Abstract
Of central importance in adapting plants of tropical origin to temperate cultivation has been selection of daylength-neutral genotypes that flower early in the temperate summer and take full advantage of its long days. A cross between tropical and temperate sorghums [Sorghum propinquum (Kunth) Hitchc.×S. bicolor (L.) Moench], revealed a quantitative trait locus (QTL), FlrAvgD1, accounting for 85.7% of variation in flowering time under long days. Fine-scale genetic mapping placed FlrAvgD1 on chromosome 6 within the physically largest centiMorgan in the genome. Forward genetic data from “converted” sorghums validated the QTL. Association genetic evidence from a diversity panel delineated the QTL to a 10-kb interval containing only one annotated gene, Sb06g012260, that was shown by reverse genetics to complement a recessive allele. Sb06g012260 (SbFT12) contains a phosphatidylethanolamine-binding (PEBP) protein domain characteristic of members of the “FT” family of flowering genes acting as a floral suppressor. Sb06g012260 appears to have evolved ∼40 Ma in a panicoid ancestor after divergence from oryzoid and pooid lineages. A species-specific Sb06g012260 mutation may have contributed to spread to temperate regions by S. halepense (“Johnsongrass”), one of the world’s most widespread invasives. Alternative alleles for another family member, Sb02g029725 (SbFT6), mapping near another flowering QTL, also showed highly significant association with photoperiod response index (P = 1.53×10 − 6). The evolution of Sb06g012260 adds to evidence that single gene duplicates play large roles in important environmental adaptations. Increased knowledge of Sb06g012260 opens new doors to improvement of sorghum and other grain and cellulosic biomass crops.
Keywords: photoperiod, flowering, conversion, FT domain, Sorghum halepense (“Johnsongrass”), single gene duplication.
Introduction
Genetic regulation of flowering is a key element of adapting plants to cultivation in different latitudes, daylength responses and/or purposes (such as seed/grain or biomass production). In the semi-arid tropical habitats to which many of the world’s most important seed/grain crops are native, flowering during “short” days of <12.5 h of light in a 24-h period coordinates gametogenesis and reproduction with generally favorable rainfall, temperature, and solar radiation (Harper 1977). Photoperiod-insensitive (“day-neutral”) mutants of many seed and grain crops that flower based on accumulation of heat units have become widely used in temperate latitudes, producing higher seed/grain yields than short-day genotypes which generally flower late in the growing season. In crops grown for biomass rather than seed/grain, flowering is generally undesirable, in sugarcane, e.g., reducing yields (Julien and Soopramanien 1976; Long 1976; Julien et al. 1978; Heinz 1987) and increasing disease susceptibility (Ricaud et al. 1980). Flowering is widely acknowledged as a significant obstacle in maximizing the cellulosic yield potential of temperate biomass crops such as Miscanthus, a close relative of sugarcane (Jakob et al. 2009).
Its small diploid genome with minimal gene duplication and substantial homozygosity make sorghum (Sorghum bicolor L. Moench.) an attractive genomic model (Paterson et al. 2009) for the Andropogoneae, tropical grasses in which biochemical and morphological specializations (“C4” photosynthesis) improve carbon assimilation at high temperatures and which include complex paleo- and/or neopolyploid genomes of important crops such as sugarcane, Miscanthus, maize and pearl millet (the latter being Paniceae). Parsimony suggests the sorghum karyotype to resemble that of the Andropogoneae common ancestor (Wang et al. 2014). During an estimated 50 My of divergence from rice, sorghum has experienced only ∼3% differential gene loss and little rearrangement (Paterson et al. 2009). With an estimated 96 My of “abstinence” from genome duplication (Wang et al. 2015), functions of sorghum genes are likely to still resemble those of the common grass ancestor. In contrast, in only ∼15 My, genome duplication and associated “fractionations” (gene and cis-regulator losses) in maize (Schnable et al. 2010; Woodhouse et al. 2010) have substantially changed gene linkage arrangements and expression patterns.
Sorghum evolution and improvement has yielded a remarkable diversity of morphologies, with genotypes of 6 m or more in height preferred in Africa as an important dual-purpose (grain + straw) crop. In contrast, genotypes of 1 m or less are widely utilized in mechanized agriculture. Introduced into USA ∼150 years ago, sorghum is grown on 8–10 million acres and has a farm-gate value of typically ∼$1 billion/year (USDA-NASS 2013). Its drought resistance provides an important “failsafe” in the agriculture of the US Southern Plains that often receive too little rain for other grains. Likewise, in arid countries of northeast and West Africa, sorghum contributes 26–39% of calories in the human diet (FAO Statistical Database on Crops 2011). In addition to its importance for food, feed, and straw, sorghum is the #2 US source of fuel ethanol and the availability of “sweet sorghums”, S. bicolor genotypes with stalk sugar concentrations approaching those of sugarcane, make it suitable for commercial-scale biofuel production from each of three feedstocks, starch, sugar and cellulose (Rooney et al. 2007).
The requirement for qualitatively different flowering regimes hinders both use of the rich diversity of tropical crop germplasm in temperate latitudes, and of high-yielding improved temperate germplasm in the tropics. For example, wild forms of sorghum and also many tropical cultivars flower only during “short” day photoperiods of ∼12.5 h daylight or less (Quinby and Karper 1945), coinciding with generally favorable rainfall, temperature, and solar radiation (Harper 1977). In temperate latitudes, such obligate short-day genotypes typically flower near the end of the growing season, by which time the majority of solar radiation has already been missed, and often with too little time to mature seeds before freezing temperatures arrive. Much higher grain yield is generally realized in temperate regions from photoperiod-insensitive (“day-neutral”) genotypes that flower much earlier and make better use of season-long solar radiation. Long-term efforts in several crops exemplified by the “Sorghum Conversion Project” (Stephens et al. 1967) have sought to render rich diversity of native tropical crop germplasm more accessible to temperate breeding programs by crossing to a temperate donor genotype and “converting” to (selecting for) day-neutral flowering while backcrossing to otherwise restore the genotype of the tropical recurrent parent. To date, in sorghum alone >700 genotypes have been converted over ∼50 years of effort, in most cases involving four generations of backcrossing each followed by two generations of selfing to identify recessive day-neutral flowering segregants.
Previously, we studied F2 progeny of a cross between S. bicolor BTx623, a day-neutral breeding line, and S. propinquum, a tropical and short-day flowering wild relative. Most of the F2 population, with at least one copy of the S. propinquum short day flowering allele, flowered in mid-September or later in the latitude where the experiment was done (30.6N; College Station, TX), consistent with the short-day flowering of the tropical S. propinquum parent. We showed a single quantitative trait locus (QTL), FlrAvgD1, to account for 85.7% of this phenotypic variation in flowering time (Lin et al. 1995), and to correspond approximately to the locus of the Triticeae photoperiodic genetic loci, Ppd1-3 and Ppd-H1 (Paterson, Lin, et al. 1995).
Here, we show by integration of positional and association genetic evidence that the short-day flowering QTL FlrAvgD1 locates on chromosome 6 (formerly linkage group D) in a 10-kb interval within the physically largest centiMorgan in the sorghum genome. This interval contains a single annotated gene that we show to partly complement the day-neutral flowering phenotype. The gene accounting for FlrAvgD1 is Sb06g012260, a member of the “Flowering locus T” (FT) family of transcription factors that is highly divergent from most known floral regulators in this family. Sb06g012260 is unique to panicoids, evolving as a single gene duplication shortly after the oryzoid (rice) – panicoid (sorghum, maize) divergence. Sb06g012260 suppresses flowering, although it is quite distant evolutionarily from other FT family members that are floral suppressors (Pin et al. 2010; Harig et al. 2012). We discuss its relationship to named sorghum flowering genes and implications of these findings for candidacy of other genes, as well as its evolution in an invasive Sorghum species that has adapted to temperate latitudes.
Results
To conduct interval mapping of flowering time in sorghum, we previously analyzed an F2 population of S. bicolor cultivar BTx623×S. propinquum using 78 RFLP loci spanning 935 cM with an average distance of 14 cM between markers (Lin et al. 1995; Paterson, Lin, et al. 1995). A QTL explaining 85.7% of phenotypic variation in flowering time, FlrAvgD1, was placed in the 21-cM interval between DNA markers pSB095 and pSB428a.
To more finely map FlrAvgD1, 34 F2 plants were selected that were putatively recombinant in the 21-cM interval between DNA markers pSB095 and pSB428a on chromosome 6. An additional 27 DNA markers were applied to pooled DNA from 50 to 150 selfed F3 progenies that were also grown in the field near College Station, Texas. About 4 of the 34 F3 families, #10, 187, 191, and 211, were excluded because the DNA marker genotypes of F2 and pooled F3 tissue were not consistent (#211), or because the FlrAvgD1 locus genotype of their F2 parents predicted from phenotypic segregation in F3 progenies was contradicted by both flanking markers and by virtually all other markers on the chromosome (#10, 187, 191). Each such inconsistency would have required a double recombination event to explain, and 3 such events among 34 progeny in a 21-cM interval are highly improbable. A few such incongruous plants were also observed in the F2 generation, and are an important example of the need for progeny testing to reliably fine-map flowering, which can be influenced by other genetic effects, temperature, and other factors such as some diseases (Quinby 1974).
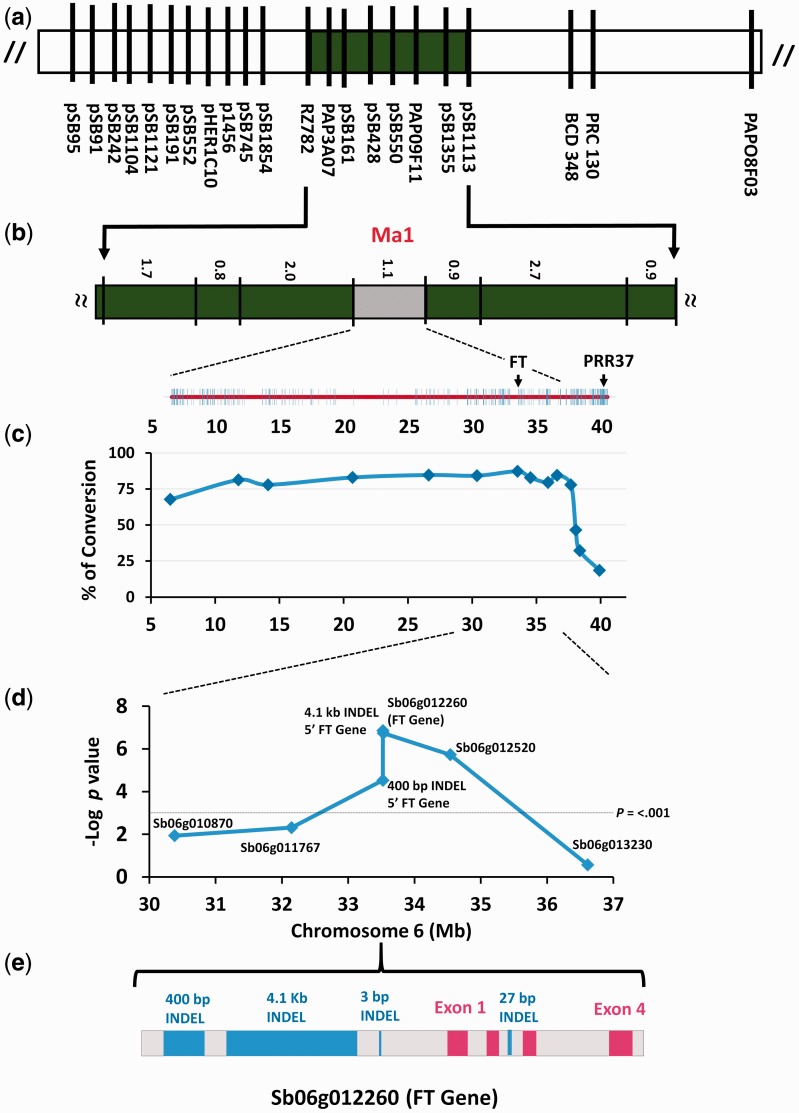
Based on progeny testing, FlrAvgD1 was placed between DNA markers pSB1113 and CDSR084, estimated to range from 0.3 to 1.1 cM apart in two different mapping populations studied (the larger distance is indicated in fig. 1a). BAC clones were identified containing each of these DNA markers, but lengthy efforts to “chromosome walk” in this region failed due to repeatedly encountering repetitive DNA near BAC ends. The sorghum genome sequence revealed this 1.1 cM interval to include 34 Mbp, thus having ∼60-fold less recombination than the genome-wide average (Paterson et al. 2009).
Fig. 1.
Genetic dissection of Ma1. (a) Linkage mapping of FlrAvgD1 to sorghum chromosome 6 (LG D; Lin et al. 1995); (b) progeny testing in 30 F3 families that were recombinant in the interval containing FlrAvgD1 delineated the locus to a 1.1-cM region including ∼400 genes; (c) analysis of 90 diverse exotic sorghums and their corresponding converted derivatives delimited FlrAvgD1 to a 4.1-Mb region including ∼63 genes. (d) Association genetics implicated Sb06g012260 in short-day flowering conferred by FlrAvgD1. (e) The day-neutral haplotype of Sb06g012260 contains three deletions in the 5′ region, one removing a CAAT box essential to many eukaryotic promoters.
To better circumscribe the location of FlrAvgD1 in the 34-Mbp target region, and to confirm that the S. propinquum-derived locus also accounts for intraspecific variation within S. bicolor (the cultigen), we utilized recombination accumulated during “conversion” of diverse short-day tropical sorghums to day-neutral flowering (Stephens et al. 1967). Sorghum conversion involves four backcrosses each followed by two generations of selfing (Stephens et al. 1967), a breeding scheme that in regions held heterozygous is expected to impart ∼3.5× more recombinational information to converted genotypes than F2 or recombinant inbred genotypes (Allard 1956). Ninety diverse exotic sorghums and their corresponding converted derivatives (supplementary table S1, Supplementary Material online) were genotyped at nine simple sequence repeat (SSR) loci distributed approximately evenly through the 34 Mbp target interval inferred for the QTL based on our S. bicolor×S. propinquum cross. The highest conversion frequency of the 90 sorghums was 87% (fig. 1c) vs. a genome-wide average of ∼10% (Lin et al. 1995), and closely coincides with FlrAvgD1, strongly supporting the hypothesis that the S. propinquum-derived QTL locus also accounts for intraspecific variation in short-day vs. day-neutral photoperiod response in S. bicolor.
In 384 diverse sorghums (Hash et al. 2008) phenotyped for “Photoperiod Response Index” (PRI: see Materials and Methods), we identified and genotyped DNA polymorphisms in annotated genes near the conversion peak. TASSEL was used to perform tests of association, employing population structure covariates and a kinship matrix for the sorghum germplasm panel based on published SSRs (Hash et al. 2008). The most significant association (P < 10 − 6) was found with Sb06g012260 (fig. 1d, with raw data in Additional data file 2), a gene containing a PEBP domain (Interpro IPR008914) shared by floral regulators in Arabidopsis (FT) (Kardailsky et al. 1999) and Oryza (Hd3a) (Kojima et al. 2002).
By comparing S. propinquum BAC sequences containing Sb06g012260 to the S. bicolor draft genome and resequencing polymorphic sites in the 384 diverse sorghums, we found two major Sb06g012260 haplotypes. One haplotype is widespread across tropical Africa, and is shared by short-day S. propinquum. The other haplotype was most abundant in accessions from South Africa, the most temperate part of the natural range of sorghum and where day-neutral flowering is common (fig. 2). The predominantly South African haplotype was characterized by four deletions, respectively, of 423 nt starting 5901 nt upstream of the Sb06g012260 transcription-start site; 4186 nt starting 4553 nt upstream; 3 nt starting 219 nt upstream; and 27 nt in the second intron of Sb06g012260 (fig. 1e). Additional indels of 2 and 7 nt (5451 and 5025 nt upstream), and three synonymous mutations in exon 1 and two in exon 2 were not analyzed in depth.
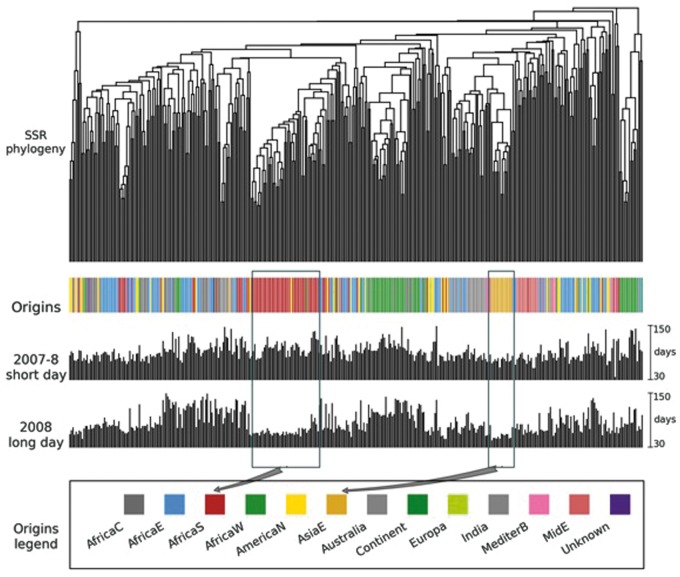
Fig. 2.
Biogeography of day-neutral flowering. Flowering of 384 diverse sorghums were compared in the 2007–2008 post-rainy (short day), and 2008 rainy (long day) seasons in peninsular India to determine “photoperiod response index” (as described in methods). Hierarchical clustering of sorghum accessions was based on distances between SSR genotypes (number of bands NOT shared). While short-day sorghums experience delayed flowering under long days (>12 h), long days accelerated flowering by ∼30 days for many genotypes from South Africa, the most temperate part of the natural range. East Asian (AsiaE) sorghums, the largest temperate-adapted group from the northern hemisphere, also had accelerated long day flowering.
Two constructs (supplementary fig. S1, Supplementary Material online) containing short-day S. propinquum Sb06g012260 alleles subcloned from a BAC and transformed into the day-neutral accession Tx430 using published methods (Howe et al. 2006) each delayed flowering of transgenic F2 progeny in long days (table 1). Widely used for sorghum transformation because of its high efficiency (Howe et al. 2006), Tx430 has day-neutral flowering but has an unusual haplotype, with deletion of seven amino acids in the 4th exon of Sb06g012260. Independent T0 transformants were selfed to produce T1 segregating progenies, then 15–24 plants from each T1 family were evaluated in the greenhouse under ambient long day conditions (at 33.95° N latitude), planting on 17 May (13 h, 58 min photoperiod) and recording the number of days to flower emergence. Plants were genotyped by PCR to determine allele state for the transgene. Among 13 transformation events carrying a ∼5 kb construct limited to Sb06g012260 and its immediate upstream elements, two conferred statistically significant delays averaging 13.1 (P = 0.03) and 24.8 days (P = 0.09), and one line showed unexpected accelerated flowering (14.1 days, P = 0.05). Among 10 independent events harboring a ∼10 kb construct spanning the entire haplotype (from Sb06g012260 through the 4,186 nt element), transgenic F2 progeny of only three showed significantly altered flowering, with delays of 4.1 (P = 0.002), 4.2 (P = 0.07) and 5.2 (P = 0.008) days.
Table 1.
Regression Analysis of Relationship of Flowering Time to Genotype for Transgenic Constructs (Transgene Events with P < 0.1 are shown).
| Construct | Event | Wild-type* | Transgenic* | Transgene effect (days) | P |
|---|---|---|---|---|---|
| pAP5.2 | 14 | 51.75 | 64.91 | 13.15 | 0.0310 |
| pAP5.2 | 16 | 61.00 | 85.82 | 24.81 | 0.0857 |
| pAP5.2 | 21 | 83.00 | 68.85 | −14.15 | 0.0463 |
| pAP10.2 | 3 | 48.29 | 52.44 | 4.16 | 0.0710 |
| pAP10.2 | 6 | 47.00 | 51.08 | 4.08 | 0.0019 |
| pAP10.2 | 7 | 49.75 | 55.00 | 5.25 | 0.0079 |
*Least-squares means.
The predominant day-neutral Sb06g012260 haplotype includes one mutation likely to alter gene function. The 3-bp deletion located 219 nt upstream of Sb06g012260 removed a predicted CAAT box, an invariant DNA sequence in many eukaryotic promoters required for sufficient transcription (Berg et al. 2002). Other elements of the haplotype appear innocuous. The 423-bp deletion removes a non-autonomous CACTA transposon; and the 4,186-nt deletion removes an open reading frame that matches a gene on chr. 7 of day-neutral sorghum (Sb07g008600, BLASTP 93.01% id, e-value 0), but with a “stop” codon in its first exon.
Sb02g029725, a family member that is more closely related to Hd3a and FT than to Sb06g012260, maps near the likelihood peak of another sorghum flowering QTL, FlrAvgB1 (Lin et al. 1995). Resequencing of this gene in the 384-member diversity panel used above (Hash et al. 2008) revealed two abundant haplotypes (resembling S. propinquum and BTx623, respectively), which showed highly significant association with PRI (P = 1.53×10 − 6). Thus, at least two members of the FT gene family are implicated in the modulation of flowering in sorghum.
Sb06g012260 is extensively diverged from known floral regulators (fig. 3). Several other sorghum, Saccharum and maize FT-containing genes occur in a clade that includes both rice Hd3a (Os06g06320.1) (Kojima et al. 2002) and Arabidopsis FT (Kardailsky et al. 1999). Indeed, Sb10g003940.1 is a colinear ortholog of Hd3a, which also formed a tandem duplicate in the rice lineage (Os06g06300: fig. 3). However, the sequence of rice Hd3a is more similar to Arabidopsis FT and another Arabidopsis homolog (AT4G20370.1) than to Sb06g012260 (fig. 3).
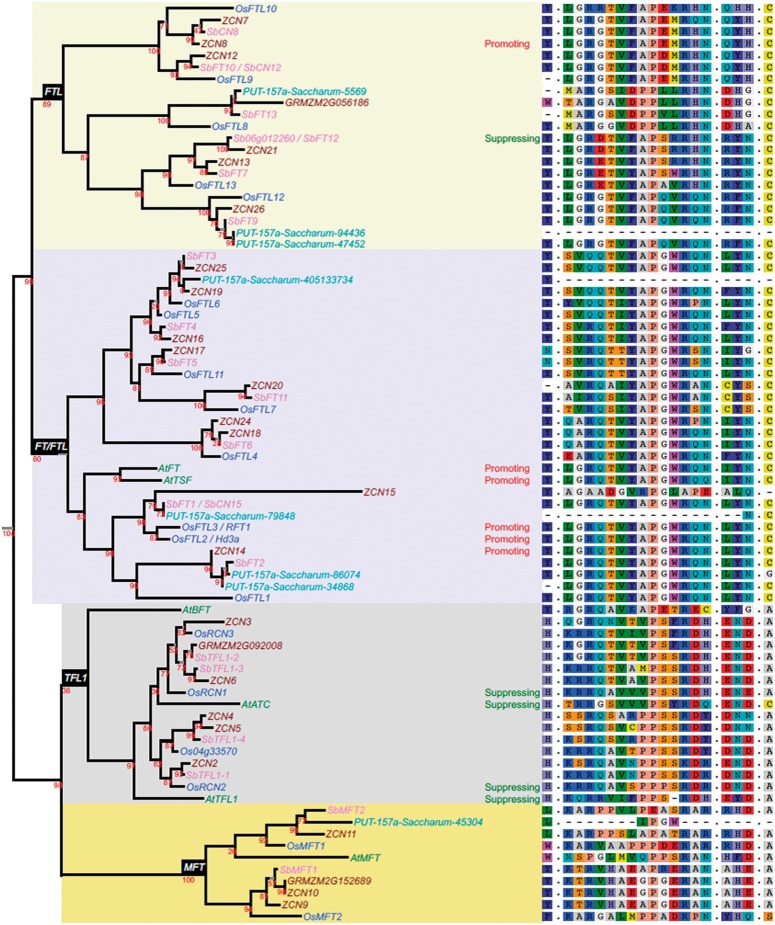
Fig. 3.
Homologs of Sb06g012260. Six homologs were found in Arabidopsis (prefixed At, including FT; Kardailsky et al. 1999), 19 in rice (Os, including Hd3a; Kojima et al. 2002), 19 in sorghum (Sb), 26 in maize (GRM or AC) and 8 in sugarcane (PUT-157a-Saccharum_officinarum), identified by BLAST from http://www.plantgdb.org/prj/ESTCluster/progress.php>PlantGDB, last accessed June 22, 2016. A phylogenetic tree of inferred homologous protein sequences was made at http://www.phylogeny.fr/, last accessed June 22, 2016, using MUSCLE for alignment and maximum-likelihood (PHYML) to determine the tree. Numbers on internal nodes indicate support values with 1000 bootstrap samples. Four distinct sub-families including FTL-like, FT/FTL-like, TFL1-like and MFT-like proteins, are highlighted on the tree. The impact on flowering (“promoting” or “suppressing”) are shown for several well studied PEBP genes (reviewed in Klintenäs et al. 2012) and for Sb06g012260/SbFT12. The panel on the right along the tree shows multiple alignments based on the Arabidopsis FT protein at amino acid position 85 at exon 2, positions 128–141 (P-loop domain), positions 150–152 and position 164 that were shown to have critical regulatory roles. For full alignments across the entire length of these proteins, see supplementary figure S4, Supplementary Material online.
An Sb06g012260 allele found at high frequency in weedy and invasive S. halepense may have contributed to its spread. Sorghum halepense (“Johnson Grass”), a tetraploid derived naturally from wild S. bicolor and S. propinquum progenitors (Paterson, Schertz, et al. 1995), has spread across much of Asia, Africa, Europe, North and South America, and Australia. Its establishment in USA is post-Columbian, by introduction as a prospective forage and/or contaminant of sorghum seedlots (McWhorter 1971). With inbreeding progenitors of tropical origin and therefore being putatively homozygous for short-day flowering, the original tetraploid S. halepense is expected to have had four copies of the short-day haplotype. Among the few Old World S. halepense accessions in the US National Plant Germplasm collection, we confirmed by PCR that PI209217 from South Africa and PI271616 from India are homozygous for the short-day haplotype, and confirmed by growouts that these two genotypes do not flower in long days.
With maximum seed production under 10.5–12 h daylengths consistent with short-day flowering, US (temperate) S. halepense nonetheless flowers and produces ample seed in photoperiods of up to 16 h (Warwick and Black 1983). We genotyped the four loci diagnostic of the short-day haplotype (423 nt, 4,186 nt, 3 nt, and intron indels) in 480 S. halepense plants sampled equally from populations in GA, TX (2), NE, and NJ described previously (Morrell et al. 2005). At the two terminal loci of the haplotype, 81.6% (423 nt indel) and 88.2% (intron indel) were homozygous for the short-day alleles, consistent with the expected genotype of their tropical progenitors. However, at the two internal loci of the haplotype (4,186 and 3 nt indels), only 1.1% and 8.0% of plants were homozygous for the short-day-associated alleles (fig. 4 and supplementary table S2, Supplementary Material online). Only 39-bp upstream from the locus of the CAAT box deletion in day-neutral S. bicolor, 85.9% of S. halepense plants have at least one copy of a 4-nt insertion that disrupts a TC-rich repeat, a cis-acting motif enriched in promoters of photoperiod-responsive genes (Mongkolsiriwatana et al. 2009). In a search of 30 Illumina-sequenced sorghum genomes, we have not found this mutation in any other members of the genus (unpublished data), suggesting that it is rare or novel. Further, at the 4,186-nt indel, 98.9% of S. halepense also have at least one allele from the day-neutral haplotype (fig. 4). Thus, it appears that an Sb06g012260 mutation in naturalized US S. halepense independent of those in S. bicolor has resulted in “replacement” of the internal portion of the short-day haplotype.
Fig. 4.
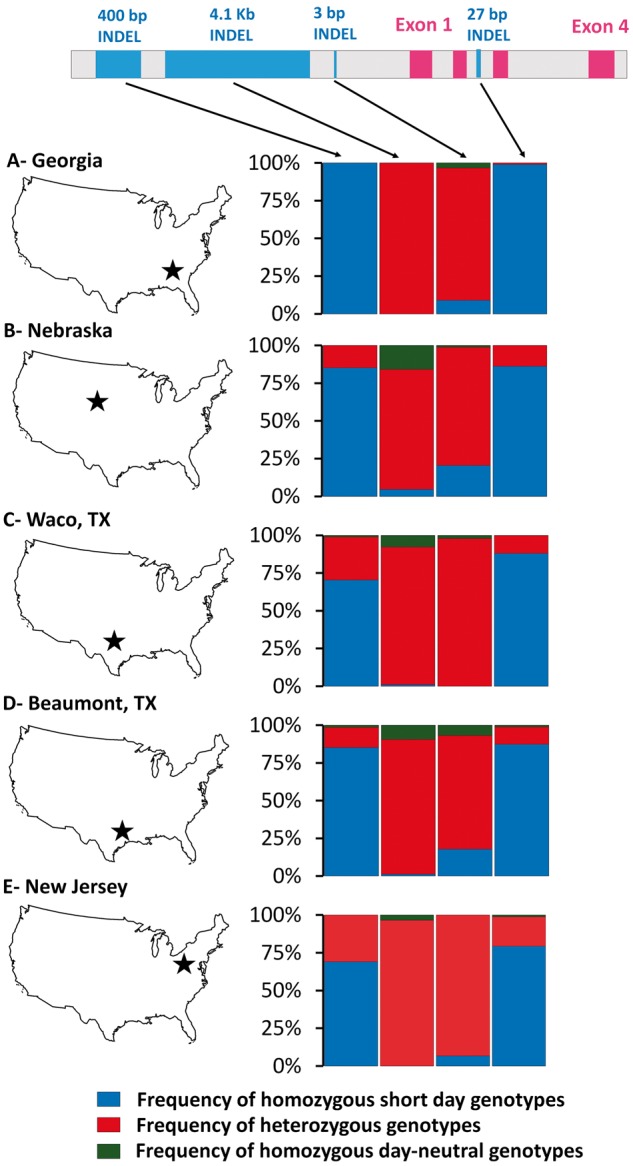
Genotype distributions of five US Johnson grass populations near the Sb06g012260 gene. Among 480 plants sampled equally from GA, TX (2), NE, and NJ populations (Morrell et al. 2005), 81.6% and 88.2% were homozygous for the short-day haplotype (blue bars) at two terminal loci (423 nt, 27 bp intron indels), but only 1.1% and 8.0% at two internal loci (4,186 and 3 nt indels). Homozygosity for day-neutral alleles (green) is nearly absent from the GA sample (from the region where Johnson grass is thought to have been introduced to USA), but exceeds 10% in the two northerly populations (NE, NJ) where day-neutral flowering would be most advantageous, and is intermediate in the two TX populations.
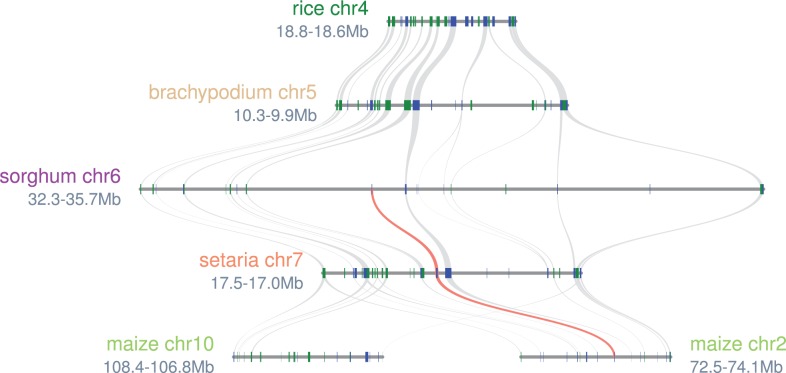
Based on several lines of evidence, Sb06g012260 appears to have evolved as a single-gene duplication (fig. 5) shortly after the oryzoid (rice) – panicoid (sorghum, maize) divergence. First, its divergence from its nearest sorghum homolog, Sb04g008320 is estimated at 40 Ma (Ks = 0.43, based on a widely-used evolutionary rate for cereal genes; Gaut et al. 1996), suggesting that it evolved after oryzoid (rice) – panicoid (sorghum, maize) divergence (consistent with gene tree topography, fig. 3). Sb04g008320 has a colinear rice ortholog (Os02g13830.1) but Sb06g012260 lacks colinear orthologs in both rice and Brachypodium distachyon, although possessing them in Setaria and maize (fig. 5). This pattern of orthology is indicative of an origin of Sb06g012260 in panicoids, shortly after divergence from a common ancestor shared with oryzoids (rice) and pooids (Brachypodium).
Fig. 5.
Microsynteny pattern around Sb06g012260 across five grasses. Sb06g012260 and associated orthologs in Setaria and maize are highlighted in red. No syntenic orthologs of Sb06g012260 can be found in rice or Brachypodium. Rectangles represent predicted gene models with colors showing relative orientations (blue: same strand, green: opposite strand). Matching gene pairs are displayed as connecting shades.
Discussion
Several independent lines of evidence including fine mapping, association genetics, mutant complementation, and evolutionary analysis all implicate a single gene, Sb06g012260, as the cause of the FlrAvgD1 QTL that accounted for 85.7% of variation in flowering time of a temperate×tropical sorghum population (Lin et al. 1995). Identification of this gene was complicated by its location in the physically largest single cM in the sorghum genome, containing ∼5% of sorghum genomic DNA and ∼1.3% (400) of genes (Paterson et al. 2009), with enrichment of retroelements and other non-genic DNA.
While both its panicoid-specific origin (fig. 5) and high-precision positional data (fig. 1) falsify the inference that Ma1 corresponds to the locus of the Triticeae photoperiodic genetic loci, Ppd1-3 and Ppd-H1 (Paterson, Lin, et al. 1995), correspondence to a major maize flowering QTL (Koester et al. 1993) remains plausible. Sb06g012260 has a maize ortholog, ZCN21 (GRMZM2G019993; fig. 3) on chr. 2, with genome duplication implying that a chr. 10 paralog should exist at ∼105 Mb, near the flowering QTL (Koester et al. 1993). However, the nearest FT homolog to this location (AC217051.3_FG006: chr. 10, 114 Mb) has a sequence so divergent from Sb06g012260 that it appears non-orthologous.
The wild-type Sb06g012260 is the first floral suppressor discovered in the “FTL” clade of the FT gene family although another clade member is a floral promoter, ZCN8 (Meng et al. 2011) (fig. 3). Despite being a floral suppressor, Sb06g012260 resembles all four functionally characterized members of the “TFL1” clade of the family (Hanzawa et al. 2005; Pin et al. 2010; Harig et al. 2012; Cao et al. 2016). Among the 23 transgene events that we evaluated, 5 showed statistically significant (0.1 or less, with 3 at 0.05 or less) delays in flowering, whereas only one showed the acceleration that might be expected from general overexpression of a florigen. The candidate mutation that removed a CAAT box upstream of the day-neutral Sb06g012260 allele would be expected to reduce gene expression, also consistent with the hypothesis that wild-type Sb06g012260 is a floral suppressor. There is growing appreciation that floral regulation and other aspects of growth and development are regulated by antagonistic actions of multiple members of FT and other gene families (Lifschitz et al. 2014). Indeed, floral activators and repressors can be only slightly differentiated from one another – exchange of a single amino acid between Arabidopsis florigen FT and floral repressor TFL1 is sufficient to exchange the functions of these opposing genes (Hanzawa et al. 2005).
The finding that mutant complementation studies of Sb06g012260 only partially recapitulated the short-day phenotype, underline the need for more work to understand the function and regulation of Sb06g012260. It is noteworthy that the overwhelming majority of transgenic lines with significant deviations from Tx430 had delayed flowering, not the general promotion of flowering that might be explained simply by overexpression of many FT-like genes (McGarry and Ayre 2012). However, it is somewhat perplexing that only one of the 23 events reached the average 24.6 (±3.5) day delay between the reference genetic stock 100M (Murphy et al. 2011) that contains Ma-1 (a dominant allele conferring short-day flowering in tropical sorghums; Quinby and Karper 1945) and Tx430 under our conditions. Shorter flowering delays than the Ma1 reference genotype 100M relative to putatively near-isogenic SM100 (Murphy et al. 2011) may indicate that some distant regulatory elements were missing from the transgene constructs and/or that its native heterochromatic chromatin environment is important to its natural function. A key element of the additional information needed is the exact timing and location of Sb06g012260 gene expression. In more than 1.7 trillion reads from (unpublished) sorghum transcriptomic data pooled across leaves, stems, and flowers of 40 diverse genotypes grown by several different investigators in different environments, we found only four reads from Sb06g012260 – two each in two different genotypes. A detailed study of 19 sorghum PEBP genes in roots, leaves (four stages), stems, shoot apices and floral heads by semiquantitative RT-PCR also detected no expression of Sb06g012260 in any tissue (Wolabu et al. 2016), reiterating a need for further investigation. Further, two of the three other sorghum members of the subclade including Sb06g012260, Sb04g008320 (SbFT7) and Sb03g002500 (SbFT13), also show no expression in any tissue, although the third member, Sb10g021790 (SbFT9) shows moderate expression in leaves (Wolabu et al. 2016). Finally, the nearest maize homolog of Sb06g012260, ZCN21, shows no discernible expression in a broad sampling of tissues (Danilevskaya et al. 2008). In contrast, Sb02g029725 (SbFT6), which we implicate above in the FlrAvgB1 sorghum flowering QTL, showed strong expression in leaves across all developmental stages and was also detected in the floral head and florets, but does not induce flowering in Arabidopsis (Wolabu et al. 2016). Other members of its subclade (SbFT3, 4, 5, and 11) all show expression but in different tissues and developmental states, and like Sb06g012260 do not show expression patterns or genic interactions consistent with being florigens (Wolabu et al. 2016).
Where does Sb06g012260 fit among the named sorghum flowering loci? Based on the high frequency at which this region has long been known (Lin et al. 1995) to be selected for during sorghum conversion, we inferred that the QTL in this region was Ma1, the sorghum maturity (flowering) locus long known to have the largest effect on phenotype (Quinby and Karper 1945). Moreover, based on relatively coarse resolution QTL data, we previously suggested correspondence of FlrAvgD1 to a homoeologous series of Triticeae photoperiodic (Ppd1-3) loci (Paterson, Lin, et al. 1995) that is now known to be PRR37 genes homologous to Sb06g014570.
Others have suggested PRR37 to be Ma1 based on positional proximity to a flowering trait mapped in genetic populations different from ours, together with its expression pattern in response to photoperiodic cues (Murphy et al. 2011). In our findings, the transition from the heterochromatic surroundings of Sb06g012260 to euchromatin, and an associated increase in recombination, coincides with a precipitous drop in levels of “conversion” at ∼37 Mb (fig. 1c). PRR37 is in a euchromatic genomic location (40.3 Mb) experiencing only ∼15% conversion (fig. 1b), only modestly above the genome-wide average of 10%. Curiously, study of 390 exotic-converted pairs genotyped at 46,062 markers (albeit with 66% missing data before imputation) suggested an introgression peak at ∼42 Mb, different from both our peak and PRR37 (Thurber et al. 2013). The fact that this region has 60-fold less recombination than the genome-wide average (fig. 1), suggests that limited recombination rather than multiple loci under selection (Thurber et al. 2013) accounts for the high frequency of introgression in this region, and indeed across much of the chromosome.
Evidence in support of PRR37 as Ma1 is further confounded by factors that were not known to its proponents (Murphy et al. 2011) or their reviewers. PRR37 was not tested by mutant complementation, but was a positional candidate that showed striking expression differences based on comparison of near-isogenic lines called 100M and SM100 that differ in PRR37 alleles (Murphy et al. 2011). We show (supplementary fig. S2, Supplementary Material online) that the short-day flowering 100M line is introgressed with not only a putatively short-day PRR37 allele but also with the short-day Sb06g012260 haplotype, based on genotyping at both the 423 and 4,186 nt indels that are on the distal side of the gene relative to PRR37. This indicates that the chromosome segment by which 100M and SM100 differ contains both PRR37 and Sb06g012260, as well as at least 82 intervening protein-coding genes and 6.75 Mb DNA (PRR37 [Sb06g014570] has an address of chr6: 40,280,414–40,290,602. Sb06g012260 has an address of chr6: 33,528,527–33,531,376). Phenotypic differences between these lines could therefore be explained by either of these two genes, interactions between the two, or other factors. Further, in the genotype that was the source of the short-day flowering allele that we mapped (S. propinquum), PRR37 is non-functional with a 2-nt insertion near the 5′ end causing 19 nonsense mutations (supplementary fig. S3, Supplementary Material online), effectively ruling out that it could confer a dominant phenotype in crosses with S. bicolor. Finally, the fine-scale mapping that we describe herein (fig. 1) now clearly separates FlrAvgD1 from PRR37.
In addition to Ma1, a more recently discovered flowering locus, Ma6, also maps to chromosome 6 very near the location of PRR37 (Brady 2006) – indeed, one report stated that PRR37 corresponded to Ma6 (Mullet et al. 2012). However, members of the same group who implicated PRR37 as Ma1 have more recently implicated a new positional candidate as Ma6 based on apparent functional polymorphism and expression differences between long-day and short-day conditions (Murphy et al. 2014) – whereas this new candidate corresponds to orthologs in several other cereals such as the rice Ghd7 gene, once again evidence from mutant complementation is lacking. The time and cost associated with sorghum transformation is clearly a hindrance to validation of candidate genes in this promising botanical model.
In partial summary, we consider the most probable scenario to be that Sb06g012260 is the cause of the Ma1 short-day flowering trait in sorghum, as supported by independent lines of evidence including fine mapping, association genetics, mutant complementation, and evolutionary analysis. Evidence in support of another nearby gene (PRR37) as Ma1 is now known to be confounded with previously unknown factors, specifically the presence of Sb06g012260 and at least 82 additional annotated genes on the crucial introgressed chromosome segment. The non-functional PRR37 allele in our population rules out the possibility that the two genes are functioning in concert to confer the trait. While this new evidence does not contradict the hypotheses that each gene independently contributes to the flowering phenotype, or that PRR37 may be Ma6 (Mullet et al. 2012), recent data implicating the sorghum ortholog of rice Grain number, plant height and heading date-7 (Ghd7) as Ma6 is more compelling (Murphy et al. 2014), albeit needing support from mutant complementation.
The evolution of Sb06g012260 from a single gene duplicate, rather than a dosage-balanced gene from whole-genome duplication, follows a trend exemplified by key members of the C4 photosynthesis pathway (Wang et al. 2009), arguably among the most important environmental adaptations in tropical grasses. WGD-duplicates are a rich potential source of genetic novelty, with longer half-lives than single gene duplicates (Lynch et al. 2001) providing more time for evolution of adaptations that warrant preservation. However, single-gene duplicates, often lacking their ancestral regulatory elements and in different chromatin environments from their ancestral gene(s), may have greater per-gene potential for the evolution of novelty as reflected by greater divergence of expression patterns than WGD-duplicates (Wang et al. 2011). An interesting hypothesis for further investigation is whether the survival of single-gene duplicates such as Sb06g012260 may be favored by location in recombinationally-recalcitrant heterochromatin, where neutral or slightly-deleterious mutations tend to survive longer than in euchromatin (Bowers et al. 2005; Paterson et al. 2009) and offer more opportunity for a mutation that confers new function to coincide with an environment that drives it to high frequency.
Recalcitrance of the Ma1 region of chromosome 6 to recombination may have contributed to the evolution of a “coadapted gene complex” (Mayr 1954). Beyond flowering, QTLs have been associated with this region affecting kernel weight (Paterson, Lin, et al. 1995), tillering, rhizomatousness and regrowth (Paterson, Schertz, et al. 1995), leaf length and width (Feltus et al. 2006), and plant stature. In particular, the Ma1 region also holds dw2, the gene of largest effect on sorghum stature (height) (Lin et al. 1995) but which has long been asserted to be separable from Ma1 by recombination. Quinby suggested that Ma1 and Dw2 were different closely-linked genes, with ca. 8% crossing over (Quinby 1974), but only evaluating 47 families based on phenotype. Based on our observation that late flowering can occasionally be a result of factors other than allelic status at the Ma1 locus and that progeny testing is necessary to validate it, such a small study must be considered tenuous. Among the 30 validated F3 families in our study, three showed different segregation patterns for flowering time and plant height. Since these 30 individuals comprised all confirmed recombinants in the region from a population of 370 individuals, this suggests a 0.5-cM linkage distance between Ma1 and Dw2 (Lin 1998). While the strongest association of allelic variation with plant height in the diversity panel was at Sb06g012260 itself (P = 0.007), we also found a marginally significant association at Sb06g007330 (P = 0.023), a putative cation efflux family protein. Genes intervening between these two loci showed no significant association with plant height. Further study is needed to determine if Sb06g007330 is dw2 – however an intriguing hypothesis for further testing is that increased height confers a competitive advantage in light interception, and alleles conferring this trait might have become abundant more quickly if co-transmitted with alleles for optimal flowering time in the native tropical environment of sorghum. Moreover, further dissection of the Ma1 region may reveal whether QTLs for other traits that have been mapped to the region (Lin et al. 1995; Paterson, Lin, et al. 1995; Paterson, Schertz, et al., 1995) are pleiotropic consequences of Ma1 or dw2, or represent additional members of a coadapted gene complex.
Potential applications of Sb06g012260 are numerous. Engineering of genotypes that silence the short-day flowering trait may render obsolete the need to laboriously “convert” tropical grasses to day-neutral flowering by twelve(!) generations of breeding (Stephens et al. 1967), potentially dramatically accelerating cross-utilization of temperate and tropical germplasm for sorghum, sugarcane, and many other crops. Targeted selection or engineering of strong floral repressor alleles in biomass crops may confer consistent high yields (Julien and Soopramanien 1976; Long 1976; Julien et al. 1978; Heinz 1987), improving the economics of cellulosic biofuel production.
Materials and Methods
RFLP and SSR mapping used published methods, with markers drawn from published maps (Bowers et al. 2003; Kong et al. 2013).
Phenotyping of F3 families was based on 50–150 selfed progenies per family, grown in the field near College Station, Texas, at ambient daylengths described in detail and with flowering dates recorded as described (Lin et al. 1995).
Association genetics used a 384-member worldwide sorghum diversity panel from ICRISAT, previously characterized with 41 SSR markers (Hash et al. 2008), evaluated in 2007 under short-day conditions (11.8–12.15 h light) and high humidity, under which short-day sorghums are expected to initiate flowering promptly. A 2008 planting was characterized by a transition from long to short-day (13.1–11.0 h) photoperiod and dry conditions, and short-day sorghums would be expected to delay flowering. Flowering time was the number of days required for 50% of the plants in a single row to flower (DFL50%). Photoperiod Response Index (PRI) was defined as the mean difference in DFL50% between the two planting seasons (i.e., PRI = DFL50%2008−DFL50%2007). Average height of the plants in the individual plots was also determined.
Resequencing used BigDye terminator chemistry, and sequences were manually checked and aligned for single nucleotide polymorphism (SNP) identification with Sequencher 4.1.
The quantity and frequency of haplotypes, and linkage disequilibrium were determined by Haplotyper 1.0, and TASSEL 2.1, respectively. TASSEL was used to perform tests of association, employing population structure covariates and a kinship matrix for the GCP/ICRISAT germplasm panel based on published SSRs (Hash et al. 2008).
Overgo hybridization (Bowers et al. 2005) was used to identify a S. propinquum BAC (Lin et al. 1999) clone (YRL11P18) that contained Sb06g012260. The BAC was sequenced to confirm the integrity of Sb06g012260 and identify restriction sites, then completely digested with PvuII to recover a 10.5-kb fragment that extends 5,220-bp upstream and 234-bp downstream from Sb06g012260. Likewise, the BAC clone was completely digested with restriction enzymes StuI and BstZ17I to recover a 5.2-kb fragment that extends 434-bp upstream and 234-bp downstream from Sb06g012260. Both fragments were cloned into transformation vector PZP211, and the clones (supplementary fig. S1, Supplementary Material online) re-sequenced to confirm the integrity of the constructs.
Transformation used published methods and vectors (Howe et al. 2006) with the 35S promoter. Independent T0 transformants were selfed to produce T1 segregating progenies, then 15–24 plants from each T1 family were evaluated in the greenhouse under ambient long day conditions (at 33.95° N latitude), recording the number of days from planting on 17 May to flower emergence. Plants were genotyped by PCR to determine allele state for the transgene.
Genotyping of the four loci diagnostic of the Ma1 haplotype (423 nt, 4186 nt, 3 nt, and intron indels) in S. halepense used 480 plants sampled equally from GA, TX (2), NE, and NJ populations described previously (Morrell et al. 2005).
Supplementary Material
Supplementary figs. S1–S4 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The late K.F. Schertz was instrumental in the early stages of this work. This work was supported by the US Department of Agriculture National Research Initiative Competitive Grants Program (00-35300-9215), the United Sorghum Checkoff Program (R0001-10), the Sun Grant Program and the USDA Biotechnology Risk Assessment Grants (BRAG) program (95-33120-1935, 2012-01658).
References
- Allard R. 1956. Formulas and tables to facilitate the calculation of recombination values in heredity. Hilgardia 24:235–278. [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. 2002. Biochemistry. 5th ed New York: W.H: Freeman. [Google Scholar]
- Bowers JE, Abbey C, Anderson S, Chang C, Draye X, Hoppe AH, Jessup R, Lemke C, Lennington J, Li Z, et al. 2003. A high-density genetic recombination map of sequence-tagged sites for sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 165:367–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Arias MA, Asher R, Avise JA, Ball RT, Brewer GA, Buss RW, Chen AH, Edwards TM, Estill JC, et al. 2005. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc Natl Acad Sci U S A. 102:13206–13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JA. 2006. Sorghum Ma5 and Ma6 maturity genes. College Station (TX: ): Texas A&M University. [Google Scholar]
- Cao K, Cui L, Zhou X, Ye L, Zou Z, Deng S. 2016. Four tomato flowering locus T-like proteins act antagonistically to regulate floral initiation. Front Plant Sci. 11:1213.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR. 2008. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146:250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO Statistical Database on Crops [Internet]. 2011. [cited 2016 Jun 22]. Available from: http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567.
- Feltus FA, Hart GE, Schertz KF, Casa AM, Brown P, Klein PE, Kresovich S, Paterson AH. 2006. Genetic map alignment and QTL correspondence between inter- and intra-specific sorghum populations. Theor Appl Genet. 112:1295–1305. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. 1996. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci U S A. 93:10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D. 2005. A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci U S A. 102:7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harig L, Beinecke FA, Oltmanns J, Muth J, Muller O, Ruping B, Twyman RM, Fischer R, Prufer D, Noll GA. 2012. Proteins from the flowering locus T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 72:908–921. [DOI] [PubMed] [Google Scholar]
- Harper J. 1977. Plant population biology. London: Academic Press. [Google Scholar]
- Hash CT, Ramu P, Folkertsma RT, Upadhyaya HD, Billot C, Rami JF, Deu M, Gardes L, Rivallan R, Li Y, et al. 2008. Diversity analysis of the sorghum global composite collection and reference set. 2008 Annual Research Meeting Generation Challege Programme, Bangkok, Thailand.
- Heinz D. 1987. Sugarcane improvement through breeding. Amsterdam: Elsevier. [Google Scholar]
- Howe A, Sato S, Dweikat I, Fromm M, Clemente T. 2006. Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep. 25:784–791. [DOI] [PubMed] [Google Scholar]
- Jakob K, Zhou F, Paterson AH. 2009. Genetic improvement of C4 grasses as cellulosic biofuel feedstocks. In Vitro Cell Dev Biol Plant 45:291–305. [Google Scholar]
- Julien M, Delaveau P, Soopramanien G, Martine J. 1978. Age, time of harvest, and environment as factors influencing differences in yield between flowering and vegetative canes. Proc Int Soc Sugar Cane Technol. 16:1771–1789. [Google Scholar]
- Julien M, Soopramanien G. 1976. The effect of flowering on yield in sugarcane. Rev Agric Sucr Ile Maurice 55:151–158. [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286:1962–1965. [DOI] [PubMed] [Google Scholar]
- Klintenäs M, Pin PA, Benlloch R, Ingvarsson PK, Nilsson O. 2012. Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol. 196:1260–1273. [DOI] [PubMed] [Google Scholar]
- Koester R, Sisco P, Stuber C. 1993. Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci. 33:1209–1216. [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43:1096–1105. [DOI] [PubMed] [Google Scholar]
- Kong W, Jin H, Franks CD, Kim C, Bandopadhyay R, Rana MK, Auckland SA, Goff VH, Rainville LK, Burow GB, et al. 2013. Genetic analysis of recombinant inbred lines for Sorghum bicolor x S. propinquum. G3 3:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Ayre BG, Eshed Y. 2014. Florigen and anti-florigen - a systemic mechanism for coordinating growth and termination in flowering plants. Front Plant Sci. 16:465.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Schertz K, Paterson A. 1995. Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 141:391–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-R. 1998. Genetic analysis and progress in chromosome walking to the sorghum photoperiodic gene, Ma1. College Station: Texas A&M. [Google Scholar]
- Lin YR, Zhu L, Ren S, Yang J, Schertz K, Paterson A. 1999. A Sorghum propinquum BAC library, suitable for cloning genes associated with loss-of-function mutations during crop domestication. Mol Breed. 5:511–520. [Google Scholar]
- Long A. 1976. A large varietal difference in cane deterioration due to flowering. Proc South Afr. Sugar Technol Assoc. 50:78–81. [Google Scholar]
- Lynch M, O’Hely M, Walsh B, Force A. 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159:1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. 1954. Change of genetic environment and evolution In: Huxley J, Hardy AC, Ford EB, editors. Evolution as a process. London: Allen & Unwin; p. 157–180. [Google Scholar]
- McGarry RC, Ayre BG. 2012. Geminivirus-mediated delivery of florigen promotes determinate growth in aerial organs and uncouples flowering from photoperiod in cotton. Plos One 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter CG. 1971. Introduction and spread of Johnsongrass in the United States. Weed Sci. 19:496. [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON. 2011. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23:942–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsiriwatana C, Pongtongkam P, Peyachoknagul S. 2009. In silico promoter analysis of photoperiod-responsive genes identified by DNA microarray in rice (Oryza sativa L.). Katsetsart J (Nat Sci.) 43:164–177. [Google Scholar]
- Morrell PL, Williams-Coplin D, Bowers JE, Chandler JM, Paterson AH. 2005. Crop-to-weed introgression has impacted allelic composition of johnsongrass populations with and without recent exposure to cultivated sorghum. Mol Ecol. 14:2143–2154. [DOI] [PubMed] [Google Scholar]
- Mullet JE, Rooney WL, Klein PE, Morishige DT, Murphy R, Brady JA. 2012. Discovery and utilization of sorghum genes (MA5/MA6) In: Office USP, editor. USA: The Texas A&M University System. USA Patent No. US 8,309793 B2: U. S. Patent Office. [Google Scholar]
- Murphy R, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE. 2011. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci U S A. 108:16469–16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RL, Morishige DT, Brady JA, Rooney WL, Yang S, Klein PE, Mullet JE. 2014. Ghd7 (Ma6) represses sorghum flowering in long days: Ghd7 alleles enhance biomass accumulation and grain production. Plant Genome 7:1–10. [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lin YR, Li ZK, Schertz KF, Doebley JF, Pinson SRM, Liu SC, Stansel JW, Irvine JE. 1995. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269:1714–1718. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Schertz KF, Lin YR, Liu SC, Chang YL. 1995. The weediness of wild plants - molecular analysis of genes influencing dispersal and persistence of Johnsongrass, Sorghum halepense (L) Pers. Proc Natl Acad Sci U S A. 92:6127–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJL, Nilsson O. 2010. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330:1397–1400. [DOI] [PubMed] [Google Scholar]
- Quinby J, Karper R. 1945. The inheritance of three genes that influence time of floral initiation and maturity date in milo. J Am Soc Agron. 37:916–936. [Google Scholar]
- Quinby JR. 1974. Sorghum improvement and the genetics of growth. College Station: Texas A&M University Press. [Google Scholar]
- Ricaud C, Autrey L, Sullivan S. 1980. Losses from the recurrence of yellow spot epiphytotics in Mauritius. Sugar Y Azucar. 75:28–29. [Google Scholar]
- Rooney WL, Blumenthal J, Bean B, Mullet JE. 2007. Designing sorghum as a dedicated bioenergy feedstock. Biofuels Bioproducts Biorefining Biofpr 1:147–157. [Google Scholar]
- Schnable JC, Springer N, Freeling M. 2010. Biased gene loss following tetraploidy in maize reflects genome dominance and ongoing selection. Proc Natl Acad Sci U S A. 108:4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens J, Miller F, Rosenow D. 1967. Conversion of alien sorghums to early combine genotypes. Crop Sci. 7:396. [Google Scholar]
- Thurber CS, Ma JM, Higgins RH, Brown PJ. 2013. Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol. 14:R68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA-NASS. 2013. Annual Crop Summary. [cited 2016 Jun 22]. Available from: www.nass.usda.gov.
- Wang X, Gowik U, Tang H, Bowers JE, Westhoff P, Paterson AH. 2009. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 10:R68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang J, Guo H, Jin D, Lee T-H, Liu T, Paterson AH. 2015. Genome alignment spanning major Poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol Plant 8:885–898. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Z, Guo H, Zhang L, Wang L, Li J, Jin D, Paterson AH. 2014. Telomere-centric genome repatterning determines recurring chromosome number reductions during the evolution of eukaryotes. New Phytol. 205:378–389. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang X, Tang H, Tan X, Ficklin S, Feltus FA, Paterson A. H. 2011. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. Plos One 6:e28150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick SI, Black LD. 1983. The biology of Canadian weeds. 61. Sorghum halepense (L.) PERS. Can J Plant Sci. 63:997–1014. [Google Scholar]
- Wolabu TW, Zhang F, Niu L, Kalve S, Bhatnagar-Mathur P, Muszynski MG, Tadege M. 2016. Three flowering locus T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 210:946–959. [DOI] [PubMed]
- Woodhouse M, Schnable J, Pedersen B, Lyons E, Lisch D, Subramaniam S, Freeling M. 2010. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homeologs. PLoS Biol. 8:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.