Abstract
The relBE family of Type II toxin-antitoxin (TA) systems have been widely reported in bacteria but none in Streptomyces. With the conserved domain searches for TA pairs in the sequenced Streptomyces genomes, we identified two putative relBE loci, relBE1sca and relBE2sca, on the chromosome of Streptomyces cattleya DSM 46488. Overexpression of the S. cattleya toxin RelE2sca caused severe growth inhibition of E. coli and S. lividans, but RelE1sca had no toxic effect. The toxicity of RelE2sca could be abolished by the co-expression of its cognate RelB2sca antitoxin. Moreover, the RelBE2sca complex, or the antitoxin RelB2sca alone, specifically interacted with the relBE2sca operon and repressed its transcription. The relBE2sca operon transcription was induced under osmotic stress, along with the ClpP proteinase genes. The subsequent in vivo analysis showed that the antitoxin was degraded by ClpP. Interestingly, the E. coli antitoxin RelBeco was able to alleviate the toxicity of S. cattleya RelE2sca while the mutant RelB2sca(N61V&M68L) but not the wild type could alleviate the toxicity of E. coli RelEeco as well. The experimental demonstration of the relBEsca locus might be helpful to investigate the key roles of type II TA systems in Streptomyces physiology and environmental stress responses.
Bacterial toxin-antitoxin (TA) system is originally identified in low copy number plasmids and shown to maintain the plasmid stability by post-segregational killing of plasmid-free daughter cells1. In recent years, bioinformatics and experimental evidence show that the type II TA modules are widely spread not only upon plasmids but also on chromosomes2,3. The TA loci typically consist of two but occasionally three tandem genes. The toxin genes invariably code for proteins, while matching antitoxin genes code for either antisense RNA or antitoxin proteins, resulting in classification as type I or type II TA loci, respectively. The chromosomal type II TA loci have been either demonstrated or hypothesized to play key roles in the stabilization of horizontally acquired genetic elements4, stress responses5, and traits in bacterial physiology such as the programmed cell death6 and persister cell formation7.
A number of functionally distinct type II TA systems have been identified by using experimental and bioinformatics approaches. As of June 2015, TADB, the web-based toxin-antitoxin database managed by our group, has collected 6,156 putative TA loci in 679 bacterial and archaeal genomes8; interestingly, 214 of the collected TA loci had been assigned to the relBE family. relBE is one of the best-documented type II TA loci with detailed reports about transcription regulation, toxin activity, antitoxin degradation and the TA complex formation9. Many of the toxins encoded on the chromosomes have been found to interfere with the protein synthesis in either a ribosome-dependent10,11 or ribosome-independent manner12,13. Under non-stress conditions, the toxicity of RelE is neutralized by the antitoxin RelB by forming a tight non-toxic RelBE complex14. The concentrations of toxin and antitoxin in the cells are regulated by the antitoxin or TA complex that is capable of repressing the transcription of the TA operon by binding specifically to the promoter. In some cases, the repression is further strengthened by conditional cooperativity15. Under environmental stress conditions, the antitoxin is degraded by cellular proteinases, such as the ATP-dependent Lon proteinases in E. coli16.
As listed in TADB, 169 out of 6,156 TA loci have been experimentally characterized, including for example 13 out of 16 type II TA pairs in E. coli K-12 MG1655, 5 out of 18 in Salmonella enterica Typhimurium LT2 and 33 out of 77 in Mycobacterium tuberculosis H37Rv8. However, there are few reports on the TA systems encoded by Streptomyces, the largest genus of Actinobacteria. To date, only one functional TA system, the yefM-yoeB locus on the S. lividans TK24 chromosome17, has been experimentally demonstrated in Streptomyces. Streptomyces are famous for producing many bioactive secondary metabolites, such as antibiotics18. Their complex life cycle that contains a vegetative and a spore stage made them excellent model organisms for studying prokaryotic differentiation19,20. The responses to nutrient limitation or other physiological stresses, including ppGpp, had been shown to play important roles in the differentiation process of Streptomyces21. It is worth noticing that ppGpp is also able to control bacterial persistence by induction of TA activity22. Availability of the complete genome sequences of Streptomyces allowed us to in silico identify the putative type II TA systems present in this genus. TADB had archived 22 putative TA loci in S. coelicolor A3(2), 27 in S. avermitilis MA-4680 and 14 in S. griseus NBRC 13350; however, none relBE family TA locus was found. We thus searched for the relBE locus in the completely sequenced Streptomyces genomes based on the conserved RelBE domains. Two putative relBE loci were obtained on the linear chromosome of Streptomyces cattleya DSM 46488.
The S. cattleya DSM 46488 strain is unusual in its ability to synthesize fluorine-containing natural products, including fluoroacetate and 4-fluorothreonine. We had completely sequenced its genome and two linear replicons were found23, a 6.3-Mb chromosome (GenBank accession no. CP003219) and a 1.8-Mb mega-plasmid (CP003229). In this study, we experimentally investigate the two putative relBE loci identified on the linear chromosome of S. cattleya DSM46488. The toxin homologous protein RelE2sca (SCATT_39270) and the antitoxin homologous protein RelB2sca (SCATT_39280) were found to be organized as an operon. The over-expression of RelE2sca resulted in the cell growth inhibition of both E. coli ΔrelBE mutant and S. lividans, and the co-expression of the cognate RelB2sca could counteract the toxicity. RelB2sca may be degraded by the ClpP proteinases under osmotic stress. Interestingly, the antitoxin RelBeco (b1564) from E. c oli K-12 MG1655 can also alleviate the toxicity of S. cattleya RelE2sca.
Results
Two putative relBE TA loci were predicted in the S. cattleya DSM46488 chromosome
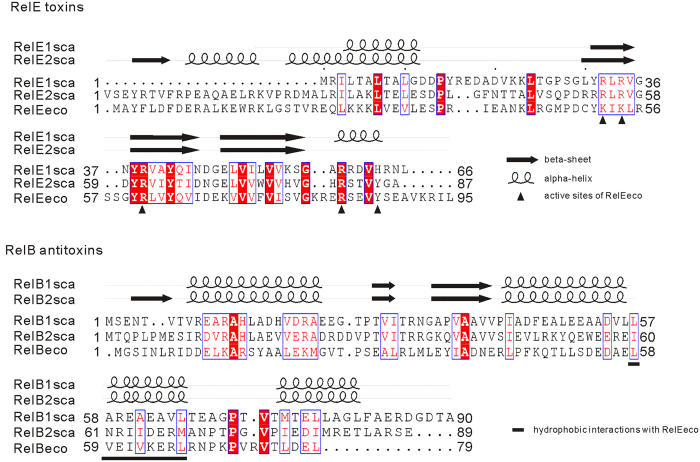
The putative type II TA pairs were detected based on typical TA features, including those of the conserved TA domains and a pair of two short genes organized as an operon structure. Thirty-three putative type II TA loci were annotated in the completely sequenced S. cattleya DSM46488 genomes (supplementary Table S1), including two relBE homologs on its linear chromosome, SCATT_20920-SCATT_20930 and SCATT_39280-SCATT_39270, named relBE1sca and relBE2sca, respectively. The S. cattleya toxin proteins RelE1sca (SCATT_20930) and RelE2sca (SCATT_39270) exhibits low sequence similarities to the well documented RelE toxin b1563 (RelEeco) of E. coli K-12 MG1655 with 23% and 28% BLASTp identities, respectively; however, four conserved motifs were found among these RelE homologs, which were predicted to form two alpha-helixes and two beta-sheets (Fig. 1). Similarly, four alpha-helixes and two beta-sheets were also present in the three cognate RelB antitoxins, RelB1sca (SCATT_20920) and RelB2sca (SCATT_39280) of S. cattleya, and RelBeco (b1564) of E. coli.
Figure 1. Amino acid sequence alignments for three RelE toxin proteins and their cognate RelB antitoxin proteins.
Putative RelBE1sca (SCATT_20920-SCATT_20930) and RelBE2sca (SCATT_39280-SCATT_39270) pairs were identified on the S. cattleya DSM46488 chromosome while the well-documented RelBE module RelBEeco (b1564-b1563) is encoded by E. coli K-12 MG1655 chromosome. Conserved residues are shown in red and by boxed. The black triangle indicates the active sites of the E. coli toxin RelEeco while the underlined sequence indicates the RelBeco conserved motif interacting with RelEeco14,30. Secondary structures were predicted by using PSIPRED43.
relE2sca and its upstream gene relB2sca were organized into an operon
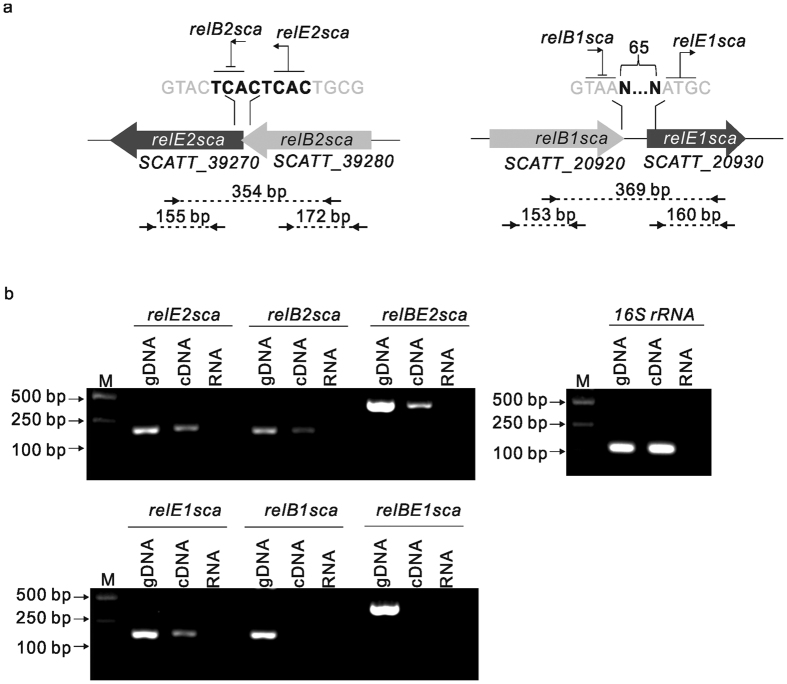
Toxin-antitoxin modules are typically organized into operons with the antitoxin genes located upstream of the toxin genes, often overlapping or with a small intergenic region between the two genes. As seen in Fig. 2a, the putative toxin gene relE2sca is overlapped by its upstream antitoxin gene relB2sca by 8 nucleotides, while relE1sca and relB1sca were separated by a 65-bp region. To determine whether the two putative relBE gene pairs were cotranscribed, respectively, and thus formed individual operons, total RNA of logarithmic phase S. cattleya DSM46488 was isolated and converted to cDNA by reverse transcriptase. The cDNA was PCR amplified by using specific primers spanning the toxin and antitoxin genes (indicated by small arrows in Fig. 2a). A band with the expected size was obtained for relBE2sca, but not for relBE1sca. mRNAs of the individual toxin genes and antitoxin genes were detected as well, and we also succeed in obtaining the expected bands except for relB1sca. Genomic DNA contamination was eliminated by using RNA without reverse transcriptase as the template. Likewise, as a positive control, bands with expected sizes were observed for 16S rRNA amplification by using cDNA and for all amplifications by using S. cattleya DSM46488 genomic DNA as the template (Fig. 2b). These data suggested that relBE2sca was organized into an operon.
Figure 2. Genetic organization of the two putative relBE loci on S. cattleya DSM46488 chromosome: relBE1sca (SCATT_20920-SCATT_20930) and relBE2sca (SCATT_39280-SCATT_39270).
(a) Scaled schematic representation of the toxin and antitoxin genes. The overlapping or separate region is shown in bold. The arrows indicate the primers used in the transcription analysis and the numbers indicate the expected size of the PCR products; (b) Transcription analysis of PCR amplification of S. cattleya relBE genes using cDNA and gene-specific primers to amplify from the 3′ end of relBs to the 5′ end of relEs (spanning both the protein-coding and the intergenic region). Lane M indicates the standard DNA size marker. Each set of the three lanes consisted of positive controls using genomic DNA as template (gDNA). PCR amplified products using cDNA prepared from log-phase S. cattleya (cDNA) and negative controls using the total RNA without reverse transcriptase (RNA). 16S rRNA was used as the positive control. The gels were cropped from the original images available at the supplementary Fig. S10.
Overexpression of S. cattleya toxin RelE2sca was lethal in E. coli and S. lividans
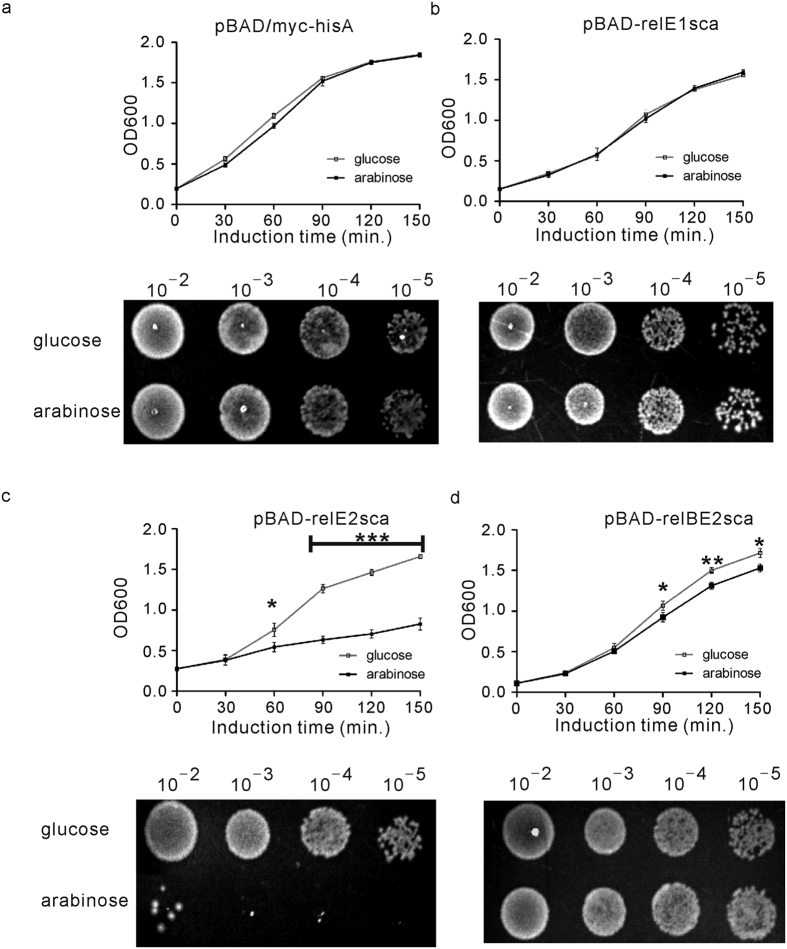
To determine whether the putative toxins RelE1sca and RelE2sca of S. cattleya were functional or not, the effect of the overexpression of their corresponding genes was assessed in E. coli MGJ5987 (MG1655 ΔmazF ΔchpB ΔrelBE Δ(dinJ-yafQ) Δ(yefM-yoeB) ΔhigBAΔ(prlF-yhaV) ΔyafNO ΔmqsRA ΔhicAB)24. The toxin genes relE1sca and relE2sca were firstly cloned into the E. coli expression vector pBAD/myc-hisA and placed under the control of arabinose inducible promoter PBAD, producing plasmids pBAD-relE1sca and pBAD-relE2sca, respectively (supplementary Table S2). Then, the growth of E. coli MGJ5987 strains containing the corresponding plasmid was used to assess the effects of RelE1sca or RelE2sca. Consequently, the growth of E. coli cells transformed with pBAD-relE2sca was strongly inhibited in the presence of 0.2% L-arabinose that induced the overexpression of relE2sca (Fig. 3c), while the normal growth was observed in the presence of 0.2% glucose. The difference in the number of E. coli colonies obtained between one-hour induction of L-arabinose and glucose was also evident when grown on Luria agar (LA) plates (Fig. 3c). This suggested that overexpression of S. cattleya toxin RelE2sca is lethal in E. coli. However, there was no significant difference in growth observed for the cells harboring pBAD-relE1sca or the blank plasmid pBAD-myc/hisA either in the presence of L-arabinose or glucose (Fig. 3a,b), indicating that RelE1sca had no toxic effect to E. coli ΔrelBE mutant under the condition studied.
Figure 3. Effect of over-expression of the putative toxin proteins RelE1sca and RelE2sca on the growth of E. coli MGJ5987 (MG1655 ∆relBE mutant).
E. coli MGJ5987 cells transformed with individual plasmids were grown in LB medium until the OD600 reached 0.2: (a) control plasmid (pBAD/myc-hisA), (b) plasmid carrying relE1sca (pBAD-relE1sca), (c) plasmid carrying relE2sca (pBAD-relE2sca) and (d) plasmid carrying the intact relBE2sca operon (pBAD-relBE2sca). Then, at time zero, 0.2% glucose (the hollow square) was added into one-half of each culture and 0.2% L-arabinose (the solid square) was added into the other half. Cell growth was monitored by measuring the OD600 every 30 minutes. The means and standard deviation of three different experiments are shown. For statistical analysis, two-way analysis of variance with Bonferroni post-tests were used to obtain P values for each time point: *P < 0.05; **P < 0.01; ***P < 0.001. 3 μl of serial dilutions of different cultures which were collected after induction by glucose or arabinose for 1 hour were spread onto the LA plate and incubated at 37 °C for 12 hours.
The toxicity of RelE2sca was predicted to be neutralized by the upstream cognate antitoxin protein RelB2sca. To balance the protein expression levels of the toxin and antitoxin, the relBE2sca region of S. cattleya was cloned into pBAD/myc-hisA, resulting in pBAD-relBE2sca (supplementary Table S2). As predicted, normal growth of E. coli MGJ5987 harboring plasmid pBAD-relBE2sca in the Luria-Bertani (LB) broth supplemented with arabinose or glucose was observed due to co-expression of RelB2sca and RelE2sca (Fig. 3d). Similarly, after induction by arabinose or glucose for one hour, the cells with different dilutions were spread onto the agar plates and showed the similar results (Fig. 3d). In addition, the direct interaction between the toxin RelE and antitoxin RelB to form the RelBE complex was examined. RelB2sca and RelE2sca-His6 were coexpressed in E. coli BL21(DE3) and co-purified. As shown in supplementary Fig. S1a, the 11.54-kDa and 10.32–kDa proteins, consistent with the expected molecular masses of the recombinant RelB2sca and RelE2sca-His6, respectively, were co-purified under native conditions. The 11.54–kDa protein (RelE2sca-His6) was purified under denaturing conditions whereas the 10.32–kDa protein (RelB2sca) was obtained from the complex under denaturing condition. The complex was finally subjected to the size-exclusion chromatography, showing that the ratio of RelB2sca and RelE2sca-His6 was 2:2 (supplementary Fig. S1b). These results all suggested that the S. cattleya toxin RelE2sca was lethal to E. coli and its toxicity could be abolished by RelB2sca. All of the results above suggested that relBE2sca might represent a functional toxin-antitoxin module while relBE1sca has no activity under the condition studied.
Results of heterologous expression experiments of RelBE2sca in Streptomyces were consistent with those in E. coli mentioned above (supplementary Fig. S2). S. lividans TK24, which was predicted to contain no homolog of RelB or RelE, was used as the host to assess the RelE2sca toxicity. The S. cattleya RelBE protein-coding regions, relE2sca, relB2sca and relBE2sca, were cloned into the Streptomyces - E. coli shuttle vector pIB139 (integrative plasmid) under the control of the constitutive promoter PermE (supplementary Table S2). Subsequently, the resulting pIB139-relE2sca was unable to be introduced into S. lividans TK24 while both pIB139-relB2sca and pIB139-relBE2sca transferred at high frequency (supplementary Fig. S2). It suggested that the expression of the toxin RelE2sca was lethal in Streptomyces but the antitoxin RelB2sca could counteract this toxic effect. Additionally, the expression of another toxin RelE1sca had no toxic effect on the growth of S. lividans TK24 under the condition studied (supplementary Fig. S2).
The relBE2sca operon was auto-regulated by the RelBE2sca TA complex
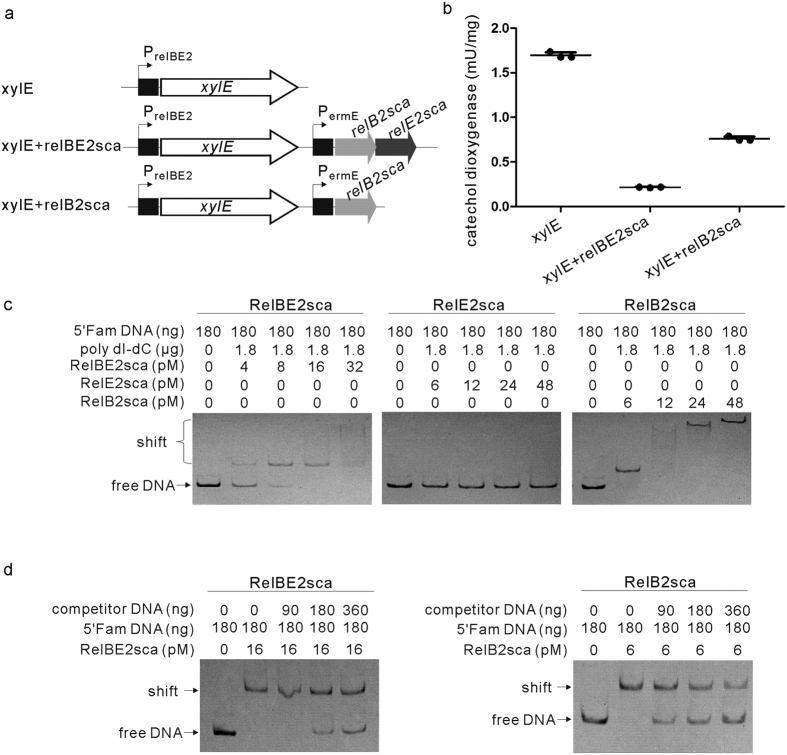
It has been reported that the transcription of the E. coli relBE operon was specifically repressed by the overexpression of its encoded TA complex or antitoxin alone. Here the reporter gene xylE (encoding catechol 2,3-dioxygenase) was used to investigate whether the transcription of S. cattleya relBE2sca operon was regulated in vivo by the RelBE2sca complex or by RelB2sca in Streptomyces25. First, the intergenic region containing the promoter region of relBE2sca of S. cattleya (PrelBE2) was amplified and placed upstream of the promoterless xylE gene to control the expression of xylE (Fig. 4a). Then, the protein-coding fragments of relBE2sca and relB2sca were amplified and placed under the control of the strong constitutive promoter PermE, respectively. Finally, cultures of S. lividans TK24 carrying the resulting plasmids were assayed for catechol dioxygenase activity. Figure 4b showed the results of three independent determinations. In the cases of the high gene expressions of the RelBE2sca complex and the antitoxin RelB2sca, catechol dioxygenase activities were consistently lower than activities obtained with XylE alone, by about 8-fold and 2-fold, respectively. It suggested that the transcription of S. cattleya relBE2sca module was inhibited by the overexpression of the antitoxin RelB2sca alone, and more efficiently by the TA complex RelBE2sca, which was in agreement with data in E. coli.
Figure 4. Auto-regulation of the S. cattleya relBE2sca operon by the RelBE2sca TA complex or the antitoxin RelB alone.
(a) Schematic representation of in vivo regulation of relBE2sca operon in S. lividans TK24 xylE, reporter gene coding for catechol 2,3-dioxygenase; PrelBE2, promoter region of S. cattleya relBE2sca; PermE, strong constitutive promoter. (b) Activity of catechol 2,3-dioxygenase was assayed by monitoring the increase in absorbance at 375 nm (A375) every 10 minutes for the S. lividans TK24 cells carrying the resulting plasmids. (c) Analysis of the interactions between PrelBE2 and RelBE2sca proteins by using EMSA in vitro. Increasing the amount of RelBE2sca-His6, RelE2sca-His6 or RelB2sca was incubated with the promoter labeled with 5′-FAM. Poly dI-dC was used to inhibit the unspecific interaction. Unbound DNA fragments were separated from protein-DNA complex by electrophoresis in 6% native PAGE gel. (d) Increasing amounts of unlabeled competitor DNA were added into the reaction system in which the amounts of His6-RelBE2sca complex or RelB2sca were set at 16 pM or 6 pM, respectively.
Furthermore, the specific interaction between the PrelBE2 region and the purified RelBE2sca-His6 complex was assessed by Electrophoretic Mobility Shift Assay (EMSA) in vitro. The 5′-FAM-labeled PrelBE2region was amplified from S. cattleya DSM46488 genomic DNA by using 5′-FAM oligonucleotides (supplementary Table S2). The retarded bands were observed when the FAM-labeled DNA probe were incubated with increasing amounts of purified RelBE2sca-His6 complex or RelB2sca alone, but not with RelE2sca alone (Fig. 4c). The signal of the retarded bands decreased with the increased unlabeled specific competitor DNA probes (Fig. 4d). In addition, the binding sites of RelBE2sca-His6 complex to the labeled probes were located precisely by DNase I footprinting (supplementary Fig. S3a). The DNA probes were the same as those used in the EMSA experiments. The DNA sequencing electropherogram showed that the 14-bp DNA fragment (GTACACCTGAGACA) upstream of the putative -35 region of PrelBE2was clearly protected by the RelBE2sca-His6 complex (supplementary Fig. S3a). This was also supported by the fact that the retarded band disappeared when the RelBE2-His6 complex was incubated with the mutant probe, in which the protected DNA sequence was deleted (supplementary Fig. S3b). Together, these observations indicated that the RelBE2sca complex repressed the transcription of the relBE2sca operon by interacting specifically with the 14-bp DNA fragment within the PrelBE2 region.
Transcription of the antitoxin relB2sca was enhanced under osmotic stress condition
To determine whether the relBE2sca operon was able to respond to the environmental stress like the typical RelBE system reported in E. coli, the transcriptional level of the antitoxin gene relB2sca was measured under osmotic stress as described in the Methods section. The relB2sca mRNA level was evaluated by a real-time quantitative PCR (qRT-PCR) assay, showing that the RelB2sca mRNA levels were increased 3 folds (Fig. 5). The result above indicated that the osmotic stress could induce the transcription of relBE2sca. In addition, the relB2sca transcription level also increased under the other environmental stresses tested, including high temperature, strong amino acid starvation, glucose starvation and antibiotic treatment (supplementary Fig. S4).
Figure 5. Transcription of the antitoxin gene relB2sca and the six clpP proteinases in S. cattleya under osmotic stress.
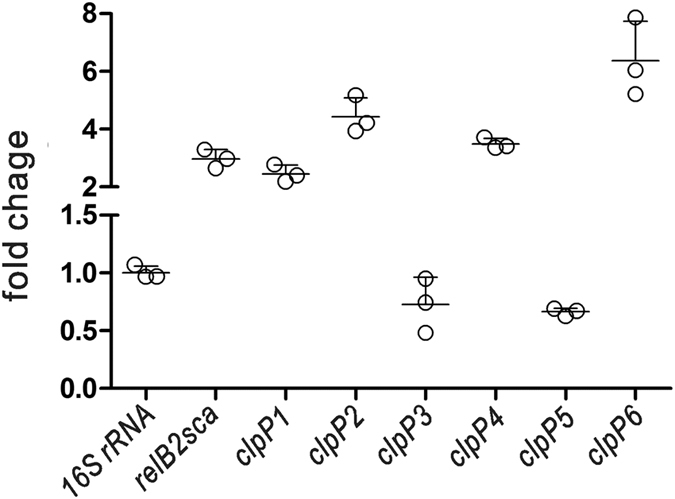
After culturing in minimal medium to log phase, S. cattleya DSM46488 cells were exposed to 0.5 M NaCl and 0.5 M sucrose for 3 hours. Cells were harvested and RNA was isolated and amplified. Gene transcription levels were measured by using real-time quantitative PCR and the 16S rRNA was used as a control. The experiments were repeated three times, and the error bars represented the standard deviation.
Antitoxin protein RelB2sca was degraded by ClpP proteinases
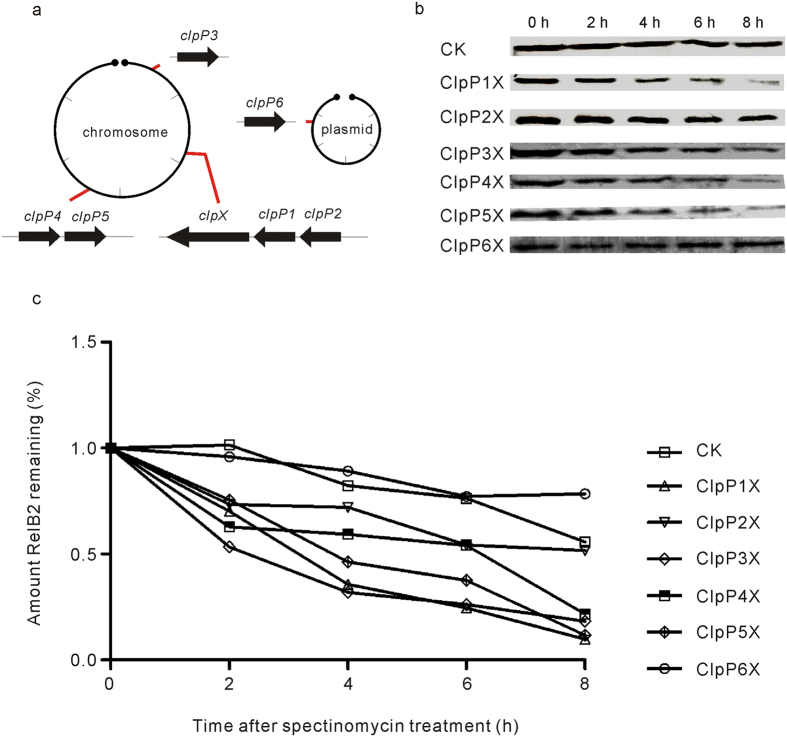
The labile antitoxin RelB in E. coli has been reported to be sensitive to the ATP-dependent proteinase Lon16, allowing the stable toxin RelE to be released and to carry out the toxic effect. Interestingly, in Synechocystis sp. PCC 6803, RelB was also found to be degraded by the ClpP proteinase with the ClpX or ClpA as the chaperon26. We thus investigated whether the S. cattleya ClpP proteinase was involved in the proteolysis of the antitoxin RelB2sca. Six putative ClpP proteinase homologs have been annotated in the S. cattleya DSM46488 genome (Fig. 6a), including five found on the linear chromosome (ClpP1, SCATT_17340; ClpP2, SCATT_17350; ClpP3, SCATT_03550; ClpP4, SCATT_32700; ClpP5, SCATT_32710) and one in the mega-plasmid (ClpP6, SCATT_p13740). One chaperone gene clpX (SCATT_17330) was also found to be clustered with the proteinase genes clpP1-clpP2. Under the osmotic stress, the transcription levels of 4 out of 6 ClpP proteinase genes increased significantly, along with the antitoxin gene relB2sca (Fig. 5 and supplemental Fig. S4). The transcription levels of clpP1, clpP2, clpP4 and clpP6 increased by 2 folds, 4 folds, 3 folds and 6 folds, respectively. As for relBE2sca, the transcription levels of ClpP proteinase genes were also determined whether they were induced by the other environmental stresses tested (supplementary Fig. S4). Diverse ClpP proteinase genes exhibited different transcription levels under these stresses. These data are in accordance with the hypothesis that the ClpPs might hydrolyze RelB2sca and participate in the activation of the relBE2sca module.
Figure 6. Intracellular stability of the S. cattleya antitoxin protein RelB2sca in the absence or presence of the proteinase ClpPnX (n = 1, 2, 3, 4, 5 or 6) detected by Western blotting.
The strains E. coli BL21(DE3)/pLysS with corresponding plasmids were grown in LB broth in the presence of 0.2% L-arabinose to induce the RelB2sca expression. Then 0.5 mM IPTG was added to induce the proteinases ClpPnX expression. One hour later, 200 μg/ml spectinomycin was added to inhibit protein synthesis. Samples were collected every two hours. The levels of the antitoxin protein RelB2sca were monitored by Western blotting analysis. (a) Six ClpP proteinase gene loci and one chaperone ClpX gene locus found on the linear chromosome and mega-plasmid of S. cattleya DSM46488. (b) RelB2sca protein band of Western blotting (CK denoted the control). (c) graph described the percentage of a signal for RelB2sca.
To determine whether the antitoxin RelB2sca was degraded by the ClpP proteinases of S. cattleya DSM46488, the relB2sca gene was firstly amplified and cloned into pBAD/myc-hisA. resulting in pBADAHis which expressed the antitoxin with C-terminal His6-tag (supplementary Table S2). The individual proteinase genes clpPs and the chaperone gene clpX were also cloned into the MCS1 and MCS2 of E. coli expression vector pRSFDuet under the control of the T7 promoter, respectively. Then, the intracellular stability of the antitoxin protein RelB2sca, which was coexpressed with ClpP1X, ClpP2X, ClpP3X, ClpP4X, ClpP5X and ClpP6X, was investigated by Western blotting analysis. The results showed that the half-life of RelB2sca coexpressed with ClpP1X, ClpP3X, ClpP4X and ClpP5X was significantly shorter than that of RelB2sca expressed alone in E. coli (Fig. 6b,c), indicating that the proteinases ClpPs of S. cattleya DSM46488 were indeed able to degrade the antitoxin RelB2sca in vivo.
Toxic effect of S. cattleya RelE2sca was alleviated by co-expression of E. coli antitoxin RelBeco
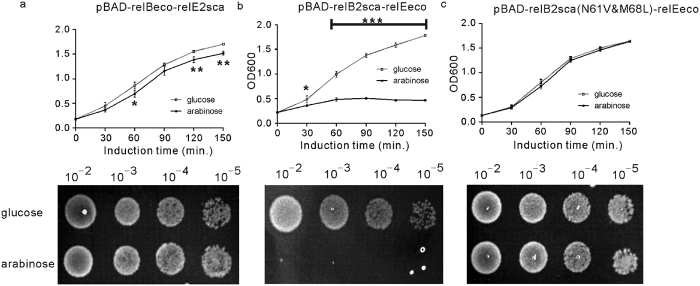
As aforementioned, the overexpression of RelE2sca could inhibit the growth of E. coli strain MGJ5987, a relBE locus defect mutant (Fig. 3c); then, we wondered whether this inhibitory effect can be neutralized by the E. coli antitoxin RelBeco. To explore this possibility, the antitoxin relBeco gene (b1564) of E. coli K-12 MG1655 was fused into the upstream region of relE2sca using SOE-PCR; subsequently, the fused fragment was cloned into the pBAD/myc-hisA vector, forming the pBAD-relBeco-relE2sca vector (supplementary Table S2). In both LB broth and LA plate, the E. coli MGJ5987 cells transformed with pBAD-relBeco-relE2sca, the co-expression plasmid of relE2sca and relBeco, grew better (Fig. 7a) than those transformed with the plasmid carrying relE2sca alone (Fig. 2b), suggesting that the E. coli antitoxin RelBeco could reduce the toxic effect of S. cattleya toxin RelE2sca. Interestingly, the toxic effect of E. coli toxin RelEeco (b1563) could not be abolished or alleviated by the S. cattleya antitoxin RelB2sca (Fig. 7b), whereas, it could be abolished by the RelB2sca mutant N61V&M68L (Fig. 7c), in which the residues Asn61 and Met68 were replaced by Val61 and Leu68, respectively. The RelB family protein has a consensus motif located at the C-terminus of their α3 helixes; the motif could be expressed as ZXnnZnnRZ, where n represents any amino acid residue, X is hydrophobic residue, and Z is hydrophobic residue with small side chain (such as Val, Ile, Leu or Ala)14. As revealed by the complex structure of E. coli RelEB, the consensus motif is critical for the interaction between RelBeco and RelEeco, via two types of interactions: salt bridge and hydrophobic interaction; the hydrophobic X and R residues are involved in the hydrophobic interactions. Formation of the RelEBeco complex will disrupt the mRNA binding and cleavage activities of RelEeco, resulting in weakened toxicity. As revealed by the sequence alignment (Fig. 1), Asn61 and Met68 of RelB2sca correspond to the X and the last Z residues of the consensus motif, respectively. Unlike the hydrophobic residues Val61 and Leu68 in RelB2eco and the RelB2sca mutant N61V&M68L, the Asn61 residue is highly hydrophilic and not suitable for the hydrophobic interaction; though Met68 is hydrophobic, it has bigger side chain compared to the Leu residue, which may further disrupt the interaction between RelE2eco and the wild type RelB2sca, leading to the complete loss of the antitoxic effect of RelB2sca.
Figure 7. Homologous RelBE toxin-antitoxin protein interactions between S. cattleya and E. coli.
Overnight cultures of E. coli MGJ5987 (MG1655 ∆relBE mutant) carrying individual plasmids were diluted 1:100 in LB broth supplemented with 50 μg/ml ampicillin: (a) pBAD-relBsco-relE2sca, (b) pBAD-relB2sca-relEeco, and (c) pBAD-relB2sca(N61V&M68L)-relEeco. When the OD600 reached to about 0.2, each culture was divided into two equals. 0.2% glucose (hollow square) or 0.2% L-arabinose (solid square) was added into the two equals, respectively. OD600 was measured every 30 min. The means and standard deviation of three different samples were present. For statistical analysis, two-way analysis of variance with Bonferroni post-tests were used to obtain P values for each time point: *P < 0.05; **P < 0.01; ***P < 0.001. 3 μl of serial dilutions of different cultures which were collected after induced by glucose or arabinose for 1 hour were spread onto the LA plate and incubated at 37 °C for 12 hours.
Discussion
Streptomyces usually inhabit soil and face a wide diversity of environmental conditions; the co-existence of various type II TA modules is hypothesized to favor the survival of Streptomyces. Based on the conserved domains of the RelBE pairs, eight putative relBE TA loci were identified on seven completely sequenced Streptomyces chromosomes (supplementary Fig. S5), including two loci in S. cattleya. The sequence similarities between the S. cattleya RelBE2sca pair and the RelBE pairs of other Streptomyces are very high (≥86% BLASTp identities); whereas, the similarity between S. cattleya RelBE2sca and RelBE1sca is relatively low (≤44% BLASTp identities). Recently, the S. lividans yefM-yoeB TA system has been characterized19; it was successfully used as the replacement of the antibiotic resistance gene, and served as a selection marker in the development of a stable plasmid expression system in Streptomyces27. The sequence similarity between the yefM-yoeB pair and the relBE modules is very low. Phylogenetic tree reconstructions also showed two distinct clades existing between the YoeB homologs and the RelE homologs in Streptomyces (supplementary Fig. S6). Remarkably, none relBE TA locus was found in the commonly used Streptomyces model strains, such as S. coelicolor A3(2), S. lividans TK24 or S. avermitilis MA-4680. Therefore, the S. cattleya relBE2sca TA pair identified in this study may be useful for developing a genetic tool to select and stabilize the plasmids, which can produce heterologous proteins in the common hosts.
As a paradigm TA system, the relBE locus has been found to be widespread. We thus examined whether the two putative relBE pairs predicted on the S. cattleya chromosome worked as functional TA modules via toxin hetero-expression and by reverse-transcriptase PCR assay. Overproduction of S. cattleya toxin RelE2sca resulted in the severe growth inhibition of E. coli MGJ5987 (MG1655 ∆10 TA) and S. lividans TK24, which were abolished by the co-expression of the cognate antitoxin RelB2sca. The E. coli RelE toxin cleaved the mRNAs displaying codon-specific manner, which cleaved the UAG faster than the other two stop codon UAA and UGA (UAG > UAA > UGA)28. It was also shown to specifically cleave the mRNA in the A site of the eukaryote ribosome29. The cleavage specificity of RelE2sca is under examination. In addition, reverse-transcriptase PCR assay showed that relBE2sca was transcribed as bicistronic mRNA. By using the xylE reporter system, the relBE2sca transcription was found to be inhibited efficiently by the overexpression of the RelBE2sca complex or the antitoxin RelB2sca. Subsequently, the EMSA and DNase I footprinting analysis suggested that the RelBE2sca complex repressed the transcription of relBE2 operon by interacting with the 14-bp DNA region of the relBE2sca promoter. These in silico analyses and experimental evidence supported that the relBE2sca locus on the S. cattleya chromosome worked as a functional relBE system of Streptomyces.
The biological significance of chromosomal RelBE systems in environmental stress response has been studied in several bacteria, especially in E. coli5. The analysis of the synonymous codon usage bias showed that the Codon Adaption Index (CAI) of relE2sca and relB2sca is 0.53 and 0.43, respectively. This indicated that the expression level of the relBE2sca module would be expected to be lower than those of the highly expressed genes, including those coding for translation elongation factors and the ribosomal proteins (supplementary Fig. S7). We found that the functional relBE2sca module of S. cattleya was induced by osmotic pressure (Fig. 5) and nutrient limitations such as amino acid and glucose starvation (supplementary Fig. S4). Further sequence analysis showed that the expression levels of the four ClpP proteinases were also increased significantly under the osmotic pressure. The antitoxin RelB was reported to be degraded by Lon proteinase but not ClpP proteinase in E. coli. There was only one heat-shock Lon family proteinase found to be encoded by S. cattleya DSM46488 genome while six ClpP proteinases found. In this study, the antitoxin RelB2sca was revealed to be degraded by the ATP-dependent ClpP proteinases in Streptomyces. However, there was much work to elucidate how ClpP proteinases degrade antitoxin under diverse environmental stresses.
All the above results suggested that relBE2sca functions in a mode similar to that of RelBE in E. coli. Besides relBE2sca, S. cattleya DSM46488 also codes for another putative relBE locus, relBE1sca. The reverse-transcriptase PCR assay failed to detect the relBE1sca pair transcription under the condition studied (Fig. 2b); interestingly, overexpression of the toxin RelE1sca had no toxic effect to E. coli MGJ5987 or S. lividans Tk24 (Fig. 3b). As revealed by the structures of RelEeco11,30 and the sequence alignment (Fig. 1), four arginine residues (including Arg54, Arg56, Arg61, and Arg81) and one tyrosine residue (Tyr85) might be important for the mRNA binding and cleavage activities of RelE2sca. Tyr85 of RelE2sca correspond to the Tyr87 in RelE2eco, which is the catalytic residue and functions as a general acid. In RelE1sca, there is a histidine residue, His63, located at the position corresponding to the catalytic Tyr residues of RelE2sca and RelE2eco. As revealed by the studies of RNase T1 and YoeB (which is also a toxin protein), the histidine residue can also catalyze the mRNA cleavage reaction, but through a mechanism different from the tyrosine residue10,11. The wild type RelE2sca is toxic; the substitution of the Tyr85 by His85 in RelE2sca resulted in the loss of toxicity (supplementary Fig. S8a), suggesting that, like RelE2eco, RelE2sca requires a Tyr residue for catalysis. Interestingly, neither the wildtype RelE1sca nor the H63Y mutant (in which the His63 was replaced by Tyr63) is toxic to E. coli (supplementary Fig. S8b). Compared to the sequences of RelE2sca and RelE2eco, RelE1sca is shorter by 24 and 23 amino acids at the N-terminus, respectively. In the RelE2eco structures, the N-terminal is composed of one β-strand (β1) and one α-helix (α1); the former forms parallel β-sheet with the last β-strand (β4), whereas the later interacts with the last helix (α3), via the formation of a strong salt bridge. Lack of these interactions may affect the overall folding of RelE1sca, especially the orientations of the last helix holding the catalytic residue, leading to the complete loss of the toxin activity. Two homologous relB-parE systems were also predicted in the Sinorhizobium melioti megaplasmid pSyme31. The first relB-parE appeared to be a functional TA system while the second pair was not; however, the RelB of the second TA system retained the antitoxin activity. Interestingly, the co-expression of the putative antitoxin RelB1sca of S. cattleya DSM46488 could alleviate slightly the E. coli MGJ5987 cell growth inhibition that was caused by the toxin RelE2sca (supplementary Fig. S9). But RelB1sca exhibited a lower degree of antitoxin activity to RelE2sca, compared to that of RelB2sca or the antitoxin RelBeco of E. coli (Fig. 7a). Therefore, it was interesting to explore deeply why the RelE1sca was not active under condition studied, which might be helpful to understand the relBE family evolution.
The toxins and their cognate antitoxins appeared to interact with high specificity. There is no toxin-antitoxin cross-interaction reported between various members of the same TA families, such as mazEF and chpAB of E. coli32, yefM-yoeB of Stahylococcus equorum33, parE-parD of Caulobacter crosotus34, paaA–parE of E. coli O157:H735 and vapBC of Mycobacteria tuberculosis36. However, the functional interaction between the ccdAB family had been detected in E. coli O157:H737. Recent research showed that the toxicity of YoeB of Streptococcus suis could be alleviated by the non-cognate antitoxin YefM of Streptococcus pneumoniae but not by YefM of E. coli38. In this study, we found that the toxic effect of S. cattleya RelE2sca was counteracted by co-expression of E. coli antitoxin RelBeco (Fig. 7a) while the toxicity of RelEeco was also abolished by the mutant RelB2sca(N61V&M68L) (Fig. 7c). It will be helpful to explore the relBE family evolution with this new finding of the cross-complementation between the toxin RelE and the heterologous antitoxin RelB from S. cattleya and E. coli, two organisms with significant evolutionary distance and important differences in their lifestyle and the chromosome structures.
In summary, here we reported a new functional RelBE TA system relBE2sca found on the S. cattleya chromosome. Overexpression of the toxin RelE2sca caused severe growth inhibition of E. coli ΔrelBE mutant and S. lividans, and the toxicity of RelE2sca could be abolished by the co-expression of RelB2sca. More interestingly, homologous toxin-antitoxin protein interactions across the species were also found; that is, the E. coli antitoxin RelBeco could alleviate the toxicity of S. cattleya RelE2sca, and the mutant RelB2sca(N61V&M68L) but not the S. cattleya wild type RelB2sca could alleviate the toxicity of E. coli RelEeco as well. It may be helpful to investigate the important regulatory role of TA loci in Streptomyces physiology, environmental stress responses, and complex secondary metabolisms such as producing fluoroacetate and 4-fluorothreonine.
Methods
Bacterial strains and growth conditions
Strains, plasmids, and oligonucleotides used in this study were listed in the supplementary Tables S1 and S2, respectively. E. coli strains were grown in LB broth medium (10 g tryptone, 5 g yeast extract and 5 g NaCl per one liter) at 37 °C and Streptomyces strains were grown in SFM medium at 30 °C, respectively, supplemented with 100 μg/ml ampicillin, 50 μg/ml apramycin, 50 μg/ml spectinomycin, 50 μg/ml kanamycin or 25 μg/ml thiostrepton as required. Conjugative transfer of DNA from E. coli ET12567/pUZ8002 to Streptomyces was performed as described by Kieser et al.39 The same amount of donor cells and recipient cells were used in each individual conjugation.
in silico identification of Type II TA loci in S. cattleya DSM46488
Nucleotide sequences and annotations of the completely sequenced S. cattleya DSM46488 chromosome and plasmid were downloaded from the NCBI RefSeq Project under the accession of NC_017586 and NC_017585, respectively. The toxin or antitoxin protein homologs were firstly obtained by HMMer-based conserved domain searches with an e-value cut-off of 0.01. The 343 hidden Markov model (HMM) profile profiles had been built from the Type II TA proteins archived by TADB. Then, the short homologs with the size of 30 - 500 a.a. were kept as TA candidates. Finally, two flanking TA candidate genes with the intergenic distances of −20 to 30 bp were paired as an operon structure, and thus predicted as a putative Type II TA locus in S. cattleya DSM46488 (supplementary Table S1).
RNA isolation, RT-PCR, and quantitative real-time quantitative PCR analysis
Total RNA samples were extracted from S. cattleya DSM46488 culturing in TSBY for 2–3 days by using Redzol reagent and the contaminated genomic DNA was eliminated by treating with RNase-free DNase I. RNA concentrations and integrity were determined by Nanodrop and agarose gel electrophoresis, respectively. Reverse transcriptase PCR (RT-PCR) was carried out using a QuantiTect Reverse Transcription Kit according to the manufacturer’s protocol. For the co-transcription assay, the genes specific primers were used for RT-PCR analysis. In addition, to assess the transcription of relB2sca and the clpP proteinases responding to the osmotic stress, S. cattleya DSM46488 was grown in TSBY medium for 2–3 days and then inoculated to the fresh minimal medium with 1:10 amount. After the cells were grown to log phase, 0.5 M sucrose and 0.5 M NaCl (final concentration) were added into the medium and the cells were kept to grow for 3 hours. Finally, total RNA was isolated as described above. Primers targeting genes relB2sca, clpP1, clpP2, clpP3, clpP4, clpP5 and clpP6 were designed for detecting the mRNA expression (supplementary Table S3). Real-time quantitative PCR was carried out with an ABI 7500 fast system. Calculations were performed by using 16S rRNA as an internal standard. The 2−∆∆CT method was used to determine the relative gene expression40. A fold change of >2 or <−0.5 between the treated and untreated cells was considered a significant difference.
Plasmid construction
Plasmids used in this study (supplementary Table S2) were constructed as the following methods by using the primers listed in the supplementary Table S3.
(i) pBAD-relE1sca, pBAD-relE2sca and pBAD-relBE2sca. Primers were used to amplify the relE1sca, relE2sca and the intact relBE2sca operon from the S. cattleya DSM46488 genomic DNA (S. cattleya gDNA), respectively. The PCR products were digested with NcoI and XhoI enzymes, which were then cloned into pBAD-myc/hisA, resulting in plasmids pBAD-relE1sca, pBAD- relE2sca and pBAD-relBE2sca.
(ii) pIB139-relE1sca, pIB139-relB1sca, pIB139-relE2sca and pIB139-relB2sca. The toxin gene relE1sca was amplified with primers IBrelE1F and IBrelE1R from the S. cattleya gDNA and then cloned into pIB139, generating pIB139-relE1sca. Similar strategy was employed to obtain pIB139-relB1sca, pIB139-relE2sca and pIB139-relB2sca.
(iii) pACYC-relB2sca-relE2scahis. The toxin gene relE2sca was amplified with primers E2exF and E2exR from the S. cattleya gDNA. The PCR product was treated with BamHI and HindIII enzymes and then cloned into the MCS1 of pACYCDuet1, generating plasmid pACYC-relE2scahis, which expressed the toxin protein with a six-histidine tag at the N-terminal. The PCR product was also digested with NdeI and XhoI and then cloned into the MCS2 of pACYC-relE2scahis, resulting in plasmid pACYC-relB2sca-relE2scahis.
(iv) pRSF-ClpP1X, pRSF-ClpP2X, pRSF-ClpP3X, pRSF-ClpP4X, pRSF-ClpP5X and pRSF-ClpP6X. The proteinase chaperone gene clpX was amplified from the S. cattleya gDNA with the primers clpXF and clpXR and then cloned into the MCS1 of pRSFDuet1 with NcoI and HindIII, producing pRSFDuet1-clpX. The proteinase gene clpP1 was amplified from the S. cattleya gDNA with primers clpP1F and clpP1R and then cloned into the MCS2 of pRSFDuet-clpX, producing plasmid pRSF-ClpP1X. The plasmids pRSF-ClpP2X, pRSF-ClpP3X, pRSF-ClpP4X, pRSF-ClpP5X and pRSF-ClpP6X were obtained with the same strategy.
Toxicity evaluation in E. coli in a liquid and solid medium
E. coli MGJ5987, a ten-TA locus defect mutant (ΔchpB, ΔrelBE, ΔdinJ-yafQ, ΔyefM-yoeB, ΔhigBA, ΔprlF-yhaV, ΔyafNO, ΔmqsRA, ΔhicAB), transformed with corresponding plasmids was grown in LB broth supplemented with 100 μg/ml ampicillin at 37 °C. When the OD600 reached to about 0.2, the cultures were divided into two equal parts. The one was grown in the presence of 0.2% glucose (inhibition of the promoter PBAD) while the other was grown in the presence of 0.2% L-Arabinose (induction of the promoter PBAD). OD600 was measured every 30 minutes to monitor the E. coli cell growth. In addition, the samples were also obtained after induction for 1 h, and 5 μl drops of different dilutions were spread onto the LA plate supplemented with 100 μg/ml ampicillin. The LA plates were incubated overnight at 37 °C.
Purification of RelE2sca-His6, RelBsca and RelBE2sca-His6
RelBE2sca-His6 complex was overproduced in E. coli BL21(DE3) transformed with pACYC-relB2sca-relE2scahis (supplementary Table S2). After induced by 0.5 mM IPTG at 37 °C for 4 hours, cells were harvested by centrifugation at 8,000 g for 10 minutes at 4 °C. The cell pellet was re-suspended in lysis buffer (25 mM Tris-HCl, pH7.4, 500 mM NaCl, 50 mM imidazole) and then sonicated and centrifuged by centrifugation at 10,000 g for 45 minutes at 4 °C to remove cell debris. The supernatant was applied to a column containing 2 ml of NTA-Ni resin. The column was washed three times with 10 ml of the buffer A (25 mM Tris-HCl, pH7.4, 500 mM NaCl, 50 mM imidazole). RelBE2sca-His6 complex was eluted with 5 ml of the buffer B (25 mM Tris-HCl, pH7.4, 500 mM NaCl, 500 mM imidazole). The overexpression of RelE2sca-His6 and RelB2sca was done as the same as the expression of RelBE2sca-His6. And the protocol described by Sterckx et al.41 was employed to purify the RelE2sca-His6 and RelB2sca from the His6-RelBE2sca complex. The purified RelBE2sca-His6 was finally subjected to size exclusion chromatography analysis.
Electrophoretic mobility shift assay
DNA probe was amplified by using PCR with FAM-labeled oligonucleotide primers (supplementary Table S3) from the S. cattleya gDNA and purified with a nucleic acid purification kit. The labeled DNA probe was incubated with different amounts of proteins in EMSA buffer (50 mM Tris-HCl, pH 7.5; 10 mM MgCl2; 1 mM DTT and 100 mM NaCl) at room temperature for 30 minutes. The mixtures were then subjected to 6% native polyacrylamide gel electrophoresis at 100 V for 1 hour at 4 °C. Unlabeled DNA probe used as a specific competitor was amplified and purified as described previously. In the competitive experiments, the labeled and unlabeled DNA probes in different ratio were mixed with the same amount of proteins in EMSA buffer, then incubated at room temperature for 30 minutes and separated by 6% native polyacrylamide gel electrophoresis. The images of gels were obtained by using Bio-Rad Molecular Imager Gel Doc XR+ System.
DNase I footprinting assay
The 5′-FAM-labeled DNA probes and the reaction system were the same as the EMSA experiments. The mixtures were treated with DNase I (0.25 units, ThermoFisher Fermentas) at room temperature for 10 minutes. The reaction was stopped by adding 0.25 μl 0.5 M EDTA and incubating in a water bath at 75 °C for 15 minutes. The treated DNA fragments were purified with the nucleic acid purification kit and eluted with 50 μl double distilled water. The purified DNA fragments were mixed with HiDi formamide and GeneScan-500 LIZ size standards, assayed with Applied Biosystems 3730XL DNA analyzer (manufactured by the Jieli Company, Shanghai). Electropherograms were analyzed using the GENEMAPPER software (Applied Biosystems).
In vivo degradation of RelB2sca in E. coli
To elucidate the degradative mechanism of RelB2sca, E. coli BL21(DE3)/pLysS were transformed with pBAD-relB2scahis and pRSFDuet-1, pBAD-relB2scahis and pRSF-ClpP1X, pBAD-relB2scahis and pRSF-ClpP2X, pBAD-relB2scahis and pRSF-ClpP3X, pBAD-relB2scahis and pRSF-ClpP4X, pBAD-relB2scahis and pRSF-ClpP5X, pBAD-relB2scahis and pRSF-ClpP6X, respectively. The intracellular stability of plasmid-encoding RelB2sca-His6 was measured using in vivo degradation experiments. E. coli cells transformed with the corresponding plasmids were grown at 37 °C until that the OD600 reached 0.5. L-arabinose was added to a final concentration of 0.2% to induce His6-relB2sca expression. After the cells induced by L-arabinose at 37 °C for 4 hours, IPTG was added to a final concentration of 0.5 mM to induce ClpP1X, ClpP2X, ClpP3X, ClpP4X, ClpP5X or ClpP6X expression for 2 hours. Then spectinomycin was added to a final concentration of 200 μg/ml to block translation. Samples (3-ml aliquots) were collected every 2 hours. Cells were pelleted by centrifugation at 4 °C and stored at −80 °C.
Preparation of protein extracts and Western blotting
Cells were re-suspended in lysis buffer (25 mM Tris-HCl, 500 mM NaCl) and then disrupted with ultrasound according to the manufacturer’s recommendations. Lysates were centrifuged at 10,000 rpm/min for 60 minutes at 4 °C. And the 6×SDS gel loading buffer was added. Then the mixture was separated by SDS-PAGE and transferred to a poly-vinylidene difluoride membrane. RelB2sca-His6 proteins were detected by using an anti-His antibody. Densitometric analysis of the scanned images from the exposed film was performed with ImageJ42.
Additional Information
How to cite this article: Li, P. et al. Identification and characterization of chromosomal relBE toxin-antitoxin locus in Streptomyces cattleya DSM46488. Sci. Rep. 6, 32047; doi: 10.1038/srep32047 (2016).
Supplementary Material
Acknowledgments
We thank Professor Kenn Gerdes of the University of Newcastle for kindly providing the E. coli MGJ5987. This work was supported by grants from the 973 program, Ministry of Science and Technology, China (2012CB721002) and the National Natural Science Foundation of China (31371261).
Footnotes
Author Contributions H.-Y.O. conceived the study. P.L. performed experiments. P.L., C.T., J.G. and Z.D. performed data analyses. P.L., M.R.O., J.G. and H.-Y.O. wrote the paper. All authors read and approved the final manuscript.
References
- Gerdes K., Rasmussen P. B. & Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proceedings of the National Academy of Sciences of the United States of America 83, 3116–3120 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R. et al. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Research 39, 5513–5525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sberro H. et al. Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Molecular Cell 50, 136–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L. & Manuel S. D. B. Bacterial toxin–antitoxin systems: more than selfish entities? PloS Genetics 5, 135–139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Christensen S. K. & Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nature Reviews Microbiology 3, 371–382 (2005). [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Amitai S., Kolodkin-Gal I., & Hazan, R. Bacterial Programmed Cell Death and Multicellular Behavior in Bacteria. Plos Genetics 2, 1518–1526 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia F. F. Kinase activity of overexpressed hipa is required for growth arrest and multidrug tolerance in Escherichia coli. Journal of Bacteriology 188, 8360–8367 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y. et al. TADB: a web-based resource for Type 2 toxin–antitoxin loci in bacteria and archaea. Nucleic Acids Research 39, 606–611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenn G. et al. Prokaryotic Toxin-Antitoxins (ed. Gerdes K.) Ch. 5, 69–92 (2013). [Google Scholar]
- Feng S. et al. YoeB-ribosome structure: A canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Research 41, 9549–9556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer C. et al. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang J., Hara H., Kato I. & Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. Journal of Biological Chemistry 280, 3143–3150 (2005). [DOI] [PubMed] [Google Scholar]
- Simanshu D., Yamaguchi Y., Park J. H., Inouye M. & Patel D. Structural basis of mRNA recognition and cleavage by toxin mazf and its regulation by antitoxin maze in Bacillus subtilis. Molecular Cell 52, 447–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bøggild A. et al. The Crystal structure of the intact E. coli RelBE toxin-antitoxin complex provides the structural basis for conditional cooperativity. Structure 20, 1641–1648 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataudella I., Trusina A., Sneppen K., Gerdes K. & Mitarai N. Conditional cooperativity in toxin-antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic Acids Research 40, 6424–6434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. K., Mikkelsen M., Pedersen K. & Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proceedings of the National Academy of Sciences of the United States of America 98, 14328 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevillano L., Díaz M., Yamaguchi Y., Inouye M. & Santamaría R. I. Identification of the first functional toxin-antitoxin system in Streptomyces. Plos One 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watve M. G., Tickoo R., Jog M. M. & Bhole B. D. How many antibiotics are produced by the genus Streptomyces? Archives of Microbiology 176, 386–390 (2001). [DOI] [PubMed] [Google Scholar]
- Lee J. S., Hah Y. C. & Roe J. H. The induction of oxidative enzymes in Streptomyces coelicolor upon hydrogen peroxide treatment. Journal of General Microbiology 139, 1013–1018 (1993). [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M. & Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. Journal of General Microbiology 129, 2257–2269 (1983). [DOI] [PubMed] [Google Scholar]
- Chater K. F. Genetics of differentiation in Streptomyces. Annual Review of Microbiology 47, 685–713 (1993). [DOI] [PubMed] [Google Scholar]
- Maisonneuve E., Castro-Camargo M. & Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150 (2013). [DOI] [PubMed] [Google Scholar]
- Zhao C. et al. Insights into fluorometabolite biosynthesis in Streptomyces cattleya DSM46488 through genome sequence and knockout mutants. Bioorganic Chemistry 44, 1–7 (2012). [DOI] [PubMed] [Google Scholar]
- Maisonneuve E., Shakespeare L. J., Jørgensen M. G. & Gerdes K. Bacterial persistence by RNA endonucleases. Proceedings of the National Academy of Sciences of the United States of America 108, 13206–13211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ingram C. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. Journal of Bacteriology 171, 6617–6624 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning D., Ye S., Liu B. & Chang J. The proteolytic activation of the relNEs (ssr1114/slr0664) toxin-antitoxin system by both proteases Lons and ClpP2s/Xs of Synechocystis sp. PCC 6803. Current Microbiology 63, 496–502 (2011). [DOI] [PubMed] [Google Scholar]
- Sevillano L., Díaz M. & Santamaría R. I. Stable expression plasmids for Streptomyces based on a toxin-antitoxin system. Microbial Cell Factories 12, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P. et al. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140 (2003). [DOI] [PubMed] [Google Scholar]
- Andreev D. et al. The bacterial toxin RelE induces specific mRNA cleavage in the A site of the eukaryote ribosome. RNA 14, 233–239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin M. A., Davis J. H. & Strobel S. A. Bacterial toxin RelE: a highly efficient ribonuclease with exquisite substrate specificity using atypical catalytic residues. Biochemistry 52, 8633–8642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders N. & Van Melderen L. Toxin-antitoxin systems as multilevel interaction systems. Toxins 6, 304–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y., Miyakawa K., Nishimura Y. & Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. Journal of Bacteriology 175, 6850–6856 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletta N., Schuster C. F. & Ralph B. Two paralogous yefM-yoeB loci from Staphylococcus equorum encode functional toxin-antitoxin systems. Microbiology 159, 1575–1585 (2013). [DOI] [PubMed] [Google Scholar]
- Fiebig A., Rojas C. M. C., Siegal-Gaskins D. & Crosson S. Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin–antitoxin systems. Molecular Microbiology 77, 236–251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallez R. et al. New toxins homologous to ParE belonging to three-component toxin–antitoxin systems in Escherichia coli O157:H7. Molecular Microbiology 76, 719–732 (2010). [DOI] [PubMed] [Google Scholar]
- Ramage H. R., Connolly L. E. & Cox J. S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PloS Genetics 5, e1000767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbaux M., Mine N., Guérout A. M., Mazel D. & Melderen L. V. Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7. Journal of Bacteriology 189, 2712–2719 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C. et al. Identification and characterization of the chromosomal yefM-yoeB toxin-antitoxin system of Streptococcus suis. Scientific Reports 5, 89–97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser Tobias, Bibb Mervyn J., Buttner Mark J., Chater Keith F. & Hopwood D. A. Practical Streptomyces Genetics. (John Inner Foundatiion, Norwich, U.K., 2000). [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Sterckx G. J. et al. An efficient method for the purification of proteins from four distinct toxin-antitoxin modules. Protein Expression & Purification 108, 30–40 (2015). [DOI] [PubMed] [Google Scholar]
- Rasband W. S. ImageJ: Image processing and analysis in Java. Astrophysics Source Code Library 2, 378 (2012). [Google Scholar]
- Buchan D. W. A., Minneci F., Nugent T. C. O., Bryson K. & Jones D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Research 41, W349–W357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.