Abstract
Aquatic organisms are increasingly exposed to lowering of environmental pH due to anthropogenic pressure (e.g. acid rain, acid mine drainages). Such acute variations trigger imbalance of fish-associated microbiota, which in turn favour opportunistic diseases. We used the tambaqui (Colossoma macropomum), an Amazonian fish tolerant to significant pH variation in its natural environment, to assess the response of fish endogenous microbiota to acute short-term acid stress. We exposed 36 specimens of tambaquis to acidic water (pH 4.0) over 2 consecutive weeks and sampled cutaneous mucus, feces and water at 0, 7 & 14 days. The 16S RNA hypervariable region V4 was sequenced on Illumina MiSeq. After two weeks of acidic exposure, fecal and skin microbiota taxonomic structures exhibited different patterns: skin microbiota was still exhibiting a significantly disturbed composition whereas fecal microbiota recovered a similar composition to control group, thus suggesting a stronger resilience capacity of the intestinal microbiota than cutaneous microbiota.
The bacteria composing the microbiota of eukaryotes have contributed to the evolution of metazoans by playing key roles in metabolic processes affecting fitness of their host1,2. Roles of endogenous microbiota have been documented in immunity3, growth4, mating preference5, behaviour6 and energy utilization7 of the host. The importance of endogenous microbiota in metazoan evolution is such that some suggest hosts and their microbiota are best understood as a single meta-organism; defined as the holobiont8. On teleostean hosts, endogenous symbiotic bacteria have been documented in the intestinal tract9,10,11, the cutaneous mucus12, and on the gill’s surface2. The microbiota found on fish body surfaces is the very first line of defence against opportunist pathogens from the environment13,14,15,16. The strength of this first line of defence (so called ‘colonisation resistance’17) may rely on the equilibrium between the relative abundance and diversity of different species of endogenous bacteria. Physiological stress of the host may promote proliferation of opportunistic pathogens via perturbation of the equilibrium between relative abundance of endogenous commensals (so called ‘dysbiosis’)18,19. A number of factors have been reported to drive dysbiosis in teleost-associated microbiota, including anoxia16, exposure to surface-acting disinfectants20 or inappropriate diet21. Variations in water physico-chemistry are likely to also drive microbial dysbiosis in fish given the permanent contact of fish with surrounding water in their environment.
In the last few years, there has been growing concern about the effect of pH decrease in water bodies22. Increased post-industrial concentrations of sulphuric and nitric oxides in the atmosphere decreases the pH of rain in urban areas23. After an acidic rainfall, the environment is temporally acidified. The resistance and resilience of ecosystems exposed to acidic rainfall relies on a multitude of abiotic and biotic characteristics from the environment: the presence of exogenous polyamines24, local soil chemistry25 or the ionic composition of the water, all of which potentially affecting the response of an ecosystem following acid precipitation. In this respect, watercourses with reduced carbonate hardness are particularly susceptible to pH drop when exposed to acid rain phenomenon, as they have a low buffering capacity.
The Amazon River is the largest drainage basin in the world26. The black water tributaries of the Amazon River have a naturally acidic pH combined with low carbonate hardness, and are thus highly sensitive to the effects of acid rain. The increasing anthropic pressure in Amazonia due to the large-scale urbanisation and industrialisation around and inside this ecosystem (e.g. the city of Manaus) could potentially affect the pH of water bodies in the near future. Freshwater capture fisheries in the Amazon represent a key component of Brazilian freshwater output (248.8 Kilotons)27. Thus, it is of major interest to assess the effect of water acidification on fish stock sustainability. Several species of Amazonian fishes are relevant models to assess the effect of acid stress, many of them being naturally exposed to two different types of water during their life cycle: white water (circumneutral pH) and black water (acidic pH ≈ 4.0–4.5). The physiological response of Amazonian fishes exposed to acid stress has been documented on the tambaqui (Colossoma macropomum), the most commercially important fish in Brazil, that migrates annually from white water to black water streams to feed during the rainy season28,29. Tambaqui exposed to acid stress experience an increase in plasma glucose, cortisol and total ammonia levels, combined with variations in transepithelial potential29. Interestingly, other studies documenting the physiological response of fish to acid stress30 show that fishes acclimate to the acid environment by modulating innate physiological parameters. Because innate physiological parameters are known to modulate both microbiota composition and activity31, the taxonomic structure of the endogenous microbiota on fish exposed to acidic pH will likewise be affected by the physiological response of its host. Perturbation of the taxonomic structure might elicit a disturbance of the bacterial functional repertory, impacting host immune defence. In addition to the host-associated physiological perturbation effects, there are potentially direct effects of the pH drop on microbiota as well, which can work synergistically with the host-associated perturbation. However, direct effects of pH drop and changes associated to host acclimatization to acidic environment are inextricable from each other. This phenomenon is best explained by the host-microbiota interdependency.
We hypothesized that exposure to short-term acid stress triggers a perturbation of the taxonomic structure of commensal microbiota (dysbiosis), potentially followed by a replacement with opportunistic bacteria from the environmental water or rare endogenous strains that are more resistant to acidic conditions. Based on Wood and co-workers29 we also hypothesized that the dysbiosis is a transitory state, eventually followed by microbiota homeostasis recovery (i.e. resilience of the microbiota (human microbiota resilience are reviewed in Lozupone et al.32)). Most importantly, a differential response of the microbiota from different ecological niches (e.g. different body sites, like cutaneous mucus versus intestines) was expected since different body sites exhibit a unique physiological response to the acid stress30. Overall, the impact of acidic stress on the taxonomic structure of the endogenous microbiota on teleosts has not been documented yet.
To address these questions, 72 juveniles of Colossoma macropomum were randomly distributed across a system of six independent tanks, and half of them were then exposed to acid stress (pH 4.0) during two consecutive weeks. Cutaneous mucus, feces and tank water were sampled at three sampling times: day 0, after one week and two weeks exposure to acidic water. Taxonomic structure of microbial communities was characterized by targeting the V4 region of the 16S gene.
Results
A total of 6 911 554 reads, assigned to 1607 OTUs with 97% identity threshold were obtained from the 85 samples. The OTUs were assigned to 525 genera, 190 families, 81 orders, 38 class and 23 phyla. OTUs with fewer than 100 reads or that only occurred in a single sample were filtered out as a step to improve accuracy and diversity33 After filtration, there were 6 072 125 reads left, assigned to 508 OTUs, with a mean of 279 OTUs per sample and an average of 77 302 reads per sample. The average Good’s coverage estimation, used to assess the sampling effort, is 98 ± 5% of estimated coverage.
Impact of pH drop on taxonomic structure
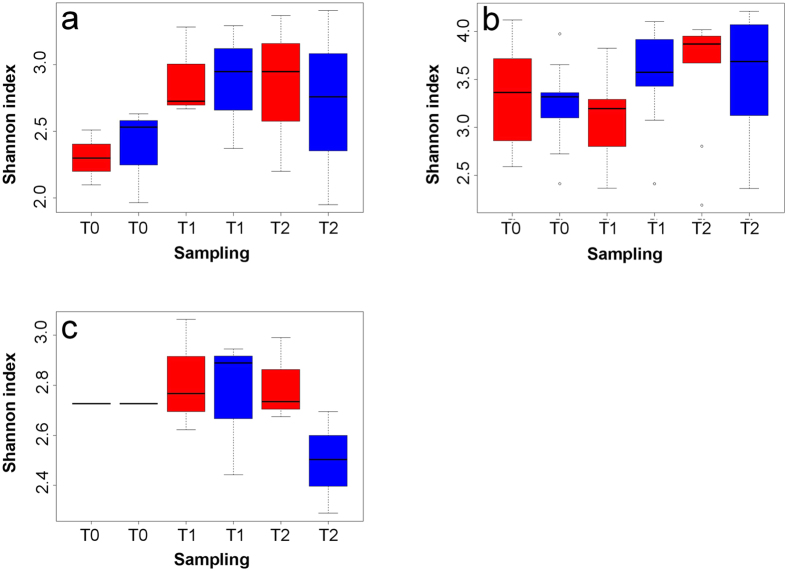
Non-parametric Shannon diversity indices for skin mucus samples showed the highest diversity (3.4 ± 0.2), which was significantly different from the diversity observed in feces (2.7 ± 0.2, p < 0.0001, df = 34, T = 5.39) and water samples (2.7 ± 0.1, p < 0.0001, df = 70, T = 7.66). There was no significant difference (p = 0.64, df = 22, T = 0.47) between diversity in feces and water samples. Finally, there was no significant variation between any sample categories (e.g. Control T0, Test T0, etc.) of the same sample type (e.g. water, feces or skin) in terms of Shannon diversity (Fig.1). Analysis of variance with permutations (PERMANOVA, 10 000 permutations34) (Table 1) revealed that the treatment factor (e.g. control or acid treatment) had a significant effect on the taxonomic structure found on the three microbial niches tested (water, feces and skin) (Table 1). At T1 (one week exposure to acid pH), the effect of the treatment was only significant in skin (p < 0.0001, R2 = 0.64) and feces samples (p < 0.002, R2 = 0.50). Although the treatment effect was still significantly different at T2 (two weeks exposure) for skin samples (p < 0.05, R2 = 0.57), it was not significantly different anymore for fecal samples (p > 0.05, R2 = 0.22). For water samples, the effect of the acid treatment was only significantly different at T2 (p < 0.002, R2 = 0.76). The tank factor showed significant difference only in skin samples at T0 and T1 (p < 0.05, R2 = 0.51). There was no significant difference between biological replicates.
Figure 1. No significant difference of alpha diversity between sampling times, for all three ecological niches tested.
The alpha diversity of bacterial communities at different sampling times is measured by non-parametric Shannon index of (a) feces samples, (b) skin mucus samples and (c) water samples. Boxes in red are test samples and in blue are control samples.
Table 1. The differential response of exposure to acid pH is niche-dependant.
| Sampling | Variable | p-value Skin mucus | p-value Feces | p-value Water |
|---|---|---|---|---|
| T0 | Biological replicate | 1 | N/A | N/A |
| Tank | 0.0441* | 1 | N/A | |
| Treatment | 0.1623 | 0.4 | N/A | |
| T1 | Biological replicate | 1 | N/A | N/A |
| Tank | 0.0002*** | 1 | 1 | |
| Treatment | 0.00009999*** | 0.001389** | 0.1 | |
| T2 | Biological replicate | 1 | N/A | N/A |
| Tank | 0.1147 | 1 | 1 | |
| Treatment | 0.0494* | 0.3014 | 0.001389** |
P-values from PERMANOVA highlight the effect of the variables: “Biological replicate” (groups of 4 fish in the same tank), “Tank” and “Treatment” (acid stress or control) on bacterial communities in three ecological niches (skin mucus, feces and water) at three sampling times. P-values in green are < 0.05. No data is available for feces and water samples for biological replicate, because only one sample was taken in each tank. No data is available for water at T0 because only one sample was taken for the whole system.
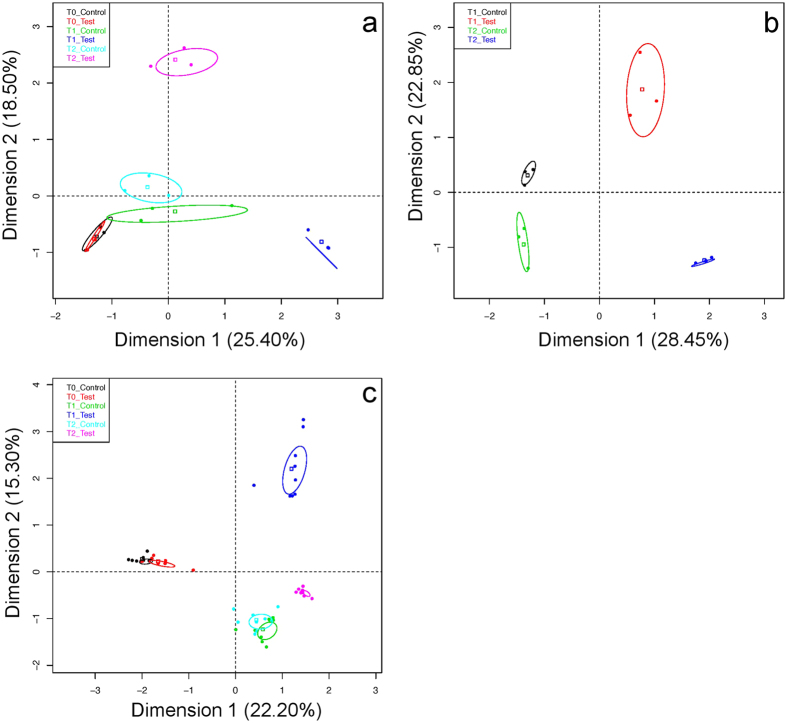
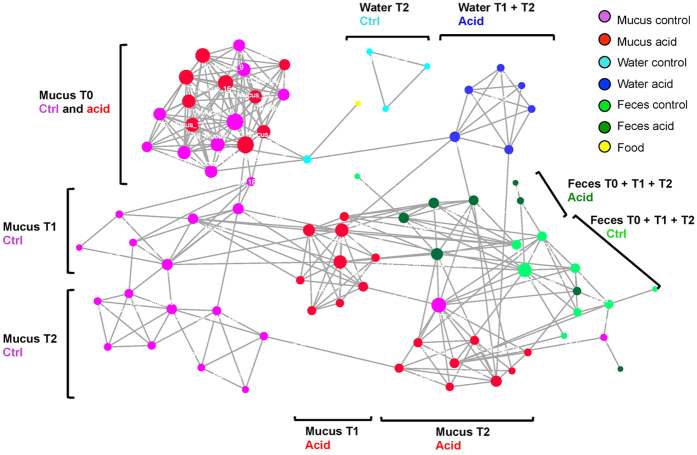
The multiple factor analysis, like the analysis of variance, revealed that acid treatment had a strong effect on the taxonomic structure of the microbiota (Fig. 2). The first two axes of the graphical representation explain 37.5% of the variance in skin samples, 43.9% in feces and 51.3% in water samples. For the three niches tested, taxonomic structure of all control samples at T0, T1 and T2 was significantly different (p < 0.05) from the structure observed in T1 and T2 test samples. Note that there is no T0 water sample in the graphical representation, as only one sample was taken at T0 for this niche (tanks were connected until T0). Control samples at T1 and T2 are significantly different (p < 0.05) for feces and water, although they are more similar than test samples at T1 and T2. Based on the number of edges (links) between samples and overall layout of sample clusters, the co-abundance network analysis (Fig. 3) showed that the taxonomic structure in all samples was mostly affected, in decreasing proportions of variance explained, by (1) the ecological niche (water, feces or skin); (2) the sampling time (T0, T1 or T2); and (3) the treatment (control or acid). The network revealed a strong inter-niche differentiation in core taxonomic structures. The layout of samples showed that pH drop impacted differentially skin and gut microbiota taxonomic structures. All skin samples clustered closely together at T0, then the network showed a strong differentiation at T1 and T2 between control and test skin samples. In this respect, there were first 59 edges (i.e. significant Spearman correlations between taxa, Bonferroni corrected p-value < 0.05) at T0 between test and control skin, then the number of edges dropped to ten edges at T1, and only one at T2. Also, there was no significant Spearman correlation (Bonferroni corrected p-value > 0.05) between T0 and T1 test skin samples. The network revealed no significant correlation between the food sample (Purina pellets) and fecal samples. As shown in the PERMANOVA (Table 1), the abundance of edges (significant Spearman correlations) in the network between skin samples at T0 confirms that biological replicates exhibited a high similarity in terms of taxonomic structure.
Figure 2. The taxonomic structure in all three niches is perturbed by pH drop.
The multi-factorial analysis (MFA) based on the variations of relative abundance of bacterial genera in regard of the combination of the: “Sampling time” (T0, T1, T2) and “Treatment” (test or control) variables. (a) feces, (b) water and (c) skin mucus samples.
Figure 3. Taxonomic structures diverge between sampling times and ecological niches.
The network of samples based on co-abundance of bacterial genera in each sample. Each dot (node) represents a sample. A link (edge) between two samples highlights a Spearman correlation index > 0.6 between the two samples and a p-value corrected with Bonferroni < 0.05. The size of each node is proportional to the number of edge that it is connected to.
Acid exposure driven taxonomic shifts
Figure 4 shows that the relative abundance of the 10 most abundant bacterial classes differed between each sampling time and ecological niche in bacterial communities exposed to acidic treatment. Interestingly, the most notable taxonomic shift is the increase of the relative abundance of the class Betaproteobacteria at T1 compared to T0 (p = 0.014) and T2 (p = 0.047) in fecal samples. At the genus level, in the tanks receiving the acid treatment, Flavobacterium composed 31.0% of the skin mucus community and 9.0% of the water community at T0. At T2, these relative abundances respectively decreased to 0.75% and 0.03%. The significant reduction of the relative abundance of Flavobacterium between T0 and T1, was 11.2% higher in test skin samples than in control (p < 0.0001) and 37.2% higher in test water samples than in control (p < 0.002).
Figure 4. There are major taxonomic structure shifts at the class level after acidic water exposure.
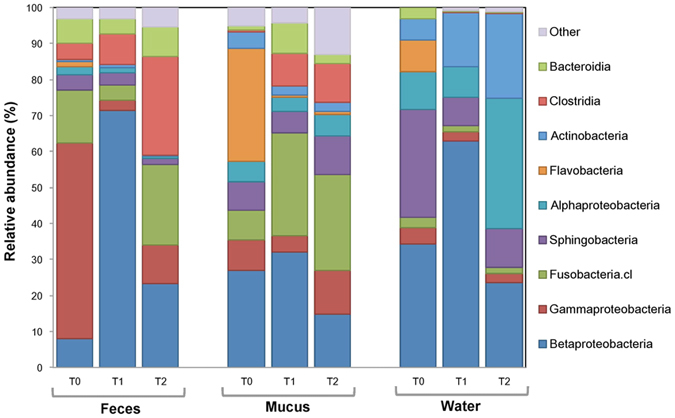
Variations of relative abundance (%) of 10 most abundant bacterial classes over sampling time and ecological niche.
In control tanks, the genus Undibacterium composed a low average of 1.48% of the microbial community in feces, 1.38% in skin and 0.08% in water. In test tanks, between T0 and T1, the relative abundance of the genus Undibacterium increased by 1652% (reaching an abundance of 14.4%) in skin samples (p < 0.01) and by 273% in fecal samples (p < 0.01). In test water samples, this genus increased from 0 detected reads (T0) to a relative abundance of 3.7% of the microbial community at T1. In test fecal samples, the relative abundance of Undibacterium further increased by 229% between T1 and T2, then reaching a relative abundance of 7.35%. For test skin samples the relative abundance of this genus decreased of 78% between T1 and T2.
In control tanks, the genus Duganella composed an average of 5.38% of the microbial community in feces, 0.53% in skin and 0.26% in water. In test tanks, between T0 and T1, the relative abundance of this genus increased significantly (p < 0.001) by 4583% (reaching a relative abundance of 21.6%) in fecal samples. In test water samples, this genus increased from 0 reads (T0) to a relative abundance of 17.11% at T1. Between T1 and T2 ( >1 week after the initial pH drop in test tanks) there was a significant decrease in the relative abundance of Duganella both in water (−96.6%, p < 0.01) and feces (−56.1%, p < 0.02).
Discussion
The results showed unambiguously that the acid pH drop had a significant effect on the structure of the bacterial communities that are closely associated with Colossoma macropomum. Structural taxonomic changes triggered by the pH occurred in all of the three microbial niches closely associated with Colosoma macropumum: surrounding water, intestine and epithelial mucus. As described on Salvelinus fontinalis16, one of the first perceivable changes on the microbiota after stress exposure is a taxonomic structure perturbation. This perturbation can act as a precursor to dysbiosis, leading to an imbalance in the equilibrium between opportunistic strains and pathogens15,16.
Each of the three microbial niches closely associated with Colosoma macropumum had a specific response to the acidic exposure. After one week of acidic exposure, there was a significant change in taxonomic structure in host-associated microbiota (i.e. epithelial mucus and fecal samples). Environmental pH has been recognized as a major driver of bacterial diversity in soil35. To add to this effect, physiological acclimatization of the host (gill structure breakdown, massive mucus production, cortisol synthesis)16,29 could have further increased the effect of acid stress on endogenous bacterial communities that are affected by (1) acid stress exposure combined with (2) a variation of available resources for growth on the host. Contrastingly, no significant change in taxonomic structure was observed in bacterioplankton at T1, highlighting the documented differentiation between host-associated microbial niches and the surrounding water36. However, we cannot rule out the possibility that important daily water changes (50% of tank volume) could have mitigated the observed perturbation on this particular niche, although pH was immediately adjusted to the target value.
No significant difference was detected in alpha diversity between control and acid exposed samples (Fig. 1). Pennanen and co-workers37 obtained similar results after exposing soil bacterial communities to simulated acid rains, suggesting that stress exposure triggers a microbial community succession (i.e a replacement of abundant taxa sensitive to acid pH by initially rare and more tolerant taxa). The microbial succession observed allowed us to detect potential stress specific biomarkers. Biomarkers (a substance produced by a living organism, or a living organism itself) are powerful and insightful tools to assess the biological state (e.g. health or perturbation) of an ecosystem38 or of a holobiont. Bacterial biomarkers in general are promising because of the short generation time of bacteria and their high sensitivity to variations in their environment making them a fast and precise tool to detect the effect of environmental perturbation on holobionts. Our results suggest that the class Betaproteobacteria, as well as the genera Flavobacterium, Duganella and Undibacterium have a great potential to be used as stress specific taxonomic biomarkers. The results revealed an increase in the relative abundance of the phylum Proteobacteria after acid stress exposure. The increase in the class Betaproteobacteria is significant (p < 0.05) in the three niche sampled in test tanks at T1. At T2, the relative abundance of this class significantly decreased (p < 0.05) from T1. These results further highlight the opportunistic nature of Proteobacteria, which was documented by Shin and co-workers (2015)39 and by Laplante and co-workers (2013)40 in acid mine drainages. Important variations were also observed for Flavobacterium, Undibacterium and Duganella genera. The genus Flavobacterium demonstrated a high sensitivity to acid pH. This is consistent with the results obtained by Suomalainen and co-workers (2005)41, who tested the effect of acid baths on rainbow trout against the opportunist pathogen Flavobacterium columnare. Results from our experiment suggest that in the future, acid baths could be a potential approach to decrease flavobacteriosis infections in tambaqui aquaculture systems, as an efficient alternative to antibiotic treatment or vaccination. On the opposite, Undibacterium and Duganella exhibited a different response than Flavobacterium to the acidic exposure: their relative abundance increased significantly the day following the pH drop (T1) and decreased significantly after stabilization of the pH (T2). These observations suggest that these taxa thrive better in perturbed or extreme environments. Interestingly, strains of Undibacterium have been documented in arsenic contaminated ground water42 and Duganella have been found in ground water beneath a uranium mine43.
The PERMANOVA reveals that fecal microbiome community structure is more resilient than cutaneous microbiota (see sampling T2 in Table 1). Gut microbiota are naturally exposed to acidic pH from gastric secretions; Payne (1978)44 has documented gastric pH as acid as 1.0 on the tilapia Tilapia guineensis. On the same species, the intestinal pH is circumneutral, highlighting the efficiency of pH buffering by alkaline secretions in the lower GI tract. Thus, given the efficiency of the intestinal pH buffering, it is possible that the dysbiosis detected in our study at T1 on fecal samples did not result directly from acute acidic exposure, but rather from host physiological stress. On the contrary, skin mucus communities likely encountered acute and prolonged acidic exposure after pH drop. Persistent dysbiosis state observed at T2 in skin mucus samples and water samples, but not in fecal samples, highlights this presumption. We cannot rule out the possibility that the taxonomic structure of the bacterial communities in skin mucus and water samples would stabilize over time. However, multiple evidence suggest that under a permanent exposure, it is unlikely that the taxonomic structure of these two particular niches will recover a taxonomic composition similar to the control group. Boutin et al.16 have shown that there was no significant resilience of the skin mucus bacterial community after a short exposition to hypoxia followed by a recovery time of 4 weeks. Laplante et al.40 have shown that there was no resilience for the water bacterial community in 5 lakes exposed to a gradient of polymetallic perturbation after a long-term exposition of 60 years.
Our results show that tambaqui exposed to acid treatment had a significantly higher ratio Firmicutes - Bacteroidetes in fecal samples than in control groups. In humans, many studies documented that a high Firmicutes / Bacteroidetes ratio could be significantly linked with obesity45. In fish models, Li et al. (2013)46 have showed that a high Firmicutes / Bacteroidetes ratio could be linked to faster growth rates of the transgenic common carp (Cyprinus carpio L.). To our knowledge, this is the first evidence that exposure to acidified water is linked to a higher Firmicutes - Bacteroidetes ratio. A long-term study needs to be exercised to assess if this ratio significantly affects the overall growth of Colosoma macropumum in an aquaculture system, as this could provide useful insight for tambaqui fish farmers in optimizing tambaqui growth rate.
The microbial community succession observed on all three ecological niches after exposure to environmental perturbation highlights the adaptation capacity of the endogenous microbial communities. As a host acclimatizes in response to a physiological stress, microbial communities at the interface between the host and the environment rapidly adapt to the modified ecosystem40,47. Interestingly, Rolli et al. (2015)48 have also shown that bacteria of root-associated microbiome can contribute to improve the adaptation of the holobiont to particular environmental conditions. Thus, it would be of great interest to assess the potential role of teleost-associated bacterial communities in facilitation of host-acclimatization to environmental stress. Indeed, as a host organism with great phenotypic plasticity might be adapted to a wide range of environmental conditions, an endogenous microbial community with substantial meta-genotypic plasticity (in this case, genotypic plasticity refers to the adaptability of the bacterial functions repertory) might confer to the holobiont a greater tolerance to fluctuating environments.
Overall, our results have shown that exposition of tambaqui to acid pH drop triggered dysbiosis in the microbiota of the three microbial niches investigated: cutaneous mucus, feces and environmental water. Furthermore, each microbial niche tested responded specifically to the acidic exposure. In this respect, our results highlight the strong resilience of tambaqui’s gut microbiota, which may be linked to its capacity to migrate from white water to black water. We have identified four potential stress specific taxonomic microbial biomarkers to assess the effect of environmental perturbation on aquatic biota: the class Betaproteobacteria, as well as the genera Flavobacterium, Duganella and Undibacterium. The results also revealed that acidic exposure had two potentially beneficial effects for tambaqui’s aquaculture: (1) a significant decrease in the relative abundance of the genus Flavobacterium responsible for flavobacteriosis infections and (2) a significant increase of the ratio Firmicutes - Bacteroidetes in feces. A long-term study is necessary to assess if the increase of this ratio improves overall growth of Colosoma macropumum. In the near future, a metagenomic approach is needed to assess whether the taxonomic structure perturbation of the tambaqui’s microbiota translates into an impairment of the microbial functional repertories dedicated to maintain the homeostasis of host physiological functions.
Methods
Fish rearing
The fish were reared in six independent 310 L FortLev tanks (3 control and 3 acid treatment tanks) at the “Laboratório de Ecofisiologia e Evolução Molecular” (LEEM) at “Instituto Nacional de Pesquisas da Amazonia” (INPA) in Manaus. Each tank contained 12 juvenile specimens of tambaqui (Ncontrol = 36, Ntest = 36, Ntotal = 72) obtained from a local fish culture station (average weight ≈40 g and length ≈15.0 cm). All of the fish were acclimated in the same conditions during six weeks before the start of the experiment: 12 h photoperiod, [dissolved O2] 7.1 ± 0.9 mg/L, water temperature 27.6 ± 0.4 °C and conductivity 48 ± 11 μmhos/cm. Every day, tanks were siphoned and 50% of the water was replaced by soft water from a local well with the following water parameters: [Ca2+] 11 μmol L−1, [Na+] 34 μmol L−1, [Cl−] 28 μmol L−1, [Mg2+] 0,8 μmol L−1, [K+] 15 μmol L−1, [dissolved organic matter] 0,9 mg C L−1, pH 6.3 and [NO2] 0–0,5 ppm. Fish were fed once per day to satiation with Purina® commercial pellets. Each fish was tagged with injection of Visible Implant Elastomer tags (Northwest Marine Technology’s) (colors = pink, green, blue) on the dorsal fin. Four fish per tank were tagged with the same color (3 colors per tank in 6 tanks, N = 18 fish groups).
Ethics statement
This project and protocol were approved by the Ethics Committee for the Use of Animals of INPA (number 026/2015 as of Dec 18th, 2015). All methods were carried out in accordance with the approved guidelines.
Sampling
Skin mucus, feces and tank water were sampled (1) after the acclimatization period at a stable pH of 6.3 (T0 = day 1); (2) after a slow pH drop from 6.3 to 4.0 (T1 = day 7) and (3) after one week exposure to pH 4.0 (T2 = day 14). The pH in experimental tanks was adjusted gradually from 6.3 to 4.0 using diluted HNO3 1 M and pH was stabilized at 6.3 in control tanks using (if needed) diluted HNO3 1 M and KOH 1 M. Skin mucus sampling was performed by gently rubbing a sterile cotton swab on ≈30% of the total surface of the right side of each fish. Skin mucus samples were pooled per tank and per color group (pink, green, blue) and were stored at −80 °C in 1.5 mL Eppendorf tubes. Feces were siphoned at the bottom of each tank. All feces samples were pooled per tank and stored at −80 °C in 1.5 mL Eppendorf tubes. Since numerous studies suggest that components of fish microbiota are shared with the bacterial community in the surrounding water21,49,50, 500 mL of tank water was sampled. Water samples were collected in each tank at T1, T2 and one water sample was collected for the whole system at T0. Water samples were filtered on 0.2 μm membranes (Nucleopore©) using a vacuum pump. Post-filtration, the membranes were stored at −80 °C.
Sample processing
DNA extraction of skin mucus samples and 0.2 μm membranes from water samples was performed using DNeasy® Blood and Tissue Kit from QIAGEN according to the manufacturer’s instructions. DNA extraction of feces samples was performed using QIamp® Fast DNA Stool Mini Kit according to the manufacturer’s instructions. Extracted DNA from feces, skin mucus and water was stored at −80 °C until amplification. DNA was amplified using universal primers specific to region V4 of the rRNA 16S gene. The hypervariable region V4 is often used to characterize thoroughly vertebrate-associated microbiota49. The forward primer S-D-Arch-0519-a-S-15 of sequence 5′CAGCMGCCGCGGTAA-3′ and the reverse primer S-D-Bact-0785-b-A-18 of sequence 5′-TACNVGGGTATCTAATCC-3′ were used. PCR reactions were performed according to the manufacturer instructions of TaKaRa Premix™ 2.0. Each reaction tube contained 5 μL of TaKaRa Premix™ 2.0 and 1 μL of each primer. Volume was completed to 11 μL with QIAGEN’s Microbial DNA Free Water and template DNA. PCR were done in triplicates to reduce PCR bias. PCR program: (1) 10 min 94 °C; (2) 1 min 94 °C; (3) 1 min 55 °C; (4) 1 min 30 sec 72 °C; (5) 10 min at 72 °C; 30 amplification cycles total. The 3 reaction replicates per sample were merged in an Eppendorf 1.5 mL post-PCR. Amplified DNA was purified with AMPure beads (Beckman Coulter Genomics) to eliminate primers, dimers, proteins and phenols. 50 μL of Microbial DNA Free Water and 10 μL of beads were added to each sample. Samples were mixed manually and incubated 5 min at RT. Using a magnetic particle concentrator (MPC), beads were concentrated on the side of the Eppendorfs and supernatant was removed. Beads were washed two times with 500 μL ethanol 70% and incubated 30 sec at RT during each wash. Supernatant was removed and beads were left to dry 5 min. Tubes were removed from the MPC and 20 μL of Microbial DNA Free Water were added for elution. DNA concentration and quality were assessed by electrophoresis on 1.5% agarose gel and by NanoDrop. After purification, samples were sequenced in the MiSeq platform from Illumina, at the LEEM. The Nextera® XT DNA Library Preparation Kit (96 samples) (Catalog number: FC-131-1096) was used for DNA library preparation. The Illumina® MiSeq Reagent Kit v2 (500-cycle; 2 × 250) (Catalog Number: MS-102-2003) was used for sequencing on MiSeq (Illumina Sequencing System). All proceedings followed the manufacturer’s protocols.
16S amplicon sequences analysis
The sequence files are available from the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra), BioProject ID: PRJNA323592. The analysis of amplicon sequences was done at the Institut de Biologie Intégrative et des Systèmes (IBIS) at Université Laval. Quality trimming of the sequences was done with sickle50. Then, the paired-end Illumina reads were merged with pandaseq51. Mothur alignment function52 was performed to align each read to 16S V4 sequence as reference template. Uchime53 was used to remove chimeric sequences. The clustering software Usearch54 was used for de novo operational taxonomic unit (OTU) picking with an identity-clustering threshold of 97%. RDP database reference taxonomy files were used to annotate OTUs55. Given the variability of the V4 region56, a bacterial genus was assigned to each OTU. OTUs with ≥100 reads total in all samples were kept for downstream analysis. Alpha diversity measures were obtained with mothur software52. Home made PERL scripts were developed to annotate OTUs taxonomy labels and to adapt the OTU table to the format requirements of mothur. The normalized OTU table was used for all analysis except for alpha diversity and Good’s index.
Shannon’s non-parametric alpha diversity index and Good’s coverage index were assessed for each sample, using mothur software52. Variance equality was assessed with F-tests on StatPlus v.5.9.92. T-tests for two-tailed distribution with unequal variance were used for non-parametric Shannon diversity indexes. Alpha diversity (see Fig. 1) was then represented in boxplots constructed with R version 3.2.1. A permutational analysis of variance (PERMANOVA) was performed with R to assess the effects of (1) the acid treatment; (2) the inter-tank variations; (3) the pooled samples (biological replicates); and (4) the time, on the variation in the taxonomic structure of the bacterial communities between samples. For each analysis of variance, 10 000 permutations were applied. To visualize the degree of similarity between samples taxonomic structures, a network of samples, based on OTU relative co-abundance between samples, was constructed with Cytoscape version 3.2.157. To construct this network, the Spearman’s correlation was calculated between each sample pair using R. A p-value, to which was applied Bonferroni correction, was determined for each Spearman’s correlation value. The significant correlation values used for the network construction had a Spearman’s correlation ≥ 0.6 and a Bonferroni corrected p-value ≤ 0.05. The nodes of the network represent the samples and the edges (i.e. connection) are attributed to significant correlation between nodes. To confirm the results obtained with PERMANOVA and the Cytoscape network, a Multiple Factor Analysis (MFA) was done with R. The MFA is similar to a Principal Components Analysis in data multidimensional visualization and it was used to simultaneously study global patterns observed in continuous (OTU normalized abundance) and categorical (e.g. time of sampling) variables. Confidence ellipses were drawn (with default settings of the plotellipses function) around each category (e.g. Control T0, Test T0, etc.) in the graphical representation. Categories with non-overlapping ellipses were considered significantly different (default confidence level of 0.95).
Additional Information
How to cite this article: Sylvain, F.-É. et al. pH drop impacts differentially skin and gut microbiota of the Amazonian fish tambaqui (Colossoma macropomum). Sci. Rep. 6, 32032; doi: 10.1038/srep32032 (2016).
Acknowledgments
We gratefully acknowledge help from all members of the Derome laboratory and of the LEEM. Thank you to Gabriel Melhado for help with fish maintenance and Daini Kochhann for tagging the fish. This research was part of the ADAPTA project at INPA and was supported by Ressources Aquatiques Québec, the NSERC Discovery grant to ND, the CAPES post-doctoral fellowship to TC, INCT ADAPTA (CNPq/FAPEAM) and INPA/MCTI grants to ALV. Thank you to FSG André Darveau (U. Laval) for travel grant to INPA.
Footnotes
Author Contributions A.L.V., N.D., F.E.S., M.L. and T.C. conceived the experiment. F.E.S. and T.C. conducted the experiment. F.E.S. and D.B.F. performed the molecular analysis. F.E.S., B.C. and M.L. analysed the results. F.E.S. and N.D. wrote the manuscript. The project was under supervision of N.D. and A.L.V. All authors reviewed the manuscript.
References
- McFall-Ngai M. et al. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America 110, 3229–3236, doi: 10.1073/pnas.1218525110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn M. S. et al. Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Frontiers in Microbiology 5. doi: 10.3389/fmicb.2014.00207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L. & Mazmanian S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology 9, 313–323, doi: 10.1038/nri2515 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. M. et al. Gut Microbiota Contributes to the Growth of Fast-Growing Transgenic Common Carp (Cyprinus carpio L.). Plos One 8, 11, doi: 10.1371/journal.pone.0064577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G. et al. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America 107, 20051–20056, doi: 10.1073/pnas.1009906107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijtza R. D. et al. Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America 108, 3047–3052, doi: 10.1073/pnas.1010529108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C. et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell 163, 1360–1374, doi: 10.1016/j.cell.2015.11.004 (2015). [DOI] [PubMed] [Google Scholar]
- Guerrero R., Margulis L. & Berlanga M. Symbiogenesis: the holobiont as a unit of evolution. International Microbiology 16, 133–144, doi: 10.2436/20.1501.01.188 (2013). [DOI] [PubMed] [Google Scholar]
- Bates J. M. et al. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Developmental Biology 297, 374–386, doi: 10.1016/j.ydbio.2006.05.006 (2006). [DOI] [PubMed] [Google Scholar]
- Rawls J. F., Mahowald M. A., Ley R. E. & Gordon J. I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–433, doi: 10.1016/j.cell.2006.08.043 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls J. F., Samuel B. S. & Gordon J. I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proceedings of the National Academy of Sciences of the United States of America 101, 4596–4601, doi: 10.1073/pnas.0400706101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin S., Sauvage C., Bernatchez L., Audet C. & Derome N. Inter Individual Variations of the Fish Skin Microbiota: Host Genetics Basis of Mutualism? Plos One 9, doi: 10.1371/journal.pone.0102649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi B. MICROBIOME The surface brigade. Nature 492, 560–561 (2012). [DOI] [PubMed] [Google Scholar]
- Sugita H. et al. Antibacterial abilities of intestinal bacteria from larval and juvenile Japanese flounder against fish pathogens. Fisheries Science 68(5), 1004–1011. doi: 10.1046/j.1444-2906.2002.00525.x (2002). [DOI] [Google Scholar]
- Boutin S., Audet C. & Derome N. Probiotic treatment by indigenous bacteria decreases mortality without disturbing the natural microbiota of Salvelinus fontinalis. Canadian Journal of Microbiology 59, 662–670, doi: 10.1139/cjm-2013-0443 (2013). [DOI] [PubMed] [Google Scholar]
- Boutin S., Bernatchez L., Audet C. & Derome N. Network Analysis Highlights Complex Interactions between Pathogen, Host and Commensal Microbiota. Plos One 8, doi: 10.1371/journal.pone.0084772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T. D. & Walker A. W. Intestinal colonization resistance. Immunology 138, 1–11, doi: 10.1111/j.1365-2567.2012.03616.x (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling C. C. & Wegner K. M. Relative contribution of reproductive investment, thermal stress and Vibrio infection to summer mortality phenomena in Pacific oysters. Aquaculture 412, 88–96, doi: 10.1016/j.aquaculture.2013.07.009 (2013). [DOI] [Google Scholar]
- Stecher B., Maier L. & Hardt W.-D. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nature Reviews Microbiology 11, 277–284, doi: 10.1038/nrmicro2989 (2013). [DOI] [PubMed] [Google Scholar]
- Mohammed H. H. & Arias C. R. Potassium permanganate elicits a shift of the external fish microbiome and increases host susceptibility to columnaris disease. Veterinary Research 46, doi: 10.1186/s13567-015-0215-y (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T. J., Smullen R. & Barnes A. C. Dietary soybean protein concentrate-induced intestinal disorder in marine farmed Atlantic salmon, Salmo salar is associated with alterations in gut microbiota. Veterinary Microbiology 166, 286–292, doi: 10.1016/j.vetmic.2013.05.009 (2013). [DOI] [PubMed] [Google Scholar]
- Orr J. C. et al. Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437, 681–686, doi: 10.1038/nature04095 (2005). [DOI] [PubMed] [Google Scholar]
- Schindler D. W. Effects of acid-rain on fresh-water ecosystems. Science 239, 149–157, doi: 10.1126/science.239.4836.149 (1988). [DOI] [PubMed] [Google Scholar]
- Velikova V., Yordanov I. & Edreva A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants - Protective role of exogenous polyamines. Plant Science 151, 59–66, doi: 10.1016/s0168-9452(99)00197-1 (2000). [DOI] [Google Scholar]
- Krug E. C. & Frink C. R. Acid-rain on acid soil - a new perspective. Science 221, 520–525, doi: 10.1126/science.221.4610.520 (1983). [DOI] [PubMed] [Google Scholar]
- Fink W. L. & Fink S. V. Central amazonia and its fishes. Comparative Biochemistry and Physiology a-Physiology 62, 13-& (1979). [Google Scholar]
- FAO, Fishery Country Profile - Brazil. (2011) Available at: http://www.fao.org/fishery/facp/BRA/en. (Accessed: 5th March 2016).
- Rocha Aride P. H., Roubach R. & Val A. L. Tolerance response of tambaqui Colossoma macropomum (Cuvier) to water pH. Aquaculture Research 38, 588–594, doi: 10.1111/j.1365-2109.2007.01693.x (2007). [DOI] [Google Scholar]
- Wood C. M. et al. Responses of an Amazonian teleost, the tambaqui (Colossoma macropomum), to low pH in extremely soft water. Physiological Zoology 71, 658–670 (1998). [DOI] [PubMed] [Google Scholar]
- Fromm P. O. Review of some physiological and toxicological responses of fresh-water fish to acid stress. Environmental Biology of Fishes 5, 79–93, doi: 10.1007/bf00000954 (1980). [DOI] [Google Scholar]
- Hevia A. et al. Human colon-derived soluble factors modulate gut microbiota composition. Frontiers in Oncology 5, doi: 10.3389/fonc.2015.00086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. A., Stombaugh J. I., Gordon J. I., Jansson J. K. & Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230, doi: 10.1038/nature11550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N. A. et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods 10, 57–U11, doi: 10.1038/nmeth.2276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecology 26, 32–46, doi: 10.1111/j.1442-9993.2001.01070.pp.x (2001). [DOI] [Google Scholar]
- Lauber C. L., Hamady M., Knight R. & Fierer N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Applied and Environmental Microbiology 75, 5111–5120, doi: 10.1128/aem.00335-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn M. S. et al. The biogeography of the atlantic salmon (Salmo salar) gut microbiome. ISME J, PMID. 26517698 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennanen T. et al. Structure of a microbial community in soil after prolonged addition of low levels of simulated acid rain. Applied and Environmental Microbiology 64, 2173–2180 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorci J. J., Paulauskis J. D. & Ford T. E. 16S rRNA restriction fragment length polymorphism analysis of bacterial diversity as a biomarker of ecological health in polluted sediments from New Bedford Harbor, Massachusetts, USA. Marine Pollution Bulletin 38, 663–675, doi: 10.1016/s0025-326x(98)90199-0 (1999). [DOI] [Google Scholar]
- Shin N.-R., Whon T. W. & Bae J.-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends in Biotechnology 33, 496–503, doi: 10.1016/j.tibtech.2015.06.011 (2015). [DOI] [PubMed] [Google Scholar]
- Laplante K., Sebastien B. & Derome N. Parallel changes of taxonomic interaction networks in lacustrine bacterial communities induced by a polymetallic perturbation. Evolutionary Applications 6, 643–659, doi: 10.1111/eva.12050 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen L. R., Throla M. & Valtonen E. T. Treatment of columnaris disease of rainbow trout: low pH and salt as possible tools? Diseases of Aquatic Organisms 65, 115–120, doi: 10.3354/dao065115 (2005). [DOI] [PubMed] [Google Scholar]
- Ghosh S. & Sar P. Identification and characterization of metabolic properties of bacterial populations recovered from arsenic contaminated ground water of North East India (Assam). Water Research 47, 6992–7005, doi: 10.1016/j.watres.2013.08.044 (2013). [DOI] [PubMed] [Google Scholar]
- Akob D. M. et al. Identification of Mn(II)-Oxidizing Bacteria from a Low-pH Contaminated Former Uranium Mine. Applied and Environmental Microbiology 80, 5086–5097, doi: 10.1128/aem.01296-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne A. I. Gut ph and digestive strategies in estuarine grey mullet (mugilidae) and tilapia (cichlidae). Journal of Fish Biology 13, 627–629, doi: 10.1111/j.1095-8649.1978.tb03476.x (1978). [DOI] [Google Scholar]
- Zhang H. et al. Human gut microbiota in obesity and after gastric bypass. Proceedings of the National Academy of Sciences of the United States of America 106, 2365–2370, doi: 10.1073/pnas.0812600106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. M. et al. Gut Microbiota Contributes to the Growth of Fast-Growing Transgenic Common Carp (Cyprinus carpio L.). Plos One 8, 11, doi: 10.1371/journal.pone.0064577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis K. M. et al. Selective progressive response of soil microbial community to wild oat roots. Isme Journal 3, 168–178, doi: 10.1038/ismej.2008.103 (2009). [DOI] [PubMed] [Google Scholar]
- Rolli E. et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environmental Microbiology 17, 316–331, doi: 10.1111/1462-2920.12439 (2015). [DOI] [PubMed] [Google Scholar]
- Werner J. J., Zhou D., Caporaso J. G., Knight R. & Angenent L. T. Comparison of Illumina paired-end and single-direction sequencing for microbial 16S rRNA gene amplicon surveys. Isme Journal 6, 1273–1276, doi: 10.1038/ismej.2011.186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N. A. & Fass J. N. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software]. Available at https://github.com/najoshi/sickle (2011).
- Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G. & Neufeld J. D. PANDAseq: PAired-eND Assembler for Illumina sequences. Bmc Bioinformatics 13, doi: 10.1186/1471-2105-13-31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied and Environmental Microbiology 75(23), 7537–7541, doi: 10.1128/aem.01541-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. & Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200, doi: 10.1093/bioinformatics/btr381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461, doi: 10.1093/bioinformatics/btq461 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Qiong, George M. Garrity, James M. Tiedje & James R. Cole. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73(16), 5261–5267, doi: 10.1128/aem.00062-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty S., Helb D., Burday M., Connell N. & Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of Microbiological Methods 69, 330–339, doi: 10.1016/j.mimet.2007.02.005 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504, doi: 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]