Abstract
Young-onset breast cancer shows certain phenotypic and etiologic differences from older-onset breast cancer and may be influenced by some distinct genetic variants. Few genetic studies of breast cancer have targeted young women and no studies have examined whether maternal variants influence disease in their adult daughters through prenatal effects. We conducted a family-based, genome-wide association study of young-onset breast cancer (age at diagnosis <50 years). A total of 602 188 single-nucleotide polymorphisms (SNPs) were genotyped for 1279 non-Hispanic white cases and their parents or sisters. We used likelihood-based log-linear models to test for transmission asymmetry within families and for maternally mediated genetic effects. Three autosomal SNPs (rs28373882, P=2.8 × 10−7; rs879162, P=9.2 × 10−7; rs12606061, P=9.1 × 10−7) were associated with risk of young-onset breast cancer at a false-discovery rate below 0.20. None of these loci has been previously linked with young-onset or overall breast cancer risk, and their functional roles are unknown. There was no evidence of maternally mediated, X-linked, or mitochondrial genetic effects, and no notable findings within cancer subcategories defined by menopausal status, estrogen receptor status, or by tumor invasiveness. Further investigations are needed to explore other potential genetic, epigenetic, or epistatic mechanisms and to confirm the association between these three novel loci and young-onset breast cancer.
Introduction
Based on recurrence risk within families, genetic factors likely have a substantial role in the etiology of young-onset breast cancer. Compared with women with no first-degree relatives with breast cancer, those with at least one affected first-degree relative are approximately three times as likely to develop the disease in their thirties, and twice as likely to develop the disease in their forties.1 Further, heritability is thought to be higher for young-onset breast cancer than for later-onset breast cancer,2 and previous studies have demonstrated that younger age at diagnosis is associated with an increased probability of having a co-twin2 or a grandmother with breast cancer.3 Mutations in genes such as BRCA1, BRCA2, CHEK2, and ATM are more common in young-onset cases,4,5 but such variants are rare and do not fully explain the disease's high heritability.6 Genetic factors may also have a role in determining the pathologic characteristics of young-onset tumors, which tend to be more aggressive and less amenable to treatment than tumors found in older women.7, 8
To date, only two genome-wide association studies (GWAS) have specifically examined single-nucleotide polymorphisms (SNPs) for associations with young-onset breast cancer.9, 10 The first of these investigations—a pilot study of 60 participants aged <45 years—did not identify any SNPs that met the Bonferroni criterion for genome-wide significance (P<5 × 10−8).10 In a larger (6993 cases, 8177 controls) and more recent study of breast cancer in women aged 51 or younger, Ahsan et al.9 reported genome-wide significant associations for 18 SNPs. All 18 were located near SNPs that had been previously identified as susceptibility variants for breast cancer across a broader age range. Given the paucity of GWAS of young-onset breast cancer, it is not yet possible to conduct pooled, consortia-based analyses at a size comparable to some of the recent breast cancer GWAS (for example, Michailidou et al.11 or Garcia-Closas et al.12).
Maternal genetic factors that influence the prenatal environment could influence the risk of young-onset breast cancer in adult daughters. We know, for example, that high birth weight is a risk factor for young-onset breast cancer.13, 14, 15 Therefore, variants that affect a child's in utero development could be related to that child's breast cancer risk later in life. Other prenatal exposures have been linked to decreased risk of breast cancer at any age, including being born to a mother with preeclampsia and being a twin.15 In addition, we previously found that family history of breast cancer in the grandmothers of affected women is skewed toward the maternal side, with the strongest asymmetry seen when the granddaughter was diagnosed between ages 45 and 54.3 Studies of maternally mediated genetic effects require a family-based design, and we know of no existing GWAS of this nature for breast cancer.
In an effort to identify variants associated with young-onset breast cancer, we conducted a family-based GWAS. We used an augmented case-parent triad design and examined the association between breast cancer diagnosed before the age of 50 and each of 602 188 SNPs. The investigation included three separate sets of genetic association tests. We first examined the influence of genetic variants carried by the affected daughter (inherited effects). Next, we examined the influence of the mother's genotype on the daughter's disease risk (maternally mediated effects). Both of these analyses included 588 961 autosomal SNPs and 13 179 X-chromosome SNPs. At last, we examined the influence of 48 mitochondrial polymorphisms on the risk of young-onset breast cancer.
Methods
Study participants
Cases were participants in The Sister Study (2003–2009) or The Two Sister Study (2008–2010) who were diagnosed with either ductal carcinoma in situ or invasive breast cancer before the age of 50 and who provided blood or saliva samples for genotyping. All participants provided written consent. The National Institute of Environmental Health Science's Internal Review Board and the Copernicus Group IRB approved both studies.
The Sister Study is a prospective cohort of 50 884 women from the United States and Puerto Rico who, at the time of enrollment, were between the ages of 35 and 74 and had a sister who had been diagnosed with breast cancer, but had not had breast cancer themselves. During follow-up, 235 Sister Study participants developed young-onset breast cancer.
For the Two Sister Study, we identified Sister Study participants who had told us in their baseline interviews that they had a sister who had been diagnosed with breast cancer within the last 4 years and before the age of 50. These unaffected Sister Study participants (n=1669) and their case sisters (n=1422) were all recruited into the Two Sister Study. Young-onset cases who consented to participate in the Two Sister Study or Sister Study were asked to forward our recruitment letter to any living parents, asking for their participation in a genetic study. The participating Two Sister Study cases and all consenting parents provided a saliva sample using the Oragene DNA self-collection kit. If the mother was unavailable, we genotyped the unaffected sister, who had already provided a blood sample when she enrolled in the Sister Study. In total, 3342 individuals were genotyped.
Genotyping analysis
DNA was extracted from saliva (80%), whole blood (19%), or blood clot (1%) and genotyped in 96-well plate batches at the Center for Inherited Disease Research at the Johns Hopkins University. Members of the same family were included on the same plate. For quality-control purposes, each plate also included at least one HapMap control sample (n=76 total) and at least one study duplicate (n=74 total).
Samples were genotyped using the Illumina HumanOmniExpressplusExome-8v1-2 array. This array includes a total of 960 697 SNPs, of which 244 707 (25%) are located in exomes. In total, 937 879 were autosomal, 22 600 were located on the X-chromosome, and 218 were mitochondrial. We uploaded all of the genotype data to dbGaP, where it is now publically available (study accession number: phs000678.v1.p1; http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000678.v1.p1). Quality control was carried out with the assistance of the Genetics Coordinating Center at the University of Washington.
Autosomal and X-chromosome SNPs were excluded from the GWAS for the following reasons: positional duplicates (n=19 948); technical filters (n=11 973); missing call rate of 2% or greater (n=6902); five or more Mendelian errors detected within trios or duos (n=4254); Hardy–Weinberg equilibrium P-value <1 × 10−6 (n=216) based on unrelated, non-Hispanic, white controls; two or more discordant calls among the 74 study duplicates (n=59); minor allele frequency <5% (n=314 987). Some additional subject-specific SNP exclusions were made based on evidence of chromosomal anomalies. After the quality-control filtering, 588 961 autosomal SNPs and 13 179 X-chromosome SNPs remained for analyses. We retained 48 of 218 genotyped mitochondrial SNPs, keeping only those variants for which there were at least 10 informative families (ie, where the mother's or daughter's genotype differed from the father's).
We excluded 11 half-sister controls who were identified as not genetically full sisters via relatedness assessment. No additional exclusions were made based on missing call rates, as all individuals had <2% missing data. The final sample included 3331 individuals (1447 cases) from 1477 families, but because parental missingness patterns differed by race/ethnicity, we limited our analyses to non-Hispanic whites. Descriptive information about the family structures for these 1296 families is included as Table 1.
Table 1. Non-Hispanic white participants included in the young-onset breast cancer genotyping analysis.
| Group description | Number of families | Number of people | Number of cases |
|---|---|---|---|
| Trios (affected sister, both parents) | 415 | 1245 | 415 |
| Sister-pairs (1 affected, 1 unaffected) | 353 | 706 | 353 |
| Affected sister and mother | 299 | 598 | 299 |
| Affected sister and father | 22 | 44 | 22 |
| Affected sister only | 108 | 108 | 108 |
| Affected sister, unaffected sister, father | 81 | 243 | 81 |
| Unaffected sister only | 11 | 11 | 0 |
| Mother only | 2 | 2 | 0 |
| Parents only | 2 | 4 | 0 |
| Unaffected sister and father | 2 | 4 | 0 |
| Tetrad (both sisters, both parents) | 1 | 4 | 1 |
| TOTAL | 1296 | 2969 | 1279 |
We used imputed genotypes to conduct fine-mapping analyses of identified susceptibility regions. This included SNPs within 20 000 base pairs of the initial hit that had imputation probabilities >95% and missing rates <5%. Pre-phasing and imputation were done using SHAPEIT (v2)16 and IMPUTE (v2),17 utilizing sequencing data from the 1000 genomes project.18 All position numbers are based on genomic reference sequence hg19.
Covariate definitions
Two Sister Study and Sister Study participants were administered the same computer-assisted telephone interview, which included questions about race, menopausal status, and a number of known or potentially breast cancer-related covariates. Women with breast cancer were asked to provide additional information about their diagnosis and for permission to access their medical records. We abstracted data on disease stage and estrogen receptor (ER) status from medical records, if available (82%), but relied on self-reported data if not. Women without data on ER status but who had been prescribed tamoxifen were presumed to be ER positive (n=2).
Although we did not directly genotype BRCA1 or BRCA2, we asked participants to report whether they had been tested for those mutations, and if so, what the results of those tests were. If an unaffected sister reported that she had a BRCA1 or BRCA2 mutation, we assumed that the affected sister did as well, unless the case had been specifically tested and found to be negative. If sisters reported discrepant races (n=16), we assigned the family to the racial category identified by the affected sister.
Statistical analysis
For the analysis of inherited effects, we used log-linear likelihood-based methods to test for transmission distortion within families.19, 20 In brief, these methods are not vulnerable to bias due to genetic population stratification and assume that if a particular SNP is associated with disease risk, then the relative frequency of the allele in affected offspring will deviate from the allele frequency expected under Mendelian inheritance. We did not assume Hardy–Weinberg equilibrium. Given complete triad data, expected frequencies follow a multinomial distribution with 15 possible outcomes and six specific mating types. Relative risks (RR) for the effect of inheriting the variant allele (relative to no copies) are estimated using Poisson regression, and P-values are calculated with a likelihood ratio test (χ2 distribution) comparing models with and without the genotype indicator variable(s). If either or both parents are unavailable, sibling genotype data are informative through use of the expectation-maximization algorithm to manage missing data.21, 22 We also employed extensions to the log-likelihood models to allow for the inclusion of case-parent dyads, case-sibling pairs, or singletons.22
To test for maternally mediated effects, we used additional extensions developed by Weinberg et al.19 and Wilcox et al.20 Here, maternal effects are assessed using the same mating type parameters and Poisson regression analysis as the inherited effects design, but with the addition of indicator variables for the mother's genotype status. In these models we controlled for the daughter's genotype.
When assessing inherited and maternally mediated genetic effects, we included analyses of X-chromosome SNPs using the parent-informed likelihood ratio test for the X-chromosome.23 This method uses information about the parental genotype and sex of the offspring to augment power.
Because daughters always inherit mitochondrial DNA from their mothers, we examined inherited mitochondrial SNP effects by calculating Z-scores for a matched-pair comparison of mothers versus fathers. If the mother was not genotyped, we imputed her genotype based on her daughter's.
Although these and other family-based methods inherently resist bias due to population stratification, this robustness may be violated if missingness itself (eg, being an unavailable father) depends on the missing genotype, conditional on the observed genotypes for the family. Based on evidence that parental missingness did vary by race in our study sample, we conducted race-specific analyses and focused on the results from non-Hispanic white families, as these comprised the largest subgroup.
We estimated RRs and association P-values for each SNP individually, assuming a log-additive genetic model. The minor allele in non-Hispanic whites was used as the index allele. We adjusted for multiple comparisons using a false-discovery rate (FDR) correction.24, 25 This was done separately for tests of inherited variants, maternally mediated effects, and mitochondrial effects. Analyses of all autosomal SNPs were conducted using LEM software (http://members.home.nl/jeroenvermunt/#Software). X-chromosome assessments were done using the ‘PIX-LRT' package in R (http://www.niehs.nih.gov/research/resources/software/biostatistics/pixlrt/index.cfm).23
For both inherited and maternally mediated effects, we conducted sensitivity analyses to examine whether the strength of the association changed when we restricted the analysis to families where the case sister had a specific sub-phenotype. Restrictive categories included were invasive breast cancer, ER-positive breast cancer, and premenopausal breast cancer. Owing to sample size concerns, we did not consider the less-common complementary categories (eg, ER-negative breast cancer). We also carried out a sensitivity analysis that excluded families known to carry BRCA1 or BRCA2 risk-related mutations (n=101 non-Hispanic, white families), reasoning that those effects could tend to overwhelm a lesser signal. When needed, we examined linkage disequilibrium (LD) patterns among the top-ranked SNPs using r2 values calculated using the 1000 genomes CEU population.18, 26
Results
Table 2 describes the women included in this GWAS. Most were non-Hispanic whites and most cases were diagnosed in their forties. The percentage of cases with ER-positive, invasive, and premenopausal breast cancer was 80, 83, and 93%, respectively.
Table 2. Characteristics of young-onset breast cancer families included in the genotyping analysis, as defined by the affected sistera: Two Sister Study (n=1242 cases; 2008–2010) and Sister Study (n=235 cases; 2003–2009).
| N (%) | |
|---|---|
| Age at diagnosis | |
| <40 | 161 (11) |
| 40–49 | 1315 (89) |
| Missing | 1 |
| Race/ethnicity | |
| Non-Hispanic white | 1296 (88) |
| Hispanic | 54 (4) |
| African-American | 85 (6) |
| Other | 40 (3) |
| Missing | 2 |
| Menopausal status at diagnosis | |
| Premenopausal (with or without hysterectomy) | 1359 (93) |
| Postmenopausal | 100 (7) |
| Missing | 18 |
| Invasive status | |
| Ductal carcinoma in situ | 239 (17) |
| Invasive | 1208 (83) |
| Missing | 30 |
| Estrogen receptor status | |
| Positive | 1146 (80) |
| Negative | 282 (20) |
| Missing | 49 |
| BRCA1/2 statusb | |
| Case carries BRCA1/2 mutation | 116 (8) |
| Case not known to have BRCA1/2 mutation | 1361 (92) |
This includes 29 families where the affected sister was not included in the genotype analysis, but other members of the family were.
Families were categorized as BRCA1 or BRCA2 mutation positive if (1) the case sister reported that she had had a positive test or (2) the case sister was not tested but the unaffected sister reported that she had had a positive test.
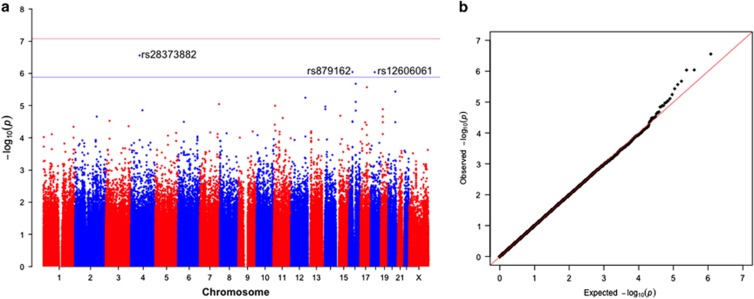
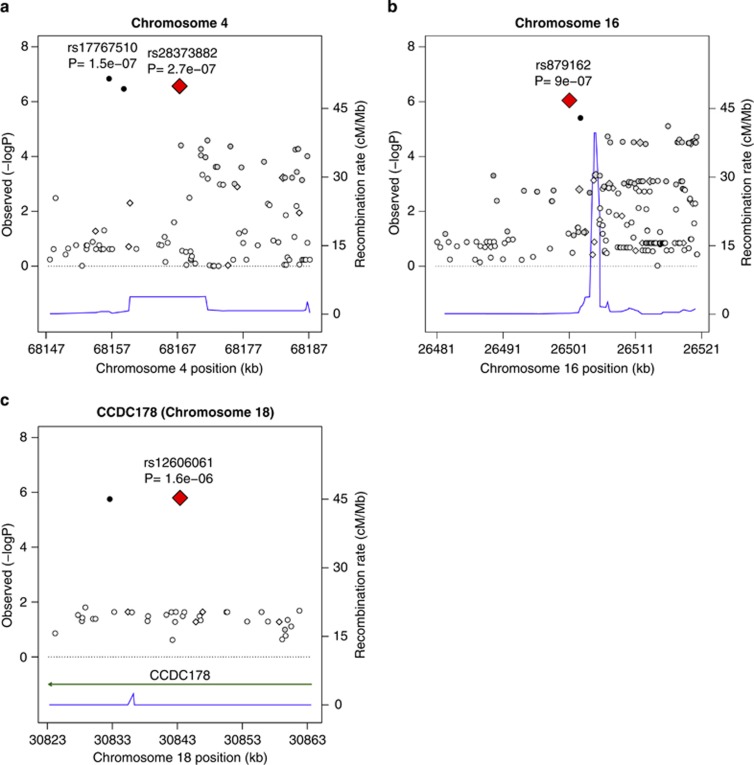
Results from our primary GWAS of inherited genetic effects among non-Hispanic whites are shown in Table 3 and Figure 1. We identified 30 320 SNPs with P<0.05, close to the number that would be expected by chance under a global null. None exceeded the cut-point for genome-wide significance based on a Bonferroni corrected two-sided P-value for 602 140 tests (8.2 × 10−8), but three had a FDR value <0.20. These were rs28373882:T>C (hg19.chr4:g.68167336:T>C; RR=1.68, 95% CI: 1.36, 2.05; P=2.8 × 10−7), rs879162:T>C (hg19.chr16:g.26501018:T>C; RR=1.50, 95% CI: 1.27, 1.76; P=9.2 × 10−7) and rs12606061:C>T (hg19.chr18:g.30843416:C>T; RR=0.63, 95% CI: 0.52, 0.76; P=9.1 × 10−7). In fine-mapping analyses of these three loci, we found that rs879162:T>C and rs12606061:C>T had the strongest associations with young-onset breast cancer within their identified regions (±20 000 base pairs), but an imputed SNP 10,738 base pairs away from rs28373882:T>C had a slightly stronger association (rs17767510:G>C, P=1.5 × 10−7) (Figure 2). These two SNPs are highly correlated with one another in our sample (r2=0.88).
Table 3. Top 20 SNPs for inherited variant effects on young-onset breast cancer in non-Hispanic white women in the Sister Study and Two Sister Study (n=1279 cases).
| Rank | SNP | Allelea | Chromosome | Positionb | Locus | MAFc | RR (95% CI) | P-value | FDR |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs28373882 | C/T | 4 | 68167336 | 4q | 0.14 | 1.68 (1.36, 2.05) | 2.8 × 10−7 | 0.17 |
| 2 | rs879162 | C/T | 16 | 26501018 | 16p | 0.22 | 1.50 (1.27, 1.76) | 9.1 × 10−7 | 0.18 |
| 3 | rs12606061 | T/C | 18 | 30843416 | CCDC178 | 0.15 | 0.63 (0.52, 0.76) | 9.1 × 10−7 | 0.18 |
| 4 | rs4784227 | T/C | 16 | 52599188 | TOX3 | 0.28 | 1.45 (1.24, 1.69) | 2.1 × 10−6 | 0.32 |
| 5 | rs8064617 | G/A | 17 | 49742142 | CA10 | 0.32 | 1.43 (1.23, 1.66) | 2.7 × 10−6 | 0.32 |
| 6 | rs169409 | T/C | 20 | 56240827 | PMEPA1 | 0.41 | 1.38 (1.20, 1.59) | 3.7 × 10−6 | 0.37 |
| 7 | rs7952757 | C/T | 12 | 114085798 | 12q | 0.32 | 1.40 (1.21, 1.62) | 5.7 × 10−6 | 0.49 |
| 8 | rs4784220 | C/T | 16 | 52535810 | TOX3 | 0.40 | 1.37 (1.19, 1.58) | 7.7 × 10−6 | 0.57 |
| 9 | rs2727565 | A/C | 7 | 151321161 | PRKAG2 | 0.27 | 0.70 (0.60, 0.82) | 9.0 × 10−6 | 0.57 |
| 10 | rs11042856 | A/G | 11 | 10519316 | AMPD3 | 0.34 | 1.39 (1.20, 1.62) | 1.0 × 10−5 | 0.57 |
| 11 | rs6572349 | T/C | 14 | 22736248 | 14q | 0.12 | 0.63 (0.52, 0.78) | 1.1 × 10−5 | 0.57 |
| 12 | rs7155927 | G/A | 14 | 22730981 | 14q | 0.13 | 0.64 (0.52, 0.78) | 1.3 × 10−5 | 0.57 |
| 13 | rs10409518 | T/C | 19 | 16606070 | CALR3 | 0.12 | 1.63 (1.30, 2.05) | 1.3 × 10−5 | 0.57 |
| 14 | rs17015190 | G/A | 4 | 90138063 | 4q | 0.14 | 0.66 (0.55, 0.80) | 1.4 × 10−5 | 0.57 |
| 15 | rs3803662 | A/G | 16 | 52586341 | TOX3 | 0.30 | 1.39 (1.20, 1.61) | 1.4 × 10−5 | 0.57 |
| 16 | rs16971851 | A/G | 15 | 33812509 | RYR3 | 0.23 | 1.41 (1.20, 1.66) | 2.1 × 10−5 | 0.73 |
| 17 | rs13430186 | G/T | 2 | 170887864 | UBR3 | 0.26 | 1.41 (1.20, 1.65) | 2.2 × 10−5 | 0.73 |
| 18 | rs875622 | A/G | 19 | 16467759 | EPS15L1 | 0.21 | 1.45 (1.22, 1.72) | 2.2 × 10−5 | 0.73 |
| 19 | rs10792551 | C/A | 11 | 70927063 | SHANK2 | 0.42 | 0.75 (0.65, 0.86) | 2.4 × 10−5 | 0.77 |
| 20 | rs9878305 | G/A | 3 | 29335426 | RBMS3 | 0.12 | 0.66 (0.54, 0.80) | 3.0 × 10−5 | 0.77 |
Minor/major allele coding; the minor allele is the index allele.
Based on genomic reference sequence hg19.
Minor allele frequency in parents.
Figure 1.
Manhattan plot (a) and quantile–quantile plot (b) of inherited variant effects on the risk of young-onset breast cancer in non-Hispanic white women. The solid line in the Manhattan plot indicates the Bonferroni cut-point for 602 140 tests SNPs (P=8.2 × 10−8) and the dashed line indicates the cut-point for a false-discovery rate correction of 0.20. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Figure 2.
Regional association plots for the top three loci for inherited effects: rs28372882 (a), rs879162 (b) and rs12606061 (c). Diamonds indicate typed SNPs (with the top hit magnified) and circles indicate imputed SNPs. SNPs in high linkage disequilibrium (r2≥0.80) with the typed hit are shown in black, whereas SNPs in moderate (0.80>r2≥0.50), low (0.50>r2≥0.20) or no linkage disequilibrium (r2<0.20) are shown in dark gray, gray, and white, respectively. The thin black line indicates the recombination rate for the identified region (in cM/Mb) and any nearby genes are shown with a bold solid line. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Although they did not meet our strict significance cut-points, three additional SNPs that were ranked in the top 20 were located in or near the TOX3 gene on chromosome 16 (rs4784227:C>T, rs4784220:T>C, and rs3803662:G>A). Two of these, rs4784227:C>T and rs3803662:G>A, are in high LD in the 1000 genomes CEU sample (r2=0.83). Two of the other top 20 SNPs, rs6572349:C>T and rs7155927:A>G on chromosome 14, were perfectly correlated (r2=1.00). None of the X-chromosome SNPs was ranked in the top 20.
Restricting the sample to families where the case had ER+, invasive, or premenopausal breast cancer did not reveal any additional noteworthy inherited SNPs (Supplementary Tables 1–3 and Supplementary Figures 1–3). However, in analyses that excluded families known to have at least one BRCA1- or BRCA2-positive individual (Supplementary Table 4 and Supplementary Figure 4), rs28373882:T>C (4q; P=7.7 × 10−8, FDR=0.05), rs4784227:C>T (TOX3; P=6.3 × 10−7, FDR=0.16) and rs879162:T>C (16p; P=8.0 × 10−7, FDR=0.16) were statistically significant at a FDR of 0.20.
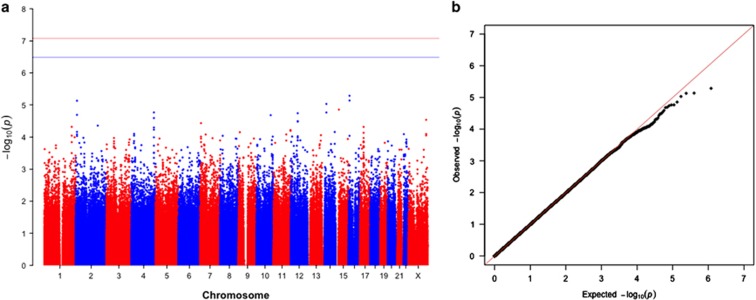
For our GWAS analysis of maternally mediated effects, we identified 29 636 SNPs with P<0.05 (Table 4, Figure 3). None of these met the criterion for genome-wide significance for a FDR of 0.20. The two highest ranked SNPs (rs12919267:C>T and rs12926526:A>G, with P-values of 5.1 × 10−6 and 7.3 × 10−6, respectively) are both located in RBFOX1 (r2=0.87 in the 1000 genomes CEU sample). Other top 20 ranked SNPs included four SNPs in or near the VEGFC gene (rs475106:G>A, rs10012721:G>T, rs4557213:A>G, and rs11131764:T>C), three SNPs in or near OR6C2 (rs6581029:A>G, rs74873135:T>C, and rs7952853:G>T), and two SNPs each in GRHL1 and NPAS3. Of these, the four VEGFC SNPS were all in high LD in the 1000 genomes CEU sample (r2≥0.90), as were the two GRHL1 SNPs (r2=1.00), with slightly lower LD seen for rs7487315:T>C and rs7952853:G>T (r2=0.81), and the two NPAS3 SNPs (r2=0.71). One X-chromosome SNP, rs6418889:T>C, was in the top 20 in the maternal effects analysis (RR=1.54, 95% CI: 1.26, 1.88; P-value=2.9 × 10−5). Sub-phenotype analyses yielded results that were not materially different, as did analyses excluding families with known BRCA1/2 mutations (Supplementary Tables 5–8 and Supplementary Figures 5–8).
Table 4. Top 20 SNPs for maternally mediated effects on young-onset breast cancer in non-Hispanic white women in the Sister Study and Two Sister Study (n=1279 cases).
| Rank | SNP | Allelea | Chromosome | Positionb | Locus | MAFc | RR (95% CI) | P-value | FDR |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs12919267 | T/C | 16 | 6922307 | RBFOX1 | 0.09 | 0.54 (0.41, 0.71) | 5.1 × 10−6 | 0.99 |
| 2 | rs12926526 | G/A | 16 | 6918341 | RBFOX1 | 0.09 | 0.56 (0.43, 0.72) | 7.3 × 10−6 | 0.99 |
| 3 | rs16867277 | C/T | 2 | 10136592 | GRHL1 | 0.07 | 0.50 (0.37, 0.69) | 7.4 × 10−6 | 0.99 |
| 4 | rs1315134 | A/G | 14 | 33827632 | NPAS3 | 0.09 | 0.52 (0.38, 0.70) | 9.3 × 10−6 | 0.99 |
| 5 | rs2355600 | G/A | 15 | 25167844 | SNRPN | 0.42 | 1.41 (1.20, 1.65) | 1.4 × 10−5 | 0.99 |
| 6 | rs11131764 | C/T | 4 | 177720359 | 4q | 0.09 | 1.90 (1.40, 2.57) | 1.7 × 10−5 | 0.99 |
| 7 | rs1315138 | A/G | 14 | 33826967 | NPAS3 | 0.12 | 0.59 (0.46, 0.76) | 1.7 × 10−5 | 0.99 |
| 8 | rs6581029 | G/A | 12 | 55686521 | 12q | 0.42 | 1.43 (1.21, 1.69) | 1.8 × 10−5 | 0.99 |
| 9 | rs7575106 | G/A | 2 | 10108037 | GRHL1 | 0.07 | 0.51 (0.38, 0.71) | 2.0 × 10−5 | 0.99 |
| 10 | rs12414450 | G/A | 10 | 112283719 | 10q | 0.24 | 0.67 (0.56, 0.81) | 2.1 × 10−5 | 0.99 |
| 11 | rs10012721 | T/G | 4 | 177659075 | VEGFC | 0.09 | 1.86 (1.38, 2.51) | 2.6 × 10−5 | 0.99 |
| 12 | rs6418889 | C/T | X | 142469312 | Xq | 0.42 | 1.54 (1.26, 1.88) | 2.9 × 10−5 | 0.99 |
| 13 | rs7487315 | C/T | 12 | 55782690 | 12q | 0.42 | 0.69 (0.57, 0.82) | 3.2 × 10−5 | 0.99 |
| 14 | rs7952853 | T/G | 12 | 55845415 | OR6C2 | 0.38 | 0.70 (0.58, 0.83) | 3.3 × 10−5 | 0.99 |
| 15 | rs1638214 | G/T | 7 | 7316977 | LOC101927354 | 0.38 | 1.42 (1.20, 1.69) | 3.7 × 10−5 | 0.99 |
| 16 | rs7608717 | A/G | 2 | 174317944 | 2q | 0.08 | 0.54 (0.40, 0.73) | 4.4 × 10−5 | 0.99 |
| 17 | rs2815220 | T/C | 1 | 217506787 | 1q | 0.22 | 0.66 (0.54, 0.81) | 4.8 × 10−5 | 0.99 |
| 18 | rs10445384 | C/T | 17 | 28182685 | SSH2 | 0.48 | 0.72 (0.61, 0.85) | 4.8 × 10−5 | 0.99 |
| 19 | rs475106 | A/G | 4 | 177656790 | VEGFC | 0.09 | 1.83 (1.35, 2.48) | 5.0 × 10−5 | 0.99 |
| 20 | rs4557213 | G/A | 4 | 177689133 | VEGFC | 0.09 | 1.77 (1.33, 2.37) | 5.9 × 10−5 | 0.99 |
Minor/major allele coding; the minor allele is the index allele.
Based on genomic reference sequence hg19.
Minor allele frequency in parents.
Figure 3.
Manhattan plot (a) and quantile–quantile plot (b) of maternal genetic effects on the risk of young-onset breast cancer in non-Hispanic white women. The solid line in the Manhattan plot indicates the Bonferroni cut-point for 602 140 tests SNPs (P=8.2 × 10−8) and the dashed line indicates the cut-point for a false-discovery rate correction of 0.20. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Four mitochondrial SNPs were associated with disease risk at P<0.05, but the smallest FDR value was 0.46 (Supplementary Table 9 and Supplementary Figure 9).
Discussion
In this family-based GWAS, three of the 602 188 SNPs (rs28373882:T>C, rs879162:T>C, and rs12606061:C>T) were associated with young-onset breast cancer after correction for multiple testing by means of a FDR. Analyses of subtype-specific families did not reveal any additional risk variants, with the exception of rs4784227:C>T (TOX3), which was associated with young-onset breast cancer after excluding families that self-identified as carrying a BRCA1 or BRCA2 mutation. Although they did not meet our cut-point for genome-wide significance, RBFOX1 and some of the other top-ranked regions from the maternal effects analysis may also be worthy of further investigation, especially as this is first GWAS to assess maternally mediated genetic effects.
None of the three loci identified in the main results has previously been linked to breast cancer or any other diseases, nor have any other nearby, correlated SNPs (within 500 000 base pairs and r2≥0.5).26, 27 rs28373882:T>C and rs879162:T>C, are intergenic (chromosomes 4q and 16p, respectively) and rs12606061:C>T is located in an intron of CCDC178, a gene of unknown function. The SNP in TOX3 (rs4784227:C>T) that was ranked fourth in the main analysis and second in the analysis that excluded the known BRCA-positive families, is a known susceptibility locus first identified in a breast cancer GWAS by Long et al.28 It has since been replicated in investigations among women of many ethnicities.29, 30, 31, 32, 33 Presence of the variant allele (T) increases the chromatin's affinity for FOXA1,34 a pioneer factor that can bind to chromatin and recruit the ER, thereby facilitating estrogen-driven transcription and cellular changes.35 The other highly ranked TOX3 SNP, rs3803662:G>A, was also originally identified in a GWAS study36 and the association was successfully replicated.37, 38, 39, 40
The top two ranked SNPs from the maternal effects analysis, rs12919267:C>T and rs12926526:A>G, are in RBFOX1, which encodes the FOX1 RNA-binding protein. The identified SNPs have no known associations with breast cancer or with the prenatal environment specifically. Of the variants in or near VEGFC, both rs11131764:T>C and rs4557213:A>G were associated with bladder cancer risk in one case-control study,41 but there are no known links to pregnancy or pregnancy-related conditions. More generally, the gene is thought to have an important role in breast cancer metastasis.42
We used log-likelihood-based methods to maximize power for our family-based design. These methods are not vulnerable to bias from population stratification and do not require an assumption of Hardy–Weinberg equilibrium. Imputation via the expectation-maximization algorithm enhanced our sample size by enabling inclusion of incompletely genotyped families. However, because there were differences in missing data frequencies by race, and inadequate numbers in the minority populations to permit ethnicity-specific stratified estimation of parental mating type parameters, we had to focus the analysis on the largest ethnic subgroup.
The families studied in the Two Sister Study are self-selected, which could lead to bias. Although we only considered sisters diagnosed within 4 years of their study enrollment, some of those who were originally identified as being eligible for inclusion may have died before we could enroll them. Others may have been too ill to participate, or deemed too ill to participate by the unaffected sister responsible for recruiting them. In addition, we found that a reported history of breast cancer in the mother was predictive of missing maternal genotype (OR=1.75, 95% CI: 1.34, 2.28). This differential missingness could have produced some bias toward the null in the assessment of maternally mediated genetic effects.
It is well known that the Bonferroni correction is too conservative, especially when the tests are positively correlated, as occurs in a GWAS where neighboring SNPs are in LD with one another.43 A FDR approach is generally less conservative than a Bonferroni cut-point and more consistent with recent guidelines described by Panagiotou et al.,44 who showed empirically that a high proportion of SNPs with P-values near 1 × 10−7 in initial GWAS are true positives. Certainly, further studies are needed to see if our results are replicable, especially given our somewhat limited sample size.
This was the third GWAS of young-onset breast cancer and the second largest. Kibriya et al.10 failed to identify any SNPs of genome-wide significance, but with only 60 cases, their power was poor. The study by Ahsan et al.9 included a much larger sample size and identified 72 SNPs that reached genome-wide significance, but all of them were located in regions that had already been identified as being associated with breast cancer over broader age ranges, so no novel loci specific to young-onset were identified.
We identified three susceptibility loci for young-onset breast cancer that have not previously been linked to breast or other cancers. These variants and genetic regions are worthy of further investigation, but considering Kibrya et al.,10 Ahsan et al.,9 and our analysis, none of the three existing young-onset breast cancer GWAS has been able to establish that there are unique genetic risk factors for young-onset breast cancer. Although the family history-based evidence and heritability evidence suggest that young-onset breast cancer has a large genetic component, it is not possible to fully explain disease heritability via identification of specific susceptibility loci. Genetic variants with effect sizes so small as to be practically undetectable might account for a large portion of heritability. However, it remains an important goal to identify and catalog any variants with noteworthy effect on disease susceptibility, and additional studies are still warranted. Investigations with larger sample sizes or pooled, consortia-based analyses may be required to detect unique susceptibility loci for young-onset breast cancer. Other genetic mechanisms, such as epigenetics, imprinting, the microbiome, gene–gene, and gene–environment interactions should also be investigated.
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, the National Institute of Environmental Health Sciences (project Z01-ES044005 to DPS and project Z01-ES102245 to CRW) and Susan G Komen for the Cure (grant FAS0703856 to CRW). We thank the Center for Inherited Disease Research, the Genetics Coordinating Center at the University of Washington (Cecelia Laurie, Quenna Wong, Sarah Nelson, and Cathy Laurie), and Jing Dong and Lauren Wilson from NIEHS for reviewing earlier drafts of this paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Collaborative Group on Hormonal Factors in Breast Cancer: Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58 209 women with breast cancer and 101 986 women without the disease. Lancet 2001; 358: 1389–1399. [DOI] [PubMed] [Google Scholar]
- Moller S, Mucci LA, Harris JR et al: The heritability of breast cancer among women in the Nordic twin study of cancer. Cancer Epidemiol Biomarkers Prev 2015; 25: 145–150. [DOI] [PubMed] [Google Scholar]
- Weinberg CR, Shi M, DeRoo LA, Taylor JA, Sandler DP, Umbach DM: Asymmetry in family history implicates nonstandard genetic mechanisms: application to the genetics of breast cancer. PLoS Genet 2014; 10: e1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung N, Battelli C, Allen B et al: Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015; 121: 25–33. [DOI] [PubMed] [Google Scholar]
- Whittemore AS, Gong G, John EM et al: Prevalence of BRCA1 mutation carriers among U.S. Non-Hispanic Whites. Cancer Epidemiol Biomarkers Prev 2004; 13: 2078–2083. [PubMed] [Google Scholar]
- Dite GS, Jenkins M, Southey M et al: Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 Germline Mutations. J Natl Cancer Inst 2003; 95: 448–457. [DOI] [PubMed] [Google Scholar]
- Anders CK, Hsu DS, Broadwater G et al: Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. J Clin Oncol 2008; 26: 3324–3330. [DOI] [PubMed] [Google Scholar]
- Anderson WF, Chen BE, Brinton LA, Devesa SS: Qualitative age interactions (or effect modification) suggest different cancer pathways for early-onset and late-onset breast cancers. Cancer Causes Control 2007; 18: 1187–1198. [DOI] [PubMed] [Google Scholar]
- Ahsan H, Halpern J, Kibriya MG et al: A genome-wide association study of early-onset breast cancer identifies PFKM as a novel breast cancer gene and supports a common genetic spectrum for breast cancer at any age. Cancer Epidemiol Biomarkers Prev 2014; 23: 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibriya MG, Jasmine F, Argos M et al: A pilot genome-wide association study of early-onset breast cancer. Breast Cancer Res Treat 2009; 114: 463–467. [DOI] [PubMed] [Google Scholar]
- Michailidou K, Hall P, Gonzalez-Neira A et al: Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 2013; 45: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Couch FJ, Lindstrom S et al: Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet 2013; 45: 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellemkjaer L, Olsen ML, Sorensen HT, Thulstrup AM, Olsen J, Olsen JH: Birth weight and risk of early-onset breast cancer (Denmark). Cancer Causes Control 2003; 14: 61–64. [DOI] [PubMed] [Google Scholar]
- Michels KB, Xue F: Role of birthweight in the etiology of breast cancer. Int J Cancer 2006; 119: 2007–2025. [DOI] [PubMed] [Google Scholar]
- Xue F, Michels KB: Intrauterine factors and risk of breast cancer: a systematic review and meta-analysis of current evidence. Lancet Oncol 2007; 8: 1088–1100. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF: A linear complexity phasing method for thousands of genomes. Nat Methods 2012; 9: 179–181. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J: A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium: an integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg CR, Wilcox AJ, Lie RT: A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet 1998; 62: 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AJ, Weinberg CR, Lie RT: Distinguishing the effects of maternal and offspring genes through studies of "case-parent triads". Am J Epidemiol 1998; 148: 893–901. [DOI] [PubMed] [Google Scholar]
- Shi M, Umbach DM, Weinberg CR: Case-sibling studies that acknowledge unstudied parents and permit the inclusion of unmatched individuals. Int J Epidemiol 2013; 42: 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg CR: Allowing for missing parents in genetic studies of case-parent triads. Am J Hum Genet 1999; 64: 1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise AS, Shi M, Weinberg CR: Learning about the X from our parents. Front Genet 2015; 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y: Controlling the false discover rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995; 57: 289–300. [Google Scholar]
- Yekutieli D, Benjamini Y: Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J Stat Plan Inference 1999; 82: 171–196. [Google Scholar]
- Broad Institute SNP Annotation and Proxy Search (SNAP) version 2.2. Available at https://www.broadinstitute.org/mpg/snap/ldsearch.php (accessed 14 October 2015).
- UCSC Genome Browser Home. Available at https://genome.ucsc.edu/ (accessed 21 September 2015).
- Long J, Cai Q, Shu XO et al: Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet 2010; 6: e1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MR, Deming-Halverson S, Cai Q et al: Evaluating 17 breast cancer susceptibility loci in the Nashville Breast Health Study. Breast Cancer 2015; 2015: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HC, Lee JY, Sung H et al: A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res 2012; 14: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KM, Cole SR, Poole C et al: Replication of breast cancer susceptibility loci in whites and African Americans using a Bayesian approach. Am J Epidemiol 2014; 179: 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueta A, Ito H, Kawase T et al: A genetic risk predictor for breast cancer using a combination of low-penetrance polymorphisms in a Japanese population. Breast Cancer Res Treat 2012; 132: 711–721. [DOI] [PubMed] [Google Scholar]
- Udler MS, Ahmed S, Healey CS et al: Fine scale mapping of the breast cancer 16q12 locus. Hum Mol Genet 2010; 19: 2507–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper-Sal lari R, Zhang X, Wright JB et al: Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 2012; 44: 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS: FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011; 43: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM et al: Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007; 447: 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Humphreys K, Heikkinen T et al: A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat 2011; 126: 717–727. [DOI] [PubMed] [Google Scholar]
- Stacey SN, Manolescu A, Sulem P et al: Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet 2007; 39: 865–869. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Kraft P et al: A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet 2009; 41: 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Ahmed S, Morrison J et al: Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 2010; 42: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Kamat AM, Aldousari S et al: Genetic variations in the transforming growth factor beta pathway as predictors of bladder cancer risk. PLoS One 2012; 7: e51758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skobe M, Hawighorst T, Jackson DG et al: Induction of tumor lymphangiogensis by VEGF-C promotes breast cancer metastasis. Nat Med 2001; 7: 192–198. [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A: Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 2008; 32: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotou OA, Ioannidis JP, Genome-Wide Significance P: What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol 2012; 41: 273–286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.