Abstract
Ohnologs –paralogous gene pairs generated by whole genome duplication– are enriched for dosage sensitive genes, that is, genes that have a phenotype due to copy number changes. Dosage sensitive genes frequently occur in the same metabolic pathway and in physically interacting proteins. Accumulating evidence reveals that functionally related genes tend to co-localize in the three-dimensional (3D) arrangement of chromosomes. We query whether the spatial distribution of ohnologs has implications for their dosage balance. We analyzed the colocalization frequency of ohnologs based on chromatin interaction datasets of seven human cell lines and found that ohnolog pairs exhibit higher spatial proximity in 3D nuclear organization than other paralog pairs and than randomly chosen ohnologs in the genome. We also found that colocalized ohnologs are more resistant to copy number variations and more likely to be disease-associated genes, which indicates a stronger dosage balance in ohnologs with high spatial proximity. This phenomenon is further supported by the stronger similarity of gene co-expression and of gene ontology terms of colocalized ohnologs. In addition, for a large fraction of ohnologs, the spatial colocalization is conserved in mouse cells, suggestive of functional constraint on their 3D positioning in the nucleus.
Keywords: ohnologs, dosage balance, spatial colocalization, copy number variation, disease-associated genes.
Introduction
Approximately 30% of the genes in the human genome are paralog pairs retained following whole genome duplication (WGD) events at the base of the vertebrate lineage. Called “ohnologs” to commemorate the work of Ohno (Ohno et al. 1968; Wolfe 2001), many of these genes function in development and transcription regulation (Bekaert et al. 2011) and have sometimes been suggested as candidates for laying the foundations of vertebrate diversity (McLysaght et al. 2002).
Accumulating evidence suggests that dosage balance constraint is a major determinant of duplicate gene retention. Under the dosage balance hypothesis, once genes have been duplicated by WGD, the subsequent loss of individual genes would result in a dosage imbalance because of insufficient gene product, thus leading to the biased retention of dosage-balanced ohnologs. Conversely, dosage-balanced genes are usually not retained after small-scale duplication (SSD), which disrupts relative dosage. Consistent with this hypothesis ohnologs have many characteristics expected of dosage sensitive genes: ohnolog pairs frequently occur in the same metabolic pathway and encode interacting proteins (Aury et al. 2006; Huminiecki and Heldin 2010; Bekaert et al. 2011; Rodgers-Melnick et al. 2012); gene knockout of an ohnolog causes more lethal phenotypes compared with the effects of knockout of SSDs (Makino et al. 2009); and ohnologs resist copy number variations (CNVs) in humans (Makino and McLysaght 2010), as well as gene duplications and losses during evolution (Birchler et al. 2005; Makino and McLysaght 2010; Makino et al. 2013). Thus the dosage sensitivity of these genes has constrained their evolution in characteristic ways, particularly in terms of evolutionary gene duplication and CNV frequency in healthy individuals.
Spatial co-localization of chromosomal regions in the 3D space of the nucleus has been linked with regulation of gene expression, either for activation or repression. Folding of chromosomes leads to high proximity and potential interactions between genes of different chromatin regions, including between genes from different chromosomes (Dixon et al. 2012; Sexton et al. 2012; Zhang et al. 2012; Jin et al. 2013). A considerable number of transcription factors regulate genes that are colocalized in the nucleus (Dai and Dai 2012). Moreover, functionally linked genes, including co-expressed genes, protein–protein interaction genes, and genes in the same pathway, have been reported to cluster together in physical proximity in both Escherichia coli and humans (Thevenin et al. 2014; Xie et al. 2015). Ancestrally-neighboring genes that have been separated by evolutionary genome rearrangements are often in spatial proximity and tend to be regulated by the same transcription factor and have similar histone modifications (Dai et al. 2014). These findings inspired us to consider whether dosage sensitive ohnolog pairs have some spatial localization features even when unlinked.
In this work, the wealth of valuable in situ Hi-C data collected for seven human cell lines was harnessed and used to generate chromatin contact matrices. With these data, we investigated the colocalization frequency between ohnolog pairs as well as SSD pairs and found frequent spatial colocalization of ohnologs but not of SSD paralogs. We compared the colocalized ohnologs with the non-colocalized ohnologs with respect to CNVs and disease-associated genes, and found that the colocalized ohnologs were less likely to experience CNVs and more likely to involve disease-associated genes. This suggests that spatially colocalized ohnolog pairs act to maintain dosage balance in the human genome.
Results and Discussion
Ohnologs Tend to Exhibit More Colocalization in the Nucleus
Colocalization of Ohnologs
We analyzed the colocalization frequency of the ohnologs identified by Makino and McLysaght (2010) based on the chromatin interactions of seven human cell lines: GM12878, HMEC, HUVEC, IMR90, K562, KBM7, and NEHK, all of which were used to generate chromatin contact maps by hiclib with default parameters (see Methods) (Imakaev et al. 2012). In order to avoid spatial proximity caused by linear gene arrangement (Dai et al. 2014; Thevenin et al. 2014), we considered only interchromosomal ohnolog pairs, excluding all intrachromosomal ohnologs (Makino and McLysaght 2010). A total of 7,923 interchromosomal ohnologs in human were identified (Makino and McLysaght 2010), of which 7,655 interchromosomal ohnologs with chromatin interaction information were used for further analysis (see Methods). The contact frequencies between ohnologs were derived from the interaction information of DNA fragments. Chromatin fragment pairs with sufficient contact frequencies (FDR <0.05) were considered to be nonrandom and spatially colocalized (see Methods) (Duan et al. 2010; Dai and Dai 2012; Dai et al. 2014).
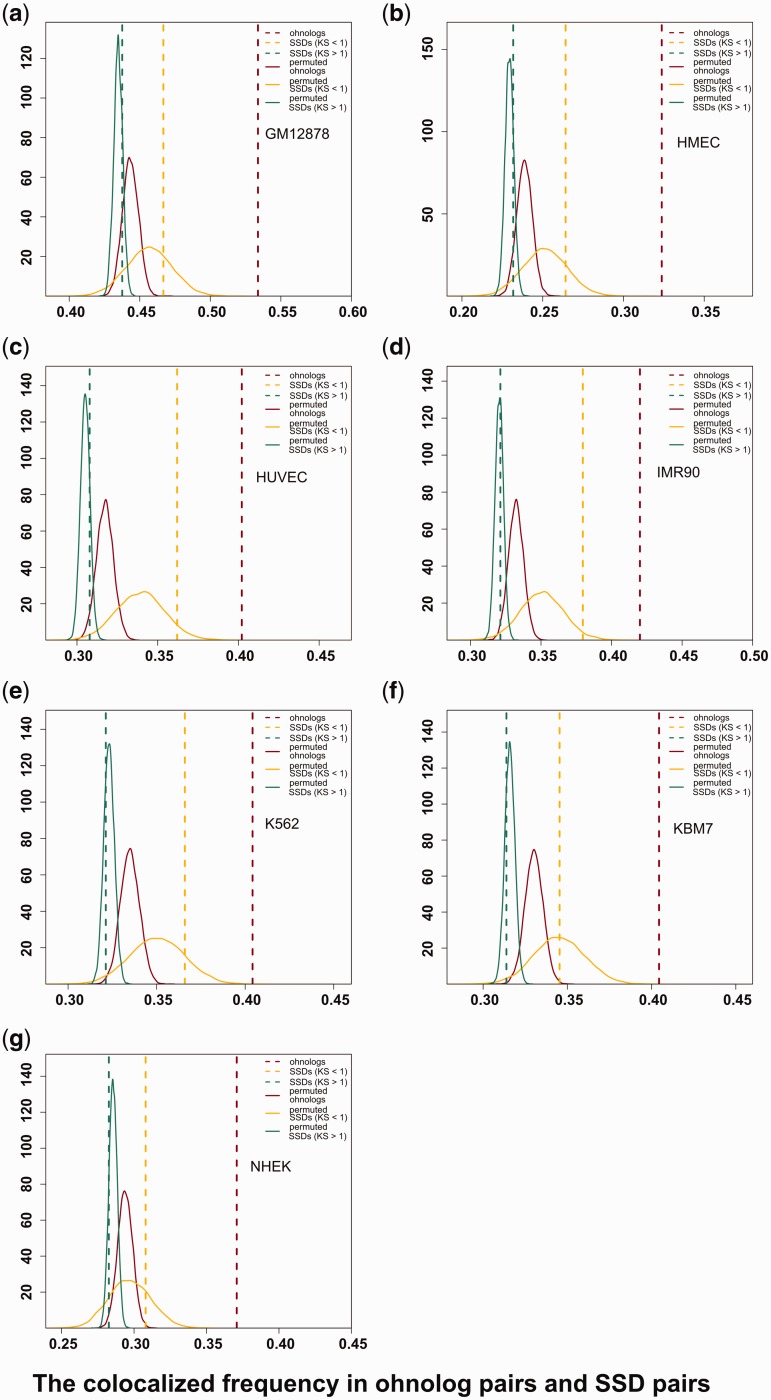
In the lymphoblastoid cell line (GM12878), we found that 4,086 of 7,655 (53.38%) interchromosomal ohnologs were colocalized (fig. 1a) and the values range from 32.37% to 42.01% colocalization for the other cell lines (fig. 1b–g and table 1). If close spatial proximity is a particular feature of ohnologs, then their colocalization frequencies should be higher than the frequencies expected by chance. To ensure that the contacts we observed here were not caused by an imbalanced chromosomal distribution of the ohnologs along the genome, the contact between ohnolog pairs were compared with those of randomized ohnolog pairs by conducting a permutation test. We generated random gene pairs equal in number to observed ohnolog pairs. In this way, we permuted ohnolog pairs 10,000 times, and found that the frequencies of colocalization for all the randomized experiments were significantly lower than those of real ohnologs in all seven cell lines (fig. 1, brown lines).
Fig. 1.
The colocalization frequency of interchromosomal ohnolog pairs and SSD pairs in seven cell lines. In each panel, the x-axis represents the percentage of colocalized interchromosomal ohnolog pairs and SSD pairs in the different cell lines. The brown, yellow, and green vertical dashed lines indicate the observed colocalization frequency for ohnologs pairs, young SSD pairs, and old SSD pairs, respectively. The curves show the colocalization frequency distributions for 10,000 permuted randomizations of the same number of pairs as in the real data.
Table 1.
Basic Information for Colocalized and Noncolocalized Ohnolog Pairs in Seven Human Cell Lines.
| Cell lines |
|||||||
|---|---|---|---|---|---|---|---|
| GM12878 | HMEC | HUVEC | IMR90 | K562 | KBM7 | NHEK | |
| Colocalized ohnolog pairs (%)a | 4086 (53.38) | 2478 (32.37) | 3078 (40.21) | 3216 (42.01) | 3094 (40.42) | 3096 (40.44) | 2839 (37.09) |
| Non-colocalized ohnolog pairs (%)a | 3569 (46.62) | 5177 (67.63) | 4577 (59.79) | 4439 (57.99) | 4561 (59.52) | 4559 (59.56) | 4816 (62.91) |
| Genes involved in colocalized ohnolog pairs | 4638 | 2984 | 3639 | 3721 | 3647 | 3698 | 3366 |
| Genes involved in non-colocalized ohnolog pairs | 4296 | 5609 | 5048 | 4877 | 4972 | 5048 | 5352 |
| Genes involved in colocalized ohnolog pairs with CNVs (%)b | 3031 (63.20) | 1882 (63.07) | 2320 (63.75) | 2125 (63.13) | 2377 (63.88) | 2293 (62.87) | 2351 (63.57) |
| Genes involved in non-colocalized ohnolog pairs with CNVs (%)c | 2878 (66.99) | 3765 (67.12) | 3398 (67.31) | 3619 (67.62) | 3280 (67.25) | 3362 (67.62) | 3394 (67.23) |
| Colocalized ohnolog pairs with GWAS disease genes (%)d | 1064 (26.04) | 664 (26.80) | 812 (26.38) | 752 (26.49) | 854 (26.55) | 831 (26.86) | 820 (26.49) |
| Non-colocalized ohnolog pairs with GWAS disease genes (%)e | 894 (23.82) | 1314 (24.13) | 1166 (24.07) | 1226 (24.11) | 1124 (23.88) | 1147 (23.75) | 1158 (23.99) |
| Colocalized ohnolog pairs with OMIM disease genes (%)d | 929 (22.74) | 558 (22.52) | 702 (22.81) | 640 (22.54) | 768 (23.88) | 696 (22.50) | 739 (23.87) |
| Non-colocalized ohnolog pairs with OMIM disease genes (%)e | 775 (20.20) | 1146 (21.05) | 1002 (20.68) | 1064 (20.93) | 936 (19.89) | 1008 (20.87) | 1025 (21.23) |
aPercentage is with respect to ohnolog pairs (the number is 7,655).
bPercentage is with respect to genes involved in colocalized ohnolog pairs.
cPercentage is with respect to genes involved in non-colocalized ohnolog pairs.
dPercentage is with respect to colocalized ohnolog pairs.
ePercentage is with respect to non-colocalized ohnolog pairs.
If spatial proximity is simply a result of sequence similarity, then all classes of paralogs should experience spatial colocalization equally. We investigated the colocalization frequency of other types of paralogs. Specifically, we wondered whether genes that had undergone SSD were also in close spatial proximity. We calculated the colocalization frequency in 25,809 SSD gene pairs, which were derived from the Ensembl database (version 79) (see Methods).
Approximate gene duplication times can be estimated based on synonymous substitution rates (KS) values, with lower and higher KS values corresponding to recent and ancient duplication events, respectively. There were 1,022 and 24,787 SSD genes for which KS was <1 (young SSDs) and ≥1 (old SSDs), respectively. The frequency of colocalization in SSD pairs was consistently lower than that of ohnologs and was similar to the random gene pairs in all seven cell lines (fig. 1, yellow and green lines). Thus spatial organization of ohnologs is not random and ohnologs retained after WGD events exhibit more colocalization in the nucleus which is absent in SSDs.
Conservation of Colocalization in Ohnologs
It is notable that SSD paralogs with lower KS (young SSDs) had a higher frequency of spatial colocalization than more divergent SSD paralogs (fig. 1). This is suggestive of cis sequence domains that regulate spatial localization becoming eroded over time. This stimulated our interest to investigate whether the interchromosomal colocalization of ohnologs is conserved in other genomes.
By using the chromatin interaction data for the mouse Lymphoblastic cell line (CH12-LX) (Rao et al. 2014), we identified 910 colocalized ohnologs and 1,768 noncolocalized ohnologs in mouse from Singh et al. (2015) (see Methods). Through comparing the colocalized ohnolog pairs between mouse CH12-LX and human GM12878, it was found that there was a large overlap of orthologous colocalized ohnologs between human and mouse. As many as 50.66% (461/910) of mouse colocalized ohnologs are orthologous gene pairs of colocalized ohnologs in human (whereas ohnologs that are noncolocalized in mouse are rarely colocalized in human (595/1,768), P < 1.48 × 10−17 Fisher’s exact test). This finding that the spatial colocalization is conserved for a large fraction of the anciently duplicated ohnologs is suggestive of functional constraint on their 3D positioning in the nucleus.
Colocalized Ohnologs Are More Dosage Sensitive
Because genes with high spatial proximity tend to be in the same functional group and have stronger regulatory linkages, we considered whether the dosage sensitivity of ohnologs is closely related to their spatial colocalization. Specifically, we hypothesized that colocalized ohnologs should be enriched for dosage-sensitive genes.
According to the dosage-balance hypothesis, either underexpression or overexpression of a dosage-sensitive gene can lower fitness; therefore, the causative mutation would be removed by purifying selection (Makino et al. 2013). Thus, CNVs of dosage-sensitive genes are deleterious and may lead to human disease or inviability (Makino and McLysaght 2010). It has been demonstrated that ohnologs are often dosage-sensitive and have fewer CNVs and more disease-associated genes (Makino and McLysaght 2010). If spatial proximity is a characteristic of dosage sensitive ohnologs, then the CNV and disease-associated gene frequencies should vary significantly between colocalized ohnologs and noncolocalized ohnologs.
Colocalized Ohnologs Are Less Likely to Experience CNV and More Likely to Be Disease-Associated
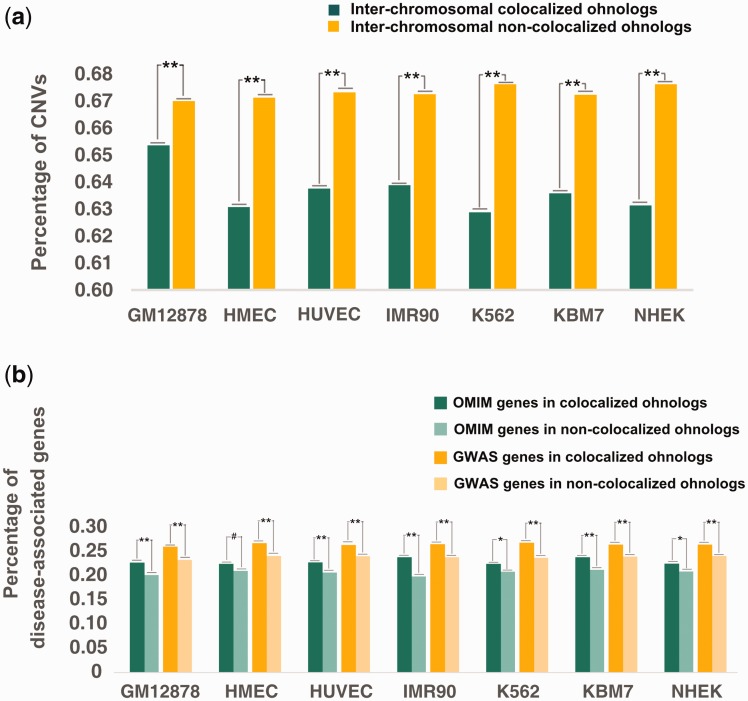
To investigate the relationship of ohnolog spatial organization and dosage balance, we compared the occurrence of CNVs between colocalized and noncolocalized ohnologs. As before, we only considered interchromosomal ohnologs. Any gene with the entire coding sequence found within a CNV region was considered to have a CNV, as per Makino and McLysaght (2010). As shown in figure 2a, in cell line GM12878, we found that (63.20%, 3,031/4,638) of the colocalized ohnologs had CNVs, and this was a significantly lower fraction than that found in noncolocalized ohnologs (66.99%, 2,878/4,296; P = 2.36 × 10−5, Fisher’s exact test). This is also true in all other cell lines (fig. 2a and table 1). Previous studies have uncovered CNV deserts in ohnolog-rich regions (Makino et al. 2013). The present results reveal that the contents of CNVs in ohnologs are influenced not only by the linear organization of ohnologs but also by their 3D nuclear organization.
Fig. 2.
Percentage of colocalized and noncolocalized ohnologs of seven cell lines that are (a) present in CNVs and (b) disease-associated genes identified in OMIM or by GWAS. P-values shown in the panels were calculated using Fisher’s exact test; ** represents P < 0.05, * represents 0.05 < P < 0.10 and # represents P > 0.10.
We retrieved 4,842 disease-associated genes from the updated “Morbidmap” database of Online Mendelian Inheritance in Man (OMIM). We also obtained genome-wide association study (GWAS)-identified disease genes from Nelson et al.’s work (Nelson, et al. 2015). The OMIM- and GWAS-derived disease genes were largely non-overlapping. We compared the colocalized and non-colocalized ohnologs with respect to the frequency of disease-associated genes in both disease gene datasets (see Methods). In GM12878, the frequency of OMIM disease genes was 22.74% in the colocalized ohnologs while this percentage drops to 20.20% in the noncolocalized ohnologs (P = 2.81 × 10−3, Fisher’s exact test, fig. 2b), and the frequency of GWAS-identified human disease genes was 26.04% in the colocalized ohnologs while this percentage drops to 23.82% in the noncolocalized ohnologs (P = 4.70 × 10−3 Fisher’s exact test, fig. 2b). This is also true in all other cell lines (fig. 2b and table 1). This observation that colocalized ohnologs are more resistant to CNVs and include more disease-associated genes than remote ohnologs is suggestive of a biological link between spatial proximity and dosage balance.
Colocalized Ohnologs Are Co-Expressed and Operate in Similar GO Categories
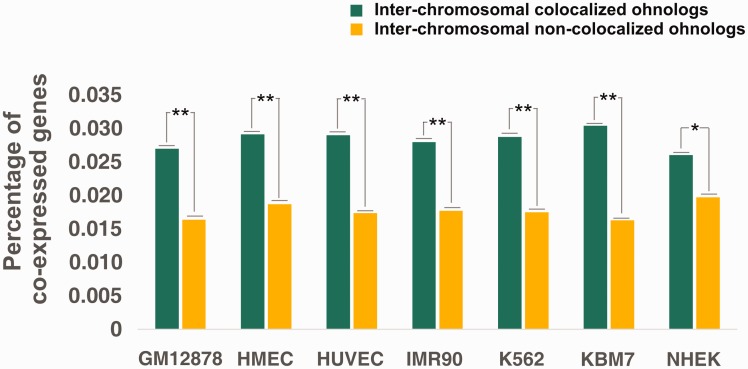
We compared the coexpression of colocalized and noncolocalized ohnologs using the human gene coexpression data derived from COXPRESdb (version 5.0; release date: 2012.08.29) (http://coxpresdb.hgc.jp) (Okamura et al. 2015). We used Pearson’s correlation coefficients (PCCs) to measure the coexpression level of the ohnolog pairs. PCC >0.4 is widely accepted as an indicator of co-expression (Pujana et al. 2007; Das et al. 2012). The coexpression frequency of colocalized ohnologs was significantly larger than that of noncolocalized ohnologs in GM12878 (P = 5.99 × 10−3, Fisher’s exact test) and in the other six cell lines (fig. 3). These results clearly show that colocalized ohnologs have a higher tendency to coexpression than noncolocalized ohnologs, consistent with previous observations of coexpression of colocalized genes (Dai et al. 2014; Xie et al. 2015).
Fig. 3.
Frequency of coexpression of colocalized and noncolocalized ohnologs of seven cell lines. P-values shown in the panels were calculated using Fisher’s exact test; ** represents P < 0.05 and * represents 0.05 < P < 0.10.
The GO-based Czekanowski–Dice distance was used to investigate the GO term similarity of colocalized and noncolocalized ohnologs (see Methods). The smaller the distance is, the more similar function the ohnologs have (Ovaska et al. 2008). The Czekanowski–Dice distance of colocalized ohnologs was significantly smaller than that of noncolocalized ohnologs tested by Wilcoxon test, indicating a smaller functional difference in colocalized ohnologs (table 2). Together, these results indicate that ohnologs in spatial proximity tend to be co-regulated, with higher coexpression levels and GO term similarity. Similar observations of greater functional overlap between ohnologs as compared with SSD paralogs in yeasts are suggestive of stoichiometric constraints (Fares et al. 2013; Vo et al. 2016), as is predicted for dosage-balanced genes. Such stoichiometric constraints are expected of dosage-balanced genes and we suggest that spatial co-localization of ohnologs in human cells is at least partly responsible for maintaining their stoichiometric balance.
Table 2.
Comparison of GO-Based Czekanowski–Dice Distance (average ± sd) for Unlinked Ohnologs.
| Cell lines |
|||||||
|---|---|---|---|---|---|---|---|
| GM12878 | HMEC | HUVEC | IMR90 | K562 | KBM7 | NEHK | |
| Colocalized ohnologs | 0.512 ± 0.066 | 0.512 ± 0.068 | 0.509 ± 0.065 | 0.506 ± 0.066 | 0.508 ± 0.066 | 0.501 ± 0.066 | 0.515 ± 0.067 |
| Noncolocalized ohnologs | 0.549 ± 0.073 | 0.538 ± 0.070 | 0.543 ± 0.072 | 0.546 ± 0.072 | 0.544 ± 0.072 | 0.548 ± 0.071 | 0.538 ± 0.071 |
| P-value (Wilcoxon test) | 6.57 × 10−8 | 3.54 × 10−4 | 1.63 × 10−6 | 5.64 × 10−8 | 3.10 × 10−7 | 2.38 × 10−11 | 4.73 × 10−3 |
Concluding Remarks
There is an increasingly clear link between complex eukaryotic transcriptional regulation and the 3D organization of eukaryotic chromosomes. In particular, genes in chromosomal regions in spatial proximity are often co-regulated (Dai and Dai 2012; Dai et al. 2014; Thevenin et al. 2014; Xie et al. 2015). Previous work has shown that ohnologs are enriched for dosage-sensitive genes which have constrained stochiometric ratios (Makino and McLysaght 2010). The analysis presented here reveals that: (1) ohnologs are not randomly distributed in 3D space, and have a higher colocalization frequency than other types of paralogs or random ohnolog pairs; and (2) colocalized ohnologs are less likely to have benign CNVs and are more likely to be human disease-associated genes as compared with spatially remote ohnologs. Thus, spatial colocalization is a characteristic of ohnologs as distinct from other paralogs and these colocalized gene pairs are more dosage sensitive than others. It is difficult to know whether the nuclear localization patterns of ohnologs predates duplication and is maintained by evolutionary constraint, or is a post-duplication adaptation to maintain dosage balance. The precise basis for dosage balance remains unclear but our observation that colocalized ohnologs maintain high co-expression and functional similarity supports the prediction that spatial colocalization is a means to achieve co-regulation and thus stoichiometric balance.
Materials and Methods
Data Sources
Unlinked Ohnologs and SSDs
A total of 9,057 ohnolog pairs in the human genome were obtained from Makino and McLysaght (2010), of which 7,923 were located on different human chromosomes and 7,655 ohnologs with chromatin interaction information were retained for subsequent analysis. Among 46,114 nonredundant duplicate pairs obtained from Ensembl v79, 39,736 SSDs remained after removing ohnolog pairs. The SSD pairs whose genes were located on the same chromosome were also removed. Finally, 26,365 SSD pairs involving 10,434 genes were derived and only 25,809 SSD pairs with chromatin interaction information were used for further analysis. A total of 2,689 ohnolog pairs in mouse were obtained from Singh et al. (2015) and 2,678 ohnologs pairs with chromatin interaction information were used for further analysis.
Chromatin Interaction Data
Chromatin interaction data from seven human cell lines were obtained from Rao et al. (2014) (GEO accession: GSE63525), which included lymphoblastoid cells (GM12878), human mammary epithelial cells (HMECs), human umbilical vein endothelial cells (HUVECs), human fetal lung cells (IMR90), epidermal keratinocytes (K562), chronic myelogenous leukemia cells (KBM7), and near haploid myelogenous leukemia cells (NHEK).
Copy Number Variants
We downloaded CNVs for the human genome from the Database of Genomic Variants (http://projects.tcag.ca/variation/, Release date: 2014-10-16). When the entire coding sequence of an ohnolog was contained within one of the CNVs, we defined the gene as a CNV gene.
Disease-Associated Genes
We extracted 4,842 disease-associated genes from the updated “Morbidmap” database of OMIM (ftp://ftp.omim.org/OMIM/morbidmap, Release 2015.11.11). GWAS-derived SNP-trait pairs were obtained from Nelson et al. (2015). After filtering the SNPs for special traits (e.g., hair color, eye color, and birth weight), 8,415 SNPs linked with 4,616 disease genes were collected.
Data Preparation and Statistical Analysis
Chromatin Interaction Derivation
For each chromatin conformation capture (Hi-C) dataset, a chromatin interaction matrix was created using the following procedures. Hi-C reads were aligned to the reference genome using Bowtie 2.1.0 (Langmead and Salzberg 2012) with default parameter settings, and an algorithm that iteratively increases the truncation length (20 bp) to maximize the yield of valid Hi-C interactions was adopted (Imakaev et al. 2012; Le et al. 2013). We retained for further analysis only pairs of reads with a unique hit of high quality (q > 30) at both ends. Subsequent analyses were performed to remove invalid read pairs. The reference genome was divided into restriction fragments by conceptually cutting them at the HindIII enzyme restriction site “AAGCTT” and this process yielded 830,194 fragments. Each read pair end was sorted into its corresponding restriction fragment. Nonligation and self-ligation products that were recognized by restriction fragment and mapping orientation were filtered (Imakaev et al. 2012). Other invalid read pairs were all removed (Diagonal, StartNearRsite, PCR amplification, random break, LargeSmallFragments, and ExtremeFragments were filtered with the default parameter settings in hiclib) (Yaffe and Tanay 2011; Imakaev et al. 2012). The quantity of the Hi-C reads after filtering in the human cell lines is listed in supplementary table S1, Supplementary Material online. We then partitioned the human genome into nonoverlapping 500 kb windows and referred to the number of filtered read pairs in the windows as the corresponding contact count of the bins (supplementary table S1, Supplementary Material online).
Calculation of Statistical Significance and FDR
To assign statistical significance to estimates of interchromosomal interactions, we used a uniform probability model to convert the observed frequencies into P-values (Duan et al. 2010; Dai et al. 2014). To do so, we counted the total number M of interchromosomal pairs of restriction fragments in the human genome. When counting these fragments, we only considered pairs in which both fragments were mapped as defined above. Assuming that the probability of observing any particular interaction is uniform, then the probability is . We then counted the total number n of observed interchromosomal interactions. The probability of observing a given interaction pair exactly k times can be calculated through a binomial distribution (Duan et al. 2010; Dai et al. 2014):
In our case, n was sufficiently large, and p was sufficiently small. Therefore, the Poisson distribution with parameter can be used as an approximation for of the binomial distribution:
We applied an FDR cutoff of 0.05 to filter the noise of the Hi-C data, and the FDR values for each map are shown in supplementary table S1, Supplementary Material online.
The contact frequencies between ohnologs were derived from the interaction information of DNA fragments. The contact of an ohnolog pair wherein one gene is located in bin i and the other gene is located in bin j was represented by the corresponding contact between bin i and bin j. These data are available upon request.
GO Term Similarity Measurement
We used the GO-based Czekanowski–Dice distance to evaluate the GO term similarity of colocalized and noncolocalized ohnologs (Ovaska et al. 2008). The Czekanowski–Dice functional distance (Dist) is defined by Equation (1):
| (1) |
where x and y denote one duplicate of unlinked ohnologs, Terms(x) and Terms(y) are the sets of their associated GO annotations, # indicates the “number of”, and Δ indicates the symmetrical difference between the two sets. The GO term information of the ohnolog pairs was obtained from the Ensembl database (version 79).
Supplementary Material
Supplementary table S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We are grateful to Professor Zhixi Su (Fudan University) for helpful discussion. The research leading to these results has received funding from the National Basic Research Program of China (973 project, Grant 2012CB721000), the National Natural Science Foundation of China (Grant 31370776) (to H.-Y. Zhang) and the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007–2013)/ERC Grant Agreement 309834 (to A.Mc.L).
References
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, et al. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171–178. [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC. 2011. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell 23:1719–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA. 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21:219–226. [DOI] [PubMed] [Google Scholar]
- Dai Z, Dai X. 2012. Nuclear colocalization of transcription factor target genes strengthens coregulation in yeast. Nucleic Acids Res. 40:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Xiong Y, Dai X. 2014. Neighboring genes show interchromosomal colocalization after their separation. Mol Biol Evol. 31:1166–1172. [DOI] [PubMed] [Google Scholar]
- Das J, Mohammed J, Yu HY. 2012. Genome-scale analysis of interaction dynamics reveals organization of biological networks. Bioinformatics 28:1873–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. 2010. A three-dimensional model of the yeast genome. Nature 465:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares MA, Keane OM, Toft C, Carretero-Paulet L, Jones GW. 2013. The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes. PLoS Genet. 9:e1003176.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huminiecki L, Heldin CH. 2010. 2R and remodeling of vertebrate signal transduction engine. BMC Biol. 8:146.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. 2012. Iterative correction of Hi-C data reveals hallmarks of chromosome organization.Nat Methods 9:999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TB, Imakaev MV, Mirny LA, Laub MT. 2013. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, Hokamp K, McLysaght A. 2009. The complex relationship of gene duplication and essentiality. Trends Genet. 25:152–155. [DOI] [PubMed] [Google Scholar]
- Makino T, McLysaght A. 2010. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 107:9270–9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, McLysaght A, Kawata M. 2013. Genome-wide deserts for copy number variation in vertebrates. Nat Commun. 4:2283.. [DOI] [PubMed] [Google Scholar]
- McLysaght A, Hokamp K, Wolfe KH. 2002. Extensive genomic duplication during early chordate evolution. Nat Genet. 31:200–204. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Tipney H, Painter JL, Shen JD, Nicoletti P, Shen YF, Floratos A, Sham PC, Li MJ, Wang JW, et al. 2015. The support of human genetic evidence for approved drug indications. Nat Genet. 47:856–860. [DOI] [PubMed] [Google Scholar]
- Ohno S, Wolf U, Atkin NB. 1968. Evolution from fish to mammals by gene duplication. Hereditas 59:169–187. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Aoki Y, Obayashi T, Tadaka S, Ito S, Narise T, Kinoshita K. 2015. COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic Acids Res. 43:D82–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovaska K, Laakso M, Hautaniemi S. 2008. Fast gene ontology based clustering for microarray experiments. BioData Min. 1:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al. 2007. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 39:1338–1349. [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Mane SP, Dharmawardhana P, Slavov GT, Crasta OR, Strauss SH, Brunner AM, Difazio SP. 2012. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 22:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148:458–472. [DOI] [PubMed] [Google Scholar]
- Singh PP, Arora J, Isambert H. 2015. Identification of ohnolog genes originating from whole genome duplication in early vertebrates, based on synteny comparison across multiple genomes. PLoS Comput Biol. 11:e1004394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenin A, Ein-Dor L, Ozery-Flato M, Shamir R. 2014. Functional gene groups are concentrated within chromosomes, among chromosomes and in the nuclear space of the human genome. Nucleic Acids Res. 42:9854–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo TV, Das J, Meyer MJ, Cordero NA, Akturk N, Wei X, Fair BJ, Degatano AG, Fragoza R, Liu LG, et al. 2016. A proteome-wide fission yeast interactome reveals network evolution principles from yeasts to human. Cell 164:310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH. 2001. Yesterday's polyploids and the mystery of diploidization. Nat Rev Genet. 2:333–341. [DOI] [PubMed] [Google Scholar]
- Xie T, Fu LY, Yang QY, Xiong H, Xu H, Ma BG, Zhang HY. 2015. Spatial features for Escherichia coli genome organization. BMC Genomics 16:37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. 2011. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet. 43:1059–1065. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. 2012. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148:908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.