Abstract
Coronary artery disease (CAD) is the leading cause of death in both the UK and worldwide. The detection of related risk factors and tracking their progress over time is of great importance for early prevention and treatment of CAD. This paper describes an information extraction system that was developed to automatically identify risk factors for heart disease in medical records while the authors participated in the 2014 i2b2/UTHealth NLP Challenge. Our approaches rely on several nature language processing (NLP) techniques such as machine learning, rule-based methods, and dictionary-based keyword spotting to cope with complicated clinical contexts inherent in a wide variety of risk factors. Our system achieved encouraging performance on the challenge test data with an overall micro-averaged F-measure of 0.915, which was competitive to the best system (F-measure of 0.927) of this challenge task.
Keywords: Risk factors, Heart Disease, Machine Learning, Rule-based Approach, Hybrid Model, Natural Language Processing, Clinical Text Mining
Graphical abstract
1. Introduction
Coronary artery disease (CAD), i.e. Coronary heart disease (CHD), is the leading cause of death in both the UK and worldwide. It is responsible for more than 73,000 deaths in the UK each year. About 1 in 6 men and 1 in 10 women die from CAD. Extensive clinical and statistical studies have identified several factors that increase the risk of CAD. The traditional risk factors for CAD are high LDL cholesterol, low HDL cholesterol, high blood pressure, family history, diabetes, smoking and obesity. The detection of related risk factors and tracking their progress over time is of great importance for early prevention and treatment of CAD.
The rapid adoption of Electronic Health Records (EHRs) in recent years has been shown to be a promising avenue for improving clinical research [13]. Despite structured information in EHRs – diagnosis codes, medications, and laboratory test results – a significant amount of medical information is still stored in narrative text format, principally in clinical notes from primary care patients. Unstructured clinical texts are widely recognized barriers for the application of clinical tools to clinical data. Natural language processing (NLP) technologies provide a solution to convert free text into structured representations that will be further re-used and re-purposed by clinical research [3].
Manual detection of heart disease risk factors from large scale medical records is prohibitively expensive, time-consuming and prone to error. Large-scale accurate risk identification therefore requires automated software that is fine-tuned to the structure of the text, the content of the medical records, and the specific requirements of a particular project. To facilitate the application of the NLP tools to the studies of heart disease, 2014 i2b2/UTHealth NLP Challenge1 Track 2 [19] was organized to comprehensively investigate the identification of related risk factors for heart disease in diabetic patients. The objective of this challenge task is to find clinical evidence from medical records, which indicates the presence and progression of diseases such as DIABETES (DM) and CORONARY ARTERY DISEASE (CAD), and associated risk factors like HYPERTENSION, HYPERLIPIDEMIA, SMOKING STATUS, OBESITY STATUS, and FAMILY HISTORY OF CAD. In addition, different categories of medications prescribed for individual diseases or risk factors are required to be recognized from the text.
The prediction of heart disease risk factors using clinical and statistical methods has been receiving much attention in the recent decade [6, 17, 28, 33]. But few published studies employed NLP techniques to investigate this research issue on the basis of textual medical records. There have been some related studies that were targeted for a number of text mining tasks such as obesity identification [24], smoking status identification [25], and medication extraction [26]. The most related work was conducted by Byrda et al. [1] where a hybrid NLP pipeline was proposed for the identification of heart failure diagnostic criteria.
This paper is the extension of our i2b2 workshop paper [31], which details our efforts to the 2014 i2b2 risk factor challenge task. A hybrid model was developed, which integrates a variety of methodological approaches, such as dictionary-based keyword spotting, rules and supervised learning, for the detection of a variety of heart disease risk factors. Our developed system achieved promising performance with an overall micro-averaged F-measure of 0.915.
The rest of the paper is organized as follows: Section 2 provides the details about the dataset used for the risk factor detection. In Section 3 we discuss the research issues in risk factor detection. Section 4 details the methods that we employ to deal with the complexity in risk detection. System performance and error analysis is reported in Section 5. Section 6 reflects related work, and our conclusions are given in Section 7.
2. Dataset
2.1. The i2b2 Corpus
The dataset used in the challenge includes discharge summaries, clinical notes and letters obtained from Partners HealthCare2. For the challenge task, a total of 1,304 medical reports for 296 patients were released to challenge participants. All records have been fully de-identified and manually annotated for disease risk factors. 790 annotated medical records (178 patients) are used as a training set, and the remaining 514 records (118 patients) are used as a test set to evaluate the performance of the participating systems.
Figure 1 gives the excerpt of a medical record with clinical evidence to denote the heart disease risk factors that a patient probably has. The annotated clinical text in Figure 1 is visualised using the Brat Annotation tool3 [18]. The text that indicates the presence of a particular risk factor (RF) is extracted as relevant evidence. Eight main risk factor categories are required to be identified from text. The distributions of eight main risk factor categories with 38 associated indicators in both training and test data are shown in Table 1. More details of the description of individual risk factors with associated indicators can be found in the i2b2 challenge annotation guideline [20].
Figure 1.

Example of clinical note with detected clinical evidence related to heart disease risk factors
Table 1.
Distributions of disease risk factors with associated indicators in both training and test data
| Risk Factor | Indicator | Training Data | Test Data |
|---|---|---|---|
|
| |||
| CAD | mention | 780 | 516 |
| event | 246 | 139 | |
| test | 79 | 59 | |
| symptom | 81 | 70 | |
|
| |||
| DIABETES | mention | 1,560 | 1,065 |
| AlC | 110 | 82 | |
| glucose | 25 | 33 | |
|
| |||
| OBESE STATUS | mention | 413 | 245 |
| BMI | 20 | 17 | |
|
| |||
| HYPERLIMIDEMIA | mention | 1,020 | 711 |
| high LDL | 33 | 29 | |
| high chol. | 9 | 11 | |
|
| |||
| HYPERTENSION | mention | 309 | 1,098 |
| high bp | 8 | 195 | |
|
| |||
| MEDICATION | ACE inhibitor | 967 | 612 |
| anti diabetes | 3 | 0 | |
| ARB | 288 | 193 | |
| aspirin | 1283 | 798 | |
| beta blocker | 1411 | 835 | |
| calcium channel blocker | 545 | 385 | |
| diuretic | 318 | 222 | |
| DPP4 inhibitors | 1 | 6 | |
| ezetimibe | 36 | 36 | |
| fibrate | 64 | 90 | |
| insulin | 634 | 395 | |
| metformin | 544 | 356 | |
| niacin | 20 | 25 | |
| nitrate | 336 | 271 | |
| statin | 1301 | 817 | |
| sulfonylureas | 471 | 288 | |
| thiazolidinedione | 124 | 61 | |
| thienopyridine | 292 | 284 | |
|
| |||
| SMOKER STUTUS | current | 58 | 33 |
| ever | 9 | 3 | |
| never | 184 | 120 | |
| past | 149 | 113 | |
| unknown | 371 | 243 | |
|
| |||
| FAMILY HISTORY | not present | 768 | 495 |
| present | 22 | 19 | |
|
| |||
| Total | 16,501 | 10,970 | |
Each risk factor category has its own set of indicators that are used to identify whether or not the disease or risk factor is present for that patient. For example, in Figure 1 the risk factor HYPERLIPIDEMIA has two indicators: (a) ‘hyperlipidemia’ → <HYPERLIPIDEMIA indicator=“mention”/> (b) ‘LDL 118’ → <HYPERLIPIDEMIA indicator=“high LDL”/>.
Moreover, each risk factor (except for SMOKING STATUS and FAMILY HISTORY) is associated with a ‘time’ attribute, i.e. when it is present, before/during/after DCT (Document Creation Time). For each medical record, the system will output a list of document-level risk factor annotations as shown in Figure 2, which are used for the final evaluation of system performance. Each annotation consists of three parts, i.e. a risk factor, a time attribute, and an associated risk indicator or medication type. In Figure 2, each risk factor annotation is supported by one particular clinical evidence instance detected from the excerpt of the clinical record in Figure 1. It is noted that sometimes one evidence instance (e.g., ‘atenolol’, and ‘hyperlipidemia’) might refer to multiple annotations with different time attributes.
Figure 2.

Document-level annotation for the risk factors detected from the free text (Figure 1)
Each risk indicator should, at minimum, have one clinical instance to support its presence. It is possible that multiple instances related to a specific indicator are found in the same medical record. Furthermore, to track the progression of heart disease, each patient has 3~5 longitudinal documents with different DCT, e.g., the file with the earlier time stamp is labelled with ‘xxx-01’ whereas the latter one with ‘xxx-02’, which allow a general timeline in the patient’s medical history.
2.2. Refining Clinical Evidence Provided in the Training Data
In the training data, the challenge organizers provided two sets of data: one is phrase-level clinical evidence found in medical records, another is document-level risk factor annotations (see Figure 2) that are generated based on the detected evidence. Each document was annotated by three different annotators in which relevant text fragments were extracted and marked as clinical evidence shown as below:
<CAD text="RCA stenting" time="during DCT" indicator="event”/>
The final document-level risk factor annotations were created by combining three sets of evidence provided by different annotators.
In the challenge task, the identification of phrase-level evidence was not required, and thus annotating phrase-level evidence was not the part of the annotation task. In fact the provided evidence set was still in its raw form, i.e. the ‘working notes’ of the annotators in support of their decision on the document-level tags. The challenge organization provided these working notes as supporting material rather than as the ground truth. As a result, to utilize this evidence for system development, we needed to refine it as follows:
- Inconsistent span boundaries in clinical evidence. The boundary of supporting evidence in the same text marked by different annotators is not consistent. For example, in the sentence (E1) below, three annotators gave different text spans to depict clinical evidence, ‘cut back his cigarettes’, ‘cigarettes to one time per week’, ‘cut back his cigarettes to one time per week’ for SMOKER STATUS.
- E1. He has cut back his cigarettes to one time per week.
Conflicted evidence. We observed that sometimes the annotators disagree with each other in terms of risk factor, associated indicator, or time attribute due to incompatible interpretations of some unclear or ambiguous contexts. For example, in the text, ‘repeat episode relived by nitro again’, one annotator considered the mention ‘nitro’ as [CAD:mention] whereas another treated it as [MEDICATION:nitrate].
Missing clinical evidence. The annotators are just required to provide, at minimum, one instance for each identified risk factor indicator. There exist some scenarios in which a document contains multiple mentions that refer to the same risk factor indicator, but the annotators only mark up one or two of them as relevant evidence.
To facilitate the detection of clinical evidence and the classification of risk factor indicators, we applied several strategies to further refine the provided annotations:
For the identical evidence instances from different annotators, replace them with a single evidence annotation.
For the evidence that the annotators disagree with regarding a risk factor indicator or time attribute, manually examine the conflicted evidence instances and select the most likely evidence as the final result. This was a subjective process. We acted as the fourth annotator and made the judgment via our understanding of the context or reference to other annotations of similar cases. About 22% of the annotations fall in this category.
Enrich the evidence set by adding more potential evidence instances that are missed by the annotators.
The newly refined i2b2 corpus contains 23,701 clinical evidence instances compared with the original 31,125 instances with noise data. The size of the corpus is reduced about 23.8% by removing the redundant or overlapped ones (6,681 instances), modifying the conflicted ones (314 instances), and adding some new ones (429 instances) for the missed instances. It took a researcher roughly 2 weeks’ time to improve the quality of the annotations.
3. Related Research Issues in Risk Factor Detection
Here we identify a number of research issues that are closely relevant to risk factor detection and require special attention during the system development.
First, evidence instances that are used to support the presence of relevant risk factors are quite different in terms of lexical, syntactic, and semantic contexts. Here we group the evidence with respect to various risk factors into three main types: (1) Token-level clinical entities (i.e. multiword phrases) (2) Sentence-level clinical facts (i.e. a clinical statement of a specific disease diagnosis) (3) Sentence-level clinical measurements (i.e. a diagnosis based on a measurement above a specified threshold), e.g., Threshold [high cholesterol]: total cholesterol of over 240. Table 2 gives three types of clinical evidence with the corresponding examples in various risk factor indicators, where abnormal clinical test values are highlighted in bold in Sentence-level Clinical Measurements.
Table 2.
Three types of clinical evidence regarding various risk factors
| Evidence Type | Risk Factor Indicator | Example |
|---|---|---|
| Token-level Clinical Entity |
CAD:mention | ‘CAD’, ‘3-vessel coronary artery disease’, ‘Coronary arteriosclerosis’ |
| DIABETES:mention | ‘DM’, ‘DMII’, ‘Diabetes mellitus’, ‘non-insulin-dependent diabetes’ | |
| OBESE:mention | ‘moderately obese’, ‘mild obesity’, ‘obesity-related conditions’ | |
| HYPERLIMIDEMIA:mention | ‘elevated cholesterol’, ‘hypercholesterolemia’, ‘hyperlipidemia’ | |
| HYPERTENSION:mention | ‘HTN’, ‘hypertension’, ‘hypertensive’ | |
| MEDICATION (+18) | ‘lisinopril’, ‘Zestril (LISINOPRIL)’, ‘Insulin 70/30 HUMAN 70-30’ | |
| Sentence-level Clinical Fact |
CAD:event | ‘stenting at that time, and subsequently had another stent placed in 2076’ |
| CAD:test | ‘The pre-intervention stenosis was 95% with 0% residual stenosis’ | |
| CAD:symptom | ‘has been getting chest pain and pressure with some shortness of breath’ | |
| SMOKER:current | ‘Smokes about a half pack per day’, ‘Tob: 2 pack/y since 30 yo’ | |
| SMOKER:ever | ‘50 pack year cigarette smoking history’, ‘History of tobacco use’ | |
| SMOKER:never | ‘Tob: denies’, ‘Nonsmoker’, ‘no history of smoking’, ‘does not drink, smoke’ | |
| SMOKER:past | ‘Tobacco: 1ppd from 14–50, quit 26 years ago’, ‘Remote h/o smoking’ | |
| FAMILY:present | ‘Mother died in her 50s of CAD’, ‘Brother – CABG with redo in his 40’s’ | |
| Sentence-level Clinical Measurement |
DIABETES:AlC | ‘Last A1c was 13.9. 5/23’, ‘HbA1c 01/13/2121 7.50’ |
| DIABETES:glucose | ‘FS: 109–160’, ‘blood sugars were found to be in the 300–400s’ | |
| OBESE:BMI | ‘BMI 31.9’, ‘BMI = 35 -> 32/33’, ‘BMI = 35’ | |
| HYPERLIMIDEMIA:high LDL | ‘Cholesterol-LDL 11/25/2115 121’, ‘LDL 196’, ‘Her last LDL was over 100’ | |
| HYPERLIMIDEMIA:high chol. | ‘cholesterol level was total 283’, ‘Cholesterol 242’, ‘cholesterol of 319’ | |
| HYPERTENSION:high bp | ‘BP: 190/102’, ‘blood pressure of 140/82’, ‘SBP 140–160mmHg’ |
Second, no single NLP technique is powerful enough to cope with a wide variety of characteristics related to different risk indicators. A hybrid approach that incorporates several NLP techniques such as machine learning, rule-based and dictionary-based keyword spotting is necessary during the system development [32].
Third, the judgments on the existence of certain risk indicators, especially the ones that are associated with the clinical conditions, e.g., glucose for DIABETES, high LDL and high chol. for HYPERLIMIDEMIA, are often not straightforward, which rely on the combination of several different clinical facts rather than a single clinical condition. Decision-making is a complicated process that requires the integration of different clinical measurements with the help of domain knowledge [9].
Fourth, some risk factor (RF) mentions are highly confusable with non-RF mentions. For example, both sentences (E2) and (E3) below contain the term ‘DM’. (E2) is a clinical fact indicating a DIABETES risk factor whereas (E3) is just some clinical testing about DIABETES. Therefore, the correct identification of ambiguous RF mentions needs to be sensitive to the linguistic context – i.e. the surrounding words – in which the cues occur.
E2. 49YOrhm w/PMH signif for CAD, Afib, DM, who presents w/RLE weakness. [DIABETES:mention]
E3. DM: diet-controlled; does not check sugars at home.
4. Methods
To participate in the i2b2 challenge, we developed a novel hybrid NLP system for evidence extraction of disease risk factors. The system framework is depicted in Figure 3, which consists of several functional components described in the following subsections.
Figure 3.
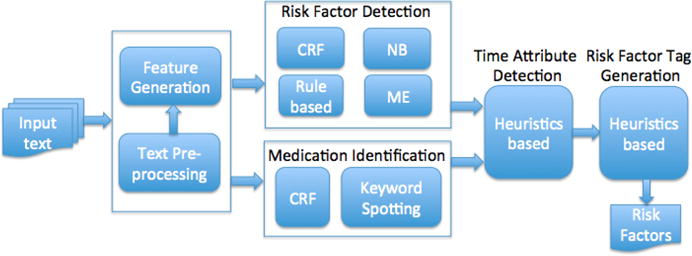
System framework for the risk factor detection task (CRF – Conditional Random Field, NB – Naive Bayes, ME – Maximum Entropy)
4.1. Text Pre-processing
Full-text medical records are first split into separate sentences. Then sentences are tokenized and specified with features characterizing tokens and token occurrences in their contexts. The syntactic information such as Part-of-speech (POS) tags, and phrase-based chunks regarding word-based tokens are extracted using Genia Tagger [23]4. Moreover, common headings of text sections (e.g., ‘Past Medical History’, ‘Medications on Admission’) are also extracted using a set of manually created regular expression rules.
4.2. Feature Generation
We systematically investigated various types of features that are extracted from the word itself and its context, and document text, including:
Token features: word lemma, POS tag, phrase chunk, orthographic information such as prefixes (e.g., the first 2–3 characters such as ‘non-’, ‘ex-’) and suffixes (e.g. the last 2–5 characters such as ‘-ide’, ‘-statin’) of words, capitalization of words, word shape information in which all upper case letters are replaced with A, all lower case letters are replaced with a, and all numbers are replaced with 0, e.g., ‘90%-treated’ → ‘00%-aaaaaaa’.
Context features: token features and their combination from three previous or following tokens of the target word.
Section features: section headings (e.g., ‘FAMILY HISTORY’) and sub-section headings (assumed to be the most recently seen mixed-case line ending with a colon) like ‘BP:’ and ‘Abdomen:’.
Domain knowledge features: word lists including immediate family members (e.g., ‘father’, ‘daughter’), frequent risk factor mentions (e.g., ‘hypercholesteremia’, ‘dyslipidemia’) or medication names (e.g., ‘simvastatin’, ‘pravastatin’), lexical cues (trigger words) indicating the occurrence of a particular risk factor indicator (e.g., ‘insulin-dependent and ‘non-insulin’ for DIABETES:mention), and smoker status keywords (e.g., ‘quit’, ‘ex-tob’). Such domain-related terms were firstly collected either from the annotated evidence instances based on the occurrence frequency or from their surrounding contexts via concordance. Then the keyword list was manually examined and filtered according to the importance of their association with each risk factor. The tf/idf (term frequency/inverse document frequency) statistics was employed to extract relevant keyword list for one specific risk indicator. The keyword lists with respect to different risk factors are available as online supplement data on the JBI web site5.
We exploited a wide range of linguistic features to capture the characteristics of different risk factor categories. The details about the feature types used for the detection of various risk indicators are given in an online data supplement on the JBI web site. In general, token, context and domain-specific features are used for both token-level clinical entities and sentence-level clinical facts whereas section features are more commonly used in sentence-level clinical measurements.
4.3. Risk Factor Detection
As described earlier, the evidence supported for the presence of risk factors can be grouped into three main categories: token-level clinical entities, sentence-level clinical facts, and sentence-level clinical measurements. Here we proposed a hybrid approach that combines several NLP techniques to handle the complexity and variations of different risk factors. Several supervised learning algorithms were employed for different types of risk factor evidence. It is shown that Conditional Random Fields (CRFs) exhibit the advantage on the recognition of word or phrase level clinical entities while Naive Bayes (NB) and Maximum Entropy (ME) are more powerful in sentence-level evidence identification. Heuristic rules are more suitable for the judgement of clinical measurements.
Table 3 provides the NLP techniques used to identify a wide variety of risk factor indicators. The details of the extraction of different evidence types are discussed below:
-
Token-level clinical entities
For this type of clinical terms, a Conditional Random Fields (CRFs) [12] based NER module that utilised optimized parameters and feature sets revealed by the training data was implemented using the CRF++ package6. Several types of features are used to build CRF-based classifiers, which include word lemma, POS tag, shallow syntactic chunk of the target word and its neighbouring words, word shape, section information, and lexical cues. Five CRF-based classifiers were built separately for the mention identification with respect to 5 different diseases, i.e. CAD, DIABETES, OBESE, HYPERLIMIDEDIA and HYPERTENSION. The CRF-based classifier for MEDICATION and three types of CAD clinical facts will be discussed in the later sections. The only difference for the building of the CRF-based classifiers is the features specific to the characteristics of one particular risk factor. Each word in a sentence is assigned one of the so-called BIO scheme tags: B (the first word of an entity mention), I (inside an entity mention), O (outside, not in an entity mention). Furthermore, a post-processing programming that makes use of heuristic rules was applied to correct possible errors, thus further improving the system performance. Some rules are intended to fix the term boundary problem, e.g., ‘Diabetes type’ → ‘Diabetes type 2’, while other rules are used to remove false positives that appear in some NEGATION assertions, e.g., ‘deny CAD’, ‘no history of CAD’. A total of 18 heuristic rules are created for the post-processing.
-
Sentence-level clinical facts
Compared with token-level clinical entity identification, the extraction of sentence-level clinical facts is quite complicated with substantial variability in expression in each risk factor category. Separate methods are proposed herein for individual risk factor categories in order to cope with the complexity of clinical fact extraction.
CAD Indicators. For sentence-level clinical facts regarding a CAD indicator, we employed three different ML-based classifiers. One is the token-level CRF-based classifier that is used to detect risk cues in the sentences. The sentence that contains one or more risk cues (e.g., ‘CABG’, ‘bypass grafting’, and ‘stent placement’ for CAD:event) is considered as relevant evidence to the targeted risk indicator. The others are two sentence-level classifiers, i.e. Naive Bayes (NB) based and Maximum Entropy (ME) based Classifiers. They were implemented using the MALLET tool7. We developed a post-processing module to combine the results from these three individual classifiers. The combination module works in a two-step way: first, it generates a list of intersection evidence instances that are predicted by two or more classifiers. Then the evidence list is extended via adding more results predicted by the best-performance classifier among the three classifiers proved at the training stage.
FAMILY Indicator. For the prediction of [FAMILY:present], a rule-based approach was employed. Sentences that contain immediate family member names (e.g., ‘mother’, ‘son’, etc.) are first located. Then relevant CAD-related keywords (e.g., ‘CAD’, ‘myocardial infarction’, ‘MIs’, ‘CABG’, etc.) are searched within a 10-token surrounding context of the family name. The optimal window size is determined by the experiments conducted on the i2b2 training data. In addition, the age of the family member is checked to see if it satisfies the required age restriction, 55 year-old for male and 65 year-old for female according to the annotation guideline of the challenge task.
SMOKER Indicators. Our approach to identify and categorize references to patient smoking in clinical reports can be divided into two stages: (a) STAGE I: we developed a sentence-level Naive Bayes (NB) classifier to identify sentences that contain smoking-related references. Documents that contain no smoking references are classified as UNKNOWN, thus being filtered out from the dataset. (b) STAGE II: for the remaining documents, the extraction engine uses linguistic analysis to associate features such as status (e.g., ‘current’, ‘ex-tob’) and time (e.g., ‘45 yrs ago’ and ‘until 8/2/81’) to smoking mentions. Four Maximum Entropy (ME) classifiers are built using these features to distinguish four SMOKER statuses, i.e. NEVER, PAST, EVER, and CURRENT, in the detected smoking-related sentences.
It is possible that several sentences referring to different SMOKER statuses are contained in the same document. For example, the sentences, ‘Smoking: 12 cig/day × 30 years’ and ‘12 pack per day hx, quit 2081’, appear in different locations of the file ‘397-01.xml’. The former implies a CURRENT status while the latter indicates a PAST status. However, it is required that there is only one SMOKER status for each clinical record. Hence, an integrated algorithm is proposed to determine the final SMOKER status with respect to a patient, which is based on the priority order: NEVER > PAST > EVER > CURRENT. The intuition for this priority order is based on the indicator ratio of these four smoker statuses in the annotated training data, NEVER(46%), PAST(37.3%), CURRENT(14.5%) and EVER(2.2%). EVER is quite rare and hard to be detected, so we put it ahead of CURRENT. After the integration, the final judgment for the two evidence instances mentioned earlier should be the PAST status.
Given a document with two or more SMOKER statuses detected, if the NEVER context is identified, the NEVER tag will be directly assigned to the record regardless of other SMOKER status. Otherwise, the system searches the PAST or EVER contexts, and the PAST has a higher priority. If the record does not contain other SMOKER-related sentence except for the CURRENT, the CURRENT tag will be highlighted.
-
Sentence-level clinical measurements
Clinical measurements conforming to certain clinical conditions are identified using several strategies:- First, a list of measurement trigger words, e.g., ‘HBAIC’ [DIABETES:AlC], ‘blood sugar’ [DIABETES:glucose], ‘CHOL’ [HYPERLIPIDEMIA:high LDL], and ‘BP’ [HYPERTENSION:high bp], are recognized from the sentences.
- Second, for the sentences that contain measurement trigger words, a number of hand-coded medical inference rules are examined to check if a clinically relevant finding could be found within the immediate context of the targeted trigger word. For example, the rule for [HYPERLIMIDEMIA:high LDL]: ‘LDL measurement of over 100 mg/dL’ and the rule for [HYPERTENSION:high bp]: ‘BP measurement of over 140/90 mm/hg’. The window size for each measurement indicator is optimized based on the performance on the training set, and it is generally set to the range of 5 words on the left and 10 words on the right.
- Third, due to terminological variations and irregularities in clinical values, e.g., ‘150/70’, ‘160’s/80’s’, ‘140s/80s’, ‘140–170s’, ‘from 220 to 230’, and ‘120–140/70–90’, an additional set of morphological rules are required to cope with orthographic variants in the measurement values.
-
Fourth, occasionally some special measurements are located in multi-line tables shown as below, which require expanding the context beyond the line, e.g., ‘GLU… 200’.
Date NA K CL CO2 BUN CRE GLU CA 07/03/93 141 4.3 104 26.6 19 1.1 200 9.4
Table 3.
NLP techniques used to detect clinical evidence in various risk factors
| Risk Factor | Indicator | Evidence type | Machine Learning
|
Rule | Dictionary | ||
|---|---|---|---|---|---|---|---|
| CRF | NB | ME | |||||
|
| |||||||
| CAD | mention | NE | ✓ | – | – | ✓ | ✓ |
| event | CF | ✓ | ✓ | ✓ | – | ✓ | |
| test | CF | ✓ | ✓ | ✓ | – | ✓ | |
| symptom | CF | ✓ | ✓ | ✓ | – | ✓ | |
|
| |||||||
| DIABETES | mention | NE | ✓ | – | – | ✓ | ✓ |
| AlC | CM | – | – | – | ✓ | – | |
| glucose | CM | – | – | – | ✓ | – | |
|
| |||||||
| OBESE STATUS | mention | NE | ✓ | – | – | ✓ | ✓ |
| BMI | CM | – | – | – | ✓ | – | |
|
| |||||||
| HYPERLIMIDEMIA | mention | NE | ✓ | – | – | ✓ | ✓ |
| high LDL | CM | – | – | – | ✓ | – | |
| high chol. | CM | – | – | – | ✓ | – | |
|
| |||||||
| HYPERTENSION | mention | NE | ✓ | – | – | ✓ | ✓ |
| high bp | CM | – | – | – | ✓ | – | |
| MEDICATION | medication (+18) | NE | ✓ | – | – | ✓ | ✓ |
|
| |||||||
| SMOKER STUTUS | current | CF | – | ✓ | ✓ | – | ✓ |
| ever | CF | – | ✓ | ✓ | – | ✓ | |
| never | CF | – | ✓ | ✓ | – | ✓ | |
| past | CF | – | ✓ | ✓ | – | ✓ | |
|
| |||||||
| FAMILY HISTORY | present | CF | – | – | – | ✓ | ✓ |
(where NE – Token-level Clinical Entity, CF – Sentence-level Clinical Fact, CM – Sentence-level Clinical Measurement)
As mentioned before, the prediction of risk indicators associated with clinical conditions is not easy, especially for [DIABETES:glucose], [HYPERLIMIDEMIA:high LDL], and [HYPERLIMIDEMIA:high chol.], which needs to consider several elements such as the number of the relevant evidence instances and the importance degree of the detected evidence (assumed to occur in some special contexts). A weight score is assigned to each predicted risk indicator, which is calculated based on the trade-off between true positive (TP) and false positive (FP) of the detected clinical measurements. The higher the weight score is, the higher likelihood for the presence of the risk factor indicator. When the total weight score of all the relevant evidence is above the assigned threshold, a warning of the corresponding risk indicator is triggered.
4.4. Identification of Medication Names
Two NLP techniques were employed for the identification of different categories of medication names. For the medication categories that are provided with enough sample instances in the training data, individual CRF-based classifiers are trained to recognize relevant medication mentions. For the medication categories that have few training samples, a dictionary-based keyword lookup is applied to distinguish drug names from the text. A list of frequent medication names is collected from the evidence instances of the training data as well as some online drug resources.
For some combination drugs (e.g., ‘Glucovance’ and ‘Zestoretic (LISINOPRIL/HYDROCHLOROTHIAZIDE)’), two different medication categories are separately identified, and are represented with the ‘type1’ and ‘type2’ attributes.
<MEDICATION time=“after DCT” type1=“sulfonylureas” typ2=“metformin”>
Finally, a rule-based post-processing step is conducted to remove some false positives, e.g., the medication names that appear in the ALLERGY section, or the NEGATION context (e.g., ‘did not take MED’), or the HYPOTHETICAL structure (e.g., ‘Consider… if/when…’).
4.5. Determination of Time Attributes
Each risk factor indicator (except for SMOKER and FAMILY_HIST) has a time attribute that reflects when the indicator occurred or was known to have existed in relation to the document creation time (DCT), i.e. the date the medical record was written. The possible values for the time attribute are continuing, before DCT, during DCT, and after DCT, where the ‘continuing’ value is actually comprised of three time tags, before DCT, during DCT, and after DCT.
A set of heuristic rules is generated based on our observations of the i2b2 training data, which is applied to determine time attributes of individual indicator types. For instance, in [E4], a temporal adverb ‘today’ is recognised from the sentence, which suggests a ‘during DCT’ attribute. In [E5], The word ‘switched from’ implies the time attribute change between different medications, i.e. from before DCT [Crestor] to continuing [Lipitor].
E4. He presented today for cath which revealed a new stenosis distal to the prior LAD stent which was patent. ([CAD:test], during DCT)
E5. He also been switched from Crestor to Lipitor due to safety concerns. ([MEDICATION:statin], before DCT → continuing)
Several types of temporal features are exploited during the heuristics-based processing of time attribute determination.
Section headings (e.g., ‘ASSESSEMENT AND PLAN’, ‘PHYSICAL EXAMINATION’, ‘PMH’)
Subsection headings ending with a colon (e.g., ‘ED Course:’, ‘PE:’, ‘VITALS:’, ‘Abdomen:’)
DATE (e.g., ‘2059-01-10’, ‘01/01/93’, ‘30Aug71’)
Sentence tense (e.g., Past, Present, and Future tense)
Regular expression template patterns (e.g., ‘change DRUG_1 to DRUG_2’, ‘catheterization/cath showed/revealed/demonstrated…’)
Location (e.g., ‘in the ED’, ‘in the hospital’)
Temporal phrases (e.g., ‘five years ago’, ‘in the past’, ‘last check’)
Temporal adjectives/adverbs (e.g., ‘previous’, ‘currently’, ‘today’, ‘yesterday’, ‘former’, ‘prior’, ‘old’)
Temporal verbs (e.g., ‘continue’, ‘discontinue’, ‘restart’, ‘stop’)
We also observed that each risk indicator is generally connected to a ‘default’ time attribute, i.e. most of the evidence instances are labelled with one particular time attribute. For example, 449 out of 458 [CAD:mention] annotations contain a ‘continuing’ time attribute. Table 4 gives the DEFAULT time attribute related to individual risk factor indicators. Therefore, our approach to time attribute determination can be considered as a processing that attempts to find those ‘exceptional’ instances with other time attributes other than the default one in each indicator category using the generated heuristics discussed above.
Table 4.
The DEFAULT time attribute for various risk factor indicators
| Risk Factor | Indicator | Continuing | Before DCT | During DCT | After DCT | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| CAD | mention | ✓ | – | – | – | ||
| event | – | ✓ | – | – | |||
| test | – | ✓ | – | – | |||
| symptom | – | ✓ | – | – | |||
|
| |||||||
| DIABETES | mention | ✓ | – | – | – | ||
| AlC | – | ✓ | – | – | |||
| glucose | – | ✓ | – | – | |||
|
| |||||||
| OBESE STATUS | mention | ✓ | – | – | – | ||
| BMI | – | – | ✓ | – | |||
|
| |||||||
| HYPERLIMIDEMIA | mention | ✓ | – | – | – | ||
| high LDL | – | ✓ | – | – | |||
| high chol. | – | ✓ | – | – | |||
|
| |||||||
| HYPERTENSION | mention | ✓ | – | – | – | ||
| high bp | – | – | ✓ | – | |||
|
| |||||||
| MEDICATION | medication (+18) | ✓ | – | – | – | ||
4.6. Generation of Risk Factor Annotations
Once the steps of clinical evidence detection and time attribute determination are completed, a set of clinical evidence annotations is automatically created. Each clinical evidence annotation (see below) includes several parts of information such as a specific risk factor, the detected evidence text, a time attribute, and a related indicator. Moreover, each evidence annotation is associated with the document ID.
<CAD text=“2cd stent was placed” time=“during DCT” indicator=“event”/>
The next step is to convert the text-fragment-level evidence annotations into the document-level risk factor annotations (recall Figure 2). It is possible that an evidence instance can be related to different time attributes regarding the same risk factor indicator. For example, an evidence annotation with the ‘continuing’ time tag should be separated to three annotations with different time attributes, before DCT, during DCT, and after DCT. Moreover, a document probably contains several evidence instances indicating the same risk factor indicator and time attribute. Here we applied a simple heuristic to create an integrated set of risk factor annotations, that is, for each clinical record, given a risk factor with a distinct indicator and a specific time attribute, e.g., ‘<CAD time=“before DCT” indicator=“test”/>’, the system looks up the evidence annotation set and checks whether there exists relevant evidence found in the document. In our current system, risk indicators supported with related evidence are treated equally – no matter how many evidence instances are found in the record. However, it is possible that the more the evidence instances are found in the text, the more confident the system should have in the prediction. It is worth exploiting in the future work.
5. Results
The risk factor identification system is evaluated using micro-average precision (P), recall (R), and F-measure (F1) [16]. As mentioned before, the output of the system is a list of document-level risk factor annotations (Recall Figure 2). The prediction is evaluated based on the ‘correctness’ of all the three tags. It is noted that clinical evidence detected from the text is NOT used for the evaluation of system performance. Micro-averaged F-measure (evaluated at the entire corpus) is used as the primary metric for the i2b2 challenge evaluation.
5.1. Overall Performance of the System
Table 5 illustrates the overall performance of the eight risk factor categories on the i2b2 test data. Our system achieved a micro-averaged F-measure of 0.915, a precision of 0.885, and a recall of 0.949 for the overall risk factor identification. Our F1-score is substantially better than the overall average (0.815) of all the participating systems in this challenge, and is not significantly different with the first-ranked system that achieved an F-measure of 0.928.
Table 5.
The overall performance of the system at the risk factor level on the i2b2 test data
| Risk Factor | #Expected | #Predicted | #Correct | Precision | Recall | F-measure |
|---|---|---|---|---|---|---|
| CAD | 784 | 817 | 630 | 0.771 | 0.803 | 0.787 |
| DIABETES | 1,180 | 1,191 | 1,094 | 0.918 | 0.927 | 0.922 |
| OBESE | 262 | 296 | 250 | 0.844 | 0.954 | 0.896 |
| HYPERLIPIDEMIA | 751 | 769 | 712 | 0.925 | 0.948 | 0.936 |
| HYPERTENSION | 1,293 | 1,347 | 1,248 | 0.926 | 0.965 | 0.945 |
| MEDICATION | 5,674 | 6,316 | 5,512 | 0.872 | 0.971 | 0.919 |
| SMOKER | 512 | 514 | 470 | 0.914 | 0.918 | 0.916 |
| FAMILY_HIST | 514 | 514 | 492 | 0.957 | 0.957 | 0.957 |
| Total | 10,970 | 11,764 | 10,408 | 0.884 | 0.948 | 0.915 |
Moreover, our system performs best on the FAMILY_HIST (0.957) and HYPERTENSION (0.945) categories, and achieves F1-scores above 0.915 for DIABETES, HYPERLIPIDEMIA, MEDICATION, and SMOKER. FAMILY_HIST is a relative easy task where only a few medical records contain the evidence of family members with CAD. The high accuracy of several CRF-based NER classifiers (discussed in the following subsection) contributes to the good performance in terms of HYPERTENSION, DIABETES, HYPERLIPIDEMIA, and MEDICATION. Poorest classification accuracy is obtained for CAD (0.787). The main reason for that is due to the difficulties in the identification of sentence-level CAD clinical facts, event, test, and symptom. The MEDICATION category that has the largest number of instances (accounting for 51.7% of the instances in the test data) obtains an F-measure of 0.919, slightly better than the overall system performance (0.916).
5.2. Performance on Individual Risk Indicators
Table 6 shows the performances of individual indicators in each risk factor category. Out of the 38 associated indicators involved in eight risk factor categories, our hybrid approach achieves macro-averaged F-measure scores above 0.8 (25 out of the 38 risk indicators) and above 0.9 (19 indicators). Our best results are achieved on the DPP4 inhibitors indicator with the ‘perfect’ score of 1.0 while the worst ones occur in trying to distinguish the [SMOKER:ever] indicator due to rare training samples. Disease mentions and most of medication names have a higher recall because of frequent term lists collected from the training data. This implies indicator-specific keyword lists play an important role in the entity identification task. In general, the more a risk indicator has training instances, the better the accuracy of the risk prediction is.
Table 6.
The overall performance of the system at the risk indicator level on the i2b2 test data
| Risk Factor | Indicator | #Expected | #Predicted | #Correct | Precision | Recall | F-measure |
|---|---|---|---|---|---|---|---|
|
| |||||||
| CAD | mention | 516 | 621 | 501 | 0.806 | 0.971 | 0.881 |
| event | 139 | 90 | 76 | 0.844 | 0.546 | 0.664 | |
| test | 59 | 85 | 43 | 0.505 | 0.728 | 0.597 | |
| symptom | 70 | 21 | 10 | 0.476 | 0.143 | 0.219 | |
|
| |||||||
| DIABETES | mention | 1,065 | 1,068 | 1,018 | 0.953 | 0.956 | 0.954 |
| AlC | 82 | 87 | 68 | 0.781 | 0.829 | 0.804 | |
| glucose | 33 | 36 | 8 | 0.222 | 0.242 | 0.231 | |
|
| |||||||
| OBESE | mention | 245 | 285 | 239 | 0.838 | 0.975 | 0.902 |
| BMI | 17 | 11 | 11 | 1.000 | 0.647 | 0.785 | |
|
| |||||||
| HYPERLIMIDEMIA | mention | 711 | 717 | 687 | 0.958 | 0.966 | 0.962 |
| high LDL | 29 | 39 | 20 | 0.512 | 0.689 | 0.588 | |
| high chol. | 11 | 13 | 5 | 0.384 | 0.454 | 0.416 | |
|
| |||||||
| HYPERTENSION | mention | 1,098 | 1,143 | 1,089 | 0.953 | 0.992 | 0.972 |
| high bp | 195 | 204 | 159 | 0.779 | 0.815 | 0.797 | |
|
| |||||||
| MEDICATION | ACE inhibitor | 612 | 674 | 583 | 0.865 | 0.952 | 0.906 |
| ARB | 193 | 211 | 193 | 0.914 | 1.000 | 0.955 | |
| aspirin | 798 | 871 | 789 | 0.906 | 0.988 | 0.945 | |
| beta blocker | 835 | 905 | 817 | 0.903 | 0.978 | 0.939 | |
| calcium channel | 385 | 414 | 368 | 0.889 | 0.955 | 0.921 | |
| blocker | 222 | 310 | 218 | 0.703 | 0.982 | 0.819 | |
| diuretic | 6 | 6 | 6 | 1.000 | 1.000 | 1.000 | |
| DPP4 inhibitors | 36 | 59 | 35 | 0.593 | 0.972 | 0.736 | |
| ezetimibe | 90 | 104 | 88 | 0.846 | 0.977 | 0.907 | |
| fibrate | 395 | 383 | 351 | 0.916 | 0.888 | 0.903 | |
| insulin | 356 | 418 | 356 | 0.852 | 1.000 | 0.920 | |
| metformin | 25 | 41 | 22 | 0.536 | 0.880 | 0.666 | |
| niacin | 271 | 369 | 268 | 0.726 | 0.989 | 0.837 | |
| nitrate | 817 | 865 | 802 | 0.927 | 0.981 | 0.953 | |
| statin | 288 | 309 | 282 | 0.912 | 0.979 | 0.944 | |
| sulfonylureas | 61 | 69 | 58 | 0.841 | 0.951 | 0.892 | |
| thiazolidinedione thienopyridine | 284 | 308 | 276 | 0.896 | 0.972 | 0.932 | |
|
| |||||||
| SMOKER | current | 33 | 51 | 30 | 0.588 | 0.909 | 0.714 |
| ever | 3 | 1 | 0 | 0.000 | 0.000 | 0.000 | |
| never | 120 | 116 | 109 | 0.939 | 0.908 | 0.923 | |
| past | 113 | 108 | 98 | 0.907 | 0.867 | 0.887 | |
| unknown | 243 | 238 | 233 | 0.979 | 0.958 | 0.969 | |
|
| |||||||
| FAMILY_HIST | not present | 495 | 489 | 481 | 0.983 | 0.956 | 0.954 |
| present | 19 | 25 | 11 | 0.440 | 0.579 | 0.500 | |
As mentioned before, we group the evidence into three main types, i.e. token-level clinical entities, sentence-level clinical facts, and sentence-level clinical measurements. As shown in Table 7, the system performs best on the token-level clinical entity recognition, and achieves a high F-measure of up to 0.929. The performances on sentence-level clinical facts and sentence-level clinical measurements are close to each other. Sentence-level clinical facts give better precision whereas sentence-level clinical measurements have better recall.
Table 7.
The performance of different types of evidence on the i2b2 test data
| Evidence Type | #Expected | #Predicted | #Correct | Precision | Recall | F-measure |
|---|---|---|---|---|---|---|
| Token-level clinical entity | 9,309 | 10,150 | 9,046 | 0.891 | 0.972 | 0.929 |
| Sentence-level clinical fact | 556 | 497 | 377 | 0.758 | 0.678 | 0.716 |
| Sentence-level clinical condition | 367 | 390 | 271 | 0.695 | 0.738 | 0.716 |
In summary, ML-based approaches exhibit the advantage in terms of the named entity recognition (NER) task. The dictionary-based keyword spotting method only works well on part of the MEDICATION categories that lack sample instances. Nevertheless, it still can be considered as an effective supplement to the ML-based NER task when there are inadequate instances for learning. For the sentence-level clinical fact extraction, the strength of the ML-based techniques heavily relies on whether or not the distinguishing features can be found in the sample instances.
The Rule-based approach to the identification of sentence-level clinical measurements performs worse than expected. The main reason for this is because the prediction of some risk factor indicators requires the merging of several different clinical facts/conditions. For example, two or more clinical evidence instances, e.g., ‘Finger blood glucose 156 two hours PP’ and ‘Glucose after eating: 200’ are required for the detection of the risk factor [DIABETES:glucose] according to the annotation guideline.
In some certain scenario, the judgment of a clinical measure (e.g., [DIABETES:glucose]) does not merely rely on the surface meaning of supporting evidence. Domain knowledge also plays an important role in the judgement of a valid clinical measurement, which increases the prediction difficulty of the system. For instance, the evidence, ‘glucose 420’, is detected in one medical record. But it is not considered as the solid indication of the risk factor [DIABETES:glucose] according to the ground truth. It implies that ‘glucose >126’ is a necessary, but certainly not a sufficient condition for [DIABETES:glucose]. More clinical knowledge is needed for the accurate judgement of some complicated risk factors, which will be exploited in our future work.
A quick glance at Table 6 suggests system performance varies with the number of the training instances available for each risk indicator. Figure 4 clearly reveals the relationship between the instance number and system performance in terms of risk indicators. When the instance number falls below 200, system performance varies greatly depending on the evidence type and the method used for the detection of the indicator. However, with the increase of the instance number, system performance obviously gets better and the predication capability tends to be more stable with high accuracy.
Figure 4.
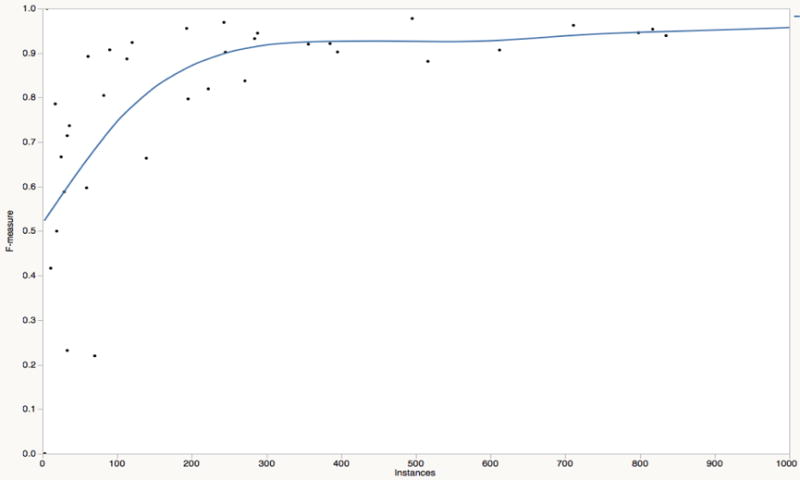
The relationship between the instance number and system performance in risk indicators
5.3. Error Analysis
As discussed before, our risk indicator prediction process in practice consists of three stages: First, relevant supporting evidence instances are identified from the text; Second, each detected evidence (except for FAMILY_HIST and SMOKER) is assigned with the appropriate time attribute tag. Finally, a set of risk factor annotations is generated based on the results from both the evidence detection and the time attribute determination. Here, we divide system error into two categories: one is the evidence-level errors, i.e. the incorrectly recognized evidence instances or missing evidence instances; the other is the time-attribute errors, i.e. a risk indicator is correctly identified, but marked with the wrong time attribute.
Of the total number of errors (1,918) made by our system, 1,356 (70.7%) are false positive (i.e. incorrect annotations) for which 920 belong to the evidence-level errors and 436 for the time-attribute errors. In the remaining 562 (29.3%) false negatives (i.e. missing tags), 480 are the evidence-level errors and 82 for the time-attribute errors.
-
Evidence-level Errors
Evidence-level errors generally can be grouped into six main categories: (1) the detection of some special terms needs to consider the surrounding contexts in some certain situation. For example, the mentions ‘CAD’ and ‘coronary artery disease’ are tagged as the [CAD:mention] indicator only in some certain contexts. (2) Cross-line evidence: the evidence spans that cross two or more lines are not supported by our current system. For example, the mention ‘coronary heart… disease’ for CAD is broken in the middle, and occurs in two different lines. (3) Token-level previously unseen evidence (e.g., ‘ischemic cardiomyopathy’ and ‘Acute coronary syndrome’) on the test data also have difficulty in being recognized by the system. (4) Misclassified SMOKER STATUS and FAMILY_HIST tags. For example, in the SMOKER category, quite a number of false positives result from the misclassification of ‘past’ and ‘unknown’ into the ‘current’ tag. (5) For the errors in terms of sentence-level clinical facts, we believe that the small number of training instances and sophisticated contexts are the primary reason that causes most of the false positives or negatives. (6) For sentence-level clinical measurements, we achieve better results with well-presented indicators (e.g., ‘A1C’, ‘BMI’, and ‘high bp’) than those of complex indicators like ‘glucose’ and ‘high chol.’ that are required where it is necessary to take into consideration.
-
Time-attribute Errors
Time-attribute annotations largely depend on the completeness and accuracy of our manually created heuristics. However, due to inadequate training instances for some time attribute tags, e.g., fewer than 10 instances for the after DCT tag regarding the [CAD:event] and [CAD:symptom] indicators, the system could not create accurate heuristics to capture the characteristics of these time attribute tags.
5.4. Lessons Learnt from the Challenge
Compared with the previous i2b2 challenges, this track task seems more complicated than before. A wide variety of clinical evidence instances are required from the text as we discussed earlier. It is important to analyse the characteristics of such evidence before the processing of the dataset. Sometimes, dividing a hard task into several small subtasks and selecting appropriate approaches to cope with each subtask will be more useful and it will facilitate the system development.
In general, the machine-learning based approaches, e.g., the identification of clinical terms and the extraction of clinic facts, require less human effort and working time than the rule-based or dictionary-based approaches. The machine can automatically learn the patterns, thus providing more flexible predictive power for a large-scale dataset. The rule-based or dictionary-based approaches have to heavily rely on the domain knowledge, even with the help of domain experts. This aspect of the work always demands more attention and time when a new dataset is provided or a domain-specific problem is tackled.
The programming modules or tools for the processing of general text information, e.g., the extraction of token features, context features, and section features, can be reusable for most of the NLP tasks. But the domain knowledge features are dependent on specific datasets or tasks.
It took roughly 6 weeks to implement a NLP system for this challenge task. A professional in computer science spent about two weeks on the refinement of the training data in order to obtain a high-quality annotation corpus. In addition, four weeks’ time was used for the system development including three weeks for the development of the rules, training classifiers, and domain-specific lexicon generation, and one week for the improvement of the overall system.
The main contribution of our work is that we proposed a hybrid approach to handle a difficult and complicated task. We exploited different NLP techniques to deal with various types of scenarios involved in the risk factor detection. We expect that our experience learnt from this challenge will be helpful for the researchers who work under the similar situations. Moreover, the quality of the annotated training data plays a role on the system performance improvement. Some experiments were conducted using a system that was built on the original annotations with noise data. The system achieved an overall F-measure of 0861. It suggests that our refined evidence instances contribute an improvement of about 5.4% in terms of the overall F-measure.
6. Related Work
In the past few years, the i2b2 organization hosted a number of NLP challenges for clinical data. Some of the previous i2b2 challenge tasks are closely related to the work of risk factor detection and address several research issues involved in this challenge task, such as the 2007 i2b2 Smoking Status Challenge [25], the 2008 i2b2 Obesity Challenge [24], the 2009 i2b2 Medication Challenge [26], the 2010 i2b2 Challenge on Concepts, Assertions, and Relations [27], and the 2012 i2b2 Temporal Relation Challenge [21].
Text-based patient medical records are a vital resource in medical research. NLP techniques, which can extract useful information from narrative clinical texts to support decision-making or represent clinical knowledge in a standardized format, are receiving much attention in the recent decade. Although various NLP approaches are proposed to address different research issues in clinical records, they can be broadly classified into two main categories: rule-based and machine learning-based. Rule-based systems usually cope with the information extraction (IE) tasks with pattern matching, regular expressions and dictionary lookup [15, 29]. They need little or no training data, but they often lack generalizability and require expertise knowledge for creating rules. Machine learning based methods [4, 8], on the other hand, can automatically learn to detect clinical patterns based on a set of examples. They are more generalizable, but require a large set of manually annotated examples. Similar to our proposed approach, several hybrid systems [14, 22, 30] were developed, which take advantage of rule-based and machine learning-based approaches to obtain better results.
Assertion status, particularly for ‘NEGATION context’, is considered in the challenge task for both disease and medication name recognition. Similar to several rule-based approaches [2, 10], we generated a number of handcrafted rules that are applied to determine the effect of negation cues on clinical concepts in the same sentence. A few systems [5, 26] used a combination of machine learning and rule-based techniques for the detection of assertion cues and the determination of assertion scopes.
The identification of temporal expressions is generally carried out by a rule-based system with a set of regular expressions. Similar to our work, Denny et al [7] used regular expressions to extract temporal expressions. Jung et al. [11] proposed a system pipeline that was based on explicit rules for temporal expression extraction and event extraction.
7. Conclusions and Future Work
In this paper, we investigated a hybrid method that can automatically identify risk factors of heart disease in clinical texts. We found that risk factor detection benefits from a wide variety of lexical, syntactic, and semantic context features as well as regular expression template patterns. Our experiments showed that the approaches combining machine-learning methods with other NLP techniques such as rule-based approaches and dictionary-based keyword spotting tend to be more robust in dealing with sophisticated clinical contexts than a single NLP approach if applied alone. Our system achieved an overall micro-average F-measure of 0.915, which was ranked the fifth place out of the 20 participating teams, and was competitive with the best model (F-measure of 0.927) of this challenge task.
This challenge provides a good opportunity to develop an information extraction system to deal with a complicated clinical task like the detection of heart disease risk factors. In this challenge, we analysed the characteristics of clinical evidence and divided it into three main types, token-level clinical entities, sentence-level clinical facts, and sentence-level clinical measurements. We believe these three evidence types could represent a majority of textual clinical information of interest. For each type of clinical evidence, we extracted relevant linguistic features and selected the appropriate approaches to handle the problems inherent in this evidence type. The lessons learned from the risk factor detection challenge task will provide valuable experience for similar clinical decision support systems in the future.
On the one side, the ML-based named entity identification might be easily adapted for the detection of a new risk indicator given a large-scale dataset in which the characteristics of the risk factor are diverse and not well formed. But the manual annotation of large training examples with pre-labelled identifiers is prohibitively expensive and time-consuming. On the other side, the rule-based clinical measure detection need little or no training data, and are less costly when a set of instances are well defined and only a few of heuristic rules could capture most of them. However, the generation of the rules requires domain knowledge from the experts. For ML-based entity identification and sentence extraction, indicator-specific linguistic features such as keyword list and morphologic features must be collected from the domain-specific area.
There are a number of areas for future work. For example, some article structure information, such as section heading and sub-heading, and the handling of lists and embedded implicit tables, is utilized for this challenge task. It is interesting to measure the usefulness of such information on the improvement of system performance. The observations on the annotated data suggest medication types are indeed associated with a particular clinical risk factor (e.g., CAD). It is also worth exploring such kind of associations to see whether they can be useful in the risk factor prediction. Due to the limited development time, we did not conduct the work about the tuning of the optimal parameters of the ML algorithms. We plan to investigate this research issue and see whether it has the potential to improve the system performance in the future. Our dictionary lookup method mainly relies on the limited keyword list directly collected from the training dataset. We intend to enrich it by making use of the existing knowledge resources such as UMLS database and web sources.
Supplementary Material
Highlights.
Risk factor detection is of great importance for the treatment of heart disease
Machine learning techniques combined with keywords and rule-based approaches
7 main heart disease risk factors with relevant medications are identified
Achieving an overall micro-averaged F-measure of 91.56%
Acknowledgments
The work was supported by funding through the Advanced Data Analysis Centre, University of Nottingham, UK. We would like to thank the i2b2 2014 challenge organizers for providing such an invaluable clinical dataset and this research opportunity. Moreover, we also thank the anonymous reviewers for the improvement of the paper quality.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Byrda RJ, Steinhublb SR, Suna J, Ebadollahi S, Stewart WF. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. J Med Inform. 2014;83(12):983–92. doi: 10.1016/j.ijmedinf.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman WW, Bridwell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 3.Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner Ö. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solution. J Am Med Inform Assoc. 2011;18(5):540–3. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark C, Good K, Jezierny L, Macpherson M, Wilson B, Chajewska U. Identifying Smokers with a Medical Extraction System. J Am Med Inform Assoc. 2008;15(1):36–9. doi: 10.1197/jamia.M2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark C, Aberdeen J, Coarr M, et al. MITRE system for clinical assertion status classification. J Am Med Inform Assoc. 2011;18(5):563–7. doi: 10.1136/amiajnl-2011-000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis M, Andrade J, Taylor C, Ignaszewski A. Cardiovascular risk factors and models of risk prediction: Recognizing the leadership of Dr Roy Dawber. BCMJ. 2010;52(7):342–8. [Google Scholar]
- 7.Denny JC, Peterson JF, Choma NN, et al. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. J Am Med Inform Assoc. 2010;17:383–8. doi: 10.1136/jamia.2010.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc. 2011;18(5):557–62. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demner-Fushman D, Chapman WW, McDonald CJ. What can Natural Language Processing do for Clinical Decision Support? J Biomed Inform. 2009;42(5):760–72. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gindl S, Kaiser K, Miksch S. Syntactical negation detection in clinical practice guidelines. Proc of MIE2008: eHealth Beyond the HorizondGet IT There. 2008:187–92. [PMC free article] [PubMed] [Google Scholar]
- 11.Jung H, Allen J, Blaylock N, Beaumont Wd, Galescu L, Swift M. Building timelines from narrative clinical records: initial results based-on deep natural language understanding. Proc of the 49th Annual Meeting of the Association for Computational Linguistics (ACL 2011) 2011:146–54. [Google Scholar]
- 12.Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labelling sequence data. Proc of the international conference on machine learning (ICML-2001) 2001:282–9. [Google Scholar]
- 13.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 14.Meystre SM, Thibault J, Shen S, Hurdle JF, South BR. Textractor: a hybrid system for medications and reason for their prescription extraction from clinical text documents. J Am Med Inform Assoc. 2010;17(5):559–62. doi: 10.1136/jamia.2010.004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra NK, Cummo DM, Arnzen JJ, Bonander J. A Rule-based Approach for Identifying Obesity and Its Comorbidities in Medical Discharge Summaries. J Am Med Inform Assoc. 2009;16(4):576–9. doi: 10.1197/jamia.M3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salton G, McGill MJ. Introduction to Modern Information Retrieval. New York: McGraw Hill; 1983. [Google Scholar]
- 17.Siontis GC, Tzoulaki I, Siontis KC, Ioannidis JPA. Comparisons of established risk prediction models for cardiovascular disease: systematic review. BMJ. 2012;344:e3318. doi: 10.1136/bmj.e3318. [DOI] [PubMed] [Google Scholar]
- 18.Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, Tsujii J. brat: a Web-based Tool for NLP-Assisted Text Annotation. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics (EACL’12) 2012:102–7. [Google Scholar]
- 19.Stubbs A, Kotfila C, Xu H, Uzuner Ö. Practical applications for NLP in Clinical Research: the 2014 i2b2/UTHealth shared tasks. The 2014 i2b2 Challenge Workshop. 2014 [Google Scholar]
- 20.Stubbs A, Uzuner Ö. Annotating Risk Factors for Heart Disease in Clinical Narratives for Diabetic Patients. J Biomed Inform. 2015 May 21; doi: 10.1016/j.jbi.2015.05.009. pii: S1532-0464(15)00089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun W, Rumshisky A, Uzuner Ö. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc. 2013;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang B, Wu Y, Jiang M, Chen Y, Denny JC, Xu H. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc. 2013;20(5):828–35. doi: 10.1136/amiajnl-2013-001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuruoka Y, Tateishi Y, Kim J, Ohta T, McNaught J, Ananiadou S. Developing a robust part-of-speech tagger for biomedical text. Advances in Informatics. 2005:382–92. [Google Scholar]
- 24.Uzuner Ö. Recognizing Obesity and Comorbidities in Sparse Data. J Am Med Inform Assoc. 2009;16(4):561–70. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uzuner Ö, Goldstein I, Luo Y, Kohane I. Identifying Patient Smoking Status from Medical Discharge Records. J Am Med Inform Assoc. 2008;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17(5):514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson PW, D’Agostino RB, Levy D. Prediction of Coronary Heart Disease Using Risk Factor Categories. Circulation. 1998;97:1837–47. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 29.Yang H. Automatic extraction of medication information from medical discharge summaries. J Am Med Inform Assoc. 2010;17(5):545–8. doi: 10.1136/jamia.2010.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Spasic I, Keane JA, Nenadic G. A Text Mining Approach to the Prediction of Disease Status from Clinical Discharge Summaries. J Am Med Inform Assoc. 2009;16(4):596–600. doi: 10.1197/jamia.M3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Garibaldi J. Automatic Extraction of Risk Factors for Heart Disease in Clinical Texts. The 2014 i2b2 Challenge Workshop. 2014 [Google Scholar]
- 32.Yang H, Willis A, Roeck Ad, Nuseibeh B. A Hybrid Model for Automatic Emotion Recognition in Suicide Notes. Biomedical Informatics Insights. 2012;5(Suppl. 1):17–30. doi: 10.4137/BII.S8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, Negishi K, Otahal P, Marwick TH. Clinical prediction of incident heart failure risk: a systematic review and meta-analysis. Open Heart. 2015;2(1):e000222. doi: 10.1136/openhrt-2014-000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


