Abstract
The bloom of genomics is revealing gene loss as a pervasive evolutionary force generating genetic diversity that shapes the evolution of species. Outside bacteria and yeast, however, the understanding of the process of gene loss remains elusive, especially in the evolution of animal species. Here, using the dismantling of the retinoic acid metabolic gene network (RA-MGN) in the chordate Oikopleura dioica as a case study, we combine approaches of comparative genomics, phylogenetics, biochemistry, and developmental biology to investigate the mutational robustness associated to biased patterns of gene loss. We demonstrate the absence of alternative pathways for RA-synthesis in O. dioica, which suggests that gene losses of RA-MGN were not compensated by mutational robustness, but occurred in a scenario of regressive evolution. In addition, the lack of drastic phenotypic changes associated to the loss of RA-signaling provides an example of the inverse paradox of Evo–Devo. This work illustrates how the identification of patterns of gene coelimination—in our case five losses (Rdh10, Rdh16, Bco1, Aldh1a, and Cyp26)—is a useful strategy to recognize gene network modules associated to distinct functions. Our work also illustrates how the identification of survival genes helps to recognize neofunctionalization events and ancestral functions. Thus, the survival and extensive duplication of Cco and RdhE2 in O. dioica correlated with the acquisition of complex compartmentalization of expression domains in the digestive system and a process of enzymatic neofunctionalization of the Cco, while the surviving Aldh8 could be related to its ancestral housekeeping role against toxic aldehydes.
Keywords: gene loss, gene coelimination, regressive evolution, evo–devo, retinoic acid, chordate.
Introduction
The recent bloom of genomic data is revealing a novel perspective of gene loss as a pervasive source of genetic variation with a great potential to generate phenotypic diversity and to shape the evolution of gene networks (Albalat and Cañestro 2016). Several fundamental questions regarding the evolutionary role of gene loss, however, still remain elusive. How do genes become dispensable and subsequently lost? How do patterns of gene loss appear to be biased rather than stochastic? What is the impact of gene loss on the evolution of the rest of surviving components of gene networks? In a simplified view, gene dispensability and the evolutionary impact of the subsequent loss of dispensable genes have been mainly associated either with the mutational robustness of biological systems (i.e., the capability of maintaining unaltered phenotypes in the face of mutations) or with changes in the functional requirements caused by the relaxation of environmental constraints (Albalat and Cañestro 2016). In the scenario of a robust genetic system, a gene can become dispensable when its loss does not imply the loss of a biological function due either to the presence of “backup genes” (e.g., redundant paralogs with overlapping functions) (Gu et al. 2003) or of “backup pathways” (e.g., alternative pathways by which the biological function can be rerouted) (Wagner 2005). Alternatively, a gene can become dispensable in a scenario of regressive evolution characterized by the loss of useless traits due to relaxation of environmental constraints, in which the loss of trait-related genes becomes neutral—for example, gene losses related to vision and pigmentation in cavefish after colonization of dark environments (Protas et al. 2006)—; or in a scenario of adaptive evolution in which the loss of the function is advantageous, and therefore positively selected—that is, the “less is more” hypothesis, classically exemplified by the advantageous losses of the human genes coding for cell receptors CCR5 and DUFFY that provide resistance to AIDS and vivax malaria, respectively (Olson 1999; Olson and Varki 2003).
Interestingly, patterns of gene loss do not appear to be stochastic, but they show clear biases depending on gene functions or genomic positions of the lost genes (reviewed in Albalat and Cañestro 2016). A special case of functional bias of gene loss is observed in species that suffer relaxation of a given biological or environmental constraint, which leads to the “coelimination” of genes that are functionally linked in distinct pathways associated with the relaxed constraint (Aravind et al. 2000; Koonin et al. 2004). The identification of patterns of gene coelimination appears therefore as a useful strategy to recognize components of functional modules within gene networks, whereas the identification of “survival genes” after the dismantling of a pathway is useful to recognize “hub” genes that cannot be lost owing their pleiotropic nature acting in multiple pathways (reviewed in Albalat and Cañestro 2016).
Most of the theoretical framework about gene dispensability and biased patterns of gene loss and survival has arisen from laboratory evolution experiments and computational models using bacteria and yeast (reviewed in Albalat and Cañestro 2016). The impact of losses affecting gene networks of multicellular eukaryotes (i.e., animals or plants), however, remains elusive owing their highly elaborated transcriptional regulatory circuits, complex signaling pathways, and frequently convoluted mechanisms of development that are less suitable for experimental or computational studies. Here, we take advantage of the availability of the deeply sequenced genomes of many chordates—including the larvacean urochordate Oikopleura dioica characterized by a high propensity to loose genes (Denoeud et al. 2010)—in order to examine in detail the evolution of an animal gene network affected by patterns of gene coelimination and survival. Thus, using the dismantling of the robust retinoic acid metabolic gene network (RA-MGN) in O. dioica as a case study, we use a multidisciplinary approach that includes not only evolutionary genomics, and phylogenetic reconstructions but also analytical biochemistry and developmental gene expression analyses, in order to provide experimental evidence supporting a plausible scenario for the gene losses in the RA-MGN and to argue about their impact on the evolution of gene networks and the phenotypic diversification.
The RA-MGN regulates the synthesis and degradation of all-trans-retinoic acid (atRA), a vitamin A derived compound that acts as a crucial signaling pathway for embryo development and adult tissue homeostasis in chordates, including humans (fig. 1) (Rhinn and Dolle 2012; Duester 2013; Cunningham and Duester 2015). The canonical pathway that regulates the synthesis of atRA during chordate embryogenesis consists of Rdh10 and Aldh1a (typically Aldh1a1, Aldh1a2, and Aldh1a3; formerly Raldhs) enzymes that catalyze two sequential oxidative reactions, first from all-trans-retinol (atROL, vitamin A) to all-trans-retinaldehyde (atRAL) and second from atRAL to atRA, respectively (Chen et al. 1995; Niederreither et al. 1999; Dalfó et al. 2002; Duester et al. 2003; Cammas et al. 2007; Belyaeva et al. 2008; Cañestro et al. 2009; Strate et al. 2009). These RA-producing enzymes—Rdh10 and Aldh1a—are coordinated with various RA-degrading enzymes of the cytochrome P450 subfamily 26 (Cyp26; typically Cyp26a, Cyp26b, and Cyp26c) to establish the spatiotemporal levels of atRA (Niederreither et al. 2002; Reijntjes et al. 2005; Schilling et al. 2012).
Fig. 1.
Schematic representation of the RA-MGN in vertebrates that establishes the levels of atRA, whose signaling regulates developmental genes (e.g., Hox1) through nuclear receptors (e.g., RAR and PPAR). The enzymes of the canonical pathway (in bold: Rdh10, Aldh1a, and Cyp26) regulate the synthesis of atRA from atROL (vitamin A) to atRAL precursors. In vertebrates, the RA-MGN is a robust system due to the presence of other redundant enzymes that bypass the canonical pathway (i.e., RdhE2, Rdh16, Adh1/4, Aldh8a1, and Cco).
The RA-MGN has been suggested to be a robust genetic system in vertebrates. In addition to the multiplicity of the enzymes in the canonical pathway, several other enzymes with redundant activities have been suggested to be able to bypass the canonical pathway and produce atRA (fig. 1). Aldh8a1, for instance, may bypass Aldh1a and catalyze the oxidation of atRAL to atRA (Lin and Napoli 2000; Lin et al. 2003; Liang et al. 2008), and RdhE2, Rdh16 and various vertebrate-specific Adhs (e.g., Adh1 and Adh4) may replace Rdh10 and catalyze atROL to atRAL oxidation (Gough et al. 1998; Jurukovski et al. 1999; Cañestro et al. 2000; Molotkov, Deltour, et al. 2002; Molotkov, Fan, et al. 2002; Cañestro et al. 2003, 2010; Lee et al. 2009; Belyaeva et al. 2012). Furthermore, atRAL can be alternatively produced by a “backup pathway” (Lindqvist and Andersson 2004) that consists of the enzymatic cleavage of dietary β-carotene by carotenoid cleavage oxygenases (Cco) enzymes, directly by β,β-carotene-15,15′-monooxygenase (Bco1) enzyme (von Lintig and Vogt 2000; Lampert et al. 2003), or indirectly, via apocarotenoids, by the β,β-carotene-9,10′-dioxygenase (Bco2) enzyme (Wang et al. 1996).
Previous work had revealed that O. dioica has lost two of the components of the canonical RA-MGN (i.e., Aldh1a and Cyp26) (Cañestro et al. 2006). However, whether O. dioica can bypass this loss and is capable of synthesizing atRA using alternative “backup” genes or pathways in a robust genetic system, remained unknown. To evaluate this possibility, in this work, we have first searched for patterns of coelimination affecting the RA-MGN, as well as survival genes of the RA-MGN that could function as redundant genes (e.g., RdhE2, Rdh16, and Aldh8a1) or alternative pathways (e.g., Cco). Second, we have investigated whether the dismantling of the RA-MGN was accompanied by the loss of biological function, that is, atRA production. Third, we have analyzed the functions of surviving genes based on their biochemical capabilities and their expression patterns during O. dioica development. And finally, we discuss possible evolutionary scenarios that may have facilitated gene losses, and whether the survival genes of the RA-MGN in O. dioica were preserved because the retention of ancestral functions or because they underwent neofunctionalization processes.
Results
Genomic Survey of the RA-MGN in O. dioica
In order to evaluate the mutational robustness of RA-MGN in O. dioica, we first investigated the existence of canonical and backup genes or pathways in this species by surveying its genome databases for all RA-MGN components known in other chordate species (fig. 1).
Rdh10 and RdhE2: Vertebrate Rdh10 (for retinol dehydrogenase 10) genes belong to the short-chain dehydrogenase/reductase (SDR)-16C family (Albalat et al. 2011; Belyaeva et al. 2015), and have been suggested to encode for the main enzyme responsible for the synthesis of atRAL from atROL during embryogenesis (Sandell et al. 2007; Belyaeva et al. 2008). RdhE2 (for epidermal retinal dehydrogenase 2) genes belong to the same SDR-16C family, and they have been shown to have the same biochemical activity (Matsuzaka et al. 2002; Belyaeva et al. 2012). Both Rdh10 and RdhE2 enzymes have the typical domains of SDR enzymes that are essential for their dehydrogenase function: 1) A Gly-rich sequence in the variable N-terminal region critical for accommodation and binding of the cofactor (Lesk 1995), 2) one acidic residue located about 20 residues downstream the Gly-rich sequence necessary for NAD(H) specificity (Wierenga et al. 1986), and 3) the Asn-Ser-Tyr-Lys tetrad that forms the active site necessary for catalysis (Albalat et al. 1992; Filling et al. 2002). BLAST searches of O. dioica genome database with Homo sapiens and Ciona intestinalis Rdh10 or RdhE2 retrieved four O. dioica genes (supplementary file S1, Supplementary Material online, CBY07137, CBY07135, CBY07673, and CBY12938; E-values < e − 50). Their best reciprocal BLAST hits (BRBH) retrieved back RdhE2, but no Rdh10, with E-values ranging from 2e − 82 to 1e − 89. In agreement with the BRBH results, phylogenetic gene trees inferred by maximum likelihood (ML) method showed that the four O. dioica genes did not group with the Rdh10 enzymes, but grouped together within the RdhE2 cluster, suggesting that O. dioica lacks an ortholog of chordate Rdh10 genes (fig. 2). The fact that Rdh10 genes had been identified in the genomes of the cephalochordate Branchiostoma floridae and the urochordate C. intestinalis (fig. 2) (Albalat et al. 2011; Belyaeva et al. 2015) indicated that the absence of an O. dioica Rdh10 was due to a gene loss in the RA-MGN of the larvacean lineage. The four O. dioica paralogs (named RdhE2a to RdhE2d) appeared to be orthologous to chordate RdhE2 genes, and they arose by gene duplications during the evolution of the larvacean lineage (fig. 2). The lineage-specific origin of O. dioica RdhE2 paralogs was further supported by the presence of several introns in conserved positions among the four O. dioica RdhE2 genes (red arrowheads in supplementary file S2A, Supplementary Material online), but absent in any other analyzed species. Sequence analysis of the four O. dioica RdhE2 enzymes showed amino acid conservation in the most important functional SDR domains (supplementary file S1, Supplementary Material online): TGAGS/NGI/LG at the Gly-rich sequence, D for NAD(H) specificity, and the NX27SX12YX3K tetrad in the active site, supporting that the O. dioica enzymes had the typical NAD(H)-dependent dehydrogenase activity.
Fig. 2.

ML phylogenetic tree of the SDR-16C Rdh10 and RdhE2 subfamilies in chordates revealing the loss of Rdh10, and the surviving and lineage-specific duplication of RdhE2 in Oikopleura dioica. The sister SDR-16C subfamily of hydroxysteroid 17-β dehydrogenases 11 and 13 (Hsd17β11/13) and the basal dehydrogenases/reductases member 3 (Dhrs3) as outgroup were included to root the tree. Scale bar indicates amino acid substitutions. Values for the approximate likelihood-ratio test (aLRT) are only shown in nodes with support values greater than 0.7. Vertebrates: Danio rerio (Dre), Gasterosteus aculeatus (Gac), Gallus gallus (Gga), Homo sapiens (Hsa), Latimeria chalumnae (Lch), and Lepisosteus oculatus (Loc), Mus musculus (Mmu), Petromizus marinus (Pmr), Xenopus tropicalis (Xtr); Urochordates: Ciona intestinalis (Cin) and Oikopleura dioica (Odi); Cephalochordates: Branchiostoma floridae (Bfl); Cnidarians: Nematostella vectensis (Nve).
Rdh16: Vertebrate Rdh16 genes (a.k.a. Rodh4) belong to SDR-9C family, and have been proposed to encode enzymes that oxidize atROL into atRAL, showing therefore a redundant activity with Rdh10 and RdhE2 (Gough et al. 1998). BLAST searches using human RDH16 as query retrieved only two O. dioica genes (CBY10263 and CBY06797) with significant similarity. The reciprocal BLAST with the O. dioica sequences did not return Rdh16 but yielded Bdh1 (for 3-hydroxybutyrate dehydrogenase type 1) gene as the best hit (E-value = 6e − 33 and 6e − 20 with human BDH1), which is another member of the SDR-9C family that catalyzes the interconversion between acetoacetate and (R)-3-hydroxybutyrate. ML phylogenetic trees corroborated the BRBH analysis (supplementary file S3, Supplementary Material online). The presence of Rdh16 pro-orthologs in ascidians and cephalochordates (Dalfó et al. 2001, 2007; Belyaeva and Kedishvili 2006) indicates that its absence in O. dioica was due again to a gene loss in the RA-MGN occurred during the evolution of the larvacean lineage.
Aldh8a1: Aldh8a1 (for aldehyde dehydrogenase 8, member a1; a.k.a. ALDH12 in human and Raldh4 in zebrafish, mouse and rat) gene encodes an enzyme capable of catalyzing the conversion of atRAL into atRA (Lin and Napoli 2000; Sima et al. 2009), being therefore functionally redundant with Aldh1a enzymes. Aldh8a1 has been found in most animal phyla, from cnidarians to mammals (Albalat et al. 2011). Its amino acid sequence is highly conserved, showing five invariant residues critical for the catalytic activity (Gly231, Gly284, Cys287, Glu391, and Phe393; numbers referred to human ALDH8A1), and 11 highly conserved residues critical for the structure and function (Arg69, Gly146, Asn155, Pro157, Gly172, Lys178, Gly255, Pro395, Gly441, Asn446, and Gly459; >95% conservation in 145 Aldh sequence comparison) (Perozich et al. 1999). BLAST searches identified one O. dioica sequence (CBY33202), which based on BRBH analysis (E-value = 0.0 with Xenopus tropicalis Aldh8a1) and ML phylogenetic tree (fig. 3) appeared to be a clear O. dioica ortholog of Aldh8a1. The high sequence conservation of O. dioica Aldh8a1 (supplementary file S1, Supplementary Material online), including the five invariant catalytic residues and 10 out of the 11 highly conserved structural and functional residues (Gly441 is Ala in O. dioica), suggested that O. dioica Aldh8a1 might have equivalent aldehyde oxidizing activity than Aldh8a1 enzymes of other chordates.
Fig. 3.
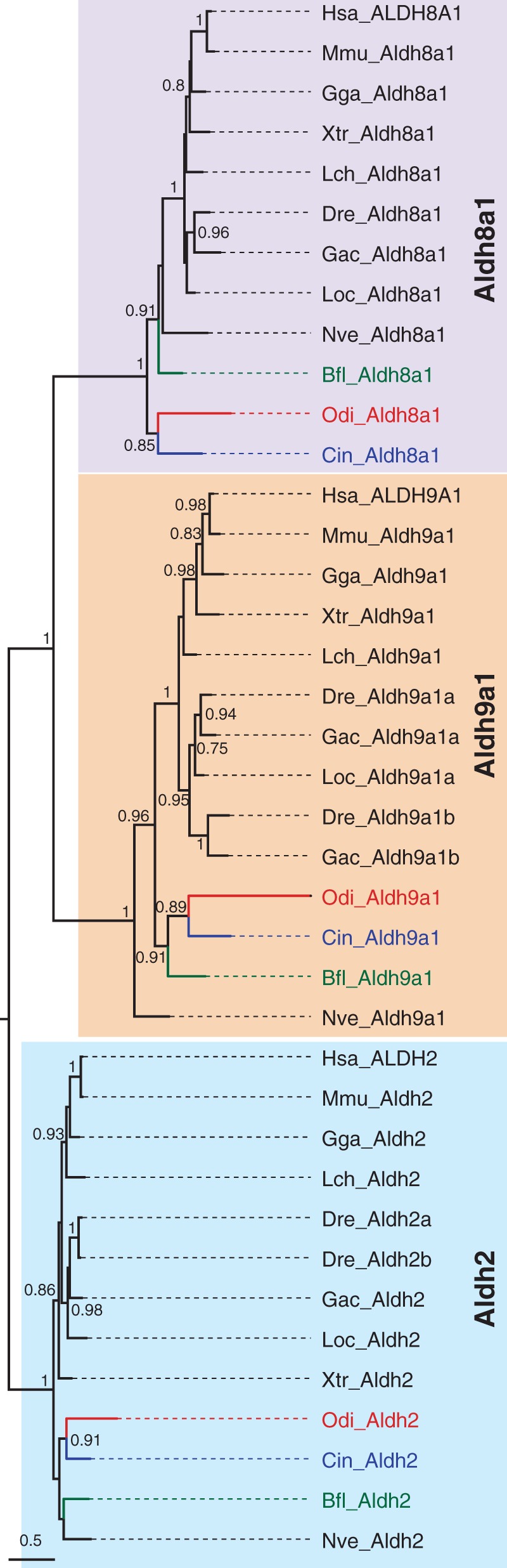
ML phylogenetic tree of the Aldh8a1 family in chordates revealing the surviving of this gene to the dismantling of the RA-MGN in Oikopleura dioica. The sister Aldh9a1 and the Aldh2 outgroup were included to root the tree. Scale bar indicates amino acid substitutions. Values for the approximate likelihood-ratio test (aLRT) are only shown in nodes with support values greater than 0.7. Abbreviations are as in figure 2.
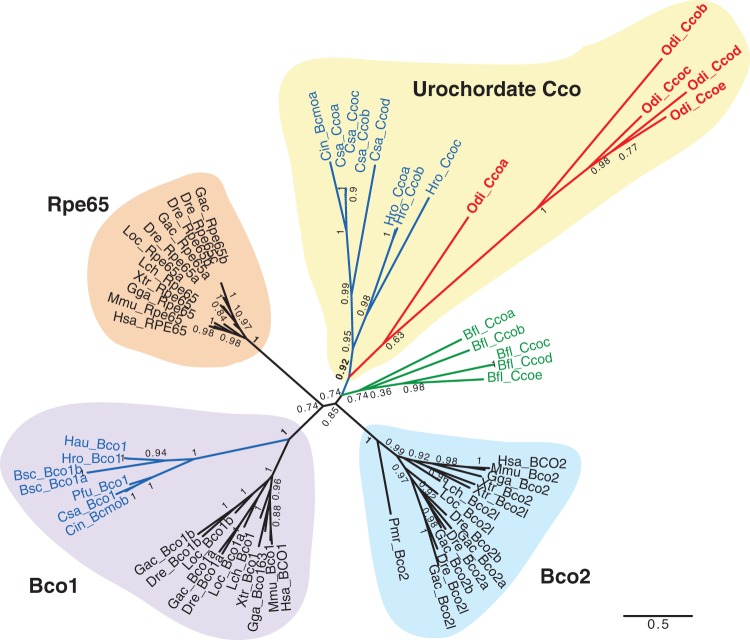
Cco (Bco1, Bco2, and Rpe65): Vertebrate β-carotene-cleaving enzymes Bco1 (for beta-carotene monooxygenase 1) and Bco2 (for beta-carotene dioxygenase 2), and retinoid isomerohydrolyzing enzyme Rpe65 (for retinal pigment epithelium-specific 65 KDa protein) belong to the Cco family. The three enzymes are involved in the production of RAL, either from the cleavage of β-carotene by Bco1 or Bco2 (Wyss 2004) or from the cleavage and isomerization of all-trans-retinyl esters by Rpe65 (Mata et al. 2004). Cco enzymes are characterized by four iron-coordinating His (His180, His241, His313, and His527) and three acidic residues that form a second coordination sphere essential for its enzymatic activity (Glu/Asp148, Glu/Asp417, and Glu/Asp469; numbers referred to human RPE65) (Sui et al. 2013). BLAST searches using human Ccos (BCO1, BCO2, and RPE65) and ascidians (C. intestinalis Bcmoa and Bcmob) identified five O. dioica genes (named Ccoa to Ccoe, CBY18123, CBY10633, CBY23475, CBY07484, and CBY07038) that were positive in BRBH analyses (E-values ranging from 2e − 47 to 5e − 97). The five O. dioica Cco showed strict conservation of the four iron-coordinating His and the three Glu/Asp acidic residues (table 1 and alignment in supplementary file S1, Supplementary Material online), clearly supporting O. dioica Ccos as potential carotenoid cleavage oxygenases. None of the O. dioica Ccos, however, contained any of the seven conserved residues responsible for fine-tuning/adaptation of the different Bco1/Bco2/Rpe65 enzymes (Poliakov et al. 2012) and none of the 13 residues considered relevant for Rpe65 enzymes (Albalat 2012) (table 1 and alignment in supplementary file S1, Supplementary Material online). The overall lack of conservation in these critical positions and the high sequence divergence besides of the seven conserved structural residues questioned that any of the five O. dioica Cco had β-carotene cleaving activity as vertebrate Bco1 or Bco2 enzymes, or an isomerohydrolyzing activity similar to vertebrate Rpe65.
The evolutionary relationship between Cco of vertebrate and nonvertebrate chordates was still unclear (Albalat 2009, 2012; Poliakov et al. 2012). In order to improve the robustness of the topology of Cco evolutionary tree of chordates of our analyses, we included in 13 new urochordate Cco sequences that we have identified after surveying the genomes of five ascidian species (sequences are provided in supplementary file S1, Supplementary Material online). ML phylogenetic analyses revealed a well-supported group of ascidian genes within the Bco1 clade, suggesting that duplications that gave rise to Bco1/Bco2/Rpe65 genes may have predated the urochordate-vertebrate split (fig. 4). None of the five O. dioica sequences, however, grouped within this ascidian/vertebrate Bco1 group, suggesting the loss of Bco1 in the O. dioica lineage. The five O. dioica Ccos grouped in a subcluster that we have named “urochordate-Cco group” (uro-Cco), which included eight Cco paralogs from three ascidian species (fig. 4). The internal tree topology of the uro-Cco group suggested that this group expanded due to recurrent independent duplications of Cco in different species within the urochordate clade. The conservation of several conserved intron positions unique to O. dioica Ccos was consistent with an origin by lineage-specific duplications (supplementary file S2B, Supplementary Material online, red arrowheads). The phylogenetic relationship between the uro-Cco group and Bco2 or Rpe65 was unclear and not well supported in the tree. The tree topology was compatible with the uro-Cco having an ancient origin, at least back to the chordate ancestor, with no surviving homolog in vertebrates, or alternatively, the uro-Cco may represent the homolog of the vertebrate Bco2 or Rpe65 families, but no clustering within those groups because of the long-branches of most urochordates Ccos (fig. 4). In either case, the five O. dioica sequences were lineage-specific duplications that represent the closest Cco members to the vertebrate Cco enzymes.
Fig. 4.
ML phylogenetic tree of the Cco family (Bco1, Rpe65, and Bco2) in chordates revealing the loss of Bco1, and the surviving and lineage specific duplication of uro-Cco paralogs in Oikopleura dioica. Scale bar indicates amino-acid substitutions. The tree is unrooted because the absence of closely related gene family that could render a reliable sequence alignment. Values for the approximate likelihood-ratio test (aLRT) are shown in nodes. Abbreviations are as in figure 2. In addition to the Cco of the ascidian Ciona intestinalis, in silico survey of the genomes of five additional ascidian species in the Aniseed database (http://aniseed.cnrs.fr/, last accessed May 2016)—Botryllus schlosseri (Bsc), Ciona savignyi (Csa), Halocynthia roretzi (Hro), Halocynthia aurantium (Hau) and Phallusia fumigata (Pfu)—allowed us to identify and include 13 new Cco sequences in the phylogenetic analysis in order to increase the robustness of tree and to clarify the position of O. dioica Cco within the uro-Cco group, characterized by multiple lineage-specific duplicated paralogs in most analyzed species.
In conclusion, our genomic survey showed that the dismantling of RA-MGN in O. dioica involved the coelimination of the main genes of the canonical metabolic pathway, that was the loss of Aldh1a, Cyp26, Rdh10, Rdh16, and Bco1 genes, while RdhE2, Aldh8a1, and some Cco genes survived the dismantling. Based on the biochemical properties of the vertebrate RdhE2, Aldh8a1, and Cco, the finding that these genes survived the RA-MGN dismantling was compatible with a hypothetical backup noncanonical pathway for atRA synthesis still present in O. dioica.
Biochemical Characterization of O. dioica Cco Paralogs
To test for the existence of such hypothetical backup pathway in O. dioica, we first investigated whether any of the O. dioica Ccos could catalyze β-carotene cleavage for atRAL production despite their sequence divergence. We used the experimental assay described in von Lintig and Vogt (2000), which has also been successfully used for ascidian Ccos (Poliakov et al. 2012). Each O. dioica Cco was heterologously expressed in the p-orange Escherichia coli strain that had been modified to synthesize and accumulate β-carotene, which yielded a yellow/orange color to bacterial pellets. In this assay, the yellow color shifted to white when β-carotene was cleaved by the induced expression of a β-carotene oxygenase enzyme (fig. 5A, compare p-orange E. coli expressing positive control (c+) mouse Bco1 (induced) and uninduced bacterial pellet). None of the O. dioica Cco did shift the color of bacterial pellets (fig. 5A), suggesting that these enzymes could not cleave β-carotene. To eliminate the possibility that the apparent lack of color shift in O. dioica Cco assays was due to a low activity of the expressed enzymes that was not evident by visual examination, we analyzed by high performance liquid chromatography (HPLC) the β-carotene content of p-orange E. coli pellets before and after the induction of Cco expression. Although expression of mouse Bco1 yielded a 9-fold reduction in the β-carotene content in comparison with the uninduced sample, no reduction was observed in any of the cultures expressing O. dioica Ccos in the assayed conditions (fig. 5B). These results suggest that in contrast to vertebrate Ccos none of O. dioica Ccos was a β-carotene-cleaving enzyme.
Fig. 5.
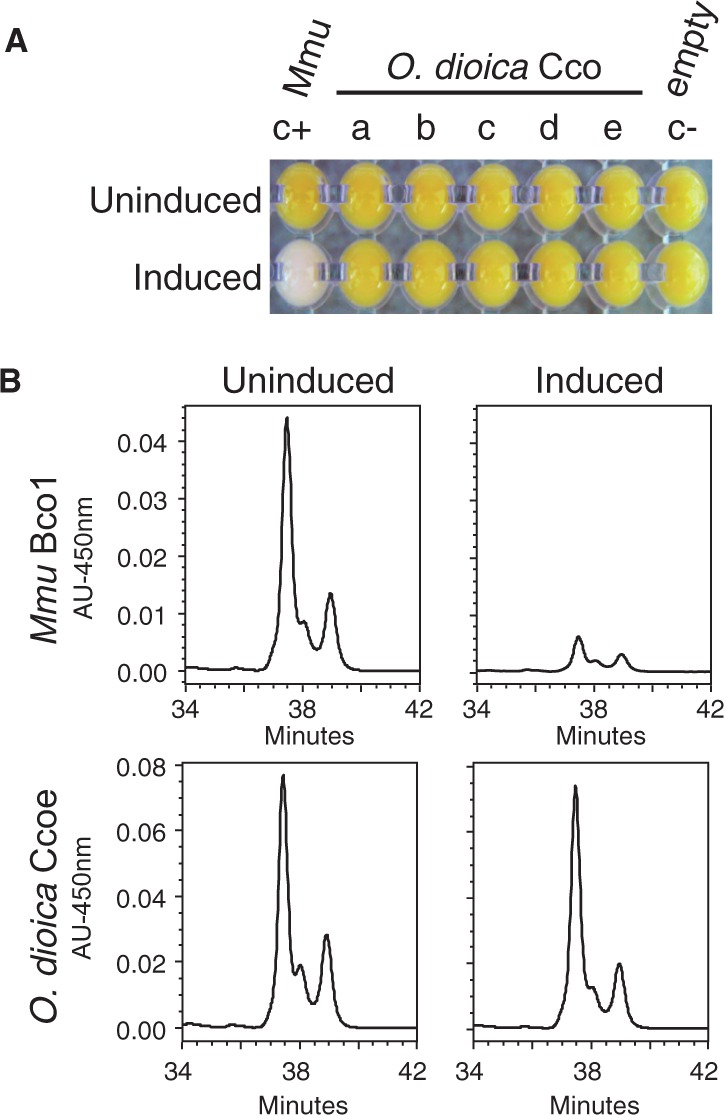
Oikopleura dioica Cco paralogs do not show β-carotene cleaving activity. (A) Induction of heterologous expression of O. dioica Cco enzymes (Ccoa to Ccoe) in p-orange Escherichia coli strain did not result in a color shift from yellow to white. In contrast, the mouse Bco1 that was used as positive control cleaved accumulated β-carotene and rendered pellets with an obvious white color in comparison to the uninduced condition. (B) HPLC analysis of β-carotene content of p-orange E. coli cultures expressing mouse Bco1 (positive control, top) and O. dioica Ccoe (bottom). While a 9-fold reduction in the β-carotene content was observed in the Mmu Bco1-expressing cultures, no reduction was observed in any of the O. dioica Cco-expressing cultures, as represented by Ccoe-induced cultures as an example. Thus, HPLC analysis supported the observation that none of the O. dioica Cco cleaves β-carotene to generate atRAL.
Retinoid and Carotenoid Content in O. dioica
Considering that O. dioica Cco did not seem to contribute to the synthesis of atRA from β-carotene cleavage, we analyzed the retinoid content of O. dioica by HPLC to test if RdhE2 and Aldh8a, or any other unidentified enzyme could still account for a noncanonical “backup” pathway for atRA synthesis in this species. In this analysis, we paid special attention to the presence of atRA, which is the most biologically active retinoid, but also to its intermediate metabolites—that is, atROL and atRAL—and carotenoids that could act as potential precursors.
Analyses of O. dioica samples from different stages, including unfertilized eggs, 7-h postfertilization (hpf) embryos, day-4 nonmature adults (at day-4, males and females were indistinguishable), and day-5 mature females and males revealed that no atRA (or 9-cis-, 11-cis-, or 13-cis-isomers) was detected in any of the samples (fig. 6A, compare the chromatograms with the “atRA standard” peak). The lower limit of reliable atRA quantification in our method was approximately 1 pmol per sample. Considering that the average weight of O. dioica sample in this study was 81 mg, we would have been able to reliable quantify the amount of RA corresponding to 12 pmol/g of wet weight, which is comparable to the concentration of atRA reported for midgestation mouse embryos (Billings et al. 2013). Since the limit of detection is much lower than the quantification limit, and we did not detect even trace amount of atRA, we conclude that O. dioica did not contain atRA at concentrations that were likely to play any role in developmental or physiological processes. The enzymes encoded by the surviving RdhE2, Cco, and Aldh8 genes might be involved, therefore, in other biological functions. Only trace amount of atROL was detected in some of the samples, indicating that vitamin A might not be abundant in O. dioica, in contrast to its higher concentrations observed in other nonvertebrate chordates (Dalfó et al. 2002). Retinyl esters, vitamin A storage form in vertebrates, were neither detected in any of O. dioica samples. Interestingly, HPLC analysis revealed the consistent presence of two peaks that may represent endogenous retinoids in O. dioica (supplementary fig. S4, Supplementary Material online). Peak 1 in the reverse phase chromatogram had elution time and absorbance spectrum similar to those of cis-RAL isomers, but it did not coelute precisely with at-, 9-cis-, 13-cis-, or 11-cis-RAL standards, leaving a possibility that it could be a double cis-isomer, or a different derivative of RAL. In normal phase chromatogram, which allowed for a better separation of cis-RAL isomers, this peak was not observed, though it could have been masked by the presence of much larger unrelated peak (peak 1 eluted at 5 min in fig. 6A). Peak 2 in figure 6A (corresponding to peak 2 in reverse phase chromatogram in supplementary fig. S5, Supplementary Material online) was present in all analyzed samples. Though its absorbance spectrum was also similar to RAL, the elution time was greatly delayed in comparison to atRAL or cis-RAL standards. This compound may also represent a derivative of RAL, such as hydroxylated forms described in invertebrates (Seki et al. 1987).
Fig. 6.
HPLC analysis of the retinoid (A) and carotenoid (B) content of Oikopleura dioica samples from different stages, including unfertilized eggs, 7-hpf embryos, day-4 nonmature adults, and day-5 mature males and day-5 mature females. (A) Oikopleura dioica extracts analyzed in normal phase HPLC. Chromatogram extracted at 350 nm. No atRA or its 9-cis-, 11-cis-, or 13-cis-isomers were detected. Unidentified peak 2 eluted at 14.2 min, may represent an endogenous retinoid, while peak 1 eluted at 5.0 min, is an unrelated compound (see text and supplementary file S4, Supplementary Material online, for details). (B) Analysis of carotenoid content in O. dioica by reverse phase HPLC. Chromatograms extracted at 450 nm showed four peaks eluted at 25.8, 28.3, 31.3, and 33.8 min in most O. dioica samples. None of the peaks appeared to be β-carotene standard. Peaks 2,3 and 4 have absorbance spectra typical for carotenoids, while peak 1 represents a different compound. Carotenoids (peaks 2, 3, and 4) likely have a dietary origin (see supplementary file S5, Supplementary Material online, for the carotenoid content of the four microalgae species used in the O. dioica diet). The inverse relative abundance of peak 1 and the carotenoid peaks at different stages suggested that peak 1 might be derived from carotenoids and that O. dioica might have the ability to actively store carotenoids in eggs, and to metabolize them throughout their life cycle. The presence of β-carotene in the dietary algae (supplementary file S5, Supplementary Material online) suggests that the absence of atRA in O. dioica was not due to a dietary deficiency of β-carotene. The absence of a β-carotene in O. dioica samples could be explained by its transformation into astaxanthin, which appears to be the one of the major carotenoids found in larvaceans (Mojib et al. 2014).
To test for the presence of potential dietary precursors of these endogenous compounds, we analyzed carotenoid content of O. dioica samples and the four algae species used in the diet to culture this organism. HPLC results revealed the presence of at least four peaks in O. dioica samples (fig. 6B), three of which had absorbance spectra typical for carotenoids (peaks 2–4), while peak 1 represented a different compound. The fact that none of the carotenoids corresponded to β-carotene (fig. 6B, compare the chromatograms with the “β-carotene standard” peak) suggested that this typical provitamin A precursor in other chordates was not used for that purpose in O. dioica, which was consistent with the loss of Bco1 and the loss of the RA-signaling in this species. Although we did not identify the nature of the detected carotenoids in O. dioica, their presence in the samples of the microalgae used to feed the animals suggested that they had a dietary origin (supplementary file S5, Supplementary Material online). The relative amount of the three carotenoid peaks detected in O. dioica varied among the analyzed samples, showing their highest level in eggs and mature females full of eggs, and gradually depleting in 7-hpf embryos, day-4 immature adults, and day-5 females (fig. 6B). On the other hand, peak 1 showed an inverse abundance relative to the three other peaks, increasing in embryos and day-4 immature animals in comparison to eggs and mature females. Oikopleura dioica, therefore, appeared to have the capability to actively store carotenoids in eggs, and to metabolize these compounds throughout their life cycle. The interdependence of the relative abundance between the different peaks suggested that they could belong to the same metabolic pathway.
Expression Analysis of O. dioica RdhE2, Aldh8a1, and Cco Genes
Having established that the surviving O. dioica Cco, RdhE2, and Aldh8a1 were not involved in the synthesis of atRA, we analyzed the expression of these genes in order to explore their possible biological functions during embryogenesis.
Cco. Most of the expression domains of the five O. dioica Cco genes were mainly, but not exclusively, observed in different compartments of the digestive system at late developmental stages (fig. 7A–U). Ccoa was the first paralog to show incipient expression in the stomach primordium by midhatchling stage (fig. 7B, black arrowhead). The signal of Ccoa became intense in the ventral part of the right stomach lobe and in the right wall of the left stomach lobe, in the region connecting both lobes by late hatchling stage (fig. 7C). By that stage, Ccoc expression signal was evident in the vertical intestine (fig. 7L). In tailshift juveniles, Ccoa expression signal became strong throughout the trunk, especially in both stomach lobes (fig. 7D), Ccob signal appeared in the posterior part of the pharynx but not in the esophagus (fig. 7H and inset), and Ccoc and Ccoe were expressed in the vertical intestine (fig. 7M and U, black arrowheads). Remarkably, by tailshift stage, all Cco genes, with the exception of Ccob, appeared to be also expressed in different fields of the oikoplastic epithelium (fig. 7D, M, Q, and U, white arrowheads). In summary, most of the expression domains of Cco genes became restricted to specific compartments of the digestive system during different developmental stages of O. dioica, suggesting that each Cco gene might have evolved distinct enzymatic activities related with a complex physiological compartmentalization of the digestive system of O. dioica.
RdhE2. Similarly to Ccos, most O. dioica RdhE2s were expressed in the digestive system at late developmental stages (fig. 8A–L). While no obvious expression was detected before hatch, by midhatchling stage, the primordium of the stomach showed the first faint expression of RdhE2b and RdhE2c (fig. 8D and G, black arrowheads). In late-hatchlings and tail-shift juveniles, the right stomach lobe expressed three out of the four RdhE2 genes (RdhE2a, RdhE2b, and RdhE2c), the midintestine expressed two of them (RdhE2a and RdhE2b), the left gastric lobe expressed Rdh2Ec and faintly RdhE2a, the vertical intestine expressed RdhE2a, and the dorsal part of the rectum expressed RdhE2c. In contrast to Cco, no expression was observed in the pharynx at any developmental stage. The signal of RdhE2d was weak and hardly distinguishable from background during all developmental stages (fig. 8J–L), with the exception of an obvious ventral domain in the oikoplastic epithelium, which also expressed RdhE2a and RdhE2b. In summary, similarly to Cco, the expression of most O. dioica RdhE2 is restricted to distinct regions of the digestive system during different developmental stages, suggesting a functional specialization for each RdhE2 enzyme, and reinforcing the idea that the digestive system of O. dioica is complex and functionally highly compartmentalized.
Aldh8a1. Aldh8a1 expression signal, in contrast to Ccos and RdhE2s, was not tissue specific but it appeared to be faintly and broadly distributed throughout the entire embryo in all analyzed developmental stages (fig. 9A–D). This ubiquitous pattern was compatible with a housekeeping role for aldehyde detoxification, which is, indeed, the role proposed for many members of superfamily of Aldh enzymes (Vasiliou and Nebert 2005). To experimentally analyze this possibility, we checked if O. dioica Aldh8a1 expression was affected by exposition to aldehydes. We treated O. dioica embryos with 0.25 µg/ml trans,trans-2,4-decadienal (DD), a model for polyunsaturated aldehydes produced by diatoms (Caldwell et al. 2002; Romano et al. 2010), and to which O. dioica might be exposed in natural environments. Aldh8a1 expression analyzed by whole-mount in situ hybridization in DD-treated O. dioica embryos before and after the hatch showed an obvious Aldh8a1 upregulation throughout the entire body of treated embryos (fig. 9E and F). These results pointed to O. dioica Aldh8a1 acting as an enzyme member of the “chemical defensome” that responds to environmental stressors with detoxification functions against biogenic or xenobiotic aldehydes.
Fig. 7.
Developmental expression patterns of Oikopleura dioica Cco paralogs. Whole-mount in situ hybridization in O. dioica late tailbubs (A, E, I, N, and R), midhatchlings (B, F, K, O, and S), late hatchlings (C, G, L, P, and T), and tailshift juveniles (D, H, M, Q, and U). Ccoa first expression signal was observed in the stomach primordium by midhatch stage (B, arrowhead), and by late-hatch stage it was strong and restricted to the right wall of the left stomach lobe, in the connection between both stomach lobes, and in the ventral part of the right stomach lobe (C). Ccob expression signal appeared as a bilateral domain in the posterior pharynx, presumptively in the peripharyngeal bands (H and dorsal view in inset). Ccoc expression signal was first observed as a faint signal in few cells near the anterior tip of the notochord (yellow arrowheads) in tailbud embryos. This notochordal domain was temporarily maintained in early hatchlings (J), together with some broad expression signal in the trunk, with special intensity in epidermal cells symmetrically (pink arrowheads) (J). Ccoc expression appeared to be temporarily downregulated in midhatchlings (K), but it became again obvious in the vertical intestine of late-hatchlings and tailshift juveniles (L and M). In tailshift juveniles, Ccoc expression signal appeared in different epithelial oikoplastic fields (M; presumptively the posterior part of the field of Fol, the middle ventral surface, the anterior crescent and the posterior rosette, white arrowheads). Ccod was the only paralog that did not show any clear expression domain in the digestive system, but it was temporally detected in late tailbuds a bilateral pair of cells adjacent near the seventh notochordal cell (N inset), and at later stages in different epithelial oikoplastic fields (Q). Finally, Ccoe expression signal appeared in the vertical intestine of tailshift juveniles (U) also labeled by Ccoc, although the onset of the former seems to be later. Large image of each panel correspond to left lateral view oriented anterior toward the left and dorsal toward the top. Inset images are dorsal views of optical cross sections at the levels of the dashed lines. Black arrowheads label expression in the digestive system, yellow in the notochord, orange in a pair of bilateral cells adjacent to the notochord, pink in the epidermis, and white in the oikoplastic epithelium. In the printed version, color codes correspond to their equivalent intensity in the grey scale. Scale bar = 20 μm.
Fig. 8.
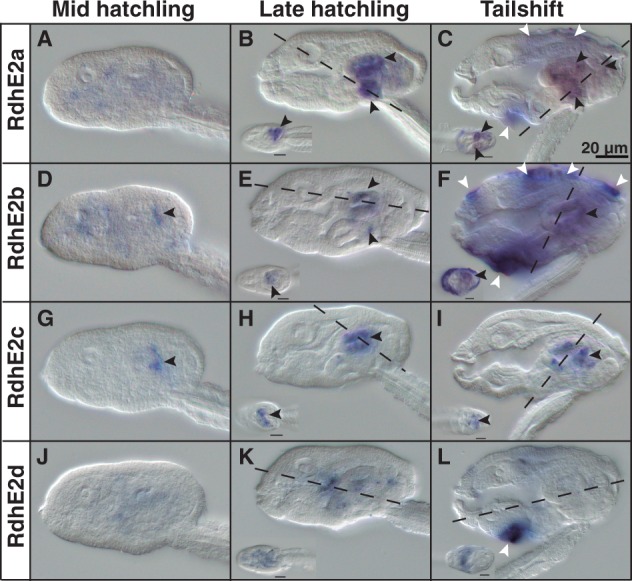
Developmental expression patterns of Oikopleura dioica RdhE2 paralogs. Whole-mount in situ hybridization in O. dioica midhatchlings (A, D, G, and J), late hatchlings (B, E, H, and K), and tailshifts (C, F, I, and L) revealed RdhE2 expression domains in the digestive system (black arrowheads) and the oikoplastic epithelium (white arrowheads). RdhE2a expression signal was strong in the right stomach lobe and midintestine, and weak in the vertical intestine and the left stomach lobe (B, C). RdhE2b expression signal was detected in the right stomach lobe and in the midintestine (E, F). RdhE2c showed the earliest expression onset of all RdhE2 paralogs in the primordium of the stomach by mid hatchling (G), and it was detected in both stomach lobes in later stages (H, I). RdhE2d was the only paralog with no clear expression in the digestive system. Similar to Cco paralogs, different fields of the oikoplastic epithelium appears to have also recruited the expression of RdhE2a, RdhE2b, and RdhE2d in different fields (e.g., the field of Fol, the middle ventral surface, the anterior crescent and the posterior rosette). Large image of each panel correspond to left lateral view oriented anterior toward the left and dorsal toward the top. Inset images are dorsal views of optical cross sections at the levels of the dashed lines. Scale bar = 20 μm.
Fig. 9.
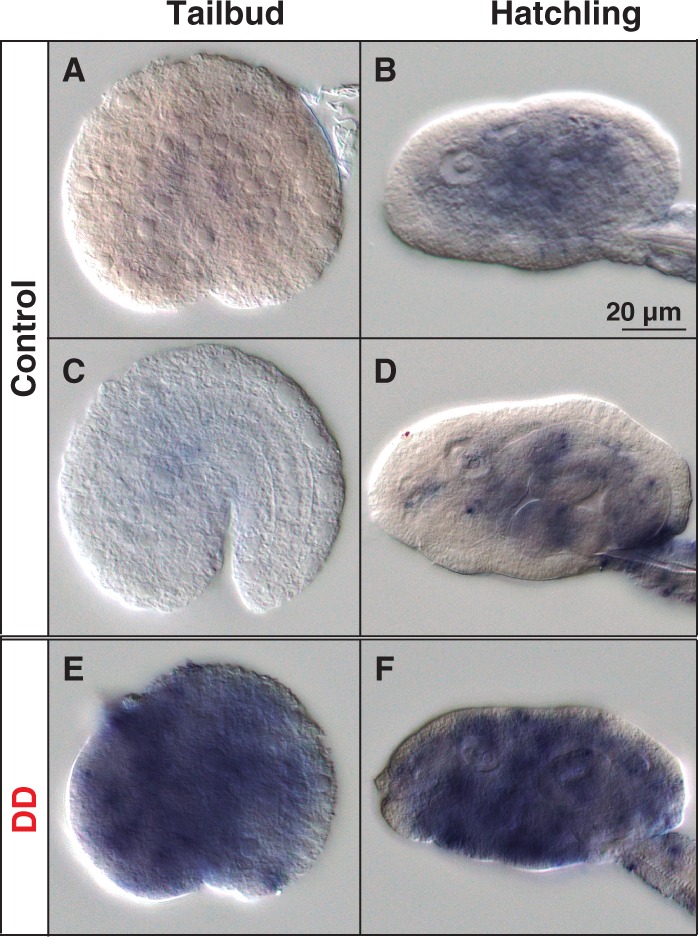
Developmental expression patterns of Oikopleura dioica Aldh8a1. Whole-mount in situ hybridization in O. dioica early tailbud stage (A and E), late tailbud stage (C) and midhatchling (B) and late-hatchling (D and F). Aldh8a1 expression signal did not show obvious tissue-specificity, but it appeared to be faintly and broadly distributed throughout the entire embryo in all analyzed developmental stages (A–D). Embryos treated with 0.25 μm/ml trans, trans-2,4-decadienal (DD), a model aldehyde for diatom-derived polyunsaturated aldehydes (PUAs), showed an obvious up-regulation of the signal throughout the embryo (E, F), suggesting a housekeeping role of the Aldh8a1 in aldehyde detoxification. Panels show left lateral views oriented anterior toward the left and dorsal toward the top. Scale bar = 20 μm.
Discussion
Gene Coelimination Pattern of the RA-MGN Associated to the Loss of Function of RA Signaling in O. dioica
Our results from HPLC analyses of retinoid content combined with those from the genome survey reveal that the loss of the ability of O. dioca to synthesize atRA was associated with a pattern of gene coelimination affecting five genes of the RA-MGN (i.e., Rdh10, Rdh16, Bco1, Aldh1a, and Cyp26) (fig. 10). This coelimination pattern suggests that the enzymes encoded by these five genes function together in a distinct, and probably specialized, pathway for atRA metabolism, and therefore, they can be considered a functional module in which the loss of one gene will likely be accompanied by the loss of the others. Interestingly, this pattern of gene coelimination does not appear to be limited to the RA-MGN, but it seems to affect other components of the regulatory gene network dependent on RA-signaling, such as the loss of nuclear receptors RAR and PPAR, which mediate RA signaling by regulating the expression of RA-target genes, or the loss of Isx (intestine-specific homeobox), a transcription factor activated by RAR that represses the expression of Bco1 (Cañestro et al. 2006 and data not shown).
Fig. 10.
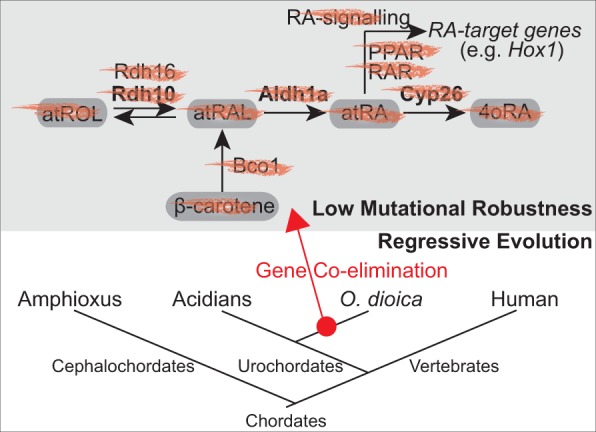
Functional biased pattern of gene loss by coelimination of genes of the retinoic acid metabolic gene network (RA-MGN). The losses of the genes Rdh10, Rdh16, Bco1, Aldh1a, and Cyp26 occurred in a context of low mutational robustness with no compensatory rerouting for the synthesis of atRA by any alternative pathway, and likely in an scenario of regressive evolution in which the loss of RA-signaling and its nuclear receptor genes (e.g., RAR, PPAR) did not imply major phenotypic changes related to RA-target genes, such as Hox1 that show the same expression pattern in Oikopleura dioica and ascidians.
Our results showing the absence of atRA (and other intermediate retinoids) in O. dioica also indicate that genes surviving the dismantling of the RA-MGN (i.e., RdhE2, Aldh8a1, and Ccos) do not form a backup pathway for atRA synthesis, at least in O. dioica. The absence of a backup pathway for at-RA synthesis suggests that the loss of the main RA-MGN genes in O. dioica did not occur in a scenario of high mutational robustness able to compensate the loss, but it was likely favored by a scenario of regressive evolution in which RA-signaling became dispensable. Several observations are consistent with the notion that RA-signaling became dispensable during the evolution of the O. dioica lineage, in which the loss of RA-MGN did not have a negative effect on its fitness, nor a drastic phenotypic impact. First, for instance, the fact that the expression pattern of O. dioica homologs of RA-target genes in other chordates (e.g., Hox1) has been preserved unaltered despite the loss of RA-signaling (Cañestro and Postlethwait 2007). Second, the disintegration of the Hox-cluster in O. dioica (Seo et al. 2004) likely relaxed the constraints to maintain the RA-signaling since this signaling requires an intact cluster to be able to regulate the temporal collinear expression of its genes (Cañestro and Postlethwait 2007; Cañestro et al. 2007; Garstang and Ferrier 2013). And third, the loss of RA-signaling did not account for a drastic remodeling of the archetypal chordate body plan in O. dioica, as it would be expected owing its crucial in anteposterior axial patterning or organogenesis in all other chordates (Nagatomo and Fujiwara 2003; Fujiwara 2006; Cañestro and Postlethwait 2007; Koop et al. 2010; Cunningham and Duester 2015). The loss of RA-signaling of O. dioica, therefore, stands as a good example to illustrate gene dispensability and patterns of gene coelimination in gene networks of multicellular eukaryotes, and provides a paradigmatic case of the “inverse paradox” of Evo–Devo, which argues that organisms might develop fundamentally similar morphologies (i.e., phenotypic unity) despite having important differences in their genetic toolkits (i.e., genetic diversity), in this case due to extensive gene losses (Cañestro et al. 2007; Albalat and Cañestro 2016). We anticipate that the study of the impact of the loss of RA-signaling on the evolution of other developmental pathways such as FGF and WNT, which are known to counteract RA-signaling in other chordates (Davidson et al. 2006; Pasini et al. 2012; Wagner and Levine 2012), will be intriguing since these developmental pathways might have been remodeled upon the loss of RA-signaling. This study will likely provide new clues about the evolution of the crosstalks between these fundamental signaling pathways.
Functions of the Genes Surviving the RA-MGN Dismantling
The absence of atRA in O. dioica indicates that the survival of Cco, RdhE2, and Aldh8a1 genes to the dismantling of the RA-MGN might be linked to other functions not related to RA synthesis.
Cco and RdhE2 Gene Family Expansions Correlate with a Complex Functional Compartmentalization of the Digestive System in O. dioica
The distinct expression of several O. dioica Cco and RdhE2 paralogs in different compartments of the digestive system (i.e., pharynx and stomach lobes) (figs. 7 and 8) suggests that the recurrent duplications affecting these gene families have allowed the evolution of a complex functional specialization of the different segments of the digestive system of O. dioica. Moreover, the differences in the temporal onset of the expression of Cco and RdhE2 paralogs likely reflect the process of differentiation of digestive cells occurring as soon as the delineation of the first organ primordia by midhatchling stage.
The endodermal expression domains of O. dioica Cco paralogs resemble the expression patterns of the ascidian C. intestinalis Bcmoa in the gills and intestine (Takimoto et al. 2006) and Bcmob in the meso-endoderm of the trunk of tailbud and larval stages (Satou et al. 2001). Bco1 and Bco2 in zebrafish (Lampert et al. 2003; Lobo et al. 2012), rodents (Kiefer et al. 2001; Raghuvanshi et al. 2015), and humans (Lindqvist and Andersson 2004) are also expressed in the digestive system, including the pharynx, liver, and gut. Bco1 shows narrow substrate specificity for cleaving β-carotene to produce RAL (Lindqvist and Andersson 2002). Bco2, in contrast, displays broad substrate specificity and can also cleave non-provitamin A carotenoids such as xanthophylls (e.g., zeaxanthin and lutein) that can serve as blue light filters and antioxidants in lipophilic environments such as in the macula of the retina improving visual acuity and protection against light damage (Krinsky and Johnson 2005; von Lintig 2010). The apparent absence of β-carotene cleaving activity in any of the O. dioica Cco set (fig. 5) and their high sequence divergence, which is reflected by long phylogenetic tree branches in comparison with other ascidian Cco (fig. 4) and by high amino acid variability at important functional positions for Bco1/Bco2/Rpe65 (table 1 in supplementary file S1, Supplementary Material online), are compatible with a process of neofunctionalization of their enzymatic activities. The expansion of the Cco family in O. dioica, as well as in some ascidians, may have facilitated its capability of metabolizing diverse carotenoids, which are especially abundant in marine environments and display great structural diversity (Liaaen-Jensen 1991; Maoka 2011). This hypothesis is supported by the detection of several unknown carotenoids, likely from a dietary origin (supplementary file S5, Supplementary Material online), already stored in eggs, which change their relative contents throughout different stages of their life cycle (fig. 6). The physiological functions of carotenoids and their derivative products, especially in marine animals, are varied and essential, including photoprotection against UV light, immunity enhancement, defense to predation by camouflage, signaling as breeding color, and normal growth, survival, and reproduction (Torrissen and Christiansen 1995; Kawakamia et al. 1998; Chew and Park 2004; Krinsky and Johnson 2005; von Lintig 2010; Maoka 2011).
The expression patterns of the RdhE2 paralogs in the digestive system of O. dioica is comparable with the ascidian expression pattern of RdhE2 in the mesoendoderm at tailbud stage (Satou et al. 2001), and with the RdhE2 expression of frogs, zebrafish, and human in the liver, stomach, and gut (Matsuzaka et al. 2002; Cheng et al. 2006; Belyaeva et al. 2012). The gastrointestinal expression and the sequence conservation of O. dioica RdhE2 enzymes suggests they might be NAD+-dependent dehydrogenases active against substrates of a dietary origin, including marine steroids and sterols that are important chemical constituents of microalgae and a major nutritional component in the diet of marine organisms (Cardozo et al. 2007). The presence of RdhE2 orthologs in cnidarians and placozoans (Albalat et al. 2011; Belyaeva et al. 2015), which are basally divergent metazoans that appear to lack RA-signaling (Cañestro et al. 2006), and the shared endodermal expression found in O. dioica and all other RdhE2 analyzed so far suggest that the digestive function might be the ancestral role of the RdhE2, and its role in RA-metabolism is an evolutionary innovation that occurred during chordate evolution (Albalat et al. 2011; Belyaeva et al. 2015).
Interestingly, none of the O. dioica Cco and RdhE2 appeared to be expressed in the CNS, notochord, or sensory organs, which contrasts with the strong neural expression of C. intestinalis Bcmoa in the photoreceptor organ of the neural complex, and Bcmob in the brain vesicle, including ocellus photoreceptor cells (Takimoto et al. 2007), or with the vertebrate RdhE2 expression in the CNS and notochord (Belyaeva et al. 2012 , 2015). The absence of any photoreceptor cell or pigment in the brain of O. dioica, in contrast to ascidians, and the lack of any atRA-metabolic role for these enzymes appear as plausible explanations accounting for the loss of neural and notochordal expression domains of O. dioica Cco and RdhE2 genes in a context of regressive evolution. The survival of these genes to the RA-MGN might be therefore due to their pleiotropic condition acting in digestive functions not related to atRA metabolism, as discussed above. Future knockdown experiments might provide further clues about the physiological roles and the evolutionary process of these duplicated enzymes.
Detoxification Role of Aldh8a
The broad expression pattern of O. dioica Aldh8a is similar to its ascidian ortholog Aldh8a, but differ from its Aldh8a ortholog in vertebrates, which is also highly expressed in the liver, intestine, and kidney (Lin et al. 2003; Marlier and Gilbert 2004; Liang et al. 2008). The biological function of Aldh8a1 is unclear, and despite a significant activity against 9-cis-RAL (but low against atRAL) has been shown in vitro (Lin and Napoli 2000; Lin et al. 2003), Aldh8a1 is most active against benzaldehyde and aliphatic aldehydes, and other metabolic aldehydes (Lin and Napoli 2000). Such substrate promiscuity and the recent finding that Aldh8a1 was one of the genes of copepods that was upregulated upon stress due to the presence of polyunsaturated aldehydes produced by blooms of diatoms (e.g., DD) (Lauritano et al. 2012) inspired us to test and discover that O. dioica Aldh81a also responds in the same way. This finding suggests, therefore, that O. dioica Aldh8a1 enzyme might play a detoxification role at least against certain polyunsaturated aldehydes similarly to other marine organisms. The presence of Aldh8a1 orthologs already in cnidarians (Albalat et al. 2011), together with the promiscuous biochemical activity of this enzyme suggest that the detoxification role observed in O. dioica might represent the ancestral function, and its contribution to the RA metabolism would be a secondary evolutionary innovation.
Conclusions
This works shows how gene loss is an important evolutionary force generating differences in the developmental genetic toolkits of different organisms. The recognition of biased patterns of gene loss due to the coelimination of set of genes is a useful strategy to identify gene network modules acting in distinct pathways related to specific biological functions. Our work demonstrates the absence of mutational robustness regarding atRA synthesis in O. dioica, suggesting, therefore that the loss of RA-signaling in O. dioica likely occurred in a scenario of regressive evolution. According to such scenario, the loss of RA-signaling did not seem to have accounted for drastic morphological or functional changes in comparison to related organisms such as ascidians in which RA-signaling still is maintained, providing an example of the inverse paradox in Evo–Devo. Our work also illustrates how the identification of genes surviving the dismantling of pathways can be a useful to recognize ancestral functions and neofunctionalizations, and to reveal the pleiotropic nature of some genes by studying their roles in a simplified context in which some functions (e.g., RA-signaling) have been lost.
Materials and Methods
Biological Material
Oikopleura dioica specimens were obtained from the Mediterranean coast of Barcelona (Catalonia, Spain). Culturing of O. dioica and embryo collections have been performed as previously described (Marti-Solans et al. 2015).
Genome Database Searches, Phylogenetic Analyses, and Intron Comparisons
Protein sequences of the RA-MGN from vertebrate H. sapiens and urochordate C. intestinalis were used as queries in BLASTp and tBLASTn searches in O. dioica genome databases (https://www.genoscope.cns.fr and http://oikoarrays.biology.uiowa.edu/Oiko, last accessed May 2016). Homologies of the O. dioica sequences were initially assessed by BRBH strategy (Wall et al. 2003). Homologies were then corroborated by phylogenetic tree analyses based on ML inferences calculated with PhyML v3.0 and automatic mode of selection of substitution model (Guindon et al. 2010) using protein alignments generated with MUSCLE and reviewed by hand (Edgar 2004). Ciona intestinalis Cco sequences were also used as queries in tBlastn searches in the genome database http://www.aniseed.cnrs.fr/aniseed (last accessed May 2016) to find Cco orthologs in five additional ascidian species (i.e., Botryllus schlosseri, Ciona savignyi, Halocynthia roretzi, Halocynthia aurantium, and Phallusia fumigata) to provide wide species representation and to increase the robustness to the phylogenetic analysis of the Cco family. Accession numbers and protein alignments for phylogenetic tree reconstructions are provided in a supplementary file S1, Supplementary Material online.
The CIWOG program (Wilkerson et al. 2009) from GECA package (Fawal et al. 2012) was used to compare the intron/exon organization of the different orthologous genes based on position and sequence conservation in the corresponding protein alignments, with no restrictions on the proportion of identical amino acids required to define a common intron. The predicted intron/exon structure of each O. dioica gene was confirmed by polymerase chain reaction (PCR) amplification, cloning, and sequencing of the corresponding cDNA or by available expressed sequence tag sequences. Briefly, total RNA was obtained from O. dioica embryos homogenized with TRI Reagent RT (MCR, RT111) using Direct-zol™ (Zymo Research, R2050). First strand cDNA was synthetized using an oligo-d(T)-anchor primer with a T7 promoter sequence at its 5′-end (table 2 in supplementary file S1, Supplementary Material online) and Superscript reverse transcriptase III (Life Technologies), and second strand was synthesized with E. coli Polymerase I (New England Biolabs). Each cDNA was amplified with gene-specific primers (table 2 in supplementary file S1, Supplementary Material online), cloned in the pCR4-TOPO vector (Invitrogen), transformed in TOP10 E. coli competent cells (Invitrogen), and sequenced using vector flanking primers. Cloned O. dioica cDNAs sequences were deposited in the Genbank under following accession numbers: Aldh8a1, KX118710; Ccoa, KX118711; Ccob, KX118712; Ccoc, KX118713; Ccod, KX118714, and Ccoe, KX118715.
Heterologous Expression and Biochemical Activity of O. dioica Ccos
To heterologously express O. dioica Cco genes, their cDNAs were amplified by two consecutive PCRs, first with a specific forward primer (table 2 in supplementary file S1, Supplementary Material online) and a reverse primer complementary to the T7 promoter sequence of the oligo-d(T)-anchor primer, and second with a specific pair of primers carrying restriction sites for AscI and NotI enzymes (table 2 in supplementary file S1, Supplementary Material online). PCR products were digested with AscI/NotI enzymes, cloned into pBAD-TOPO vector (Invitrogen) previously modified for containing three extra restriction sites (AscI, SpeI, and NotI), and transformed in a β-carotene-producing E. coli strain kindly provided by Dr Johannes von Lintig (von Lintig and Vogt 2000). The β-carotene-producing E. coli strain transformed with each O. dioica Cco cDNAs was inoculated in 25 ml of LB medium in 100-ml flasks, and grown at 28 °C in the dark until reached a 600 nm-absorbance of 0.8. Expression of each Cco was then induced by adding 0.1% w/v of L-arabinose for 20 h. The β-carotene cleaving activity of the recombinant proteins in the E. coli cultures was first evaluated by visual examination of the shift of the color of the bacterial pellets harvested by centrifugation—mouse Bco1 was used as positive control β-carotene cleavage Cco enzyme for comparison (von Lintig and Vogt 2004)—, and later confirmed by HPLC analysis of the β-carotene content in the bacterial pellets.
Analysis of Carotenoid and Retinoid Content by HPLC
Oikopleura dioica unfertilized eggs (96.1 mg, ∼35,000 eggs), 7-hpf embryos (52.3 mg, ∼31,000 embryos), day-4 nonmature adults (97.6 mg, ∼2,000 individuals), day-5 mature males (70.3 mg, ∼200 individuals), day-5 mature females (90.5 mg, ∼280 individuals), and pellets of the four algae species used in O. dioica diet –Isochrysis sp (11.1 mg), Chaetoceros calcitrans (22.9 mg), Rhinomonas reticulate (24.2 mg) and Synechococcus sp. (101.9 mg)—grown as described in (Marti-Solans et al. 2015)—were frozen at −80 and stored until analyses. For the analysis of O. dioica and algal samples, the extraction procedure described in Napoli and Horst (1998) and Belyaeva et al. (2009) was modified as follows to allow for the extraction of different classes of retinoids from the same sample: samples were homogenized on ice in 0.5 ml of PBS, mixed with 0.35 ml of methanol and extracted with 2.5 ml of hexane:dichloromethane (4:1). The upper organic phase was collected in a siliconized glass tube and dried. Ethanol (0.65 ml) containing 0.025 N potassium hydroxide was added to the aqueous phase, and the extraction was repeated with 3 ml of hexane. The upper hexane phase was pooled with the first extraction. The aqueous phase was acidified by the addition of 40 μl of 4 N hydrochloric acid, and extracted again with 3 ml of hexane. The hexane phase was collected and pooled with the previous extractions, and dried. The residue was reconstituted in 120 μl of the mobile phase and 100 μl were analyzed by normal phase HPLC. Fractions of elute containing peaks of interest were collected, pooled, dried, reconstituted in 50 μl of mobile phase, and analyzed by reverse phase HPLC. Normal and reverse phase HPLC analysis was performed essentially as described in Adams et al. (2014).
For the analysis of β-carotene content in bacterial culture, heterologous expression of O. dioica Cco enzymes was performed as described in the section Heterologous Expression and Biochemical Activity of O. dioica Ccos. Bacterial pellets were obtained from 12.5 ml of the cultures, rinsed with PBS, homogenized in 0.5 ml of PBS, mixed with 1 ml of ethanol, and extracted twice with 4 ml of hexane. Hexane extractions were pooled, dried, and analyzed by reverse phase HPLC as described in Adams et al. (2014).
Gene Expression Analysis by Whole-Mount In Situ Hybridization
Fragments of O. dioica genes were PCR amplified and cloned to synthesize gene-specific riboprobes (table 2 in supplementary file S1, Supplementary Material online). Whole-mount in situ hybridization on fixed embryos was performed as previously described (Bassham and Postlethwait 2000; Cañestro and Postlethwait 2007) with minor modifications. To promote permeabilization, embryos were incubated with 1% DMSO and 0,2% Tween-20 in PBS for 30 min before prehybridization, and washed with a PBT solution with 0.2% Tween-20 in the posthybridization.
Pharmacological Treatments
Avoiding light exposure, 5 µl of trans,trans-2,4-decadienal (DD; Sigma-Aldrich W313505) were diluted in 200 µl of DMSO (Sigma-Aldrich D8418) to prepare a stock solution of 15 mg/ml inferred from optical density at 282 nm following the Beer-Lambert law, and it was stored at 4 °C until used. Working solution of DD (0.03 mg/ml) was made by diluting the stock 1:500 in 0.2 µM-filtered sterilized seawater (sSW). Eggs were preincubated for 10 min before fertilization in 0.25 µg/ml of DD in a volume 900 µl of sSW (50 µl of DD-working solution in 6 ml of sSW). Ten minutes after fertilization, excess of sperm was washed by transferring the zygotes into a new 5.1 ml of 0.25 µg/ml DD solution in sSW in 50 mm-diameter glass dishes. Control embryos were incubated in 0.0017% DMSO solution. Embryos were maintained at 19 °C for 3 h and 20 min postfertilization (until late tailbud stage) or for 9 h 45 min postfertilization (until late hatchling stage) and fixed for whole-mount in situ hybridization analysis.
Supplementary Material
Supplementary files S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Johannes von Lintig for kindly sharing the β-carotene-producing E. coli strain and mouse Bco1 construct, and to D. Buj and Professor D. Chourrout for its technical assistance and scientific advise at the beginning of the project. The authors also thank the team members of the lab for thoughtful discussions on gene loss. They also thank to F. Palau and the staff of the Club Nàutic de Coma-ruga, and to G. Muñoz and the Badalona City Hall for facilitating us the access for animal collections. To J. Guinea and the staff of the Scientific and Technological Centers (CCiT) at the Facultat de Biologia of the Universitat de Barcelona for providing seawater. This work was supported by National Institutes of Health (grant AA12153), Ministerio de Ciencia e Innovación (grant number BFU2010-14875), and by Generalitat de Catalunya (grant number SGR2014-290).
References
- Adams MK, Belyaeva OV, Wu L, Kedishvili NY. 2014. The retinaldehyde reductase activity of DHRS3 is reciprocally activated by retinol dehydrogenase 10 to control retinoid homeostasis. J Biol Chem. 289:14868–14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R. 2009. The retinoic acid machinery in invertebrates: ancestral elements and vertebrate innovations. Mol Cell Endocrinol. 313:23–35. [DOI] [PubMed] [Google Scholar]
- Albalat R. 2012. Evolution of the genetic machinery of the visual cycle: a novelty of the vertebrate eye? Mol Biol Evol. 29:1461–1469. [DOI] [PubMed] [Google Scholar]
- Albalat R, Brunet F, Laudet V, Schubert M. 2011. Evolution of retinoid and steroid signaling: vertebrate diversification from an amphioxus perspective. Genome Biol Evol. 3:985–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet. 17:379–391. [DOI] [PubMed] [Google Scholar]
- Albalat R, Gonzàlez D, Atrian S. 1992. Protein engineering of Drosophila alcohol dehydrogenase. The hydroxyl group of Tyr152 is involved in the active site of the enzyme. FEBS Lett. 308:235–239. [DOI] [PubMed] [Google Scholar]
- Aravind L, Watanabe H, Lipman DJ, Koonin EV. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci U S A. 97:11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham S, Postlethwait J. 2000. Brachyury (T) expression in embryos of a larvacean urochordate, Oikopleura dioica, and the ancestral role of T. Dev Biol. 220:322–332. [DOI] [PubMed] [Google Scholar]
- Belyaeva OV, Chang C, Berlett MC, Kedishvili NY. 2015. Evolutionary origins of retinoid active short-chain dehydrogenases/reductases of SDR16C family. Chem Biol Interact. 234:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva OV, Johnson MP, Kedishvili NY. 2008. Kinetic analysis of human enzyme RDH10 defines the characteristics of a physiologically relevant retinol dehydrogenase. J Biol Chem. 283:20299–20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva OV, Kedishvili NY. 2006. Comparative genomic and phylogenetic analysis of short-chain dehydrogenases/reductases with dual retinol/sterol substrate specificity. Genomics 88:820–830. [DOI] [PubMed] [Google Scholar]
- Belyaeva OV, Lee SA, Adams MK, Chang C, Kedishvili NY. 2012. Short chain dehydrogenase/reductase rdhe2 is a novel retinol dehydrogenase essential for frog embryonic development. J Biol Chem. 287:9061–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva OV, Lee SA, Kolupaev OV, Kedishvili NY. 2009. Identification and characterization of retinoid-active short-chain dehydrogenases/reductases in Drosophila melanogaster. Biochim Biophys Acta. 1790:1266–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings SE, Pierzchalski K, Butler Tjaden NE, Pang XY, Trainor PA, Kane MA, Moise AR. 2013. The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J. 27:4877–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell GS, Olive PJ, Bentley MG. 2002. Inhibition of embryonic development and fertilization in broadcast spawning marine invertebrates by water soluble diatom extracts and the diatom toxin 2-trans,4-trans decadienal. Aquat Toxicol. 60:123–137. [DOI] [PubMed] [Google Scholar]
- Cammas L, Romand R, Fraulob V, Mura C, Dolle P. 2007. Expression of the murine retinol dehydrogenase 10 (Rdh10) gene correlates with many sites of retinoid signalling during embryogenesis and organ differentiation. Dev Dyn. 236:2899–2908. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Albalat R, Postlethwait JH. 2010. Oikopleura dioica alcohol dehydrogenase class 3 provides new insights into the evolution of retinoic acid synthesis in chordates. Zool Sci. 27:128–133. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Catchen JM, Rodríguez-Marí A, Yokoi H, Postlethwait JH. 2009. Consequences of lineage-specific gene loss on functional evolution of surviving paralogs: ALDH1A and retinoic acid signaling in vertebrate genomes. PLoS Genet. 5:e1000496.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañestro C, Godoy L, Gonzàlez-Duarte R, Albalat R. 2003. Comparative expression analysis of Adh3 during arthropod, urochordate, cephalochordate and vertebrate development challenges its predicted housekeeping role. Evol Dev. 5:157–162. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Hjelmqvist L, Albalat R, Garcia-Fernàndez J, Gonzàlez-Duarte R, Jörnvall H. 2000. Amphioxus alcohol dehydrogenase is a class 3 form of single type and of structural conservation but with unique developmental expression. Eur J Biochem. 267:6511–6518. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Postlethwait JH. 2007. Development of a chordate anterior-posterior axis without classical retinoic acid signaling. Dev Biol. 305:522–538. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Postlethwait JH, Gonzàlez-Duarte R, Albalat R. 2006. Is retinoic acid genetic machinery a chordate innovation? Evol Dev. 8:394–406. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Yokoi H, Postlethwait JH. 2007. Evolutionary developmental biology and genomics. Nat Rev Genet. 8:932–942. [DOI] [PubMed] [Google Scholar]
- Cardozo KH, Guaratini T, Barros MP, Falcao VR, Tonon AP, Lopes NP, Campos S, Torres MA, Souza AO, Colepicolo P, et al. 2007. Metabolites from algae with economical impact. Comp Biochem Physiol C Toxicol Pharmacol. 146:60–78. [DOI] [PubMed] [Google Scholar]
- Chen H, Namkung MJ, Juchau MR. 1995. Biotransformation of all-trans-retinol and all-trans-retinal to all-trans-retinoic acid in rat conceptal homogenates. Biochem Pharmacol. 50:1257–1264. [DOI] [PubMed] [Google Scholar]
- Cheng W, Guo L, Zhang Z, Soo HM, Wen C, Wu W, Peng J. 2006. HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev Biol. 294:482–496. [DOI] [PubMed] [Google Scholar]
- Chew BP, Park JS. 2004. Carotenoid action on the immune response. J Nutr. 134:257S–261S. [DOI] [PubMed] [Google Scholar]
- Cunningham TJ, Duester G. 2015. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 16:110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalfó D, Albalat R, Molotkov A, Duester G, Gonzàlez-Duarte R. 2002. Retinoic acid synthesis in the prevertebrate amphioxus involves retinol oxidation. Dev Genes Evol. 212:388–393. [DOI] [PubMed] [Google Scholar]
- Dalfó D, Cañestro C, Albalat R, Gonzàlez-Duarte R. 2001. Characterization of a microsomal retinol dehydrogenase gene from amphioxus: retinoid metabolism before vertebrates. Chem Biol Interact. 130-132:359–370. [DOI] [PubMed] [Google Scholar]
- Dalfó D, Marqués N, Albalat R. 2007. Analysis of the NADH-dependent retinaldehyde reductase activity of amphioxus retinol dehydrogenase enzymes enhances our understanding of the evolution of the retinol dehydrogenase family. FEBS J. 274:3739–3752. [DOI] [PubMed] [Google Scholar]
- Davidson B, Shi W, Beh J, Christiaen L, Levine M. 2006. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 20:2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Henriet S, Mungpakdee S, Aury JM, Da Silva C, Brinkmann H, Mikhaleva J, Olsen LC, Jubin C, Cañestro C, et al. 2010. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330:1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. 2013. Retinoid signaling in control of progenitor cell differentiation during mouse development. Semin Cell Dev Biol. 24:694–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G, Mic FA, Molotkov A. 2003. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem Biol Interact. 143-144:201–210. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawal N, Savelli B, Dunand C, Mathe C. 2012. GECA: a fast tool for gene evolution and conservation analysis in eukaryotic protein families. Bioinformatics 28:1398–1399. [DOI] [PubMed] [Google Scholar]
- Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jörnvall H, Oppermann U. 2002. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem. 277:25677–25684. [DOI] [PubMed] [Google Scholar]
- Fujiwara S. 2006. Retinoids and nonvertebrate chordate development. J Neurobiol. 66:645–652. [DOI] [PubMed] [Google Scholar]
- Garstang M, Ferrier DE. 2013. Time is of the essence for ParaHox homeobox gene clustering. BMC Biol. 11:72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough WH, VanOoteghem S, Sint T, Kedishvili NY. 1998. cDNA cloning and characterization of a new human microsomal NAD+-dependent dehydrogenase that oxidizes all-trans-retinol and 3alpha-hydroxysteroids. J Biol Chem. 273:19778–19785. [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421:63–66. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Jurukovski V, Markova NG, Karaman-Jurukovska N, Randolph RK, Su J, Napoli JL, Simon M. 1999. Cloning and characterization of retinol dehydrogenase transcripts expressed in human epidermal keratinocytes. Mol Genet Metab. 67:62–73. [DOI] [PubMed] [Google Scholar]
- Kawakamia T, Tsushimab M, Katabamia Y, Mine M, Ishida A, Matsuno M. 1998. Effect of b,b-carotene, b-echinenone, astaxanthin, fucoxanthin, vitamin A and vitamin E on the biological defense of the sea urchin Pseudocentrotus depressus. J Exp Mar Biol Ecol. 226:165–174. [Google Scholar]
- Kiefer C, Hessel S, Lampert JM, Vogt K, Lederer MO, Breithaupt DE, von Lintig J. 2001. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J Biol Chem. 276:14110–14116. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. 2004. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5:R7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop D, Holland ND, Semon M, Alvarez S, de Lera AR, Laudet V, Holland LZ, Schubert M. 2010. Retinoic acid signaling targets Hox genes during the amphioxus gastrula stage: insights into early anterior-posterior patterning of the chordate body plan. Dev Biol. 338:98–106. [DOI] [PubMed] [Google Scholar]
- Krinsky NI, Johnson EJ. 2005. Carotenoid actions and their relation to health and disease. Mol Aspects Med. 26:459–516. [DOI] [PubMed] [Google Scholar]
- Lampert JM, Holzschuh J, Hessel S, Driever W, Vogt K, von Lintig J. 2003. Provitamin A conversion to retinal via the beta,beta-carotene-15,15'-oxygenase (bcox) is essential for pattern formation and differentiation during zebrafish embryogenesis. Development 130:2173–2186. [DOI] [PubMed] [Google Scholar]
- Lauritano C, Carotenuto Y, Miralto A, Procaccini G, Ianora A. 2012. Copepod population-specific response to a toxic diatom diet. PLoS One 7:e47262.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Belyaeva OV, Kedishvili NY. 2009. Biochemical characterization of human epidermal retinol dehydrogenase 2. Chem Biol Interact 178:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesk AM. 1995. NAD-binding domains of dehydrogenases. Curr Opin Struct Biol. 5:775–783. [DOI] [PubMed] [Google Scholar]
- Liaaen-Jensen S. 1991. Marine carotenoids: recent progress. Pure Appl Chem. 63:1–12. [Google Scholar]
- Liang D, Zhang M, Bao J, Zhang L, Xu X, Gao X, Zhao Q. 2008. Expressions of Raldh3 and Raldh4 during zebrafish early development. Gene Expr Patterns. 8:248–253. [DOI] [PubMed] [Google Scholar]
- Lin M, Napoli JL. 2000. cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and identification of a rat ortholog, ALDH12. J Biol Chem. 275: 40106–40112. [DOI] [PubMed] [Google Scholar]
- Lin M, Zhang M, Abraham M, Smith SM, Napoli JL. 2003. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 278:9856–9861. [DOI] [PubMed] [Google Scholar]
- Lindqvist A, Andersson S. 2002. Biochemical properties of purified recombinant human beta-carotene 15,15'-monooxygenase. J Biol Chem. 277:23942–23948. [DOI] [PubMed] [Google Scholar]
- Lindqvist A, Andersson S. 2004. Cell type-specific expression of beta-carotene 15,15'-mono-oxygenase in human tissues. J Histochem Cytochem. 52:491–499. [DOI] [PubMed] [Google Scholar]
- Lobo GP, Isken A, Hoff S, Babino D, von Lintig J. 2012. BCDO2 acts as a carotenoid scavenger and gatekeeper for the mitochondrial apoptotic pathway. Development 139:2966–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maoka T. 2011. Carotenoids in marine animals. Mar Drugs. 9:278–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlier A, Gilbert T. 2004. Expression of retinoic acid-synthesizing and -metabolizing enzymes during nephrogenesis in the rat. Gene Expr Patterns. 5:179–185. [DOI] [PubMed] [Google Scholar]
- Martí-Solans J, Ferrández-Roldán A, Godoy-Marín H, Badia-Ramentol J, Torres-Águila NP, Rodríguez-Marí A, Bouquet JM, Chourrout D, Thompson EM, Albalat R, et al. 2015. Oikopleura dioica culturing made easy: a low-cost facility for an emerging animal model in EvoDevo. Genesis 53:183–193. [DOI] [PubMed] [Google Scholar]
- Mata NL, Moghrabi WN, Lee JS, Bui TV, Radu RA, Horwitz J, Travis GH. 2004. Rpe65 is a retinyl ester binding protein that presents insoluble substrate to the isomerase in retinal pigment epithelial cells. J Biol Chem. 279:635–643. [DOI] [PubMed] [Google Scholar]
- Matsuzaka Y, Okamoto K, Tsuji H, Mabuchi T, Ozawa A, Tamiya G, Inoko H. 2002. Identification of the hRDH-E2 gene, a novel member of the SDR family, and its increased expression in psoriatic lesion. Biochem Biophys Res Commun. 297:1171–1180. [DOI] [PubMed] [Google Scholar]
- Mojib N, Amad M, Thimma M, Aldanondo N, Kumaran M, Irigoien X. 2014. Carotenoid metabolic profiling and transcriptome-genome mining reveal functional equivalence among blue-pigmented copepods and appendicularia. Mol Ecol. 23:2740–2756. [DOI] [PubMed] [Google Scholar]
- Molotkov A, Deltour L, Foglio MH, Cuenca AE, Duester G. 2002. Distinct retinoid metabolic functions for alcohol dehydrogenase genes Adh1 and Adh4 in protection against vitamin A toxicity or deficiency revealed in double null mutant mice. J Biol Chem. 277:13804–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A, Fan X, Deltour L, Foglio MH, Martras S, Farres J, Pares X, Duester G. 2002. Stimulation of retinoic acid production and growth by ubiquitously expressed alcohol dehydrogenase Adh3. Proc Natl Acad Sci U S A. 99:5337–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatomo K, Fujiwara S. 2003. Expression of Raldh2, Cyp26 and Hox-1 in normal and retinoic acid-treated Ciona intestinalis embryos. Gene Expr Patterns. 3:273–277. [DOI] [PubMed] [Google Scholar]
- Napoli JL, Horst RL. 1998. Quantitative analyses of naturally occurring retinoids In: Redfern, editor. Methods Mol Biol. Totowa, NJ: Humana Press; 29–40. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dolle P. 2002. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet. 31:84–88. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, Chambon P. 1999. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 21:444–448. [DOI] [PubMed] [Google Scholar]
- Olson MV. 1999. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 64:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MV, Varki A. 2003. Sequencing the chimpanzee genome: insights into human evolution and disease. Nat Rev Genet. 4:20–28. [DOI] [PubMed] [Google Scholar]
- Pasini A, Manenti R, Rothbacher U, Lemaire P. 2012. Antagonizing retinoic acid and FGF/MAPK pathways control posterior body patterning in the invertebrate chordate Ciona intestinalis. PLoS One 7:e46193.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozich J, Nicholas H, Wang BC, Lindahl R, Hempel J. 1999. Relationships within the aldehyde dehydrogenase extended family. Protein Sci. 8:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov E, Gubin AN, Stearn O, Li Y, Campos MM, Gentleman S, Rogozin IB, Redmond TM. 2012. Origin and evolution of retinoid isomerization machinery in vertebrate visual cycle: hint from jawless vertebrates. PLoS One 7:e49975.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. 2006. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 38:107–111. [DOI] [PubMed] [Google Scholar]
- Raghuvanshi S, Reed V, Blaner WS, Harrison EH. 2015. Cellular localization of beta-carotene 15,15' oxygenase-1 (BCO1) and beta-carotene 9',10' oxygenase-2 (BCO2) in rat liver and intestine. Arch Biochem Biophys. 572:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijntjes S, Blentic A, Gale E, Maden M. 2005. The control of morphogen signalling: regulation of the synthesis and catabolism of retinoic acid in the developing embryo. Dev Biol. 285:224–237. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dolle P. 2012. Retinoic acid signalling during development. Development 139:843–858. [DOI] [PubMed] [Google Scholar]
- Romano G, Miralto A, Ianora A. 2010. Teratogenic effects of diatom metabolites on sea urchin Paracentrotus lividus embryos. Mar Drugs. 8:950–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, Rey JP, Ma JX, Staehling-Hampton K, Trainor PA. 2007. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev 21:1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Takatori N, Yamada L, Mochizuki Y, Hamaguchi M, Ishikawa H, Chiba S, Imai K, Kano S, Murakami SD, et al. 2001. Gene expression profiles in Ciona intestinalis tailbud embryos. Development 128:2893–2904. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Nie Q, Lander AD. 2012. Dynamics and precision in retinoic acid morphogen gradients. Curr Opin Genet Dev. 22:562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Fujishita S, Ito M, Matsuoka N, Tsukida K. 1987. Retinoid composition in the compound eyes of insects. Exp Biol. 47:95–103. [PubMed] [Google Scholar]
- Seo HC, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, Flaat M, Weissenbach J, Lehrach H, Wincker P, et al. 2004. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431:67–71. [DOI] [PubMed] [Google Scholar]
- Sima A, Parisotto M, Mader S, Bhat PV. 2009. Kinetic characterization of recombinant mouse retinal dehydrogenase types 3 and 4 for retinal substrates. Biochim Biophys Acta. 1790:1660–1664. [DOI] [PubMed] [Google Scholar]
- Strate I, Min TH, Iliev D, Pera EM. 2009. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development 136:461–472. [DOI] [PubMed] [Google Scholar]
- Sui X, Kiser PD, Lintig J, Palczewski K. 2013. Structural basis of carotenoid cleavage: from bacteria to mammals. Arch Biochem Biophys. 539:203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto N, Kusakabe T, Horie T, Miyamoto Y, Tsuda M. 2006. Origin of the Vertebrate Visual Cycle: III. Distinct Distribution of RPE65 and b-carotene 15,15'-Monooxygenase Homologues in Ciona intestinalis. Photochem Photobiol. 82:1468–1474. [DOI] [PubMed] [Google Scholar]
- Takimoto N, Kusakabe T, Tsuda M. 2007. Origin of the vertebrate visual cycle. Photochem Photobiol. 83:242–247. [DOI] [PubMed] [Google Scholar]
- Torrissen O, Christiansen R. 1995. Requirements for carotenoids in fish diets. J Appl Ichthyol. 11:225–230. [Google Scholar]
- Vasiliou V, Nebert DW. 2005. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2:138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J. 2010. Colors with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annu Rev Nutr. 30:35–56. [DOI] [PubMed] [Google Scholar]
- von Lintig J, Vogt K. 2000. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving b-carotene to retinal. J Biol Chem. 275:11915–11920. [DOI] [PubMed] [Google Scholar]
- von Lintig J, Vogt K. 2004. Vitamin A formation in animals: molecular identification and functional characterization of carotene cleaving enzymes. J Nutr. 134:251S–256S. [DOI] [PubMed] [Google Scholar]
- Wagner A. 2005. Distributed robustness versus redundancy as causes of mutational robustness. Bioessays 27:176–188. [DOI] [PubMed] [Google Scholar]
- Wagner E, Levine M. 2012. FGF signaling establishes the anterior border of the Ciona neural tube. Development 139:2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall DP, Fraser HB, Hirsh AE. 2003. Detecting putative orthologs. Bioinformatics 19:1710–1711. [DOI] [PubMed] [Google Scholar]
- Wang XD, Russell RM, Liu C, Stickel F, Smith DE, Krinsky NI. 1996. Beta-oxidation in rabbit liver in vitro and in the perfused ferret liver contributes to retinoic acid biosynthesis from beta-apocarotenoic acids. J Biol Chem. 271:26490–26498. [PubMed] [Google Scholar]
- Wierenga RK, Terpstra P, Hol WG. 1986. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 187:101–107. [DOI] [PubMed] [Google Scholar]
- Wilkerson MD, Ru Y, Brendel VP. 2009. Common introns within orthologous genes: software and application to plants. Brief Bioinform. 10:631–644. [DOI] [PubMed] [Google Scholar]
- Wyss A. 2004. Carotene oxygenases: a new family of double bond cleavage enzymes. J Nutr. 134:246S–250S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.