:73–79. doi: 10.3109/08916934.2014.962024. [DOI] [PubMed] [Google Scholar]
- 5.Fox DA. Citrullination: a specific target for the autoimmune response in rheumatoid arthritis. J Immunol. 2015;195:5–7. doi: 10.4049/jimmunol.1501021. [DOI] [PubMed] [Google Scholar]
- 6.Scinocca M, Bell DA, Racape M, Joseph R, Shaw G, McCormick JK, et al. Antihomocitrullinated fibrinogen antibodies are specific to rheumatoid arthritis and frequently bind citrullinated proteins/peptides. J Rheumatol. 2014;41:270–279. doi: 10.3899/jrheum.130742. [DOI] [PubMed] [Google Scholar]
- 7.van Venrooij WJ, Pruijn GJM (2014) How citrullination invaded rheumatoid arthritis research. Arthritis Res Ther 16 [DOI] [PMC free article] [PubMed]
- 8.Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling S, Cline EN, Haug TS, Fox DA, Holoshitz J. Citrullinated calreticulin potentiates rheumatoid arthritis shared epitope signaling. Arthritis Rheum. 2013;65:618–626. doi: 10.1002/art.37814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harlow L, Rosas IO, Gochuico BR, Mikuls TR, Dellaripa PF, Oddis CV, et al. Identification of citrullinated hsp90 isoforms as novel autoantigens in rheumatoid arthritis-associated interstitial lung disease. Arthritis Rheum. 2013;65:869–879. doi: 10.1002/art.37881. [DOI] [PubMed] [Google Scholar]
- 11.Van Steendam K, Tilleman K, De Ceuleneer M, De Keyser F, Elewaut D, Deforce D. Citrullinated vimentin as an important antigen in immune complexes from synovial fluid of rheumatoid arthritis patients with antibodies against citrullinated proteins. Arthritis Res Ther. 2010;12:R132. doi: 10.1186/ar3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Beers JJBC, Schwarte CM, Stammen-Vogelzangs J, Oosterink E, Bozic B, Pruijn GJM. The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein E, myeloid nuclear differentiation antigen, and beta-actin. Arthritis Rheum. 2013;65:69–80. doi: 10.1002/art.37720. [DOI] [PubMed] [Google Scholar]
- 13.Pruijn GJ. Citrullination and carbamylation in the pathophysiology of rheumatoid arthritis. Front Immunol. 2015;6:192. doi: 10.3389/fimmu.2015.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao G, Wang D, Gu J, Shen Q, Gross SS, Wang Y. Neutral loss of isocyanic acid in peptide CID spectra: a novel diagnostic marker for mass spectrometric identification of protein citrullination. J Am Soc Mass Spectrom. 2009;20:723–727. doi: 10.1016/j.jasms.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, 3rd, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 16.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, et al. Development of criteria for the classification and reporting of osteoarthritis. Arthritis Rheum. 1986;29:1039–1049. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 17.Huang CC, Yan SH, Chen D, Chen BC, Zhao NW. Application of on-line nanoLC-IT-TOF in the identification of serum beta-catenin complex in mice scald model. PLoS One. 2012;7:e46530. doi: 10.1371/journal.pone.0046530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneko N, Yamamoto R, Sato T-A, Tanaka K. Identification and quantification of amyloid beta-related peptides in human plasma using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90:104–117. doi: 10.2183/pjab.90.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsubara T, Mita A, Minami K, Hosooka T, Kitazawa S, Takahashi K, et al. PGRN is a key adipokine mediating high fat diet-induced insulin resistance and obesity through IL-6 in adipose tissue. Cell Metab. 2012;15:38–50. doi: 10.1016/j.cmet.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Li XJ, Xu M, Zhao XQ, Zhao JN, Chen FF, Yu W, et al. Proteomic analysis of synovial fibroblast-like synoviocytes from rheumatoid arthritis. Clin Exp Rheumatol. 2013;31:552–558. [PubMed] [Google Scholar]
- 21.Shoda H, Fujio K, Shibuya M, Okamura T, Sumitomo S, Okamoto A, et al. Detection of autoantibodies to citrullinated BiP in rheumatoid arthritis patients and pro-inflammatory role of citrullinated BiP in collagen-induced arthritis. Arthritis Res Ther. 2011;13:R191. doi: 10.1186/ar3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T, Shibazaki A, Ono R, Kaisho T. HSP70 mediates degradation of the p65 subunit of nuclear factor kappaB to inhibit inflammatory signaling. Sci Signal. 2014;7:356. doi: 10.1126/scisignal.aaa5291. [DOI] [PubMed] [Google Scholar]
- 23.Noort AR, Tak PP, Tas SW. Non-canonical NF-kappa B signaling in rheumatoid arthritis: Dr Jekyll and Mr Hyde? Arthritis Res Ther. 2015;17:15. doi: 10.1186/s13075-015-0527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Headland S, Norling L, Patel H, Dalli J, Kelly S, Pitzalis C, et al. (2014) A novel mechanism for protecting the arthritic joint: microparticles deliver Annexin A1 into cartilage. FASEB J 28
- 25.Yang YH, Morand E, Leech M. Annexin A1: potential for glucocorticoid sparing in RA. Nat Rev Rheumatol. 2013;9:595–603. doi: 10.1038/nrrheum.2013.126. [DOI] [PubMed] [Google Scholar]
- 26.Patel HB, Kornerup KN, Sampaio AL, D’Acquisto F, Seed MP, Girol AP, et al. The impact of endogenous annexin A1 on glucocorticoid control of inflammatory arthritis. Ann Rheum Dis. 2012;71:1872–1880. doi: 10.1136/annrheumdis-2011-201180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Mechin MC, et al. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007;56:3541–3553. doi: 10.1002/art.22983. [DOI] [PubMed] [Google Scholar]
- 28.Slade DJ, Horibata S, Coonrod SA, Thompson PR. A novel role for protein arginine deiminase 4 in pluripotency: the emerging role of citrullinated histone H1 in cellular programming. Bioessays. 2014;36:736–740. doi: 10.1002/bies.201400057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrade F, Darrah E, Gucek M, Cole RN, Rosen A, Zhu XM. Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum. 2010;62:1630–1640. doi: 10.1002/art.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1136 kb)