Abstract
Basan syndrome is a rare autosomal dominant genodermatosis, characterized by rapidly healing congenital acral bullae, congenital milia and lack of fingerprints. A mutation in the SMARCAD1 gene was recently reported to cause Basan syndrome in one family. Here, we present a large Chinese family with Basan syndrome; some patients presented with hyperpigmentation and knuckle pads in addition to previously reported clinical manifestations. We used genome-wide linkage analysis and whole-genome sequencing (WGS) to identify the pathogenic gene in this unique pedigree. Genome-wide linkage analysis successfully mapped the candidate gene to 4p15.31-4p14 and 4q13.2-4q23. The maximal LOD score was 3.01. WGS in one patient identified a splice variant (c.378+1G>T) in the SMARCAD1 gene (NG_031945.1) that was confirmed by Sanger sequencing. Co-segregation of the variant was confirmed in this pedigree. The same variant was recently found to be associated with isolated adermatoglyphia (ADG) in another family, suggesting that this variant is causative for both Basan syndrome and autosomal dominant ADG (OMIM 136 000). This indicates that ADG and Basan syndrome may be the phenotypic variants of the same disease. Further studies should be performed to elucidate the pathogenic mechanisms induced by this variant. This report increases the phenotypic spectrum of Basan syndrome and furthers our understanding of the genetic basis of this disease. Our results also highlight the power of combining WGS and genome-wide linkage analysis in identifying causative genes in pedigrees with a genetic disorder.
INTRODUCTION
Basan syndrome is a rare genetic skin disease with autosomal dominant inheritance that is characterized by rapidly healing congenital acral bullae, congenital milia and lack of fingerprints.1 Other phenotypes include contractures of digits, hypohidrosis, palmoplantar keratoderma (PPK) and nail dystrophy.2 Nine pedigrees with Basan syndrome have been reported worldwide.3, 4, 5, 6, 7, 8, 9, 10 Recently, Marks et al.10 identified SMARCAD1 (OMIM 612761) as a promising candidate gene for Basan syndrome in an American family. This gene encodes a member of the SNF subfamily of helicase proteins.9 Variants in the SMARCAD1 gene were previously reported in patients with adermatoglyphia (ADG, OMIM 136000), which is characterized by the congenital absence of epidermal ridges.11, 12 To date, five variants (c.378+1G>T, c.378+1G>A, c.378+2T>C, c.378+3A>T and c.378+5G>C) have been reported in patients with ADG and Basan syndrome. These variants were all found in splice sites of a short, skin-specific isoform of the gene.10, 11, 12
In this paper, we describe a large pedigree with Basan syndrome and reveal the causative gene through genome-wide linkage analysis and whole-genome sequencing (WGS).
SUBJECTS AND METHODS
Patients
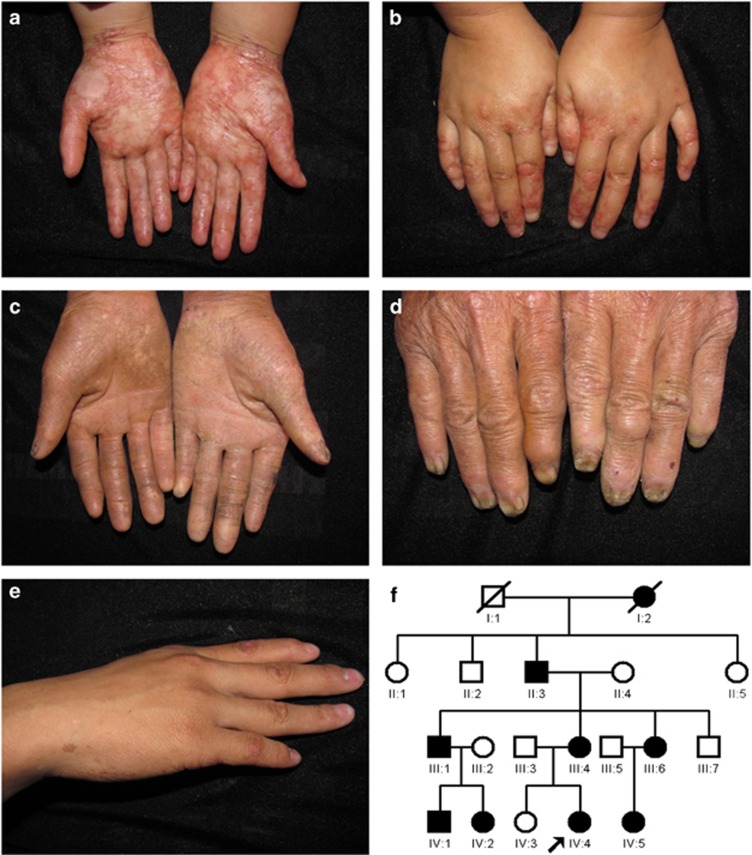
We investigated a 3-year-old girl with Basan syndrome from the Henan province of China. She had acral bullae on the fingers and soles of the feet at birth, which were completely healed by two months of age. Milia were prominent on the forehead, nose and around the mouth. Upon examination, her hands had a leather-like texture and transverse palmar creases were observed with increased fine wrinkling (Figure 1a). She had knuckle pads on the dorsal side of her hands (Figure 1b). She lacked sweating on her hands and feet. The hair, eyebrows, eyelashes, teeth and nails were all normal. Her mother and cousin were also affected, with knuckle pads on the dorsal side of their hands and hyperpigmented macules on the hands and feet (Supplementary Figures 1a and b). In total, eight individuals were affected in this family and all affected individuals presented with similar clinical manifestation (Figures 1c–e) (Table 1). The pedigree of the family is shown in Figure 1f.
Figure 1.
(a) Leather-like texture, absence of fingerprints and transverse palmar crease with increased fine wrinkling of hands (IV:4). (b) Knuckle pads on the dorsal aspect of the hands (IV:4). (c) Contractures of digits and palmoplantar keratoderma (II:3). (d) Nail dystrophy (II:3). (e) Hyperpigmented macules on the dorsal aspect of hand (III:6). (f) Pedigree of the family. The arrow denotes the proband.
Table 1. Clinical manifestations of this pedigree with Basan syndrome.
| Skin blistering | Congenital milia | Lack of fingerprints | Contractures of digits | Hypohidrosis | PPK | Nail dystrophy | Hyperpigmentation | Knuckle pads | |
|---|---|---|---|---|---|---|---|---|---|
| II:3 | + | + | + | Severe | + | Severe | + | + | Mild |
| III:1 | + | + | + | Moderate | + | Moderate | − | + | Mild |
| III:4 | + | + | + | Severe | + | − | − | + | Severe |
| III:6 | + | + | + | Mild | + | − | − | − | − |
| IV:1 | + | + | + | Mild | + | − | − | + | Severe |
| IV:2 | + | + | + | Mild | + | − | − | + | Moderate |
| IV:4 | + | + | + | Mild | + | − | − | − | Severe |
| IV:5 | + | + | + | Mild | + | − | − | − | Severe |
Abbreviation: PPK, palmoplantar keratoderma.
Genomic DNA was extracted from peripheral blood samples of the eight affected and eight unaffected family members. Informed consent was obtained from all participants and approval was obtained from the ethics committee of the Shanghai Jiaotong University School of Medicine and was conducted in accordance with the principles of the Declaration of Helsinki.
Genome-wide linkage analysis
Approximately 200 ng of genomic DNA from each of the 16 participants was genotyped using an Illumina Infinium HumanOminiZhongHua-8 whole genome array (Illumina, San Diego, CA, USA). Genome-wide linkage analysis was performed with a total of 900 015 SNP markers. SNPs with a call rate of <90% and monomorphic or non-Mendelian-transmitted markers were removed. A total of 11 152 informative autosomal SNPs within an average genetic distance of 0.25 cM were retained in the linkage analysis. Multipoint parametric linkage analysis was performed using MERLIN program version 1.1.2 software (Ann Arbor, MI, USA). A fully penetrance autosomal dominant model was used with a rare disease frequency of 0.0001. Critical recombination events of the pedigree were also determined through haplotype construction using the MERLIN software.
Exome sequencing and whole-genome sequencing
We performed exome capture using Agilent SureSelect Human All Exon Kits (Agilent, Santa Clara, CA, USA) according to the manufacturer's instructions. Sequencing was performed on a HiSeq 2000 platform with read lengths of 100 bp. The mean coverage depth for each sample was 100 ×. The sequencing reads were aligned with the NCBI human reference sequence.
For whole-genome sequencing, paired-end DNA libraries were prepared according to the manufacturer's instructions (Illumina Truseq Library Construction (Illumina). Genomic DNA (1.5 μg) was sheared into 350 bp fragments in a Covaris S220 sonicator (Woburn, MA, USA). DNA libraries were sequenced on an Illumina Hiseq X according to the manufacturer's instructions to generate paired-end 150 bp reads. Variants with a minor allele frequency (MAF) of >1% were filtered in the dbSNP databases and variants with a MAF>0.5% were filtered in the 1000 Genomes databases (1000 Genomes Project Consortium). Only SNPs occurring in exons or in canonical splice sites were further analyzed. The functional relevance of the detected variants was assessed using SIFT and PolyPhen-2programs. Variants with SIFT scores of <0.05 and PolyPhen-2 scores of >0.95 for PolyPhen-2 were regarded as functionally relevant.
Sanger sequencing
We designed primers flanking all coding exons and intron–exon boundaries of SMARCAD1 (NG_031945.1) using Premier 5.0 software (Palo Alto, CA, USA). All PCR products were purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced using an ABI PRISM 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA). The SMARCAD1 variant c.378+1G>T was submitted to the Leiden Open Variation Database (LOVD) at http://databases.lovd.nl/shared/individuals/00057917.
RESULTS
Linkage analysis
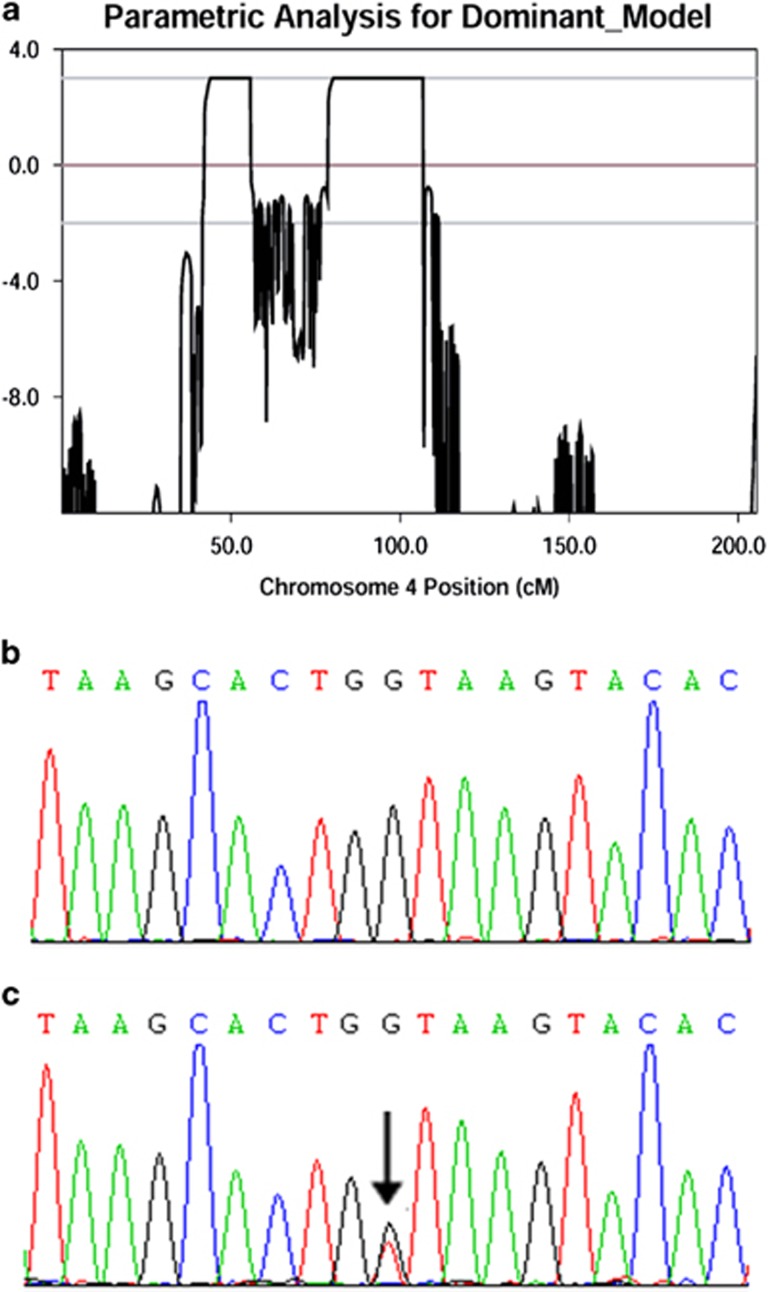
The affected patients in this pedigree had acral bullae on the fingers and soles of the feet at birth, which followed a similar manifestation as epidermolysis bullosa simplex (EBS, OMIM 131900). For this reason, we excluded mutations in the keratin 5 gene (KRT5, OMIM 148040). After confirming the exclusion of KRT5 mutations by Sanger sequencing, we performed a genome-wide linkage scan using SNP markers. LOD scores were calculated under the assumption of autosomal dominant inheritance with a penetrance of 100%. Linkage and haplotype analyses mapped the pathogenic gene in this family to two loci. One mapped region was 4p15.31-4p14, located between rs1378946 and rs1435378 (Supplementary Figure 2a). The other region was 4q13.2-4q23, located between rs6840820 and rs6818863 (Supplementary Figure 2b). Maximal LOD scores were all 3.01 (Figure 2a) (Supplementary Table 1). No other locus of suggestive linkage was detected.
Figure 2.
(a) Output of chromosome 4 multipoint LOD score analysis. (b) Normal sequence of the SMARCAD1 gene. (c) Mutation of the SMARCAD1 gene in this family. The black arrow indicates mutation.
Exome sequencing and whole-genome sequencing
We performed exome sequencing in one affected individual and found no functional variants in the mapped regions. WGS using the Illumina HiSeq XTen platform with an average coverage of 27.94 reads per base identified 3 832 626 variants (SNVs and short indels). After filtering of the identified heterozygous variants through the 1000 Genomes Project, HapMap 8 and dbSNP135 databases, two variants remained within the mapped region. These included the splice variant c.378+1G>T in a splice site of the SMARCAD1 gene (RefSeq NG_031945.1) and a duplication of GGC at position 328 330 (c.328_330dup) in exon 1 of the ANKRD17 gene (RefSeq NM_032217.4). We found that the splice variant c.378+1G>T was not covered by whole-exome sequencing.
Sanger sequencing
The splice variant in SMARCAD1 was confirmed by conventional sequencing and co-segregated with the phenotype in this family (Figures 2b and c). It was not found in any of the 100 healthy controls, suggesting it is an uncommon variant with functional relevance. The non-frameshift variant found in the ANKRD17 gene did not co-segregate with the phenotype in this family.
DISCUSSION
ADG is a rare heritable disorder, which may present as an isolated feature,3, 12 but is often characterized by additional manifestations, such as skin blisters, congenital milia, hypohidrosis, nail dystrophy, palmoplantar keratoderma and contractures of digits.1, 2, 4, 5, 10 Some complex syndromes also present similar symptoms to ADG, such as Naegeli–Franceschetti–Jadassohn syndrome, dermatopathia pigmentosa reticularis or Basan syndrome.1, 13, 14, 15 In this study, some patients presented with hyperpigmentation and knuckle pads in addition to previously reported clinical manifestations. This report has increased the phenotypic spectrum of Basan syndrome.
Only five variants in the SMARCAD1 gene have been identified in patients with isolated ADG or Basan syndrome so far (c.378+1G>T, c.378+1G>A, c.378+2T>C, c.378+3A>T and c.378+5G>C).10, 11, 12 These variants disrupt a single conserved donor splice site and are predicted to result in haploinsufficiency by reducing protein translation in affected patients.12 In this pedigree with Basan syndrome, we detected a recurrent splice variant, which has been reported in a Swiss family with isolated autosomal dominant ADG.12 Affected individuals in this family presented with rapidly healing congenital acral bullae, congenital milia, lack of fingerprints and other clinical features. This suggests that ADG and Basan syndrome may be phenotypic variants of the same disease.
Marks et al.10 reported a splice variant (c.378+3A>T) in a family with Basan syndrome and seven individuals were affected over four generations. These patients presented with congenital absence of fingerprints and a history of facial milia during the newborn period; but only three affected members had rapidly healing skin blisters, and two affected individuals had palmoplantar callosities and lacked sweating on their hands and feet. In this Chinese family, we report skin blistering, congenital milia, lack of fingerprints, contractures of digits and hypohidrosis in all eight patients; but only II:2 and III:1 presented with PPK and only II:2 had nail dystrophy. Five affected individuals (II:1, III:1, III:4, IV:1 and IV:2) presented with hyperpigmentation on the dorsal side of the hands. Excepting III:6, all patients presented with knuckle pads (Table 1). These results indicate that the same variant leads to different phenotypes even within the same family, suggesting that environmental factors such as sunlight, pressure, ethnic differences, modifier genes or polymorphic sequences of SMARCAD1 may play an important role. Further investigation is required to elucidate the pathological mechanisms leading to the phenotypes associated with the skin-specific isoform of SMARCAD1 in this family.
In conclusion, we have reported a splice variant in the skin-specific isoform of the SMARCAD1 gene that is associated with Basan syndrome. To our knowledge, this represents the second family with a splice variant that is causative for Basan syndrome. This report has advanced our understanding of the genetic basis of Basan syndrome and enlarges the phenotypic spectrum of this disease. The ongoing identification of causative variants may provide further insight into the mechanisms underlying the development of this disorder.
Acknowledgments
We thank all subjects for their ongoing participation in this study. This work was supported by a grant from the National Nature Science Foundation of China (81472867) and a grant from the Research Fund for the Doctoral Program of Higher Education (20130073120014).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Basan M: Ectodermal dysplasia. Missing papillary pattern, nail disorders and furrows on 4 fingers. Arch Klin Exp Dermatol 1965; 222: 546–557. [PubMed] [Google Scholar]
- Gagey-Caron V, Stalder JF, Barbarot S: Basan's syndrome: Congenital absence of dermatoglyphs and milia. Ann Dermatol Venereol 2009; 136: 419–421. [DOI] [PubMed] [Google Scholar]
- Burger B, Fuchs D, Sprecher E, Itin P: The immigration delay disease: adermatoglyphia-inherited absence of epidermal ridges. J Am Acad Dermatol 2011; 64: 974–980. [DOI] [PubMed] [Google Scholar]
- Baird HW 3rd: Kindred showing congenital absence of the dermal ridges (fingerprints) and associated anomalies. J Pediatr 1964; 64: 621–631. [DOI] [PubMed] [Google Scholar]
- Límová M, Blacker KL, LeBoit PE: Congenital absence of dermatoglyphs. J Am Acad Dermatol 1993; 29: 355–358. [DOI] [PubMed] [Google Scholar]
- Luna PC, Larralde M: Profuse congenital familial milia with absent dermatoglyphics (Basan's syndrome): description of a new family. Pediatr Dermatol 2012; 29: 527–529. [DOI] [PubMed] [Google Scholar]
- Reed T, Schreiner RL: Absence of dermal ridge patterns: genetic heterogeneity. Am J Med Genet 1983; 16: 81–88. [DOI] [PubMed] [Google Scholar]
- Rutter KJ, Judge MR: Profuse congenital milia in a family. Pediatr Dermatol 2009; 26: 62–64. [DOI] [PubMed] [Google Scholar]
- Rowbotham SP, Barki L, Neves-Costa A et al: Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell 2011; 42: 285–296. [DOI] [PubMed] [Google Scholar]
- Marks KC, Banks WR 3rd, Cunningham D, Witman PM, Herman GE: Analysis of two candidate genes for Basan syndrome. Am J Med Genet A 2014; 164A: 1188–1191. [DOI] [PubMed] [Google Scholar]
- Nousbeck J, Sarig O, Magal L et al: Mutations in SMARCAD1 cause autosomal dominant adermatoglyphia and perturb the expression of epidermal differentiation-associated genes. Br J Dermatol 2014; 171: 1521–1524. [DOI] [PubMed] [Google Scholar]
- Nousbeck J, Burger B, Fuchs-Telem D et al: A mutation in a skin-specific isoform of SMARCAD1 causes autosomal-dominant adermatoglyphia. Am J Hum Genet 2011; 89: 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugassy J, Itin P, Ishida-Yamamoto A et al: Naegeli-Franceschetti-Jadassohn syndrome and dermatopathia pigmentosa reticularis: two allelic ectodermal dysplasias caused by dominant mutations in KRT14. Am J Hum Genet 2006; 79: 724–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimer WL 2nd, Brauner G, James WD: Dermatopathia pigmentosa reticularis: a report of a family demonstrating autosomal dominant inheritance. J Am Acad Dermatol 1992; 26: 298–301. [DOI] [PubMed] [Google Scholar]
- van Steensel MA, Lemmink HH: A missense mutation in KRT14 causing a dermatopathia pigmentosa reticularis/Naegeli-Franceschetti-Jadassohn phenotype. J Eur Acad Dermatol Venereol 2010; 24: 1116–1117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.