Abstract
This paper presents a natural language processing (NLP) system that was designed to participate in the 2014 i2b2 De-identification Challenge. The challenge task aims to identify and classify seven main Protected Health Information (PHI) categories and 25 associated sub-categories. A hybrid model was proposed which combines machine learning techniques with keyword-based and rule-based approaches to deal with the complexity inherent in PHI categories. Our proposed approaches exploit a rich set of linguistic features, both syntactic and word surface-oriented, which are further enriched by task-specific features and regular expression template patterns to characterize the semantics of various PHI categories. Our system achieved promising accuracy on the challenge test data with an overall micro-averaged F-measure of 93.6%, which was the winner of this de-identification challenge.
Keywords: Protected Health Information (PHI), De-identification, Hybrid Model, Natural Language Processing, Clinical Text Mining
Graphical abstract
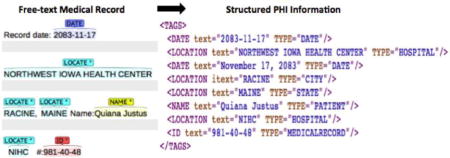
1. Introduction
Narrative clinical texts of patient medical records that contain rich clinical information (e.g., disease treatment and medication information) are gaining increasing recognition as an important component of clinical studies and many medical applications such as disease treatment and decision-making. To protect patient privacy and facilitate the dissemination of patient-specific data, it is required that Protected Health Information (PHI) should be removed from medical records before they are publicly available for non-hospital researchers. De-identification is a step that removes or replaces all the sensitive information while keeping the records otherwise intact.
The 2014 i2b2 De-identification Challenge Task1 [14] is to identify and extract various types of PHI data from clinical free-texts like patient discharge summaries, clinical notes and letters. The data released for this task consists of 1304 medical records with respect to 296 patients, of which 790 records (178 patients) are used for training, and the remaining 514 records (118 patients) for testing. The medical records are a fully annotated gold standard set of clinical narratives as shown in Figure 1. The PHI categories are grouped into seven main categories with 25 associated sub-categories. The distributions of PHI categories in the training and test sets are shown in Table 1.
Figure 1.
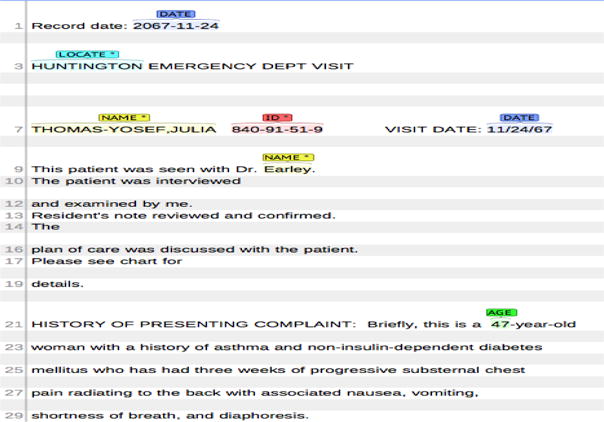
Example of clinical record with annotated PHI categories
Table 1.
Distributions of PHI categories in the training and test corpora
| PHI Category | Sub-category | Training Data | Test Data |
|---|---|---|---|
|
| |||
| DATE | DATE | 7,495 | 4,980 |
|
| |||
| NAME | DOCTOR | 2,877 | 1,912 |
| PATIENT | 1,315 | 879 | |
| USERNAME | 264 | 92 | |
|
| |||
| AGE | AGE | 1,233 | 764 |
|
| |||
| CONTACT | PHONE | 309 | 215 |
| FAX | 8 | 2 | |
| 4 | 1 | ||
| URL | 2 | 0 | |
|
| |||
| ID | MEDICALRECORD | 611 | 422 |
| IDNUM | 261 | 195 | |
| DEVICE | 7 | 8 | |
| BIOID | 1 | 0 | |
| HEALTHPLAN | 1 | 0 | |
|
| |||
| LOCATION | HOSPITAL | 1,437 | 875 |
| CITY | 394 | 260 | |
| STATE | 314 | 190 | |
| STREET | 216 | 136 | |
| ZIP | 212 | 140 | |
| ORGANIZATION | 124 | 82 | |
| COUNTRY | 66 | 117 | |
| LOCATION-OTHER | 4 | 13 | |
|
| |||
| PROFESSION | PROFESSION | 234 | 179 |
|
| |||
| Total | 17,389 | 11,462 | |
It is noted that in this dataset, each patient has 3~5 documents with different Document Creation Time (DCT), which allow a general timeline present in the patient’s medical history. The sets of longitudinal patient records are named with the combination of patient ID and document order ID, e.g., the files, ‘100-01.xml’ and ‘100-02.xml’ denote the first and second timeline record for the patient with ID ‘100’.
2. Related Research Issues in De-identification
Here we discuss a number of research issues that arise from the analysis of the i2b2 de-identification training data, and need to be dealt with during the system development.
First, due to terminological variations and irregularities in PHI terms, PHI term identification that is resolved on the basis of token level remains a challenging task. For example, the tokens ‘T-Th-Sa’ and ‘TThSa’ in fact consist of three different DATE mentions, ‘T’ [Tuesday], ‘Th’ [Thursday] and ‘Sa’ [Saturday]. The token ‘3041023MARY’ contains two different PHI category mentions, i.e. ‘3041023’ for the MEDICALRECORD, and ‘MARY’ for the HOSPITAL.
Second, in some well-formed categories like DATE, AGE, USERNAME, PHONE, ZIP, and MEDICALRECORD, a number of regular expression template patterns can be generated to capture the characteristics of such categories. However, due to lexical variations and the nonstandard ‘free’ forms used by the doctors, e.g., ‘37 yoM’, ‘37 yo Male’, ‘37 yo M’, ‘37yoM’, ‘37 y.o.m’, an additional set of morphological rules are required to cope with orthographic variants in PHI mentions.
Third, the seven main categories of PHI entities are quite different, each exhibiting distinct characteristics in lexicon, syntax, semantics, and discourse descriptions. Due to the wide variety and complexity of features inherent in different categories, a hybrid model coupled with several NLP techniques such as machine-learning approaches, keyword-based and rule/pattern-based methods, is more appropriate in this challenge task than a single language model.
Fourth, resolving ambiguity is another challenging task for the detection of PHI entities, which includes the ambiguity of PHI terms with non-PHI terms. For example, ‘9/12’ can be regarded as either a DATE instance or a medical test value, or the ambiguity between different PHI categories (i.e. inter-PHI ambiguity) such as whether the term ‘40’s’ should be considered as an AGE entity or a DATE entity (depending on context).
Fifth, we observed that quite a number of PHI mentions explicitly or implicitly correlate to each other in the challenge corpus. Several entities co-occur in a coordination-structured expression, such as ‘GQ/NV/whalen’ for different DOCTOR names and ‘EDVISITˆ84091519ˆThomas-yosef, Juliaˆ09/21/68ˆKEMPER, SYLVAN’ for the mentions in different PHI categories. Moreover, coreference relations among different mentions in the HOSPITAL, PATIENT, and DOCTOR categories are also worth investigating for the purpose of improving the accuracy of PHI recognition. For example, the terms, ‘Homestead Hospital’, ‘Homestead’, and ‘HH’ all refer to the same HOSPITAL.
Sixth, it is noticed that some PHI terms frequently appear in different timeline documents regarding the same patient, because the patient is likely to visit the same HOSPITAL or DOCTOR throughout his/her medical history. To uncover the relations among PHI terms across different timeline documents is another interesting issue to explore.
In the following sections, we will discuss how we address these research issues during system development and how the de-identification task benefits from making use of various types of relations between PHI terms discovered in the challenge corpus.
3. Methods
We developed an automated system to detect, at the token level, PHI instances from full-text medical records. The system diagram is shown in Figure 2. The system consists of four major functional process modules, which are briefly described below.
Figure 2.
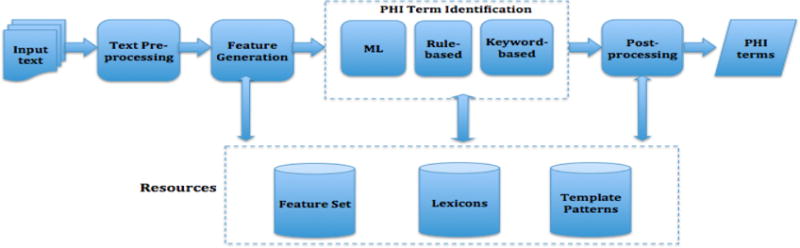
System Diagram for the De-identification Task
3.1. Text Pre-processing
This process is composed of several text pre-processing steps like sentence splitting, tokenization, POS Tagging, and shallow parsing in order to obtain word lemmas, part-of-speech (POS) tags, and syntactic chunks used for the machine learners. Moreover, a few document-level features such as section heading (e.g., PAST MEDICAL HISTORY, MEDICATIONS, PHYSICAL EXAMINATION) and sentence position (e.g., the beginning or the end of the record) are also extracted using a set of manually-crafted rules.
3.2. Feature Generation
We extracted a wide variety of linguistic features, both syntactic and word surface-oriented, which attempt to characterize the semantics of PHI terms. The feature set is further enriched by a set of task-specific features and regular expression template features extracted from the training data. The features used for the PHI classification are grouped into the following main categories:
Token Features: This type of feature includes word lemma, Part-of-Speech (POS) tag, and chunk tag of the target word, which are obtained from the Genia Tagger2.
Contextual Features: The combined features for word lemma, POS tag, and chunk tag of the neighboring tokens (within a 3-word context window of the target word) are also considered.
Orthographic Features: The features characterize word form information, e.g., capitalization (INIT-CAP, ALL-CAPS, CAPS-MIX), digit (HAS-DIGIT, ALL-DIGIT, DIGIT-PUNCTUATION, REAL-NUM, ALPHA-NUM), and special punctuation marks like ‘-’, ‘/’, ‘:’, and ‘.’ (HAS-PUNCT). In addition, regular expression template patterns (See Table 2) that describe common surface characteristics of well-structured terms in the categories, e.g., DATE, USERNAME, AGE, PHONE, MEDICALRECORD, IDNUM, ZIP, are generated. It is noted that each regular expression template pattern is treated as one orthographic feature for machine learning in different PHI categories.
Discourse Features: The features that indicate the position of the sentence in the text, the closest section heading as well as the sentences starting with some strong PHI-related contextual cues like ‘Transcribed by’, ‘CC:’, and ‘Dictated by’.
Task-specific Features: Several task-related term lists are collected, which include the full names and acronyms of US states (e.g., ‘New York’ and ‘NY’), English names of different countries (e.g., ‘Spain’) and their languages (e.g., ‘Spanish’), the full names and associated abbreviations regarding week (e.g., ‘Tuesday’, ‘Tu’), month (e.g., ‘January’, ‘Jan’) and season (e.g., ‘Winter’). Moreover, lexical cues with respect to individual PHI categories are also taken into account as an important type of task-specific features. Lexical cues are trigger words that indicate the occurrence of a particular PHI category, e.g., ‘Dr.’, ‘MD’ for DOCTOR, and ‘street’ and ‘road’ for STREET. Such lexical cues are directly collected from the surrounding contexts of the target PHI terms and are filtered according to their occurrence frequency and the importance associated with each PHI category. The tf-idf-statistics was employed to extract relevant keyword lists in terms of different PHI categories in the training corpus.
Table 2.
Part of regular expression template patterns
| Regular Expression Template | Example | |
|---|---|---|
| DATE | yyyy-mm-dd, yyyy, yy-yy [m | mm]-[d | dd]-[yy | yyyy] [m | mm]/[d | dd]/[yy | yyyy] [m | mm]/[yy | yyyy] m/’yy, ‘yy, [yy | yyyy] ‘s, [yy | yyyy]s [m | mm].[d | dd].yy [d | dd] [Alpha-mm] [yy | yyyy] [Alpha-mm] [yyyy] |
2059-01-10, 2014, 65–68, 01-01-93, 1-4-76, 5-20-75, 11-4-1983, 4-8-2056 01/02/2092, 3/12/2092, 10/02/93, 2/4/82 01/01, 1/07, 3/2133, 12/1965 2/’95, ‘63, 60’s, 2060’s, 54s, 2060s 10.25.78, 3.23.64 10-Feb-2011, 5-March-2054, 30Aug71, 10 Feb 2011 Sep-1976, April 2072, April of 2011, Oct. ‘74 |
| USERNAME | Alpha[2–3]Digit[1–3] | XO71, RM5, MU830, vmf47, yd42 |
| AGE | DigitAlpha Digit year old Digit [month | week | mos] Digit [s |’s] Digit[y]Digit[m] |
87yo, 73y.o., 70y/o, 77ym 72-year-old, 57-yr-old, 72 year old, 72year old 22 months, 6 weeks, 9 mos 20s, 60 s, 40’s 49y7.7m |
| PHONE | Digit{3} Digit{3} Digit{4} Digit{3} Digit{4} Digit{3} Digit{3} Digit{4} Digit{5} |
123-102-2039, 503 155-7742, (842) 544-2703, 895.376.3157, 942 077 9578 120-2345-120 829-1293 48125, 5-6394, #24104 |
| MEDICALRECORD | Digit{6–7} Digit{3–4}Aphpa{1–2}Digit{4–5} Alpha{2–4}Digit{3–8} |
9290228, 94482315, 7-351769, 7-9689009, 981-40-48, 948-59-58-0, 879 20 76, 481 85 43 7 4824E7560, 4894U89067, 2593:A79604 CK759182, EO25834097, HW988, NXO 2-359 |
| IDNUM | Digit{1–2}[−]Digit{6–8} Digit{3–8} DigitAlpha AlphaDigit AlphaDigit[/]Digit Alpha[:]Alpha[:]Digit Alpha[:]Digit[:]Digit |
3-638570, 2-22566361, 33-437857, 36-33387396 224, 23755, 2048395, 28618552 5-0269342 GC, 204QY ZOXU6, YRUK 7 BE249/3185, CG993/95284, pf 0760305 CCL:FG:1879, PC:RP: 1660 DE:38535 :70, CH:18306 :0077 |
| ZIP | Digit{5} Digit{5}[−]Digit{4} |
12151 19741-6273 |
3.3. A Hybrid Model for PHI Term Identification
We treat the de-identification problem as the identification and classification of PHI terms at the token level. For the PHI categories with sufficient amount of available training data, we employed a machine-learning (ML) algorithm named Conditional Random Fields (CRFs), implemented by the CRF++ package3. Each word token in a sentence is assigned one of the so-called BIO scheme tags: B (the first word of a PHI entity mention), I (inside an entity mention), O (outside, not in an entity mention). Several CRFs-based PHI classifiers are created, each of which is targeted for the sub-categories under one particular main PHI category. The PHI categories that use the CRFs algorithm can be found in Table 3, discussed in the following subsection.
Table 3.
NLP approaches used in individual PHI categories at different processing stages
| Category | Sub-category | Term Identification
|
Post-processing
|
|||
|---|---|---|---|---|---|---|
| ML (CRFs) | Rule/Pattern | Keywords | Term Extraction | Trusted PHI | ||
|
| ||||||
| DATE | DATE | ✓ | ✓ | ✓ | ✓ | ✓ |
|
| ||||||
| NAME | DOCTOR | ✓ | – | ✓ | ✓ | ✓ |
| PATIENT | ✓ | – | ✓ | – | ✓ | |
| USERNAME | ✓ | ✓ | – | – | – | |
|
| ||||||
| LOCATION | HOSPITAL | ✓ | – | ✓ | – | ✓ |
| CITY | ✓ | – | ✓ | – | – | |
| STATE | ✓ | – | ✓ | – | – | |
| STREET | ✓ | – | ✓ | – | – | |
| ZIP | ✓ | ✓ | – | – | – | |
| ORGANIZATION | ✓ | – | ✓ | – | – | |
| COUNTRY | ✓ | – | ✓ | – | – | |
| LOCATION-OTHER | – | – | ✓ | – | – | |
|
| ||||||
| AGE | AGE | ✓ | ✓ | ✓ | ✓ | – |
|
| ||||||
| ID | MEDICALRECORD | ✓ | ✓ | ✓ | ✓ | ✓ |
| IDNUM | ✓ | ✓ | ✓ | – | – | |
| DEVICE | – | ✓ | ✓ | – | – | |
| BIOID | – | ✓ | – | – | – | |
| HEALTHPLAN | – | – | ✓ | – | – | |
|
| ||||||
| CONTACT | PHONE | ✓ | ✓ | ✓ | – | ✓ |
| FAX | – | ✓ | – | – | – | |
| – | ✓ | – | – | – | ||
| URL | – | – | ✓ | – | – | |
|
| ||||||
| PROFESSION | PROFESSION | ✓ | – | ✓ | – | – |
As described earlier, we exploited a wide range of linguistic features to capture the characteristics of different PHI categories. The details about the feature types used for the recognition of individual categories are given in an online data supplement available on the JBI web site. A total of 220.1m features are extracted from different PHI categories, that is, 0.6m features for DATE, 4.0m features for NAME, 106m features for LOCATION, 2.0m features for AGE, 3.3m features for ID, 2.0m features for CONTACT, and 2.2m features for PROFESSION, respectively. The LOCATON category generates the most features due to multiple sub-categories.
Moreover, for the PHI categories (e.g., FAX, EMAIL, DEVICE, BIOID, etc.) with few sample instances, keyword spotting and rule-based approach are more appropriate methods to detect PHI-related phrases in the text. Keyword list and PHI-related regular expression patterns are manually generated from the training data.
3.4. Post-processing
At the stage of post-processing, several methods are used either to correct the errors at the term identification stage or to find more potential PHI candidates:
-
Token-level entity extraction from identified PHI markups
As discussed before, due to irregularities and non-standard forms in PHI terms, some PHI terms in certain categories are required to further process and are extracted from the PHI-related tokens labeled by the PHI classifiers. For example, [DATE]: ‘MWFS’ → ‘M’, ‘W ‘, ‘F’, ‘S’; [AGE]: ‘70yoM’ → ‘70’; [MEDICALRECORD]: ‘MR:6746781’ → ‘ 6746781’; [DOCTOR]: ‘GQ/NV/whalen’ → ‘GQ’, ‘NV’, ‘Whalen’.
-
Generation of trusted PHI terms
Similar to other de-identification work [15], we also create a trusted PHI term list to help find more PHI terms that are missed at the earlier stage of term identification. A trusted PHI term is a highly unambiguous term associated with just one PHI category. It is assumed that all the occurrences of a trusted PHI term in a medical record will be considered as TRUE positives and be assigned with a valid label to the associated PHI category. The trusted PHI terms are determined using several strategies:- If a term matches a reliable template pattern identified from the training data, e.g., the DATE pattern ‘yyyy-mm-dd’, this term is considered as a trusted term to the target PHI category. This strategy is applied to the well-represented PHI categories, such as DATE, AGE, PHONE, and MEDICALRECORD.
- If a string matches a regular expression pattern that indicates a strong association to one or more PHI categories, then all the PHI-related terms contained in the matching string are considered as trustable ones. For instance, In the string ‘EDVISITˆ84091519ˆThomas-yosef, Juliaˆ09/21/68ˆKEMPER, SYLVAN’, all the terms ‘84091519’, ‘Thomas-yosef, Julia’, ‘09/21/68’, and ‘KEMPER, SYLVAN’ are treated as trusted PHI terms for the MEDICALRECORD, PATIENT, DATE, and DOCTOR categories, respectively.
- If a term is recognized as a valid PHI mention for more than one time in the same medical record, we assume that this is a trusted PHI term. This strategy is employed just for the PATIENT, DOCTOR, and HOSPITAL categories.
- If Term A (e.g., ‘Xai Dunn’) and Term B (e.g., ‘X. Dunn’) are identified as the same PHI category, and these two terms are coreferent to each other, both terms should be regarded as trusted PHI terms. This method is used in the PATIENT, DOCTOR, and HOSPITAL categories. Coreference relations among PHI terms were predicted by the adapted coreference resolution system [20] developed for the 2011 i2b2 Coreference Challenge.
-
Extension of the trusted PHI term list
We extend the existing trusted PHI term list by applying a set of permutation rules based on the observations in the training data:- In PATIENT and DOCTOR categories, several additional names can be generated based on the full name of a person, e.g., ‘Harlan Valdez’ → {‘Harlan Valdez’, ‘Valdez, Harlan’, ‘H. Valdez’, ‘Valdez’}
- In the HOSPITAL category, more candidates are created by removing some informative words from long multi-word terms (generally 4 or more words) with the suffix like ‘Center’, ‘Hospital’, and ‘Clinic’. For example, ‘Atlantic North Rehabilitation Center’ → {‘Atlantic North Rehabilitation Center’, ‘Atlantic North Rehabilitation’, ‘Atlantic North’, ‘ANRC’}
-
Find more potential PHI candidates using trusted PHI terms
In the generated trusted PHI term list, each trusted term is associated with a specific PHI category and the file name in which it is detected. Given a trusted PHI term, all the occurrences of this term in the same document will be marked as a valid candidate with respect to the associated PHI category. For the PATIENT, DOCTOR, and HOSPITAL categories, the search will be expanded to other relevant timeline documents regarding the same patient to find more unidentified PHI terms.
In summary, to handle the complicated characteristics inherent in various PHI subcategories, we proposed a hybrid model that explores several NLP approaches to uncover as many potential PHI terms as possible in narrative clinical texts. The approaches used in individual PHI sub-categories at different processing stages are summarized in Table 3.
4. Evaluations, Results, and Discussions
4.1. Evaluation Measures
De-identification performance is evaluated using precision (P), recall (R), and F-measure (F) at both token and entity level. Entity-level measures give credit to the predicted entity mentions of multiple words that match the ground truth whereas token-level measures consider the correction of each token (i.e. word) in a mention separately. Entity-level performance is studied via a strict evaluation and a relaxed evaluation. The strict metric denotes the exact string match of the predicted entity mention against the gold standard, while the relaxed metric means the approximate string matching that allows for some leeway (1–2 characters) at the end of a mention string.
All the evaluations are performed against the gold standard of the I2B2 test data. System performance is conducted based on two types of PHI categories:
I2B2 PHI categories: All the seven main PHI categories and 25 associated sub-categories as discussed before.
HIPAA PHI categories: the I2B2 PHI categories that are compliant to the Administrative Simplification Regulations promulgated under the Health Insurance Portability and Accountability Act (HIPAA). HOSPITAL and PROFESSION categories are excluded.
4.2. Overall Performance of the System
Overall performance of the developed system is measured using Micro and Macro evaluation matrices:
Micro: all the tags are evaluated at the entire corpus level
Macro: the tags are first evaluated at the document level, and then the scores are averaged across the corpus
We evaluated overall system performance on distinguishing PHI from non-PHI. All PHI categories are put together and treated as an overall PHI category. Precision, recall and F-measure are separately calculated based on the overall PHI category. Table 4 demonstrates that the performance at the token level is generally better than that of the entity level. Part of the reason for that is due to the inexact string matching in terms of multi-word PHI terms, especially for the PATIENT, DOCTOR, HOSPITAL, and STREET categories containing a high number of multi-word terms. The slight difference in performance between the strict entity level and the relaxed entity level implies that the system is capable of extracting the entity name that is contained within a token, e.g., ‘79yo’ → ‘79’.
Table 4.
The overall performance of both i2b2 and HIPAA PHI categories on the i2b2 test set
| Token Level | Strict Entity Level | Relaxed Entity Level | |||||
|---|---|---|---|---|---|---|---|
| Micro | Macro | Micro | Macro | Micro | Macro | ||
| I2B2 PHI Categories | Precision | 0.9806 | 0.9815 | 0.9645 | 0.9653 | 0.9665 | 0.9668 |
| Recall | 0.9414 | 0.9414 | 0.9092 | 0.9156 | 0.9111 | 0.9171 | |
| F-measure | 0.9611 | 0.9611 | 0.9360 | 0.9398 | 0.9380 | 0.9413 | |
| HIPAA PHI Categories | Precision | 0.9889 | 0.9864 | 0.9763 | 0.9751 | 0.9784 | 0.9768 |
| Recall | 0.9629 | 0.9599 | 0.9390 | 0.9425 | 0.9411 | 0.9441 | |
| F-measure | 0.9570 | 0.9730 | 0.9573 | 0.9585 | 0.9594 | 0.9602 | |
It is not surprising that the HIPAA PHI categories generally perform better than the I2B2 PHI categories when the non-HIPAA categories such as HOSPITAL and PROFESSION are excluded from the evaluation. The Non-HIPAA categories exhibit more difficulty in terms of PHI term identification compared with the HIPAA-compliant categories (see the discussion in Section 4.3).
4.3. Performance on Individual PHI Categories
In addition to overall system evaluation on the detection of PHI entity names, we evaluated the system for its ability to recognize the exact category of PHI. Tables 5 and 6 provide the performances of seven main PHI categories and their associated sub-categories. Among the main PHI categories, DATE performs best with the highest F-measure of 0.9764. It is followed by AGE, CONTACT, and NAME categories that are not significantly different from each other in F-measure (above 0.94). PROFESSION performs worst, which suffers from a lack of training examples and the fact that no informative features were found in the training data.
Table 5.
The overall performance of main i2b2 PHI categories on the i2b2 test set at the strict entity level
| Category | #Expected | #Predicted | #Correct | Precision | Recall | F-measure |
|---|---|---|---|---|---|---|
| DATE | 4,980 | 4,877 | 4,812 | 0.9867 | 0.9663 | 0.9764 |
| NAME | 2,883 | 2,726 | 2,643 | 0.9696 | 0.9168 | 0.9424 |
| LOCATION | 1,813 | 1,523 | 1,380 | 0.9061 | 0.7612 | 0.8273 |
| AGE | 764 | 728 | 707 | 0.9712 | 0.9254 | 0.9477 |
| ID | 625 | 611 | 573 | 0.9378 | 0.9168 | 0.9272 |
| CONTACT | 218 | 208 | 201 | 0.9663 | 0.9220 | 0.9437 |
| PROFESSION | 179 | 132 | 107 | 0.8106 | 0.5978 | 0.6881 |
| Total | 11,462 | 10,805 | 10,423 | 0.9645 | 0.9092 | 0.9360 |
Table 6.
The overall performance of i2b2 PHI sub-categories on the i2b2 test set at the strict entity level
| Category | Sub-category | #Expected | #Predicted | #Correct | P | R | F |
|---|---|---|---|---|---|---|---|
|
| |||||||
| DATE | DATE | 4,980 | 4,877 | 4,812 | 0.9867 | 0.9663 | 0.9764 |
|
| |||||||
| NAME | DOCTOR | 1,912 | 1,808 | 1,758 | 0.9723 | 0.9195 | 0.9452 |
| PATIENT | 879 | 830 | 797 | 0.9602 | 0.9067 | 0.9327 | |
| USERNAME | 92 | 88 | 88 | 1.0000 | 0.9665 | 0.9778 | |
|
| |||||||
| LOCATION | HOSPITAL | 875 | 807 | 727 | 0.9009 | 0.8309 | 0.8644 |
| CITY | 260 | 214 | 184 | 0.8598 | 0.7077 | 0.7764 | |
| STATE | 190 | 167 | 154 | 0.9222 | 0.8105 | 0.8627 | |
| STREET | 136 | 134 | 132 | 0.9851 | 0.9706 | 0.9778 | |
| ZIP | 140 | 136 | 136 | 1.0000 | 0.9714 | 0.9855 | |
| ORGANIZATION | 82 | 35 | 25 | 0.7143 | 0.3049 | 0.4274 | |
| COUNTRY | 117 | 28 | 22 | 0.7857 | 0.1880 | 0.3034 | |
| LOCATION-OTHER | 13 | 1 | 0 | 0.0000 | 0.0000 | 0.0000 | |
|
| |||||||
| AGE | AGE | 764 | 728 | 707 | 0.9712 | 0.9254 | 0.9477 |
|
| |||||||
| ID | MEDICALRECORD | 422 | 427 | 412 | 0.9649 | 0.9763 | 0.9706 |
| IDNUM | 195 | 182 | 158 | 0.8681 | 0.8103 | 0.8382 | |
| DEVICE | 8 | 3 | 3 | 1.0000 | 0.3750 | 0.5455 | |
|
| |||||||
| CONTACT | PHONE | 215 | 203 | 199 | 0.9803 | 0.9256 | 0.9522 |
| FAX | 2 | 2 | 1 | 0.5000 | 0.5000 | 0.5000 | |
| 1 | 1 | 1 | 1.0000 | 1.0000 | 1.0000 | ||
|
| |||||||
| PROFESSION | PROFESSION | 179 | 132 | 107 | 0.8106 | 0.5978 | 0.6881 |
For I2B2 PHI sub-categories, USERNAME, STREET, and EMAIL are the best performers in terms of precision with a perfect score of 1.0. USERNAME, STREET, ZIP, and MEDICALRECORD give the best recall up to 0.97. DATE, USERNAME, STREET, ZIP, and MEDICALRECORD are the top five categories with respect to F-measure. All of them have an F-measure of over 0.975.
In general, the categories (e.g., DATE, AGE, USERNAME, PHONE, ZIP, and MEDICALRECORD) that heavily rely on regular expression template patterns perform well, and all achieve scores above 0.9 in terms of both precision and recall. It means that regular expression template patterns, when combined with other orthographic features, can be quite effective in predicting these PHI categories. Moreover, lexical trigger words play a crucial role in entity detection of the PATIENT, DOCTOR, and STREET categories, and help these categories to achieve high F-measures above 0.93.
Performances are relatively poor for some sub-categories within the LOCATION category due to the lack of enough training samples and the complexity of term expression (e.g., HOSPITAL). For PROFESSION, poor performance is partly caused by the presence of relatively infrequent and broadly defined training examples. Our keyword list for PROFESSION was directly collected from the training corpus, which contains merely 187 words. Many general occupation keywords (e.g., ‘veteran’, ‘cashier’, ‘instructor’) are missed in the current keyword list, and thus could not be recognized in the test corpus.
Moreover, the system has difficulty in recognizing the categories that have few examples in the training data, and fails to recognize most of the entity names in such categories as ORGANIZATION and COUNTRY. For example, some infrequent country or country-related names like ‘trinidad and tobago’, ‘Puerto Rican’, ‘Kazakhstani’, and ‘Kazakhstan’ are not identified by our system. The main reason for that is due to the insufficiency of our collected COUNTRY list from which such country terms are excluded.
LOCATION-OTHER performs worst among all the PHI sub-categories. The identification of LOCATION-OTHER heavily relies on a keyword list that is directly generated from limited training examples. However, out manual examination of both training and test samples reveals that most of the test instances are unseen in the training data.
To investigate how the keyword-based matching relies on the completeness and comprehensiveness of the related term lists, we enriched two sets of keyword lists for both COUNTRY and PROFESSION by making use of some existing knowledge resources (e.g., UMLS database) or web sources after the challenge competition. For COUNTRY, country/region names and related language and nationality information were collected. A new list of 1252 country-related terms was created, which resulted in 72 missing country names being detected from the test data. The F-measure of COUNTRY was significantly increased from 0.3034 to 0.8818. Some misspelled or irregular country names, e.g., ‘Ghanna’, ‘Khazakhstani’, and ‘Equadorian’ could not be identified by the system.
We also extended the PROFESSION list to a total of 1252 terms, which help find 23 more potential terms in the test data, and thus increase the F score by 9.1%. However, the system still has some difficulty in recognizing some ambiguous terms (e.g., ‘mapping’, ‘intern’ and ‘banking’) and imprecise/relaxed terms (e.g., ‘managing production’, and ‘commercial diving’).
4.4. The Impact of Post-processing
As discussed previously, we employed two strategies at the post-processing stage in order to further improve the overall system accuracy: one is to extract PHI terms from the PHI-related tokens, and another is to discover more potential candidates using trusted PHI terms. To gain insight into the strength of the post-processing module, we compared system performance on some specific PHI sub-categories both before and after post-processing, to determine whether the de-identification system could benefit from the post-processing stage, as shown in Table 7.
Table 7.
System comparison of two stages: Before Post-processing vs. After Post-processing
| Sub-category | Before Post-processing | After Post-processing | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-measure | Precision | Recall | F-measure | |
| DATE | 0.9845 | 0.8643 | 0.9204 | 0.9867 | 0.9663 | 0.9764 |
| DOCTOR | 0.9701 | 0.8674 | 0.9158 | 0.9723 | 0.9195 | 0.9452 |
| PATIENT | 0.9682 | 0.8826 | 0.9234 | 0.9602 | 0.9067 | 0.9327 |
| HOSPITAL | 0.8288 | 0.7539 | 0.7895 | 0.9009 | 0.8309 | 0.8644 |
| AGE | 0.7183 | 0.6529 | 0.6840 | 0.9712 | 0.9254 | 0.9477 |
| MEDICALRECORD | 0.9446 | 0.9314 | 0.9379 | 0.9649 | 0.9763 | 0.9706 |
| PHONE | 0.9873 | 0.9070 | 0.9454 | 0.9803 | 0.9256 | 0.9522 |
In general, the token-level PHI term extraction has a slight impact on system performance due to limited mention instances except for the AGE sub-category that has a quite number of mentions appearing in the forms like ‘78yo’ or ‘77yM’. As expected, the trusted PHI term method shows excellent performance in terms of recall improvement when this step results in more new PHI terms found in the records. Table 7 illustrates that the best results are achieved in the AGE subcategory, raising accuracy considerably after conducting the post-processing step with Precision, Recall and F-measure of 0.25, 0.27 and 0.26, respectively. The HOSPITAL sub-category is the second best with substantial F-measure improvement up to 0.08. Other sub-categories also have an improvement in the range 0.1 ~ 0.5 in terms of F-measure.
4.5. Error Analysis
As shown in Table 8, we performed a detailed error analysis for system output, and grouped the errors into several broad classes:
-
Class label errors (misclassification/false positives)
This kind of error occurs when a term that originally belongs to Category A is wrongly assigned with the class label of Category B. Most such errors fall into inter-PHI ambiguity instances. It is expected that DOCTOR and PATIENT are highly ambiguous to each other due to the similarity in terms of name form. 17 PATIENT names are wrongly identified as DOCTOR whereas 8 DOCTOR names are labeled as PATIENT. Among all the main categories, LOCATION is the most ambiguous category in which several sub-categories (e.g., COUNTRY, CITY, ORGANIZATION, and HOSPITAL) are easily ambiguous to other sub-categories. For example, 18 COUNTRY names are incorrectly assigned to the STATE, CITY, and DOCTOR categories respectively.
-
Missing tag errors (false negatives)
In a total of 1,161 errors produced by the system in the test dataset, 69.7% of the errors (880 false negatives) are missing tags. Missing tag errors can fall into the following main categories:- In DOCTOR and PATIENT, quite a number of single-word person names (e.g., ‘talbot’ and ‘ray’) are much harder to detect compared with full names that have two or more words due to the lack of the context (i.e., surrounding words and characters) and morphology of the words.
- The system still has difficulty in identifying short terms, especially abbreviations in HOSPITAL and DOCTOR (e.g., ‘WA’ and ‘DG’), which are ambiguous to non-PHI medical test terms.
- Rare-frequency terms that do not conform to the generated regular expression template patterns in certain categories cannot be recognized by the system, e.g., ‘RICO,HELEN U’ [DOCTOR].
- Unseen terms in the test data are another source of hard instances in PHI term detection, especially for the terms in the CITY, COUNTRY, ORGANIZATION, and PROFESSION categories.
-
Spurious tag errors (false positives)
Spurious tag errors are mainly caused by partial matches of long multi-word terms, e.g., ‘September 13, 2070’ [DATE] and ‘House Of Calvary Hospital’ [HOSPITAL]. A few of cross-line mentions, e.g., ‘Peter … Vaderberg’ and ‘April 12, … 2091’, are also not correctly recognized. Moreover, a number of non-PHI medical terms, such as ‘Sacred Heart’, ‘TDK’, and ‘Nutrition Clinic’ are marked with wrong PHI class labels like DOCTOR and HOSPITAL.
Table 8.
The confusion matrix of our model on the i2b2 test set at the strict entity level
| Output | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||||||
| Key | Dt | Dct | Pt | Un | Hpt | Ct | Stt | Str | Zip | Org | Ctr | Lct | Age | Mdr | Idn | Dv | Ph | Fx | Em | Prf | Missing | Total |
|
| ||||||||||||||||||||||
| Dt | 4812 | 1 | 1 | 106 | 4980 | |||||||||||||||||
| Dct | 1758 | 8 | 1 | 1 | 144 | 1912 | ||||||||||||||||
| Pt | 17 | 797 | 65 | 879 | ||||||||||||||||||
| Un | 2 | 88 | 2 | 92 | ||||||||||||||||||
| Hpt | 1 | 727 | 7 | 1 | 139 | 875 | ||||||||||||||||
| Ct | 1 | 184 | 5 | 1 | 1 | 68 | 260 | |||||||||||||||
| Stt | 9 | 154 | 27 | 190 | ||||||||||||||||||
| Str | 132 | 4 | 136 | |||||||||||||||||||
| Zip | 136 | 2 | 2 | 140 | ||||||||||||||||||
| Org | 9 | 1 | 25 | 47 | 82 | |||||||||||||||||
| Ctr | 2 | 6 | 10 | 22 | 77 | 117 | ||||||||||||||||
| Lct | 1 | 0 | 12 | 13 | ||||||||||||||||||
| Age | 707 | 57 | 764 | |||||||||||||||||||
| Mdr | 412 | 2 | 8 | 422 | ||||||||||||||||||
| Idn | 6 | 158 | 31 | 195 | ||||||||||||||||||
| Dv | 3 | 5 | 8 | |||||||||||||||||||
| Ph | 1 | 199 | 1 | 14 | 215 | |||||||||||||||||
| Fx | 1 | 1 | 0 | 2 | ||||||||||||||||||
| Em | 1 | 0 | 1 | |||||||||||||||||||
| Prf | 107 | 72 | 179 | |||||||||||||||||||
| Spurious | 65 | 28 | 25 | 0 | 69 | 5 | 3 | 2 | 0 | 4 | 5 | 0 | 20 | 9 | 20 | 0 | 1 | 0 | 0 | 25 | 281 | |
| Total | 4877 | 1898 | 830 | 88 | 807 | 214 | 167 | 134 | 136 | 35 | 28 | 1 | 728 | 427 | 182 | 3 | 203 | 2 | 1 | 132 | 880 | 11462 |
(where Dt=DATE, Dct=DOCTOR, Pt=PATIENT, Un=USERNAME, Hpt=HOSPITAL, Ct=CITY, Stt=STATE, Str=STREET, Org=ORGANIZATION, Ctr=COUNTRY, Lct=LOCATION-OTHER, Mdr=MEDICALRECORD, Id= IDNUM, Dv=DEVICE, Ph=PHONE, Fx=FAX, Em=EMAIL, Prf=PREFESSION)
5. Related Work
As manual de-identification can no longer keep up with the tremendous growth in the use of Electronic Health Records (EHR) for clinical research, automatic algorithms have received much attention in recent years. Although many automated systems adopt different approaches to de-identify specific PHI types, the main techniques used to detect PHI terms can be classified into two groups of methodologies: rule/pattern based and machine learning based.
Rule/pattern based methods [2, 7, 9, 12, 13] typically make use of dictionaries and hand-coded rules to match PHI patterns in the texts. They need little or no training data, and can be easily modified (e.g., by adding new rules or adapting existing rules for new data structure). However, they usually require additional data curation or annotation by experienced domain experts and have limitations. The generated rules practically contain already known domain-related knowledge and patterns, which provide inflexible predictive power for a large-scale dataset in which new unknown knowledge is created or added over time, and thus new rules and the minor or major modification of existing rules are required.
Machine learning-based approaches can automatically recognize PHI patterns based on statistical learning of the characteristics of data using different ML algorithms such as Conditional Random Fields (CRFs) [1, 3, 8, 9], Maximum Entropy [16], Support Vector Machines (SVM) [18], and Decision Trees [10, 15]. But they require manual annotation of large training examples with pre-labeled identifiers, which are prohibitively expensive and time-consuming.
Ferrández et al. [5] compared and evaluated system performance of five text de-identification systems “out-of-the-box” using a corpus of VHA Clinical documents. Uzuner et al. [17] summarized several de-identification systems that participated in the 2006 i2b2 de-identification challenge. Similar to our work, Ferrández et al. [6] implemented a best-of-breed (BoB) automated text de-identification system that takes advantage of rule-based and machine learning-based approaches to obtain better results. Deleger et al. [4] conducted de-identification experiments on a large-scale clinical corpus that consists of a wide variety of clinical notes (over 22 different types) to examine the accuracy and generalizability of NLP approaches under the situation of heterogeneous document sources. They found that the performance of the automatic system competes with that of the human annotators, and there is little impact of automated de-identification on subsequent information extraction tasks. More details of de-identification techniques and system analysis can be found in the research review paper by Meystre et al. [11].
Our de-identification work differs from relevant previous work in two aspects. Firstly, regular expression templates play several roles during the PHI detection process. Not only do they function as distinguishing features in both machine learning and rule/pattern approaches, but also they are used to help find more potential instances in the post-processing step. Secondly, we exploit several useful syntactic and semantic relations at the entity level (e.g., coordination and co-reference relations between entities) or document level (e.g., the timeline present in the patient’s medical history) in order to discover more trusted PHI terms, thus improving the system recall.
6. Conclusions
In this paper, we introduced a de-identification system that was designed to recognize and classify Protected Health Information (PHI) present in free-text medical records. We proposed a hybrid model that combines machine learning technique with other NLP approaches such as keyword-based and rule-based approaches to cope with the complexity inherent in various PHI categories. A rich set of linguistic features are extracted to characterize the semantics of a variety of PHI categories, which are enriched by task-specific features as well as regular expression template patterns. At the post-processing step, a trusted PHI term set that is generated by making use of various types of relations between PHI terms is used to further improve the system accuracy. Our developed system achieved an overall micro-average F-measure of 0.936, which was ranked first in this de-identification challenge.
The results reported here show that the proposed hybrid approach is capable of accurately identifying PHI terms from text. However, a number of interesting issues remain to be resolved. One of the research issues is how to distinguish short PHI terms (e.g., abbreviations) from other non-PHI terms in medical records. A possible solution is to create lists of non-PHI terms such as common words and UMLS terms useful for determining ambiguous PHI short terms. In addition, it is interesting to investigate whether our system can be easily extended to detect PHI mentions with good performance on more heterogeneous document sources to assess generalizability across clinical documents. Finally, to facilitate the public to use this de-identification system, we plan to implement it as a web service on our university web server for public access in the future.
Supplementary Material
Highlights.
A model to automatically detect protected health information in clinical texts
Machine learning techniques combined with keywords and rule-based approaches
7 main PHI categories with 25 associated sub-categories are identified
Achieving an overall micro-averaged F-measure of 93.6%
The winner of 2014 i2b2 de-identification challenge task
Acknowledgments
The work was supported by funding through the Advanced Data Analysis Centre, University of Nottingham, UK. The author would like to thank the 2014 i2b2 challenge organizers for providing such an invaluable clinical dataset and this research opportunity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aberdeen J, Bayer S, Yeniterzi R, et al. The MITRE Identification Scrubber Toolkit: design, training, and assessment. Int J Med Inform. 2010;79:849–59. doi: 10.1016/j.ijmedinf.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Beckwith BA, Mahaadevan R, Balis UJ, Kuo F. Development and evaluation of an open source software tool for deidentification of pathology reports. BMC Med Inform Decis Mak. 2006;6:12. doi: 10.1186/1472-6947-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benton A, Hill S, Ungar L, et al. A system for de-identifying medical message board text. BMC Bioinformatics. 2011;12(Suppl 3):S2. doi: 10.1186/1471-2105-12-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deleger L, Molnar K, Savova G, et al. Large-scale evaluation of automated clinical note de-identification and its impact on information extraction. J Am Med Inform Assoc. 2013;20:84–94. doi: 10.1136/amiajnl-2012-001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrández O, South BR, Shen S, Friedlin FJ, Samore MH. Evaluating current automatic de-identification methods with Veteran’s health administration clinical documents. BMC Med Res Methodol. 2012;12:109. doi: 10.1186/1471-2288-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrández O, South BR, Shen S, Friedlin FJ, Samore MH, Meystre SM. BoB, a best-of-breed automated text de-ideintification system for VHA clinical documents. J Am Med Inform Assoc. 2013;20:77–83. doi: 10.1136/amiajnl-2012-001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedlin FJ, Mcdonald CJ. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc. 2008;15(5):601–10. doi: 10.1197/jamia.M2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner J, Xiong L. HIDE: an integrated system for health information DE-identification. The 21st IEEE International Symposium on Computer-Based Medical Systems. 2008;2008:254–9. [Google Scholar]
- 9.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol. 2004;121:176–8. doi: 10.1309/E6K3-3GBP-E5C2-7FYU. [DOI] [PubMed] [Google Scholar]
- 10.McMurry AJ, Fitch B, Savova G, Kohane IS, Reis BY. Improved de-identification of physician notes through integrative modeling of both public and private medical text. BMC Med Inform Decis Mak. 2003;13:112. doi: 10.1186/1472-6947-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol. 2010;10:70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison FP, Li L, Lai AM, Hripcsak G. Repurposing the clinical record: can an existing natural language processing system de-identify clinical notes? J Am Med Inform Assoc. 2009;16:37–9. doi: 10.1197/jamia.M2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neamatullah I, Douglass MM, Lehman LH, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak. 2008;8(1):32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stubbs A, Xu H, Kotfila C, Uzuern O. Practical applications for NLP in Clinical Research: the 2014 i2b2/UTHealth shared tasks. The 2014 i2b2 Challenge Workshop. 2014 [Google Scholar]
- 15.Szarvas G, Farkas R, Busa-Fekete R. State-of-the-art Anonymization of Medical Records Using an Iterative Machine Learning Framework. J Am Med Inform Assoc. 2007;14(5):574–80. doi: 10.1197/j.jamia.M2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taira RK, Bui AA, Kangarloo H. Identification of patient name references within medical documents using semantic selectional restrictions. Proc AMIA Symp. 2002;2002:757–61. [PMC free article] [PubMed] [Google Scholar]
- 17.Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc. 2007;14(5):550–63. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uzuner Ö, Sibanda TC, Luo Y, Szolovits P. A de-identifier for medical discharge summaries. Artificial intelligence in medicine. 2008;42(1):13–35. doi: 10.1016/j.artmed.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wellner B, Huyck M, Mardis S, et al. Rapidly Retargetable Approaches to De-identification in Medical Records. J Am Med Inform Assoc. 2007;14(5):550–63. doi: 10.1197/jamia.M2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Wills A, de-Roeck A, Nuseibeh B. A System for Coreference Resolution in Clinical Documents. 2011 i2b2/VA/Cincinnati Medical NLP Challenge workshop. 2011 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


