Abstract
Identification of prognostic gene expression signatures may enable improved decisions about management of breast cancer. To identify a prognostic signature for breast cancer, we performed DNA methylation profiling and identified methylation markers that were associated with expression of ER, PR, HER2, CK5/6 and EGFR proteins. Methylation markers that were correlated with corresponding mRNA expression levels were identified using 208 invasive tumors from a population-based case-control study conducted in Poland. Using this approach, we defined the Methylation Expression Index (MEI) signature that was based on a weighted sum of mRNA levels of 57 genes. Classification of cases as low or high MEI scores were related to survival using Cox regression models. In the Polish study, women with ER-positive low MEI cancers had reduced survival at a median of 5.20 years of follow-up, HR=2.85 95%CI=1.25-6.47. Low MEI was also related to decreased survival in four independent datasets totaling over 2500 ER-positive breast cancers. These results suggest that integrated analysis of tumor expression markers, DNA methylation, and mRNA data can be an important approach for identifying breast cancer prognostic signatures. Prospective assessment of MEI along with other prognostic signatures should be evaluated in future studies.
Introduction
DNA methylation of CpG dinucleotides within promoter regions of tumor suppressor genes is an early, stable, heritable event that may be associated with loss of mRNA and protein expression and contribute to breast carcinogenesis [1]. Accordingly, analysis of DNA methylation profiling in conjunction with expression of mRNA and protein may identify genes that associated with the biology and clinical behavior of breast cancers and aid in clinical management [2-4].
Several recent studies employing candidate gene approaches [5-6] or profiling strategies [7-18] have identified DNA methylation markers that are associated with breast cancer characteristics or clinical outcomes. These studies demonstrate that DNA methylation is more frequent in estrogen receptor (ER)-positive compared with ER-negative tumors, suggesting that DNA methylation may play a more important role in shaping the biology and behavior of ER-positive cancers [7,9,11]. To date, most efforts to identify prognostic signatures for breast cancer have relied on mRNA profiling, but recent studies suggest that integrated analyses using multiple molecular platforms may be promising [19,7,20-21].
Although many immunohistochemical (IHC) signatures have been described to classify molecular subtypes of breast cancer, the five IHC markers that we used have shown important etiologic and survival differences for breast cancer [2,22-23]. We reasoned that evaluation of methylation profiles using these five-key IHC markers could provide new insights on epigenetic markers related to these important pathologic types..
Materials and Methods
Study Population
Subjects were selected from a population-based breast cancer case-control study, the Polish Breast Cancer Study (PBCS) [24]. The PBCS included 2386 cases and 2502 age and study site matched controls, between ages 20 and 74 years who resided in Warsaw or Łódź, Poland from 2000-2003. Breast cancer pathology was reviewed centrally to provide standardized classification. We selected 227 PBCS cases with snap frozen and formalin-fixed paraffin embedded tumor tissues obtained prior to treatment. Primary treatment (chemotherapy, radiation therapy, and hormone therapy) and vital status for up o 10 years after diagnosis (mean = 7.8 years, S.D. = 2.1), were collected through review of medical records and national databases in Poland.
Assessment of protein markers in TMAs
To develop a prognostic signature for breast cancer based on mRNA expression, we performed a cross-platform analysis using a multi-step strategy. Specifically, DNA methylation profiling data were first filtered based on their significant relationship with protein expression of ER, progesterone receptor (PR), human epidermal Growth factor receptor 2 (HER2), cytokeratin 5/6 (CK5/6), and epidermal Growth factor receptor 1 (EGFR) by immunohistochemical (IHC) analysis using data from the PBCS, a population-based case-control study.
Fixed tissues were prepared as tissue microarrays (TMAs) and evaluated for expression of ER, PR, HER2, EGFR or CK 5/6 using Automated Quantitative Analysis (AQUA) as previously described elsewhere [25,22]. Results of AQUA were highly correlated with results of IHC performed on TMAs and assessed visually and staining of whole sections performed for clinical management [25]. AQUA or IHC data was used to define tumors that were positive for each individual protein marker: ER, PR, HER2, EGFR and CK 5/6 and the scores for each individual marker were combined to approximate correspondence to the intrinsic molecular subtypes, including luminal (ER or PR positive), HER2 overexpression, and basal (negative for ER, PR and HER 2 with expression of CK 5/6 or EGFR) as previously described [25,22].
DNA Isolation and Methylation Analysis of Breast Cancer Tissues
DNA and RNA were isolated by standard protocols simultaneously from 30 mg tissue samples. For DNA, 30 mg of frozen tissue was shaved into 400 μl digestion buffer (0.1 M NaCl, 0.01 M Tris, pH 8.0, 0.025 M EDTA, pH 8.0, and 0.5% SDS) containing 0.1 mg/ml proteinase K. Samples were incubated overnight at 50°C with gentle rocking and purified using phenol:chloroform:isoamyl alcohol (25:24:1) and PhaseLock Gel (heavy) tubes for DNA extraction. DNA quality and quantity was assessed using agarose gel electrophoresis and PicoGreen dsDNA Quantitation Kit (Molecular Probes, Eugene, OR). Upon bisulfite conversion, DNA methylation status of 25,578 CpG probes was assayed using the Illumina Methylation27 bead-array (Illumina, San Diego CA) using the manufacturer's protocol. Data were extracted from the scanned arrays using Bead Studio. Of the 227 cases selected 226 had methylation data.
RNA Isolation, labeling, and microarray hybridization
Frozen tumor samples were stored in liquid nitrogen (−196°C) prior to nucleic acid extraction. RNA was isolated using 350 microliters TRIzol reagent (Invitrogen, Carlsbad, CA) and purified on Qiagen RNAeasy Mini columns per manufacturer protocols. RNA quantity and integrity was assessed using Nanodrop Spectrophotometry (Thermo Scientific, Waltham, MA) and 6000 Nano LabCip Kit on Agilent 2100 BioAnalyzer (Agilent, Santa Clara, CA), respectively.
Two-hundred fifty nanograms of input RNA was amplified and labeled using the Illumina TotalPrep RNA Amplification kit (Applied Biosystems/Ambion, Austin, TX). The biotin-labeled cRNAs were quantitated using RiboGreen RNA Quantitation reagent (Molecular Probes, Eugene, OR) and 750 ng was hybridized to Illumina HumanRef-8 v2 Expression BeadChip microarrays (Illumina, San Diego, CA). BeadChips were scanned in an Illumina scanner. Data were deposited with NCBI under GSEXXXXX. Of the 227 cases selected, 208 had usable mRNA expression data.
Data Analysis
The representativeness of cases with regards to tumor characteristics or risk factors included in the analysis was compared with the entire set of cases of the original PBCS population using Chi-squared or Fischer's exact tests.
Methylation data were processed in the R statistical environment. Analysis of mRNA expression data was processed using the lumi package in Bioconductor (R). These methylation markers were correlated with mRNA profiling data to define a Methylation Expression Index (MEI) composed of 57 genes whose expression was correlated (typically negatively) with DNA methylation.
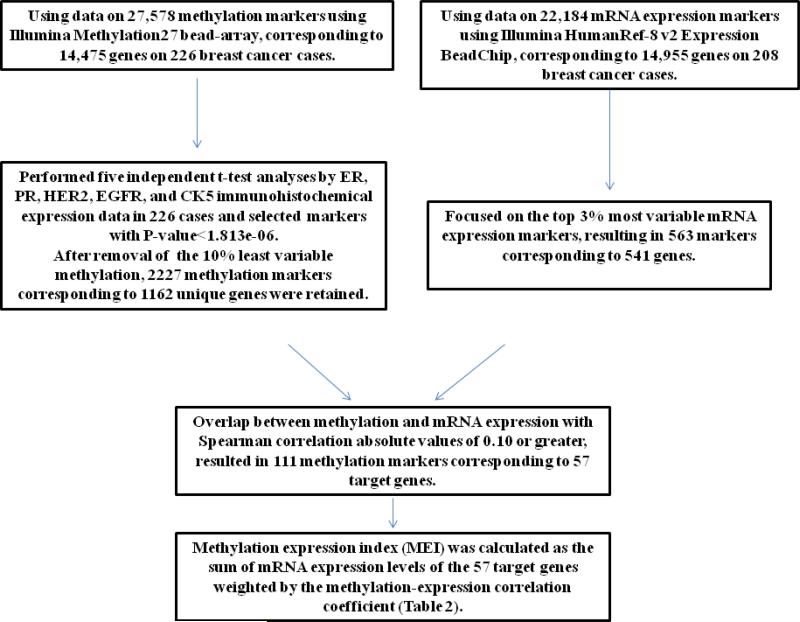
To develop a prognostic signature, we classified tumors as negative or positive for five IHC markers, ER, PR, HER2, EGFR, and CK5, as previously described [22]. Using the methylation data from the Illumina Infinium Human Methylation27 BeadChip, we selected markers related to IHC protein expression, using a Bonferroni adjustment threshold of p <1.8 e-06. Through this selection process we identified 2227 methylation markers in 1162 genes, which also had corresponding mRNA expression data (Figure 1). In a separate analysis, to identify mRNA markers that had substantial variation, we selected the 3% most variable mRNA expression probes from the Illumina HumanRef-8 v2 Expression BeadChip, which identified 563 probes corresponding to 541 genes. To we selected 3% most variable genes based on gene expression and intersected them with the 1162 genes. The intersection resulted in 65 genes. We further selected 57 genes that had spearman correlation absolute values of 0.1 or greater.
Figure 1.
Analysis strategy to define the 57 gene Methylation Expression Index (MEI), using data from 27,578 methylation markers and 22,184 mRNA expression markers from the Polish breast cancer study
We then determined the overlap between the 1162 methlylation marker genes associated with IHC marker expression, and the 541 genes with variable mRNA expression data, which identified 65 genes. Because we were interested in methylation markers that could influence mRNA expression, we determined the Spearman correlations coefficients between methylation and mRNA expression among the 65 genes. We further restricted the gene list to those with an absolute Spearman correlation rho equal or greater than 0.1 (for genes with multiple probes we calculated the average), which resulted in 57 genes with corresponding mRNA expression data available in the Polish study (Table 1).
Table 1.
Summary of the methylation markers and whether they are CpG islands significantly associated with tumor IHC marker expression at p< 1.813e-06
| IHC marker | count | notCPGI | CPGI |
|---|---|---|---|
| ER | 922 | 574 | 348 |
| PR | 646 | 394 | 252 |
| HER2 | 203 | 127 | 76 |
| EGFR | 286 | 167 | 119 |
| CK5 | 124 | 64 | 60 |
MEI scores were related to demographic, prognostic and treatment variables using Chi-squared or Fischer's exact tests. We assessed the relationship between MEI and overall survival among 208 women with breast cancer who had available mRNA expression data. The Kaplan-Meier (KM) method stratified by ER-status was used to generate survival curves for categories of the MEI [26]. Hazard ratios (HR) and 95% confidence intervals (CI) associated with methylation status adjusted for age (in five-year categories), tumor size (<2cm vs >2cm) , grade (well/moderately differentiated vs poorly differentiated), and node status (positive vs negative), were estimated using Cox proportional hazard models [27]. We checked for violations of the proportional hazards assumption for methylation variables and covariates by using Schoenfeld residuals. Interaction with MEI and ER was assessed using an interaction term in Cox models. Analyses were performed using Stata/SE v11.2 for Windows (College Station, TX) and the R statistical environment.
Replication datasets
To validate associations between MEI and breast cancer survival, we analyzed mRNA expression data from four independent datasets. The first consisted of a combined dataset of cDNA microarrays from 337 breast cancer patients (88 ER-negative and 249 ER-positive) ages 26-62 from the Netherlands Kanker Instituut (NKI) [28,4]. A second dataset included expression data from GEO GSE6532 Affymetrix Human Genome U133A microarray assays from 414 breast cancer patients ages 24- 88 available at the National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/geo/ [29]. In this dataset, 277 patients were treated with tamoxifen 5 of which were ER-negative, while the remaining 137 patients were untreated, 86 of which were ER-positive. The third dataset is the TCGA breast cancer dataset [19] and used data on 532 patients (407 of which were ER-positive) ages 26-90 with both gene expression and survival data. The fourth dataset was from METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) with expression data on 1992 patients of which 1508 were ER-positive (referred to as BT2000) [20]. Gene expression data were weighted by the correlation of methylation with expression determined in the PBCS dataset. The correlations provided a way to combine gene expression so that a gene with less correlation had less contribution to the MEI.
Result
Development of Methylation Express Index (MEI)
Cases included in this study ranged in age from 28-75 years, with a median age of 55 years. The percentage of cases positive for IHC markers of interest was 72% for ER, 49% for PR, 10% for HER2, and 18% for CK5/6 or EGFR (Supplemental Table 1). Patient demographic and tumor characteristics for the 227 cases selected for analysis were generally similar to the remaining cases in this population, except that tumors were larger (53% of profiled > 2.1 cm vs. 42% not profiled; p=0.002), more frequently ER positive (72% profiled vs 65% not profiled; p=0.02), and more often node positive (profiled 46% vs. 36% non-profiled p=0.008, Supplemental Table 1). Additional analysis restricted to these 208 showed similar results but use of the full set of 226 samples with methylation profiling data. 93% of the methylation markers found by the 208 samples were also found by the 226 samples, however to increase the power of methylation markers we selected relevant markers from the full set of 226 samples.
The MEI signature we developed based on these 57 genes was defined as the sum of the mRNA expression values of the 57 genes weighted by the Spearman correlation coefficient, which we refer to as the MEI (see Methods). Of the 57 genes, 54 showed a negative correlation between DNA methylation and mRNA expression (Table 2, Supplemental Table 3). The 57 genes identified through the selection process are related to a broad range of molecular functions based on Gene Ontology (GO), including binding, catalytic activity, enzyme and receptor activities and other processes.
Table 2.
Weights based on 208 breast cancer cases from the Polish breast cancer study for the 57 genes identified from methylation analysis that have corresponding mRNA expression data and used as weighting factors for calculation of prognostic signature
| Gene | Gene name | weights |
|---|---|---|
| DNALI1 | dynein, axonemal, light intermediate chain 1 | 0.7370 |
| CALML5 | calmodulin-like 5 | 0.6845 |
| HCLS1 | hematopoietic cell-specific Lyn substrate 1 | 0.6748 |
| SERPINA5 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 | 0.6397 |
| PDZK1 | PDZ domain containing 1 | 0.6325 |
| AGR3 | anterior gradient 3 | 0.6307 |
| PLAT | plasminogen activator, tissue | 0.6215 |
| SCUBE2 | signal peptide, CUB domain, EGF-like 2 | 0.6146 |
| KYNU | kynureninase | 0.6116 |
| SCNN1A | sodium channel, non-voltage-gated 1 alpha subunit | 0.5683 |
| SPOCK2 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 | 0.5480 |
| REEP6 | receptor accessory protein 6 | 0.5400 |
| STC2 | stanniocalcin 2 | 0.5338 |
| C8orf4 | chromosome 8 open reading frame 4 | 0.5319 |
| CAP2 | CAP, adenylate cyclase-associated protein, 2 (yeast) | 0.5118 |
| CFB | complement factor B | 0.5039 |
| C1orf64 | chromosome 1 open reading frame 64 | 0.5024 |
| IL20 | interleukin 20 | 0.4972 |
| CD247 | CD247 molecule | 0.4967 |
| FABP7 | fatty acid binding protein 7, brain | 0.4952 |
| CCND1 | cyclin D1 | 0.4827 |
| AKR7A3 | aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) | 0.4678 |
| TFF1 | trefoil factor 1 | 0.4575 |
| TAP1 | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | 0.4360 |
| ZG16B | zymogen granule protein 16B | 0.4226 |
| MMP7 | matrix metallopeptidase 7 (matrilysin, uterine) | 0.3514 |
| HMGCS2 | 3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) | 0.3425 |
| CAMK2N1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 | 0.3393 |
| GZMA | granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) | 0.3298 |
| ATP6V1B1 | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 | 0.3214 |
| GLIPR2 | GLI pathogenesis-related 2 | 0.3054 |
| PRSS8 | protease, serine, 8 | 0.2737 |
| MLPH | melanophilin | 0.2733 |
| C10orf116 | adipogenesis regulatory factor | 0.2715 |
| THRSP | thyroid hormone responsive | 0.2706 |
| CRYAB | crystallin, alpha B | 0.2598 |
| ALOX5AP | arachidonate 5-lipoxygenase-activating protein | 0.2439 |
| IFITM3 | interferon induced transmembrane protein 3 | 0.2414 |
| CD248 | CD248 molecule, endosialin | 0.2359 |
| CXCL12 | chemokine (C-X-C motif) ligand 12 | 0.2344 |
| SERPINA3 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 0.2328 |
| CILP | cartilage intermediate layer protein, nucleotide pyrophosphohydrolase | 0.2233 |
| CST3 | cystatin C | 0.2154 |
| HLA-DRA | major histocompatibility complex, class II, DR alpha | 0.2039 |
| SCGB3A1 | secretoglobin, family 3A, member 1 | 0.1925 |
| S100A7 | S100 calcium binding protein A7 | −0.1892 |
| SLPI | secretory leukocyte peptidase inhibitor | 0.1823 |
| GP1BB | glycoprotein Ib (platelet), beta polypeptide | −0.1781 |
| S100A9 | S100 calcium binding protein A9 | 0.1761 |
| CCL8 | chemokine (C-C motif) ligand 8 | −0.1726 |
| SMOC2 | SPARC related modular calcium binding 2 | 0.1738 |
| TFF3 | trefoil factor 3 (intestinal) | 0.1617 |
| MSLN | mesothelin | 0.1611 |
| MUC1 | mucin 1, cell surface associated | 0.1539 |
| PRSS23 | protease, serine, 23 | 0.1518 |
| TMEM119 | transmembrane protein 119 | 0.1026 |
| PI15 | peptidase inhibitor 15 | 0.1024 |
*These genes had more than one methylation marker associated with the mRNA expression target and the average of the sites was used to calculate the weighted Rho value.
MEI score association with death among ER-positive breast cancer cases in the PBCS
MEI scores approximated a normal distribution (median value = 168.172). We classified tumors as high/low MEI using the median value as the cutpoint, to evaluate its association with tumor characteristics and prognosis. Low MEI was associated with tumors that were poorly differentiated, ER-negative, PR-negative, and HER2-postive (p=<0.001), Supplemental Table 3.
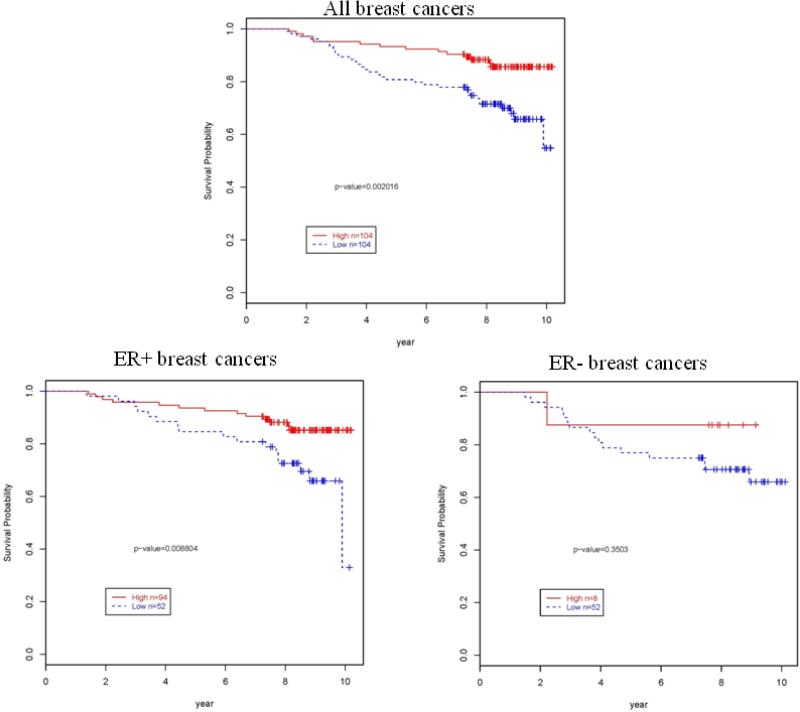
Survival analysis based on the classification of the MEI as high/low showed a significant association with survival overall; however, analysis stratified by ER status demonstrated that a significant association was restricted to ER-positive tumors. Among ER-positive breast cancer cases, low MEI was significantly associated with an increased risk of death in KM curves (P=0.0088, Figure 2) and in multivariable models adjusted for age, tumor size, node status, grade, primary hormone treatment and secondary radiation treatment (HR=2.85 95%CI=1.25-6.47, Table 3).
Figure 2.
Kaplan-Meir curves by low and high MEI overall survival for 208 breast cancer cases from the Polish breast cancer study
Table 3.
Hazard ratios (HR) and 95% confidence intervals (CI) between low and high MEI among ER-positive breast cancers and risk of death in the Polish breast cancer study
| Person years | Alive | Dead | HR* | 95%CI | P-value | HR** | 95%CI | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| MEI | |||||||||
| High | 697.82 | 81 | 13 | 1.00 | 1.00 | ||||
| Low | 375.49 | 35 | 17 | 2.59 | 1.22-5.49 | 0.01 | 2.85 | 1.25-6.47 | 0.01 |
| Invasive grade | |||||||||
| Well/moderately differentiated | 927.94 | 102 | 25 | 1.00 | 1.00 | ||||
| Poorly differentiated | 145.37 | 14 | 5 | 1.55 | 0.58-4.16 | 0.38 | 0.98 | 0.34-2.80 | 0.97 |
| Tumor size, CM | |||||||||
| 0.1-2.0 | 582.56 | 63 | 17 | 1.00 | 1.00 | ||||
| 2.1+ | 490.75 | 53 | 13 | 1.39 | 0.75-2.57 | 0.29 | 0.95 | 0.43-2.07 | 0.89 |
| Nodal status | |||||||||
| Negative | 564.70 | 65 | 13 | 1.00 | 1.00 | ||||
| Positive | 508.61 | 51 | 17 | 1.70 | 0.79-3.65 | 0.17 | 1.79 | 0.79-4.03 | 0.16 |
| Hormone therapy- primary treatment | |||||||||
| No | 226.89 | 23 | 10 | 1.00 | 1.00 | ||||
| Yes | 846.41 | 93 | 20 | 0.51 | 0.23-1.14 | 0.10 | 0.42 | 0.18-0.99 | 0.05 |
| Radiation therapy -secondary treatment | |||||||||
| No | 771.11 | 86 | 18 | 1.00 | 1.00 | ||||
| Yes | 302.20 | 30 | 12 | 2.66 | 1.34-5.30 | 0.005 | 2.93 | 1.34-6.41 | 0.01 |
Cox-proportional hazard ratios adjusting for age in 5-year intervals
Cox-proportional hazard ratios adjusting for age in 5-year intervals, tumor size, node status, grade, treatment regimen, and MEI
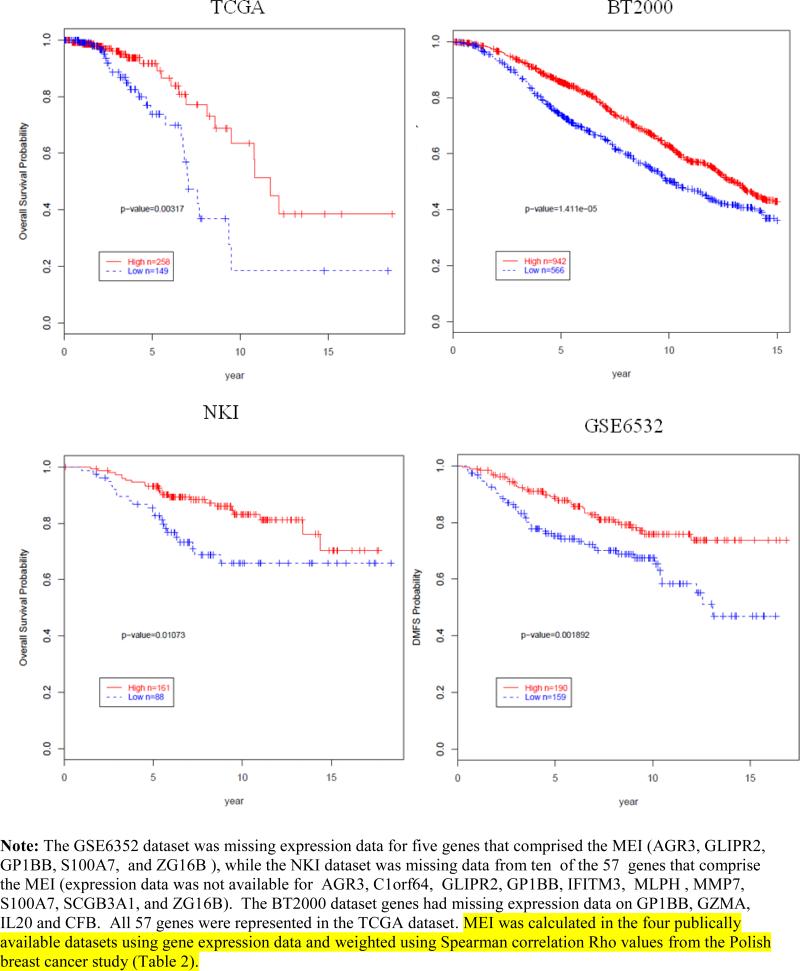
Validation of the MEI score and survival among ER-positive breast cancer cases
We generated a high/low categorical variable of MEI based on the median value of the MEI in each of four published mRNA expression datasets and evaluated its relationship with clinical outcomes. Among ER-positive breast cancer cases, low MEI was significantly associated with an increased risk of death (TCGA dataset P=0.003, BT2000 dataset P=1.4×10−5, NKI dataset P=0.01); and increased risk of distant metastases (GSE6532 dataset P=0.0018) (Figure 3). Tests for interaction of the MEI and ER by dataset showed significant interaction only in TCGA (P=0.02) and not in the other datasets (PBCS, P=0.37; NKI, P=0.52; BT2000, P=0.06; GSE6532, P=0.81). Evaluation of an unweighted MEI showed similar results to the weighted MEI in the datasets PBCS, TCGA and GSE6532, however, weighting of the markers gave more consistent findings (See Supplemental Figure 1),
Figure 3.
Kaplan-Meir curves and low and high MEI among ER-positive breast cancers and association with overall or disease metastasis free survival in four independent datasets
Discussion
In this report, we describe the development of the MEI, a prognostic signature consisting of a weighted combinatorial score of mRNA levels corresponding to 57 genes. These genes were selected because they showed significant relationships between DNA methylation levels and expression of key protein markers in the Polish Breast Cancer Study. In this study, women with ER-positive cancers and low MEI demonstrated reduced survival, and similar results were found in four large independent datasets. These results provide further support for integrated multi-platform molecular analyses to characterize tumor biology and search for prognostic signatures for breast cancer.
Although the MEI is novel, our findings are consistent with other published data. We found that the frequency of DNA methylation was higher in ER-positive as compared with ER-negative breast cancers, similar to prior studies [7,9,11,30]. Two MEI genes are included in other well-known prognostic gene expression signatures, including MLPH in the PAM50 panel and SCUBE2 in the Oncotype DX [2,31]. DNA methylation has also been identified in histologically normal tissues surrounding breast cancers, potentially implicating methylation as an early mechanism in carcinogenesis [32]. Accordingly, identifying panels of frequently methylated genes in breast cancers and surrounding tissues related to critical changes in mRNA and protein expression may provide markers of early detection and molecular targets for prevention or treatment. Analysis not using the weights showed similar results to the weighted sum of the MEI, showing the signature to be robust and support these genes to potentially have some important biological function.
Our analysis independently identified 55 genes that are not in well-known gene expression signatures (PAM50, OncotypeDx, and MammaPrint), thus are specific to the MEI. These genes might provide further clues into the biology of aggressive ER-positive breast cancers. Some of the MEI-specific genes are well-known cancer associated-genes, including CCND1, MUC1, MMP7 , C10orf116, and KYNU, which are involved in DNA repair, cell proliferation and cancer metastasis, and have been noted to be related to aggressive breast cancers [33-44].
Strengths of our study include: 1) analysis of samples from a population-based case-control study, with detailed epidemiologic data and long term follow-up and treatment data from medical records for these cases; 2) use of high density profiling arrays; 3) availability of detailed pathology and IHC data; and 4) validation of our findings in four independent datasets. Our study was limited by small numbers of less common subtypes, including ER-negative breast cancers. Although, some differences were found between the pathologic characteristics of Polish cases included in the analysis and the entire case group, confirmation of the prognostic significance of the MEI in independent datasets reduces concerns about generalizability. In addition, data were lacking from some MEI markers in the validation sets, but findings replicated nonetheless, suggesting that the signature is robust.
In conclusion, our analysis demonstrated that an integrated biomarker discovery approach in which DNA methylation profiling is integrated with mRNA expression and IHC is promising for discovering prognostic signatures for breast cancer. Specifically, discovery approaches that identify genes which show important associations at the DNA, RNA and protein levels may improve the specificity of signatures and limit the identification of chance findings. Future prospective studies are needed to assess the performance of the MEI along with other signatures to determine how best to employ these tools to direct patient care.
Supplementary Material
References
- 1.Laird PW, Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. Spec No. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J, Cheang MC, Mardis ER, Perou CM, Bernard PS, Ellis MJ. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16(21):5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. doi:1078-0432.CCR-10-1282 [pii] 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. doi:NEJMoa041588 [pii] 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 4.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. doi:10.1038/415530a415530a [pii] [DOI] [PubMed] [Google Scholar]
- 5.Veeck J, Esteller M. Breast cancer epigenetics: from DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia. 2010;15(1):5–17. doi: 10.1007/s10911-010-9165-1. doi:10.1007/s10911-010-9165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21(35):5462–5482. doi: 10.1038/sj.onc.1205606. doi:10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 7.Bediaga NG, Acha-Sagredo A, Guerra I, Viguri A, Albaina C, Ruiz Diaz I, Rezola R, Alberdi MJ, Dopazo J, Montaner D, de Renobales M, Fernandez AF, Field JK, Fraga MF, Liloglou T, de Pancorbo MM. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010;12(5):R77. doi: 10.1186/bcr2721. doi:bcr2721 [pii] 10.1186/bcr2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Kushi LH, Kwan ML, Wiencke JK. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6(7):e1001043. doi: 10.1371/journal.pgen.1001043. doi:10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedeurwaerder S, Desmedt C, Calonne E. Largest ever DNA methylation dataset for breast cancer completed. Expert Rev Mol Diagn. 2011;11(5):470. [PubMed] [Google Scholar]
- 10.Fackler MJ, Umbricht CB, Williams D, Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P, Visvananthan K, Marks J, Ethier S, Gray JW, Wolff AC, Cope LM, Sukumar S. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71(19):6195–6207. doi: 10.1158/0008-5472.CAN-11-1630. doi:0008-5472.CAN-11-1630 [pii] 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LG, Shen R, Seshan V, Mo Q, Heguy A, Baylin SB, Ahuja N, Viale A, Massague J, Norton L, Vahdat LT, Moynahan ME, Chan TA. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3(75):75ra25. doi: 10.1126/scitranslmed.3001875. doi:3/75/75ra25 [pii] 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flanagan JM, Cocciardi S, Waddell N, Johnstone CN, Marsh A, Henderson S, Simpson P, da Silva L, Khanna K, Lakhani S, Boshoff C, Chenevix-Trench G. DNA methylome of familial breast cancer identifies distinct profiles defined by mutation status. Am J Hum Genet. 2010;86(3):420–433. doi: 10.1016/j.ajhg.2010.02.008. doi:S0002-9297(10)00090-X [pii] 10.1016/j.ajhg.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill VK, Ricketts C, Bieche I, Vacher S, Gentle D, Lewis C, Maher ER, Latif F. Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer Res. 2011;71(8):2988–2999. doi: 10.1158/0008-5472.CAN-10-4026. doi:0008-5472.CAN-10-4026 [pii] 10.1158/0008-5472.CAN-10-4026. [DOI] [PubMed] [Google Scholar]
- 14.Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jonsson G, Olsson H, Borg A, Ringner M. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010;12(3):R36. doi: 10.1186/bcr2590. doi:bcr2590 [pii] 10.1186/bcr2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamalakaran S, Varadan V, Giercksky Russnes HE, Levy D, Kendall J, Janevski A, Riggs M, Banerjee N, Synnestvedt M, Schlichting E, Karesen R, Shama Prasada K, Rotti H, Rao R, Rao L, Eric Tang MH, Satyamoorthy K, Lucito R, Wigler M, Dimitrova N, Naume B, Borresen-Dale AL, Hicks JB. DNA methylation patterns in luminal breast cancers differ from non luminal subtypes and can identify relapse risk independent of other clinical variables. Mol Oncol. 2011;5(1):77–92. doi: 10.1016/j.molonc.2010.11.002. doi:S1574-7891(10)00126-2 [pii] 10.1016/j.molonc.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Lee KM, Han W, Choi JY, Lee JY, Kang GH, Park SK, Noh DY, Yoo KY, Kang D. Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum Mol Genet. 2010;19(21):4273–4277. doi: 10.1093/hmg/ddq351. doi:ddq351 [pii] 10.1093/hmg/ddq351. [DOI] [PubMed] [Google Scholar]
- 17.Ronneberg JA, Fleischer T, Solvang HK, Nordgard SH, Edvardsen H, Potapenko I, Nebdal D, Daviaud C, Gut I, Bukholm I, Naume B, Borresen-Dale AL, Tost J, Kristensen V. Methylation profiling with a panel of cancer related genes: association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol Oncol. 2011;5(1):61–76. doi: 10.1016/j.molonc.2010.11.004. doi:S1574-7891(10)00128-6 [pii] 10.1016/j.molonc.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van der Auwera I, Yu W, Suo L, Van Neste L, van Dam P, Van Marck EA, Pauwels P, Vermeulen PB, Dirix LY, Van Laere SJ. Array-based DNA methylation profiling for breast cancer subtype discrimination. PLoS One. 2010;5(9):e12616. doi: 10.1371/journal.pone.0012616. doi:10.1371/journal.pone.0012616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. doi:nature11412 [pii] 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. doi:nature10983 [pii] 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kristensen VN, Lingjaerde OC, Russnes HG, Vollan HK, Frigessi A, Borresen-Dale AL. Principles and methods of integrative genomic analyses in cancer. Nat Rev Cancer. 2014;14(5):299–313. doi: 10.1038/nrc3721. doi:nrc3721 [pii] 10.1038/nrc3721. [DOI] [PubMed] [Google Scholar]
- 22.Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, Hewitt SM, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Cartun R, Mandich D, Rymkiewicz G, Ligaj M, Lukaszek S, Kordek R, Garcia-Closas M. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007;16(3):439–443. doi: 10.1158/1055-9965.EPI-06-0806. doi:16/3/439 [pii] 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 23.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. doi:295/21/2492 [pii] 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Closas M, Brinton LA, Lissowska J, Chatterjee N, Peplonska B, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Blair A, Kalaylioglu Z, Rymkiewicz G, Mazepa-Sikora D, Kordek R, Lukaszek S, Sherman ME. Established breast cancer risk factors by clinically important tumour characteristics. Br J Cancer. 2006;95(1):123–129. doi: 10.1038/sj.bjc.6603207. doi:6603207 [pii] 10.1038/sj.bjc.6603207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherman ME, Rimm DL, Yang XR, Chatterjee N, Brinton LA, Lissowska J, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Cartun R, Mandich D, Rymkiewicz G, Ligaj M, Lukaszek S, Kordek R, Kalaylioglu Z, Harigopal M, Charrette L, Falk RT, Richesson D, Anderson WF, Hewitt SM, Garcia-Closas M. Variation in breast cancer hormone receptor and HER2 levels by etiologic factors: a population-based analysis. Int J Cancer. 2007;121(5):1079–1085. doi: 10.1002/ijc.22812. doi:10.1002/ijc.22812. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 27.Cox DR. Regression models and life-tables (with discussion). J R Stat Soc Ser C Appl Stat Series B. 1972;(34):187–220. [Google Scholar]
- 28.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. doi:10.1056/NEJMoa021967 347/25/1999 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, Klijn JG, Larsimont D, Buyse M, Bontempi G, Delorenzi M, Piccart MJ, Sotiriou C. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. doi:25/10/1239 [pii] 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 30.Killian JK, Bilke S, Davis S, Walker RL, Jaeger E, Killian MS, Waterfall JJ, Bibikova M, Fan JB, Smith WI, Jr., Meltzer PS. A methyl-deviator epigenotype of estrogen receptor-positive breast carcinoma is associated with malignant biology. Am J Pathol. 2011;179(1):55–65. doi: 10.1016/j.ajpath.2011.03.022. doi:S0002-9440(11)00338-5 [pii] 10.1016/j.ajpath.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gianni L, Zambetti M, Clark K, Baker J, Cronin M, Wu J, Mariani G, Rodriguez J, Carcangiu M, Watson D, Valagussa P, Rouzier R, Symmans WF, Ross JS, Hortobagyi GN, Pusztai L, Shak S. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol. 2005;23(29):7265–7277. doi: 10.1200/JCO.2005.02.0818. doi:JCO.2005.02.0818 [pii] 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 32.Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, Centeno B, Weber F, Leu YW, Shapiro CL, Eng C, Yeatman TJ, Huang TH. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res. 2006;12(22):6626–6636. doi: 10.1158/1078-0432.CCR-06-0467. doi:12/22/6626 [pii] 10.1158/1078-0432.CCR-06-0467. [DOI] [PubMed] [Google Scholar]
- 33.Sandhu R, Rivenbark AG, Mackler RM, Livasy CA, Coleman WB. Dysregulation of microRNA expression drives aberrant DNA hypermethylation in basal-like breast cancer. Int J Oncol. 2014;44(2):563–572. doi: 10.3892/ijo.2013.2197. doi:10.3892/ijo.2013.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roll JD, Rivenbark AG, Sandhu R, Parker JS, Jones WD, Carey LA, Livasy CA, Coleman WB. Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Exp Mol Pathol. 2013;95(3):276–287. doi: 10.1016/j.yexmp.2013.09.001. doi:S0014-4800(13)00104-4 [pii] 10.1016/j.yexmp.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Stefansson OA, Jonasson JG, Olafsdottir K, Hilmarsdottir H, Olafsdottir G, Esteller M, Johannsson OT, Eyfjord JE. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics. 2011;6(5):638–649. doi: 10.4161/epi.6.5.15667. doi:15667 [pii] 10.4161/epi.6.5.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matuschek C, Bolke E, Lammering G, Gerber PA, Peiper M, Budach W, Taskin H, Prisack HB, Schieren G, Orth K, Bojar H. Methylated APC and GSTP1 genes in serum DNA correlate with the presence of circulating blood tumor cells and are associated with a more aggressive and advanced breast cancer disease. Eur J Med Res. 2010;15:277–286. doi: 10.1186/2047-783X-15-7-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada N, Nishida Y, Tsutsumida H, Hamada T, Goto M, Higashi M, Nomoto M, Yonezawa S. MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res. 2008;68(8):2708–2716. doi: 10.1158/0008-5472.CAN-07-6844. doi:68/8/2708 [pii] 10.1158/0008-5472.CAN-07-6844. [DOI] [PubMed] [Google Scholar]
- 38.Zrihan-Licht S, Weiss M, Keydar I, Wreschner DH. DNA methylation status of the MUC1 gene coding for a breast-cancer-associated protein. Int J Cancer. 1995;62(3):245–251. doi: 10.1002/ijc.2910620303. [DOI] [PubMed] [Google Scholar]
- 39.Yu Z, Wang L, Wang C, Ju X, Wang M, Chen K, Loro E, Li Z, Zhang Y, Wu K, Casimiro MC, Gormley M, Ertel A, Fortina P, Chen Y, Tozeren A, Liu Z, Pestell RG. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun. 2013;4:2812. doi: 10.1038/ncomms3812. doi:ncomms3812 [pii] 10.1038/ncomms3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowling VH. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene. 2010;29(6):930–936. doi: 10.1038/onc.2009.368. doi:onc2009368 [pii] 10.1038/onc.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu T, Niu Y, Feng Y, Niu R, Yu Y, Lv A, Yang Y. Methylation of CpG islands of p16(INK4a) and cyclinD1 overexpression associated with progression of intraductal proliferative lesions of the breast. Hum Pathol. 2008;39(11):1637–1646. doi: 10.1016/j.humpath.2008.04.001. doi:S0046-8177(08)00157-3 [pii] 10.1016/j.humpath.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Sizemore ST, Sizemore GM, Booth CN, Thompson CL, Silverman P, Bebek G, Abdul-Karim FW, Avril S, Keri RA. Hypomethylation of the MMP7 promoter and increased expression of MMP7 distinguishes the basal-like breast cancer subtype from other triple-negative tumors. Breast Cancer Res Treat. 2014;146(1):25–40. doi: 10.1007/s10549-014-2989-4. doi:10.1007/s10549-014-2989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Zhou XG, Zhou XY, Zhu C, Ji CB, Shi CM, Qiu J, Guo XR. Overexpression of C10orf116 promotes proliferation, inhibits apoptosis and enhances glucose transport in 3T3-L1 adipocytes. Mol Med Rep. 2013;7(5):1477–1481. doi: 10.3892/mmr.2013.1351. doi:10.3892/mmr.2013.1351. [DOI] [PubMed] [Google Scholar]
- 44.Simpson NE, Lambert WM, Watkins R, Giashuddin S, Huang SJ, Oxelmark E, Arju R, Hochman T, Goldberg JD, Schneider RJ, Reiz LF, Soares FA, Logan SK, Garabedian MJ. High levels of Hsp90 cochaperone p23 promote tumor progression and poor prognosis in breast cancer by increasing lymph node metastases and drug resistance. Cancer Res. 2010;70(21):8446–8456. doi: 10.1158/0008-5472.CAN-10-1590. doi:0008-5472.CAN-10-1590 [pii] 10.1158/0008-5472.CAN-10-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.