Abstract
The analytical aspects of measuring hydrogen exchange by mass spectrometry are reviewed. The nature of analytical selectivity in hydrogen exchange is described followed by review of the analytical tools required to accomplish fragmentation, separation, and the mass spectrometry measurements under restrictive exchange quench conditions. In contrast to analytical quantitation that relies on measurements of peak intensity or area, quantitation in hydrogen exchange mass spectrometry depends on measuring a mass change with respect to an undeuterated or deuterated control, resulting in a value between zero and the maximum amount of deuterium that could be incorporated. Reliable quantitation is a function of experimental fidelity and to achieve high measurement reproducibility, a large number of experimental variables must be controlled during sample preparation and analysis. The method also reports on important qualitative aspects of the sample, including conformational heterogeneity and population dynamics.
Keywords: Deuterium, fragment separation method, protein conformation, protein dynamics, biopharmaceutical
1. Introduction
The following review is meant to encompass the principle analytical aspects of hydrogen exchange (HX) mass spectrometry (MS). All readers are highly encouraged to familiarize themselves with the early descriptions of the fragment separation method (1–5) wherein large peptides or proteins were labeled with an isotope of hydrogen, enzymatic digestion fragmented the labeled protein into smaller pieces, chromatographic separation was used to resolve the pieces, and some sort of isotope-selective detection was used to quantify the labeling. These studies in the late 1970s and early 1980s formed the foundation for the integrated method of HX MS. After MS began to be used for the analysis of large peptides and whole proteins, the first MS measurements of deuterium incorporation into proteins were made (6–8). Two subsequent reports (9, 10) described the integration of direct MS detection into the fragment separation method that was described in 1979. Many of the analytical challenges that were already anticipated and apparent in these early experiments of 1979 have been addressed in the years since 1993. These analytical aspects are the primary focus of this article.
The theory and practice of HX MS has been reviewed many times. The reader is directed to the many reviews from the past 10 years as a starting point for familiarization with this topic [e.g., (11–26)]. Several historical accounts of the early days of HX measurements, not limited to detection by mass spectrometry, are also highly recommended (27–31). In light of these many other descriptions, the details of the actual hydrogen exchange reaction, the mechanisms of exchange, and fundamental principles that govern how protein structure is connected to exchange will not be covered in great detail in this article. While various chemical moieties exchange with various rates, the backbone amide hydrogen position is the one that is used the most for studies of proteins and protein conformation (9, 32–35). The location of the label within the protein for all subsequent discussions is the hydrogen located at backbone amide positions. Several other theoretical aspects, including the restraints placed on analysis that are intimately connected to the analytical method, must be made clear in order that the justification for the analytical approach is obvious. Such reviews of theory will be accomplished in turn, with each section that follows.
2. Selectivity in measuring hydrogen exchange
In hydrogen exchange, hydrogen in the protein is exchanged for an isotope of hydrogen in the solvent (liquid or gas) (34–40). In some experiments, these parameters are reversed and the protein starts out heavily labeled (e.g., deuterated) and hydrogen from solvent replaces deuterium in the protein. As with any analytical measurement, there must be selectivity – the ability to detect/distinguish a given analyte in a complex mixture without interference from other components in the mixture/matrix – to distinguish deuterium from hydrogen, or distinguish tritium from hydrogen. The matrix signal may originate from other hydrogens in the protein molecule, hydrogen in the solvents, or hydrogens in other components of the solution (buffer molecules, etc.) (1). For example, a peptide of 10 residues that has 30% of its backbone amide hydrogens replaced with deuterium will have a background signal of 70% from the backbone amide hydrogens that are not deuterated. As long as the selectivity is high enough, these two isotopes of hydrogen and their respective positions in the peptide can be distinguished. Selectivity in detecting hydrogen exchange can be afforded by exploiting any chemical property that is unique to the three isotopes of hydrogen, as summarized in [TABLE 1].
TABLE 1.
Distinguishing chemical properties of the isotopes of hydrogen used to detect hydrogen exchange
| Chemical property | Details | Detection Method | Example Refs. |
|---|---|---|---|
| Density | Hd = 1.000 g mL−1 Dd = 1.106 g mL−1 Td = 1.215 g mL−1 |
Density gradient tube | (36, 38) |
| Radioactivity | H,D half-life = stable Thalf-life = 12.3 yr |
Scintillation counting | (1, 2, 5, 42–44) |
| Absorption | Blue shift upon deuteration, measure at 220–230 nm | Ultraviolet spectroscopy | (47) |
| Absorption | e.g., downshift of amide II band from ~1550 cm−1 to 1450 cm−1 upon deuteration | Infrared spectroscopy | (39, 48, 49) |
| Nuclear spin/Magnetic resonance | D silent in 1H NMR; H spin = ½ D spin = 1 | NMR spectroscopy | (51–55) |
| Nuclear scattering | Scattering amplitudes of H and D of are opposite sign | Neutron diffraction | (50) |
| Mass | Mass (natural abundance) H=1.0078 Da (99.9885%) D=2.0141 Da (0.0115%) T=3.0160 Da (trace) |
Mass spectrometry | (6, 9, 10, 58–60) |
The selectivity of mass spectrometry can be classified as “moderate” for typical fragment separation methods and “high” in single-residue experiments using MS/MS, as described further below. Other detection methods are exquisitely selective – such as nuclear magnetic resonance (NMR), for example, where signal from deuterium at a single backbone amide hydrogen can be observed in a field of hundreds of other hydrogens. Much older methods, such as density measurements or spectrophotometric methods (UV and FTIR) [TABLE 1], do not have such selectivity and therefore much larger amounts of change (more deuteration) must be realized before reliable detection is possible. With low selectivity methods, the location of deuterium to single amino acids is not possible. Note also that single molecules are never detected (see Section 4.2 below on population measurements).
Historically speaking, density was the first selectivity property exploited to measure exchange, as described by Linderstrøm-Lang and colleagues (36, 38, 41). Scintillation counting (42–44) was an obvious way to measure exchange as tritium is radioactive. The fragment separation method of Rosa/Richards (1) used tritium labeling and scintillation counting as did subsequent studies using fragment separation (2, 5). With the development of other more modern analytical tools, the use of radioactivity as the selectivity agent for exchange measurements decreased. The isotope effects with tritium are larger than those with deuterium (44–46) and the complications (including associated costs) of production and disposal of the radioactive isotope, particularly in modern times, greatly reduced the use of tritium as a labeling agent. It was shown than deuteration can be monitored by far UV absorption (47), absorption in the infrared (39, 48, 49) or with neutron diffraction (50). Many examples of exploiting the magnetic resonance properties of the hydrogen isotopes to provide selectivity exist [e.g., (51–55)]. While very selective, the time required to take an NMR measurement, unless quenching methods are used (56, 57), and size restrictions and concentration requirements generally preclude NMR as a method for analysis of large and complicated protein systems. Each amide hydrogen in HX NMR must be assigned and spectral overlap in large proteins can reduce the selectivity for individual amide hydrogens and thereby compromise the spatial resolution.
Selectivity in mass spectrometry measurements of HX arises from the mass differences between the isotopes, the ability to measure the mass of peptides to very high accuracy, and the ability with MS/MS to locate individual atoms in molecules. Early mass spectrometry measurements of isotope exchange were done indirectly using the Linderstrøm-Lang method (37, 38) followed by MS of the solvent. Deuterated protein was incubated in pure H2O, deuterium from the protein exchanged back out into the H2O, small samples were taken of this H2O solution after various amount of exchange time and converted to a gas by reduction of the water in the presence of a platinum-zinc mixture (58, 59). MS detection of the isotope ratio was done with a Thomson-Houston THN 202 mass spectrometer. This HX MS indirectly measured the deuterium level on proteins and it was not until much later that deuterated molecules were directly introduced into the mass spectrometer, by both fast-atom bombardment (9, 60, 61) and then by electrospray (6, 7, 10). It was clear from the first direct measurements that the mass difference afforded by the isotopes supplies sufficient selectivity to distinguish deuterated versus undeuterated species. What was also obvious was that the selectivity was not high enough to distinguish which amide hydrogens were labeled in species that had more than one possible position. It was proposed (6, 7) that MS/MS or digestion were needed to localize the deuterium. Much work has been done in the last ~25 years to address this issue, the so-called spatial resolution problem. While adaptation of the Rosa and Richards fragmentation method (1) to mass spectrometry detection (9) increased spatial resolution from whole proteins to smaller peptides, there has been a strong desire to increase resolution even more. This topic will be addressed in Section 3 below.
The fragment separation method relies on a chromatographic step to separate the fragments for individual interrogation by the detection method, a mass spectrometer in the case of HX MS implementation. MS, and in particular electrospray, is ideally suited for a chromatographic step prior to ionization and thus detection of exchange by MS seemed destined or predetermined to be a method even before MS was used to do it. However, many analytical challenges are introduced by the inclusion of chromatography prior to MS. While there are some challenges in the analysis that arise from the mass spectrometer itself (e.g., ionization temperature, mass accuracy, etc.), the majority of problems come from all the steps that precede the actual MS measurement. To address these challenges, a review of the components of a typical fragmentation separation method HX MS experiment is needed.
3. Analytical tools required
What is required in any hydrogen exchange method is to use the selectivity of the analytical method at hand to measure which positions have become deuterated, and at what rate. With mass spectrometry, this means measuring a mass change upon deuteration. Measuring a mass change in HX sounds much more straightforward than it is – it is actually quite an analytical challenge. The major reason for the challenge when using MS as the detection system is that the label itself is labile (37, 39). As rapidly as deuterium exchanges into a protein placed into a 100% D environment, it can exchange back out of the protein if placed into a 100% H environment [FIGURE 1]. While the labeling aspects are simplified by the spontaneity of the labeling reaction, i.e. it occurs naturally in water and physiological pH, this spontaneity also causes major analytical problems in the liquid chromatography (LC) and MS steps. If one wishes to measure the instantaneous level of deuterium in a protein, one must prevent changes to the amount of label during the analysis (1, 5). The label must not disappear during the analysis (back-exchange) nor must more labeling happen during analysis (forward-exchange). In addition to the reviews of HX MS mentioned in Section 1 above, reviews and descriptions of back- and forward-exchange can be found in Refs. (6, 9, 11, 62–64). To deal with the labile nature of hydrogen exchange, the two choices are to not allow back- or forward-exchange at all [FIGURE 1, point 1], or to limit it greatly by “quenching” [FIGURE 1, point 2].
Figure 1.
Hydrogen exchange is spontaneous. In this diagram, hydrogen or deuterium at backbone amide positions are represented by blue or red balls, respectively. When a protein with all hydrogen is placed in a solvent with a different isotope (here at left, D2O), exchange occurs spontaneously. If a partially deuterated protein were to suddenly find itself in a pure H2O solvent, the deuterium in the protein exchanges back to the solution (right). Options for measuring the amount of labeling include 1) measure deuteration before the protein is placed in pure H2O, or (2) slow the labeling (with quench conditions) to limit-the back-exchange in H2O and allow time to make the measurement.
To not allow any back- and forward-exchange, the liquid water solvent must be rapidly removed at the exact moment desired (the labeling time-point) by either going into organic solvent (either pure or high percentage) where there is no/reduced solvent hydrogen that could exchange with the protein sample (65) or by going immediately into the gas phase (66) where there is a greatly reduced local concentration of hydrogen that could exchange. The problem with both of these strategies is that proteins are not necessarily happy in organic solvent or in the gas phase and steps may be required after labeling – such as digestion – which must occur in liquid water. Additionally, some back-exchange is desirable, almost essential for data interpretation, such as that of deuterium incorporated into various side chains (9). A primary alternative is to slow the exchange reaction with conditions that limit the back- and forward-exchange as much as possible, that is, to quench. This quenching step is not modern, and has been used since the very beginning in HX with scintillation counting or UV-Vis measurement (1, 5), HX NMR (56, 57), and HX MS (9, 10).
The theory of quenching is essential to understand. Because the exchange reaction itself is both acid and base catalyzed, with the cross-over point between the two regimes occurring around pH 2.5–3.0, one can label at pH 7.0–8.0 where there are high rates of exchange and then slow the exchange orders of magnitude by adjusting to acidic pH (44, 65, 67–69). In the pH range 2.5–3.0, the intrinsic rate of exchange is the slowest for the average backbone amide hydrogen, with variability in intrinsic exchange introduced by the local environment, including the sequence (70–73). Reduction in temperature also slows the exchange (20, 44, 73). From the first reports of the fragment separation method (1–5), therefore, pH 2.5 and a temperature of 0 °C were used to slow (i.e., quench) exchange for analysis. While a pH of 2.5 is optimal for analysis, temperatures lower than 0 °C provide even more reduction in exchange [e.g., recent examples (74, 75)].
The constraints of HX quench conditions introduce the majority of the analytical challenges to the HX MS method. Bearing quench conditions in mind, the next several sections will address three areas of analytical technology needed to make the measurements, how each is influenced by the restraints of quench conditions, and examples of how some constraints can be overcome. The sections are: fragment generation, separation, and MS analysis.
3.1 Fragment generation
The fragments generated in the fragment separation method must be created under quench conditions. For digestion with a protease, there must be enzymatic activity under quench conditions, meaning pH 2.5 and low temperature. There are a limited number of enzymes that fulfill these requirements, including porcine pepsin [EC 3.4.23.1] (1, 2, 5, 9), aspergillopepsin [protease type XIII, EC 3.4.23.18] (76–78), rhizopuspepsin [protease type XVIII, EC 3.4.23.21] (1, 76, 78–80), plasmepsin [EC 3.4.23.39] (81), aspartic proteases from the Antarctic rock cod (82) or the rice field eel (Monopterus albus Zuiew) (83), and nepenthesin [EC 3.4.23.12] from plants (84, 85). Digestion with immobilized enzymes, also first described by Rosa and Richards in 1979 (1), has been described multiple times [e.g., (85–89)]. Immobilized enzymes that are packed into columns for online digestion are preferable to digestion with enzyme in solution because the relative enzyme to substrate ratio can be much higher with immobilized material and no free enzyme is introduced into the subsequent separation and MS steps, further enhancing selectivity by eliminating background signals that are not of interest.
Pepsin is the most commonly used HX MS enzyme and as a result, the most characterized. It is known that some care must be taken with pepsin, which while very active in acid is irreversibly inactivated should the pH rise above pH 5 (90, 91). Pepsin has some preference for what sequences it will cleave (92–94), but the molten globule conformation of a protein in acid may significantly contribute to the digestion pattern observed, more so than the amino acids on either side of the cleavage point. The addition of denaturants (e.g., guanidine hydrochloride, urea) and reducing agents (e.g., TCEP, DTT) can improve digestion (7, 95–99) by changing the conformation in acid to one more favorable to the protease. While digestion is unpredictable based only on sequence, digestion reproducibility can be very high given identical experimental conditions and a group of very reproducible peptides emerges when the same protein is digested many times (83). Peptic peptides are not necessarily ideal for electrospray and may exhibit a wide variety of intensities with less than ideal sequences for good ionization and multiple charging (83).
The original report of the fragment separation method (1) described the idea of increasing the spatial resolution of the exchange data by the analysis of smaller pieces, and that single-residue resolution could be possible if there were enough pieces that overlapped. They also exploited the use of multiple enzymes, that being pepsin and the acid protease from Rhizopus chinensis. Other reports over the years described using overlapping fragments to improve spatial resolution [e.g. (63, 77, 100, 101)] and a recent renaissance (102–104) has revived the idea but with the use of much improved computational and analytical tools. Using overlapping fragments is not without problems (105) so caution must be exercised.
3.2 Separation
Quench-conditions – aimed at retaining as much label as possible – restrict separations to high speed, low pH and at low temperature. High chromatographic speed degrades chromatographic performance because there is not enough time for sufficient equilibration between mobile and stationary phases. Low temperature results in poor mass transfer, further degrading chromatographic performance. Luckily, peptide separations can be accomplished well at low pH so there is at least one LC/MS variable in favor of HX MS.
For mass spectrometry analysis of chromatographic effluent, the most ideal scenario (i.e., high peak capacity case), is when the chromatography sequentially presents each peptide to the electrospray source for ionization and detection. Poor-efficiency separation (i.e., low peak capacity case) deviates from this ideality and presents multiple peptides to the source simultaneously. The resulting mass spectra can become quite complex, especially in the case of large proteins, and valuable data can be lost if too much overlap occurs. Conventional HPLC peptide chromatography has greatly advanced from what it was in the 1980s (5). At present, proteins smaller than ~30–40 kDa generally present few problems during traditional HPLC separation (1×50mm C18 column, 3.5 μm particles) with gradients under 10 minutes because there are not that many peptides to be separated which are not also resolved from one another in the mass spectrometer. When larger more complicated systems are analyzed, sometimes presenting thousands of amino acids of unique sequence and therefore hundreds of peptides, co-elution becomes a major issue; species overlap in the mass spectra, thereby inhibiting data analysis and interpretation. When many species co-elute, ion suppression effects can reduce the MS signal of some species. The dynamic range of the mass spectrometer can then become an issue when very strongly responding species are present along with verily poorly responding species [examples shown in (83)].
In many separation scenarios, the complexity of the digestion which must be separated relates to unique sequence. For example, in HX MS analysis of large viral capsids such as brome mosaic virus (106) or the P22 capsid (107), or the chaperonins GroEL/ES (108), there are many identical copies of each subunit. Upon digestion, the amount of unique sequence is low because each identical monomer produces identical peptides (e.g. for brome mosaic virus, the total size of the assembled capsid is 3.6 mega Daltons but the unique sequence of the monomer, from which all peptides are drawn, is only 20 kilo Daltons). Therefore, chromatography is only challenged with separating a small number of unique peptides and is relatively trivial. In contrast, much more complex systems with large amounts of unique sequence [e.g. (109, 110)] present chromatographic challenges wherein many hundreds of unique peptides must often be separated with the same peak capacity used for much simpler digestions.
Another separations complication is analysis of proteins present at low concentration relative to undesirable proteins present in vast excess, such as studies of the folding substrates of GroEL/ES (111, 112). In the case of the later, dynamic range considerations prevent easy analysis of the comparably weak desired signals of the substrate protein in the presence of the very strong, undesirable signals of the chaperonins. In the analysis of membrane proteins, lipids, detergents or other components of the lipid mimetics may also compromise separation efficiency and MS response. These matrix components must be removed or isolated to enable optimal peptide signals, as has been reported using several strategies (113, 114).
One solution to overwhelmed peak capacity, matrix effects, dynamic range issues, and the overall complexity of digesting a lot of unique sequence is to limit the number of species being digested and therefore introduced to the separation and mass spectrometer. For analysis of protein complexes, this could mean only digesting the members of the complex for which HX information is desired while the members that are not of interest are discarded. Accomplishing rapid chromatographic separation of proteins (for example using HPLC, SEC, etc.) to remove the undesired proteins under quench conditions is challenging [e.g., (111)]. Other more attractive strategies include: 1) immobilization of one member of the complex prior to labeling and digestion or 2) affinity capture after quench. An example of immobilization methods is the use of biotin:streptavidin, as initially reported (115) and more recently described [e.g., (116–118)]. While the second approach, affinity capture/extraction, is an ideal method to fish out what is desired from what is not desired, most affinity associations either do not hold together in low pH quench conditions or the kinetics of association are such that they are incompatible with the HX MS workflow. More work in this area is needed.
In the absence of better separation or reduction to the complexity of the digestion, overlapping peptide signals have be deconvoluted by the mass spectrometer itself when there is high mass spectrometric resolving power [e.g., (119–123)] or with software methods [e.g., (104, 120, 124–127)]. These approaches, while valuable, do not solve the underlying issue of poor analytical performance under quench conditions. With the field of HX MS pushing towards analysis of very large and complex systems, the total peak capacity of the separation step in HX MS must be improved. Pure peaks with high signal-to-noise ratio are also desirable for other manipulations including MS/MS (see next section) – if not enough precursor ions are available, fragment ion detection will be poor.
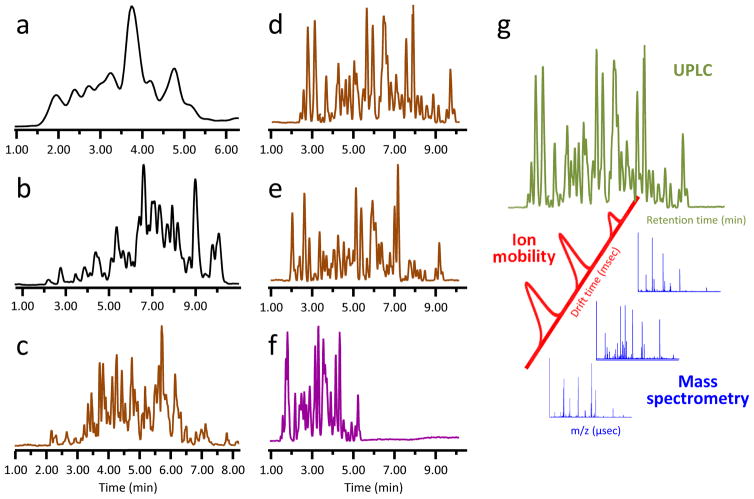
What can solve the problem of separations in quench conditions is separation methodology that has higher performance under quench conditions. To improve mass transfer at low temperature, separations with larger surface area at higher pressure can be used (128). Chromatography with sub-2-micron particles is vastly superior at 0 °C to that of more conventional separations at 0 °C with particles >3.0–3.5 microns in diameter (129). However, when the particle size is smaller, backpressure is higher (near 8,000 psi at 0 °C, flow rate 40 μL/min, 1×100mm column, 1.7 μm diameter particles) and pumps that can withstand higher pressure must be used. In 2004, the Waters Corporation commercialized (trademark UPLC) such pumps, along with columns that use 1.7 micron particles (130). There are now many instrument vendors that provide commercial systems compatible with small diameter separation particles and higher backpressure. UPLC separation for HX MS was first described in 2006 (129), with additional reports and improvements in the years following [e.g., (109, 131–133)]. Examples comparing separation under quench conditions for various particle diameters are shown in [FIGURE 2]. An added benefit of using smaller particles is that the gradient time can be made shorter without compromising separation quality, resulting in higher deuterium recovery (129, 132), or the flow rate can be increased to further enhance chromatographic efficiency if pumps capable of higher backpressure are utilized.
Figure 2.
Improving HX MS chromatographic separations at 0 °C. Panels (a–c) are taken from Ref. (132), with permission, and correspond to separation of a peptic digestion of a 52 kD protein with 20, 3.5, and 1.7 μm diameter particles, respectively. Panels (d–g) are separations of peptic digestion of monoclonal antibody (IgG) using a 1×50 mm HSS T3 1.8 μm column and a 5–35% water:acetonitrile gradient with changes to gradient time, flow rate, and backpressure: (d) 12 minute separation, 65 μL/min., backpressure 8000 psi; (e) 12 minute separation, 100 μL/min., backpressure 12000 psi; (f) 6 minute separation, 100 μL/min., backpressure 12000 psi, with addition of ion mobility separation. Separation with >2 μm particles (black traces, panels a,b) is inferior to separations with sub-2 μm particles (brown traces, panels c,d,e). Separation with both chromatography and ion mobility (purple trace, panel f) greatly enhances peak capacity. (g) Ion mobility separations require milliseconds and fit nicely between the time scale of liquid chromatography (minutes) and that of time-of-flight MS detection (microseconds).
Another way to improve peak capacity is to incorporate orthogonal separations, or add other separation modalities to the reversed-phase LC separation. However, the addition of more separation(s) must not come at the expense of more separation time and more deuterium loss. Including ion mobility spectrometry (IMS) increases the overall resolving power of LC/MS [e.g., (134–136)] by adding an additional separation step based on collisional cross section and charge, but does so without any increase in the analysis time. IMS separation, which happens in milliseconds, fits nicely between LC separation where peaks are seconds wide and TOF MS where each TOF pulse is microseconds [FIGURE 2g]. IMS and its uses for HX MS has been explored (137, 138). New developments in IMS have led to great improvements in the total peak capacity and chromatographic efficiency possible in HX MS. An example of using it in HX MS is in the analysis of a sample where higher deuterium recovery is obtained by significantly shortening the gradient program [FIGURE 2e,f]; overlapping species in the shorter LC step are resolved in the IMS step and there is no compromise to the quality of the MS data that are recorded. Future developments that include other types of separation modalities could also improve peak capacity.
3.3 MS analysis
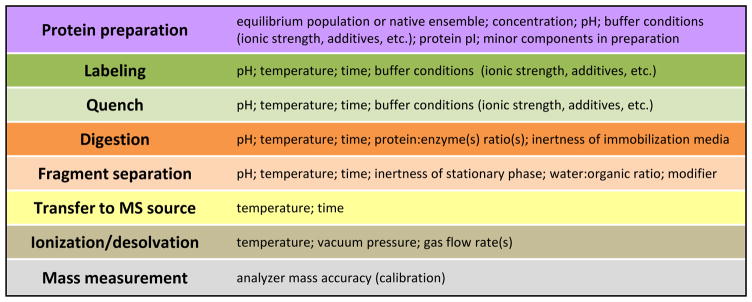
There are many mass spectrometers capable of making the measurement of mass increase as a result of deuteration. A survey of the HX MS literature over the past 20 years shows that instruments from all vendors and in many configurations have been used. The mass spectrometry, the final step in the HX MS workflow [FIGURE 3] is the most straightforward of all steps overall (see also Section 4.1). A number of new features of mass spectrometers make the measurements easier, less time-consuming, and raise the upper limit of sample complexity that can be analyzed.
Figure 3.
Variables in HX MS. Each step of the experiment from earliest (top) to latest (bottom) is in a different colored box with the variables associated with that step indicated to the right.
Good measurements of deuterium incorporation come from instruments with a number of characteristics that are now commonplace. Relatively cool ionization sources with good desolvation are desirable as less deuterium is lost during the ionization process. The source conditions need to be set for low in-source fragmentation to prevent identification of “peptides” that were not, in fact, produced by enzymatic digestion. An instrument should have good ion transmission and high duty cycle resulting in low limits of detection. As there is a wide range of ion intensity for peptic fragments (see Sect 3.2 above), detectors with high dynamic range provide the best results and most reliable, total sequence coverage. A mass analyzer with high mass accuracy, and good overall calibration stability (for example, with lock mass calibration methods) is desired so there is no drift in measured m/z during a long day or more of data acquisition. Since the centroid m/z value of an isotope distribution is generally the final outcome (9, 61, 139), not the pattern of the isotopes, even a single quadrupole mass analyzer will work if that is all that is available. However, it is certainly beneficial to have high (20,000 or more) mass resolution, if available, especially for charge state assignment and peptide identification.
Where the other bells and whistles of modern peptide and protein mass spectrometry come in handy is in integrated ion-mobility for increasing peak capacity (Section 3.2), peptide identification using data independent fragmentation methods (140), and the push toward single amino acid resolution via fragmentation by electron transfer/capture dissociation (ETD/ECD) wherein deuterium scrambling is greatly reduced, as described by Rand, Jorgensen and colleagues [reviewed in (141)] and others [e.g., (142–145)]. ETD/ECD fragmentation may be of whole, intact proteins just deuterated (top-down) or of deuterated peptides produced by enzymatic digestion (middle-down). Top-down methods [e.g., (75, 146, 147)], where deuterium quantity is measured in fragments of the protein produced within the mass spectrometer, offer the possibility of single amino acid resolution without protein digestion and chromatography (and the resulting deuterium losses that accompany both), but are not without complications. Incomplete fragmentation, poor ion statistics (i.e., weak signals hard to detect), hydrogen scrambling, difficulty in isolating single peptide species in crowded fields of ions (isolation window too wide), and the potential for retaining deuterium at side-chain positions are all issues that must be resolved prior to such methods becoming mainstream.
4. Quantitation of deuterium
Quantitation in HX MS is distinct from most types of MS quantitation experiments where quantity is related to peak height, peak area, or intensity with respect to an internal standard. Rather, the quantitation in HX MS arises from mass change with respect to an undeuterated (or totally deuterated) control. The amount of mass change that is measured can report on protein conformation and dynamics, i.e. more exchangeable regions unprotected from exchange will have a higher deuterium level and so forth. However, a mass change can also arise from changes to the analysis conditions (6, 9, 61). FIGURE 4a describes an example scenario. If, for example, pH was improperly controlled and was 0.25 pH units too high in one sample versus in another, the m/z measured would be higher (FIGURE 4a, red spectrum) than the properly-controlled-pH sample [FIGURE 4a, green spectrum]. An analogous situation from other types of MS quantitation would be if a random error from improper handling of samples during extraction, for example in small molecule quantitation with GC/MS, led to lower than appropriate peak height with respect to a standard, a concentration value for the small molecule could be reported as lower than it really was. For HX MS methodology errors based on altered conditions that change the measured m/z, if the experimenter was unaware of the altered conditions, they may falsely attribute the higher mass (red spectrum versus green spectrum, FIGURE 4a) to a feature of protein conformation, binding, etc., effectively fooling the experimenter into thinking changes originating from poor experimental control are real. As it turns out, most all of the challenges in MS measurements of deuterium levels come not from the mass spectrometer, but from the steps prior to the mass analysis. Section 4.1 on reproducibility addresses some of these issues. Quantitation also depends on the shape of the isotope distribution, which reports on the population of molecules during labeling, as discussed in Section 4.2
Figure 4.
Quantitation of deuterium by mass change. (a). An undeuterated spectrum (black) is compared to deuterated spectra where the increase in mass may be small (grey spectrum), intermediate (green spectrum) or large (red spectrum). Improper control of experimental conditions could produce an isotope cluster with an average m/z higher or lower than would be found with proper control of conditions, in which case changes could be falsely attributed to changes in the protein that were in fact due to undesirable changes in experimental conditions. (b) in EX1 kinetics, there can be a broadened isotope cluster which may resolve to two distributions (blue spectra): a higher-mass envelope representing a more unfolded/unprotected species and a lower-mass envelope representing a more folded/protected species. Quantitation in EX1 kinetics can be done by finding the centroid of each distribution (gray or orange bars) or the centroid of the entire distribution (purple bar).
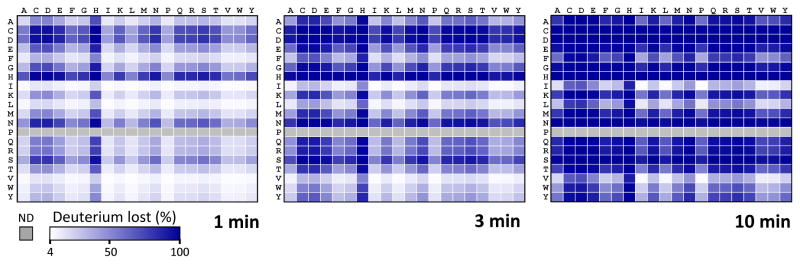
The range of quantitation for forward exchange (deuterium replacing hydrogen) in each peptide extends from zero (undeuterated reference) to the theoretical maximum amount of deuterium that could possibly be incorporated if every backbone amide hydrogen of every amino acid were deuterated (totally deuterated reference). When there is back-exchange during analysis, it is nearly impossible to observe the totally deuterated form. The theoretical maximum takes into account that proline has no backbone amide hydrogen and therefore does not contribute to the maximum value. The other major consideration is the N-terminal end of each peptide. Digesting a protein into peptides creates an unprotected amide on the N-terminus of each peptide (72, 73). This N-terminal amide hydrogen has a very fast exchange rate and invariably exchanges with the solvent soon after it is created during enzymatic cleavage. During LC/MS in H2O, after labeling in D2O, this N-terminal position will quickly lose any deuterium it may have incorporated making it appropriate to subtract this position from the theoretical number of available backbone amide hydrogens that could exist in a particular peptide (1, 9). Subtracting also the penultimate, or second residue, of each peptide from consideration is also sometimes warranted because of the potential for fast back-exchange and loss of label in this position. As various sequence combinations lead to different amounts of back-exchange, due to primary structure effects, the Bai/Molday factors (72, 73) can be used to calculate the exchange rate constants for all 400 hundred sequence combinations of the first two amino acids of peptides [FIGURE 5]. Sequences involving certain pairs of Ile, Val, Ala, Tyr, Trp and Leu mostly retain deuterium at the second position after even 10 minutes in H2O quench conditions while pairs involving certain combinations of Asp, His, Cys, Asn and Gln easily lose deuterium at the second position even after 1 minute of quench conditions. A wise strategy is to consider the sequence of each peptide, known from MS identification, and apply a smart algorithm that deducts deuterium for consideration from the second position if warranted based on sequence, quench pH and temperature, and the amount of time exposed to the quench isotope (usually H2O). With modern computation methods, this strategy can be implemented automatically during data processing.
Figure 5.
Loss of deuterium incorporated at the penultimate backbone amide hydrogen position of peptides, as calculated using (73). All 400 combinations of the twenty common amino acids at the N-terminus (along the top of each panel) and penultimate position (vertically along the side) are shown. Deuterium loss after 1 min, 3 min or 10 min at pH 2.5 and 0 °C is colored using the gradient scale indicated. Note that proline has no backbone amide hydrogen (colored light grey).
4.1 Measurement reproducibility
The sources of variability in an HX MS experiment were summarized in Sect 3.3, FIGURE 3 [also recently discussed in (22)]. Each experimental parameter at each step of the experiment must be under control in order that the information that is obtained [FIGURE 4a] reports on the protein and not on the methodology. While small changes in pH, time and temperature may at first consideration seem trivial and inconsequential to the experiment, they can in fact [FIGURE 6] make changes to the measured mass that are much larger than the error of most peptide mass determinations (±0.01 Da or less). The inclusion of sample handling robotics [e.g., (121)] enhances reproducibility. Robots can assist in sample preparation, labeling, and subsequent injection into LC/MS systems.
Figure 6.
Examples of how changes to various experimental parameters affect the deuterium level of peptic peptides. Three random sequences were chosen (blue, MYSLCEQTVNFK; red, QCSVFMTNYEKL; green, IHGASDFWVWER) and the deuterium level after forward exchange, 100% H to D (a, b) or back-exchange, 100% D to H, (c, d) was calculated using (73) for various conditions. (a) labeling at 25 °C for 1 second at variable pH (x-axis). (b) Same as panel a but labeling for 2 seconds. (c) Back-exchange for 5 minutes at 0 °C with variable quench pH. (d) Back-exchange for 5 minutes at pH 2.5 with variable quench temperature. Differences in deuterium levels at specific conditions are highlighted with numbers in colored boxes.
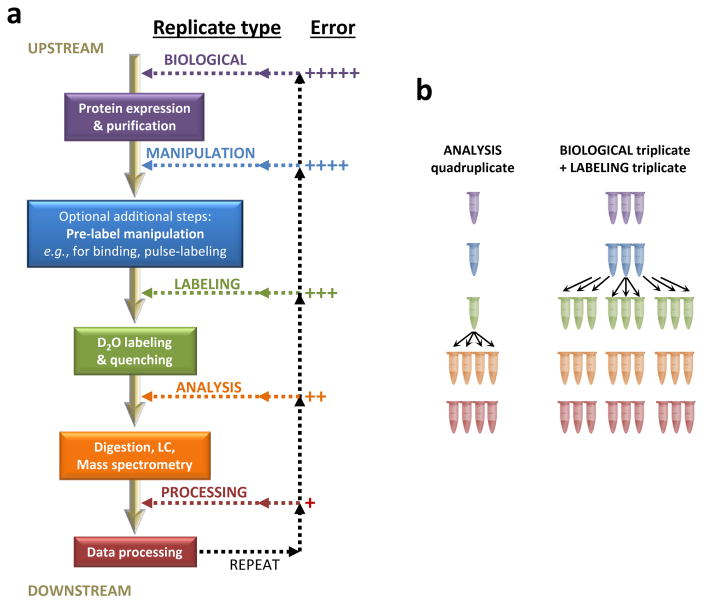
As each of the experimental parameters contributes to variability, overall global experimental reproducibility is an aggregate of these many parameters. The type of experimental replication is a key factor when describing error because it changes the number of variables that contribute to the overall reproducibility [FIGURE 7]. We here define five classes of HX MS replicates: biological, manipulation, labeling, analysis and processing. We do not wish to confuse these terms with interday or intraday variability because such variability may also be a function of the type of replicate, not just the frequency with which it was performed. For example, measuring the interday variability by taking repeated measurements over a 3 month period for a sample that was prepared once and frozen in aliquots is quite different from measuring the intraday variability of 3 samples of protein made from three distinct labelings of isolates from 3 separate cell culture pellets. The most error arises in performing biological replicates because there are more parameters that can vary [see also FIGURE 3] while the least error arises in processing replicates. Biological replicates are a much better representation of the biological system as they include all the variability that comes with recombinant protein expression and purification. Low overall error in a biological replicate has more meaning than low overall error in an analysis replicate.
Figure 7.
The five main types of HX MS experimental replication. (a) The major steps of the HX MS experiment are shown down the central spine with colored boxes. Replication can be one of five types, shown with the proposed nomenclature (colored lines and text). Some experiments (e.g., binding, pulse-labeling) require a(n) additional step(s) (manipulation, blue box and shading) just before labeling in which sample volumes, concentrations and other conditions are manipulated; simple, one-protein HX MS generally does not have a manipulation step. The final relative error for each type of replicate, as determined by the spread of the replicate data points during final data interpretation, is shown at the left (+++++ is most error, + is least error). (b) Examples of two types of replication experiments. In the analysis quadruplicate, a single sample of protein is labeled and quenched, and the quenched material is divided into four separate tubes for four independent LC/MS analyses and processing. In the other example, protein is overexpressed/isolated three independent times and then divided into three separate aliquots per biological preparation for independent labeling, analysis and processing. A total of nine replicates comprises this final data set.
Detailed measurements from several laboratories (18, 148, 149) have explored the error of HX MS experiments [reviewed in (22)]. Some of these reports describe labeling replicates or analysis replicates, which should not be confused with biological replicates that bear higher error. A valuable practice is to provide more experimental details about how replication was performed, the criteria for similarity attribution, etc. [e.g., (109, 149–152)]. Without such details, error cannot be compared between various types of experiments with different kinds of replication. An understanding of error helps gauge the significance of measured differences between states, or forms of a protein that are being labeled [e.g., see Ref. (153)]. When strict tests of statistical significance are applied, it should be done along with common sense. For example, in a protein of 100 amino acids if a mass difference of 1 Dalton was found in a peptide of 10 amino acids for a bound versus free form of the protein, all statistical tests may indicate this is a real and statistically meaningful difference in that 10 amino acid peptide. The question to be answered next is does a change in deuteration of one amide position in a protein of 100 amino acids mean something biologically significant to the protein. The answer may depend on many factors, not necessarily all of them rooted in analytical chemistry and statistics.
4.2 Qualitative aspects – sample homogeneity and conformational mixtures
Protein molecules in solution are in an equilibrium of interconverting states that are not all identical [the native state ensemble (154, 155)]. This conformational heterogeneity introduces variability in HX MS measurements because what is measured is the sum of all populations of molecules in the sample, an aggregate of each of the different co-existing conformational forms. When the mass spectra are first taken, the shape, the width, the number, and the intensity of each isotope cluster reports on the conformational heterogeneity of the sample that was labeled. An example is shown in [FIGURE 4b] where two populations are apparent: a lower-m/z population that is more protected from exchange than a higher m/z population that has become more deuterated. Often, what is extracted from isotope clusters is the center of mass, or first moment, of each isotopic distribution (9, 61, 139). Unusual shapes, isotope cluster widths and distributions can report on interesting protein dynamics, conformational heterogeneity and interconverting states (8, 156–159). Perturbing the native state ensemble (with buffer conditions, concentration, pH, and temperature) may change the average deuterium incorporation and therefore both the mass and pattern of the isotope clusters that are measured. Care must be exercised to ensure that the analytical conditions do not cause the experimenter to falsely attribute peak shape to protein conformation and dynamics, as can be the case when carryover occurs from sample to sample (160, 161).
Deuterium quantitation in situations where there is conformational heterogeneity in the labeled sample is not the same as when there is a single, homogenous conformation. Mixed populations during pulse-labeling experiments of protein folding [e.g., (8, 111, 112, 162)] or in continuous labeling of proteins with obvious EX1 kinetics [e.g., (157–159, 163, 164)] can be handled in a number of ways [FIGURE 4b]. It is best to present the spectra in such situations to illustrate the heterogeneity. The quantity of label in each individual population [FIGURE 4b, gray and orange bars] can be extracted by fitting peaks (156, 162) or the entire isotope distribution [FIGURE 4b purple bar] can be processed to produce peak width plots (158) or bubble plots (127). Particularly in the case of mixed populations displaying heterogeneous isotope distributions, attention to the experimental details of the data processing steps is crucial to ensure reproducibility and validity of the final conclusions. Biological replicates [FIGURE 7] are essential in cases of conformational heterogeneity as the ensemble of protein molecules being labeled may be very sensitive to the experimental conditions, particularly those of the initial buffer [FIGURE 3, purple].
Acknowledgments
This work is supported in part by a grant from the National Institutes of Health (R01-GM101135) and by a research collaboration with the Waters Corporation.
Footnotes
Disclosure statement
JRE is a paid consultant of the Waters Corporation which has a commercial HX MS system for sale.
Literature cited
- 1.Rosa JJ, Richards FM. An experimental procedure for increasing the structural resolution of chemical hydrogen-exchange measurements on proteins: application to ribonuclease S peptide. J Mol Biol. 1979;133:399–416. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- 2.Englander SW, Calhoun DB, Englander JJ, Kallenbach NR, Liem RK, et al. Individual breathing reactions measured in hemoglobin by hydrogen exchange methods. Biophys J. 1980;32:577–89. doi: 10.1016/S0006-3495(80)84991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosa JJ, Richards FM. Hydrogen exchange from identified regions of the S-protein component of ribonuclease as a function of temperature, pH, and the binding of S-peptide. J Mol Biol. 1981;145:835–51. doi: 10.1016/0022-2836(81)90318-1. [DOI] [PubMed] [Google Scholar]
- 4.Rosa JJ, Richards FM. Effects of binding of S-peptide and 2′-cytidine monophosphate on hydrogen exchange from the S-protein component of ribonuclease S. J Mol Biol. 1982;160:517–30. doi: 10.1016/0022-2836(82)90311-4. [DOI] [PubMed] [Google Scholar]
- 5.Englander JJ, Rogero JR, Englander SW. Protein hydrogen exchange studied by the fragment separation method. Anal Biochem. 1985;147:234–44. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katta V, Chait BT. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun Mass Spectrom. 1991;5:214–17. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 7.Katta V, Chait BT. Hydrogen/deuterium exchange electrospray ionization mass spectrometry: a method for probing protein conformational changes in solution. J Am Chem Soc. 1993;115:6317–21. [Google Scholar]
- 8.Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Detection of transient protein folding populations by mass spectrometry. Science. 1993;262:896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–31. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson RS, Walsh KA. Mass spectrometric measurement of protein amide hydrogen exchange rates of apo- and holo-myoglobin. Protein Sci. 1994;3:2411–18. doi: 10.1002/pro.5560031224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier CS, Deinzer ML. Protein conformations, interactions, and H/D exchange. Methods Enzymol. 2005;402:312–60. doi: 10.1016/S0076-6879(05)02010-0. [DOI] [PubMed] [Google Scholar]
- 12.Hamuro Y, Weber PC, Griffin PR. High-throughput analysis of protein structure by hydrogen/deuterium exchange mass spectrometry. Methods Biochem Anal. 2005;45:131–57. [PubMed] [Google Scholar]
- 13.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–70. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 14.Tsutsui Y, Wintrode PL. Hydrogen/deuterium exchange-mass spectrometry: a powerful tool for probing protein structure, dynamics and interactions. Curr Med Chem. 2007;14:2344–58. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- 15.Brier S, Engen JR. Hydrogen exchange mass spectrometry: Principles and capabilities. In: Chance M, editor. Mass spectrometry analysis for protein-protein interactions and dynamics. New York: Wiley-Blackwell; 2008. pp. 11–43. [Google Scholar]
- 16.Yan X, Maier CS. Hydrogen/deuterium exchange mass spectrometry. Methods Mol Biol. 2009;492:255–71. doi: 10.1007/978-1-59745-493-3_15. [DOI] [PubMed] [Google Scholar]
- 17.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–72. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev Proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–34. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 20.Engen JR, Wales TE, Shi X. Hydrogen Exchange Mass Spectrometry for Conformational Analysis of Proteins. In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. Wiley; 2011. [DOI] [Google Scholar]
- 21.Brock A. Fragmentation hydrogen exchange mass spectrometry: a review of methodology and applications. Protein Expr Purif. 2012;84:19–37. doi: 10.1016/j.pep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Iacob RE, Engen JR. Hydrogen exchange mass spectrometry: are we out of the quicksand? J Am Soc Mass Spectrom. 2012;23:1003–10. doi: 10.1007/s13361-012-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Percy AJ, Rey M, Burns KM, Schriemer DC. Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry-a review. Anal Chim Acta. 2012;721:7–21. doi: 10.1016/j.aca.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 24.Jaswal SS. Biological insights from hydrogen exchange mass spectrometry. Biochim Biophys Acta. 2013;1834:1188–201. doi: 10.1016/j.bbapap.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Cao J, Burke JE, Dennis EA. Using hydrogen/deuterium exchange mass spectrometry to define the specific interactions of the phospholipase A2 superfamily with lipid substrates, inhibitors, and membranes. J Biol Chem. 2013;288:1806–13. doi: 10.1074/jbc.R112.421909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei H, Mo J, Tao L, Russell RJ, Tymiak AA, et al. Hydrogen/deuterium exchange mass spectrometry for probing higher order structure of protein therapeutics: methodology and applications. Drug Discov Today. 2014;19:95–102. doi: 10.1016/j.drudis.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards FM. Linderstrom-Lang and the Carlsberg Laboratory: The view of a postdoctoral fellow in 1954. Protein Sci. 1992;1:1721–30. doi: 10.1002/pro.5560011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schellman JA, Schellman CG. Kaj Ulrick Linderstrom-Lang (1896–1959) Protein Sci. 1997;6:1092–100. doi: 10.1002/pro.5560060516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Englander SW, Mayne L, Bai Y, Sosnick TR. Hydrogen exchange: the modern legacy of Linderstrom-Lang. Protein Sci. 1997;6:1101–9. doi: 10.1002/pro.5560060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldwin RL. In memoriam: Reflections on Fred Richards (1925–2009) Protein Sci. 2009;18:682–85. [Google Scholar]
- 31.Baldwin RL. Early days of protein hydrogen exchange: 1954–1972. Proteins. 2011;79:2021–6. doi: 10.1002/prot.23039. [DOI] [PubMed] [Google Scholar]
- 32.Klotz IM, Frank BH. Deuterium--Hydrogen Exchange in Amide N--H Groups. J Am Chem Soc. 1965;87:2721–8. doi: 10.1021/ja01090a033. [DOI] [PubMed] [Google Scholar]
- 33.Klotz IM. Molecular aspects of hydrogen-deuterium exchange in macromolecules. J Colloid Interface Sci. 1968;27:804–17. doi: 10.1016/0021-9797(68)90114-8. [DOI] [PubMed] [Google Scholar]
- 34.Englander SW, Downer NW, Teitelbaum H. Hydrogen exchange. Annu Rev Biochem. 1972;41:903–24. doi: 10.1146/annurev.bi.41.070172.004351. [DOI] [PubMed] [Google Scholar]
- 35.Woodward CK, Hilton BD. Hydrogen exchange kinetics and internal motions in proteins and nucleic acids. Annu Rev Biophys Bioeng. 1979;8:99–127. doi: 10.1146/annurev.bb.08.060179.000531. [DOI] [PubMed] [Google Scholar]
- 36.Hvidt A, Linderstrom-Lang K. Exchange of hydrogen atoms in insulin with deuterium atoms in aqueous solutions. Biochim Biophys Acta. 1954;14:574–5. doi: 10.1016/0006-3002(54)90241-3. [DOI] [PubMed] [Google Scholar]
- 37.Linderstrom-Lang K. Deuterium Exchange Between Peptides and Water. Chem Soc (London) 1955:1–20. Spec. Pub. 2. [Google Scholar]
- 38.Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 39.Ottesen M. Methods for measurement of hydrogen isotope exchange in globular proteins. Methods of Biochemical Analysis. 1971;20:135–68. doi: 10.1002/9780470110393.ch5. [DOI] [PubMed] [Google Scholar]
- 40.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 41.Hvidt A, Johansen G, Linderstrom-Lang K. Laboratory manual of analytical methods of protein chemistry. 1960;2:103. [Google Scholar]
- 42.Leach SJ, Springell PH. Tritium-hydrogen exchange in studies of protein structure. Australian J Chem. 1962;15:350–64. [Google Scholar]
- 43.Englander SW. A Hydrogen Exchange Method Using Tritium and Sephadex: Its Application to Ribonuclease. Biochemistry. 1963;2:798–807. doi: 10.1021/bi00904a030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Englander SW, Poulsen A. Hydrogen-tritium exchange of the random chain polypeptide. Biopolymers. 1969;7:379–93. [Google Scholar]
- 45.Gregory RB, Carbo L, Percy AJ, Rosenberg A. Water catalysis of peptide hydrogen isotope exchange. Biochemistry. 1983;22:910–17. doi: 10.1021/bi00273a031. [DOI] [PubMed] [Google Scholar]
- 46.Connelly GP, Bai Y, Jeng M-F, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins: Struct Funct Genet. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 47.Englander JJ, Calhoun DB, Englander SW. Measurement and calibration of peptide group hydrogen-deuterium exchange by ultraviolet spectrophotometry. Anal Biochem. 1979;95:517–24. doi: 10.1016/0003-2697(79)90693-6. [DOI] [PubMed] [Google Scholar]
- 48.Blout ER, De Loze C, Asadourian A. The Deuterium Exchange of Water-soluble Polypeptides and Proteins as Measured by Infrared Spectroscopy. J Am Chem Soc. 1961;83:1895–900. [Google Scholar]
- 49.Parker FS, Bhaskar KR. Infrared Studies of Hydrogen-Deuterium Exchange in Biological Molecules. Appl Spectrosc Rev. 1970;3:91–142. [Google Scholar]
- 50.Kossiakoff AA. Protein dynamics investigated by the neutron diffraction-hydrogen exchange technique. Nature. 1982;296:713–21. doi: 10.1038/296713a0. [DOI] [PubMed] [Google Scholar]
- 51.Woodward CK, Rosenberg A. Studies of hydrogen exchange in proteins. VI. Urea effects on ribonuclease exchange kinetics leading to a general model for hydrogen exchange from folded proteins. J Biol Chem. 1971;246:4114–21. [PubMed] [Google Scholar]
- 52.Wagner G, Wuthrich K. Amide proton exchange and surface conformation of the basic pancreatic trypsin inhibitor in solution. J Mol Biol. 1982;160:343–61. doi: 10.1016/0022-2836(82)90180-2. [DOI] [PubMed] [Google Scholar]
- 53.Barksdale AD, Rosenberg A. Acquisition and interpretation of hydrogen exchange data from peptides, polymers, and proteins. Methods Biochem Anal. 1982;28:1–113. doi: 10.1002/9780470110485.ch1. [DOI] [PubMed] [Google Scholar]
- 54.Roder H, Wuthrich K. Protein folding kinetics by combined use of rapid mixing techniques and NMR observation of individual amide protons. Proteins. 1986;1:34–42. doi: 10.1002/prot.340010107. [DOI] [PubMed] [Google Scholar]
- 55.Englander SW, Mayne L. Protein folding studied using hydrogen-exchange labeling and two-dimensional NMR. Annu Rev Biophys Biomol Struct. 1992;21:243–65. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]
- 56.Paterson Y, Englander SW, Roder H. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science. 1990;249:755–59. doi: 10.1126/science.1697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang YZ, Paterson Y, Roder H. Rapid amide proton exchange rates in peptides and proteins measured by solvent quenching and two-dimensional NMR. Protein Sci. 1995;4:804–14. doi: 10.1002/pro.5560040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nonnenmacher G, Coursaget J. Utilizing deuterium in study of exchangeable hydrogen atoms of proteins and application to ribonuclease. J Chim Phys. 1969;66:616–18. [Google Scholar]
- 59.Nonnenmacher G, Viala E, Thiery JM, Calvet P. A deuterium-hydrogen exchange study of inhibitor-induced conformational changes in ribonuclease A. Eur J Biochem. 1971;21:393–9. doi: 10.1111/j.1432-1033.1971.tb01482.x. [DOI] [PubMed] [Google Scholar]
- 60.Sethi SK, Smith DL, McCloskey JA. Determination of active hydrogen content by fast atom bombardment mass spectrometry following hydrogen-deuterium exchange. Biochem Biophys Res Commun. 1983;112:126–31. doi: 10.1016/0006-291x(83)91806-5. [DOI] [PubMed] [Google Scholar]
- 61.Thévenon-Emeric G, Kozlowski J, Zhang Z, Smith DL. Determination of amide hydrogen exchange rates in peptides by mass spectrometry. Anal Chem. 1992;64:2456–8. doi: 10.1021/ac00044a027. [DOI] [PubMed] [Google Scholar]
- 62.Engen JR, Smith DL. Investigating the higher order structure of proteins. Hydrogen exchange, proteolytic fragmentation, and mass spectrometry. Methods Mol Biol. 2000;146:95–112. doi: 10.1385/1-59259-045-4:95. [DOI] [PubMed] [Google Scholar]
- 63.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 64.Hoofnagle AN, Resing KA, Ahn NG. Practical methods for deuterium exchange/mass spectrometry. Meth Mol Biol. 2004;250:283–98. doi: 10.1385/1-59259-671-1:283. [DOI] [PubMed] [Google Scholar]
- 65.Leichtling BH, Klotz IM. Catalysis of hydrogen-deuterium exchange in polypeptides. Biochemistry. 1966;5:4026–37. [Google Scholar]
- 66.Suckaw D, Shi Y, Beu SC, Senko MW, Quinn JP, et al. Coexisting stable conformations of gaseous protein ions. Proc Natl Acad Sci U S A. 1993;90:790–93. doi: 10.1073/pnas.90.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berger A, Linderstrom-Lang K. Deuterium exchange of poly-DL-alanine in aqueous solution. Arch Biochem Biophys. 1957;69:106–18. doi: 10.1016/0003-9861(57)90478-2. [DOI] [PubMed] [Google Scholar]
- 68.Hvidt A. A Discussion of the pH Dependence of the Hydrogen-Deuterium Exchange of Proteins. C R Trav Lab Carlsberg. 1964;34:299–317. [PubMed] [Google Scholar]
- 69.Eigen M. Proton transfer, acid-base catalysis, and enzymatic hydrolysis. Part I: elementary processes. Angewandte Chemie. 1964;3:1–19. [Google Scholar]
- 70.Klotz IM, Mueller DD. Local environment effects on hydrogen--deuterium exchange. Biochemistry. 1969;8:12–6. doi: 10.1021/bi00829a003. [DOI] [PubMed] [Google Scholar]
- 71.Woodward CK, Rosenberg A. Oxidized RNase as a protein model having no contribution to the hydrogen exchange rate from conformational restrictions. Proc Natl Acad Sci U S A. 1970;66:1067–74. doi: 10.1073/pnas.66.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11:150–8. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 73.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venable JD, Okach L, Agarwalla S, Brock A. Subzero temperature chromatography for reduced back-exchange and improved dynamic range in amide hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2012;84:9601–8. doi: 10.1021/ac302488h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan J, Zhang S, Parker CE, Borchers CH. Subzero Temperature Chromatography and Top-Down Mass Spectrometry for Protein Higher-Order Structure Characterization: Method Validation and Application to Therapeutic Antibodies. J Am Chem Soc. 2014 doi: 10.1021/ja507880w. in press. [DOI] [PubMed] [Google Scholar]
- 76.Cravello L, Lascoux D, Forest E. Use of different proteases working in acidic conditions to improve sequence coverage and resolution in hydrogen/deuterium exchange of large proteins. Rapid Commun Mass Spectrom. 2003;17:2387–93. doi: 10.1002/rcm.1207. [DOI] [PubMed] [Google Scholar]
- 77.Englander JJ, Del Mar C, Li W, Englander SW, Kim JS, et al. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc Natl Acad Sci U S A. 2003;100:7057–62. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang HM, Kazazic S, Schaub TM, Tipton JD, Emmett MR, Marshall AG. Enhanced Digestion Efficiency, Peptide Ionization Efficiency, and Sequence Resolution for Protein Hydrogen/Deuterium Exchange Monitored by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal Chem. 2008;80:9034–41. doi: 10.1021/ac801417d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fukumoto J, Tsuru D, Yamamoto T. Studies on Mold Proteases. Part I. Purification, crystallization and some enzymatic properties of acid protease of Rhizopus chinensis. Agr Biol Chem. 1967;31:710–17. [Google Scholar]
- 80.Rey M, Man P, Brandolin G, Forest E, Pelosi L. Recombinant immobilized rhizopuspepsin as a new tool for protein digestion in hydrogen/deuterium exchange mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:3431–8. doi: 10.1002/rcm.4260. [DOI] [PubMed] [Google Scholar]
- 81.Marcoux J, Thierry E, Vives C, Signor L, Fieschi F, Forest E. Investigating alternative acidic proteases for H/D exchange coupled to mass spectrometry: plasmepsin 2 but not plasmepsin 4 is active under quenching conditions. J Am Soc Mass Spectrom. 2010;21:76–9. doi: 10.1016/j.jasms.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 82.Brier S, Maria G, Carginale V, Capasso A, Wu Y, et al. Purification and characterization of pepsins A1 and A2 from the Antarctic rock cod Trematomus bernacchii. Febs J. 2007;274:6152–66. doi: 10.1111/j.1742-4658.2007.06136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahn J, Cao MJ, Yu YQ, Engen JR. Accessing the reproducibility and specificity of pepsin and other aspartic proteases. Biochim Biophys Acta. 2013;1834:1222–9. doi: 10.1016/j.bbapap.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rey M, Yang M, Burns KM, Yu Y, Lees-Miller SP, Schriemer DC. Nepenthesin from monkey cups for hydrogen/deuterium exchange mass spectrometry. Mol Cell Proteomics. 2013;12:464–72. doi: 10.1074/mcp.M112.025221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kadek A, Mrazek H, Halada P, Rey M, Schriemer DC, Man P. Aspartic protease nepenthesin-1 as a tool for digestion in hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2014;86:4287–94. doi: 10.1021/ac404076j. [DOI] [PubMed] [Google Scholar]
- 86.Ehring H. Hydrogen exchange/electrospray ionization mass spectrometry studies of structural features of proteins and protein/protein interactions. Anal Biochem. 1999;267:252–9. doi: 10.1006/abio.1998.3000. [DOI] [PubMed] [Google Scholar]
- 87.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–8. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 88.Busby SA, Chalmers MJ, Griffin PR. Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled columns. Int J Mass Spectrom. 2007;259:130–39. [Google Scholar]
- 89.Ahn J, Jung MC, Wyndham K, Yu YQ, Engen JR. Pepsin immobilized on high-strength hybrid particles for continuous flow online digestion at 10,000 psi. Anal Chem. 2012;84:7256–62. doi: 10.1021/ac301749h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang JL, Edelman GM. Fluorescent probes for conformational states of proteins. IV. The pepsinogen-pepsin conversion. J Biol Chem. 1971;246:1185–91. [PubMed] [Google Scholar]
- 91.Fruton JS. A history of pepsin and related enzymes. Q Rev Biol. 2002;77:127–47. doi: 10.1086/340729. [DOI] [PubMed] [Google Scholar]
- 92.Powers JC, Harley AD, Myers DV. Subsite specificity of porcine pepsin. Advances in experimental medicine and biology. 1977;95:141–57. doi: 10.1007/978-1-4757-0719-9_9. [DOI] [PubMed] [Google Scholar]
- 93.Palashoff MH. MS thesis. Northeastern University; Boston: 2008. Determining the specificity of pepsin for proteolytic degestion; p. 78. [Google Scholar]
- 94.Hamuro Y, Coales SJ, Molnar KS, Tuske SJ, Morrow JA. Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun Mass Spectrom. 2008;22:1041–6. doi: 10.1002/rcm.3467. [DOI] [PubMed] [Google Scholar]
- 95.Woods VL., Jr 6,291,189 USA Patent No. 2001
- 96.Yan X, Zhang H, Watson J, Schimerlik MI, Deinzer ML. Hydrogen/deuterium exchange and mass spectrometric analysis of a protein containing multiple disulfide bonds: Solution structure of recombinant macrophage colony stimulating factor-beta (rhM-CSFbeta) Protein Sci. 2002;11:2113–24. doi: 10.1110/ps.0204402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hamuro Y, Coales SJ, Southern MR, Nemeth-Cawley JF, Stranz DD, Griffin PR. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J Biomol Tech. 2003;14:171–82. [PMC free article] [PubMed] [Google Scholar]
- 98.Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR. Characterization of IgG1 conformation and conformational dynamics by hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2009;81:2644–51. doi: 10.1021/ac802575y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang HM, McLoughlin SM, Frausto SD, Tang H, Emmett MR, Marshall AG. Simultaneous reduction and digestion of proteins with disulfide bonds for hydrogen/deuterium exchange monitored by mass spectrometry. Anal Chem. 2010;82:1450–4. doi: 10.1021/ac902550n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abzalimov RR, Kaltashov IA. Extraction of local hydrogen exchange data from HDX CAD MS measurements by deconvolution of isotopic distributions of fragment ions. J Am Soc Mass Spectrom. 2006;17:1543–51. doi: 10.1016/j.jasms.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 101.Althaus E, Canzar S, Ehrler C, Emmett MR, Karrenbauer A, et al. Computing H/D-exchange rates of single residues from data of proteolytic fragments. BMC Bioinformatics. 2010;11:424. doi: 10.1186/1471-2105-11-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mayne L, Kan ZY, Chetty PS, Ricciuti A, Walters BT, Englander SW. Many overlapping peptides for protein hydrogen exchange experiments by the fragment separation-mass spectrometry method. J Am Soc Mass Spectrom. 2011;22:1898–905. doi: 10.1007/s13361-011-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fajer PG, Bou-Assaf GM, Marshall AG. Improved sequence resolution by global analysis of overlapped peptides in hydrogen/deuterium exchange mass spectrometry. J Am Soc Mass Spectrom. 2012;23:1202–8. doi: 10.1007/s13361-012-0373-3. [DOI] [PubMed] [Google Scholar]
- 104.Lindner R, Lou X, Reinstein J, Shoeman RL, Hamprecht FA, Winkler A. Hexicon 2: automated processing of hydrogen-deuterium exchange mass spectrometry data with improved deuteration distribution estimation. J Am Soc Mass Spectrom. 2014;25:1018–28. doi: 10.1007/s13361-014-0850-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sheff JG, Rey M, Schriemer DC. Peptide-column interactions and their influence on back exchange rates in hydrogen/deuterium exchange-MS. J Am Soc Mass Spectrom. 2013;24:1006–15. doi: 10.1007/s13361-013-0639-4. [DOI] [PubMed] [Google Scholar]
- 106.Wang L, Lane LC, Smith DL. Detecting structural changes in viral capsids by hydrogen exchange and mass spectrometry. Protein Sci. 2001;10:1234–43. doi: 10.1110/ps.100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tuma R, Coward LU, Kirk MC, Barnes S, Prevelige PE., Jr Hydrogen-deuterium exchange as a probe of folding and assembly in viral capsids. J Mol Biol. 2001;306:389–96. doi: 10.1006/jmbi.2000.4383. [DOI] [PubMed] [Google Scholar]
- 108.Zhang Q, Chen J, Kuwajima K, Zhang HM, Xian F, et al. Nucleotide-induced conformational changes of tetradecameric GroEL mapped by H/D exchange monitored by FT-ICR mass spectrometry. Sci Rep. 2013;3:1247. doi: 10.1038/srep01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Houde D, Demarest SJ. Fine details of IGF-1R activation, inhibition, and asymmetry determined by associated hydrogen /deuterium-exchange and peptide mass mapping. Structure. 2011;19:890–900. doi: 10.1016/j.str.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 110.Vadas O, Dbouk HA, Shymanets A, Perisic O, Burke JE, et al. Molecular determinants of PI3Kgamma-mediated activation downstream of G-protein-coupled receptors (GPCRs) Proc Natl Acad Sci U S A. 2013;110:18862–7. doi: 10.1073/pnas.1304801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen J, Walter S, Horwich AL, Smith DL. Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nat Struct Biol. 2001;8:721–8. doi: 10.1038/90443. [DOI] [PubMed] [Google Scholar]
- 112.Georgescauld F, Popova K, Gupta AJ, Bracher A, Engen JR, et al. GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding. Cell. 2014;157:922–34. doi: 10.1016/j.cell.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hebling CM, Morgan CR, Stafford DW, Jorgenson JW, Rand KD, Engen JR. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal Chem. 2010;82:5415–9. doi: 10.1021/ac100962c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rey M, Mrazek H, Pompach P, Novak P, Pelosi L, et al. Effective removal of nonionic detergents in protein mass spectrometry, hydrogen/deuterium exchange, and proteomics. Anal Chem. 2010;82:5107–16. doi: 10.1021/ac100171m. [DOI] [PubMed] [Google Scholar]
- 115.Baerga-Ortiz A, Hughes CA, Mandell JG, Komives EA. Epitope mapping of a monoclonal antibody against human thrombin by H/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein. Protein Sci. 2002;11:1300–8. doi: 10.1110/ps.4670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Coales SJ, Tuske SJ, Tomasso JC, Hamuro Y. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:639–47. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- 117.Ling JM, Silva L, Schriemer DC, Schryvers AB. Hydrogen-deuterium exchange coupled to mass spectrometry to investigate ligand-receptor interactions. Methods Mol Biol. 2012;799:237–52. doi: 10.1007/978-1-61779-346-2_15. [DOI] [PubMed] [Google Scholar]
- 118.Jensen PF, Jorgensen TJ, Koefoed K, Nygaard F, Sen JW. Affinity capture of biotinylated proteins at acidic conditions to facilitate hydrogen/deuterium exchange mass spectrometry analysis of multimeric protein complexes. Anal Chem. 2013;85:7052–9. doi: 10.1021/ac303442y. [DOI] [PubMed] [Google Scholar]
- 119.Zhang Z, Li W, Logan TM, Li M, Marshall AG. Human recombinant [C22A] FK506-binding protein amide hydrogen exchange rates from mass spectrometry match and extend those from NMR. Protein Sci. 1997;6:2203–17. doi: 10.1002/pro.5560061015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Palmblad M, Buijs J, Hakansson P. Automatic analysis of hydrogen/deuterium exchange mass spectra of peptides and proteins using calculations of isotopic distributions. J Am Soc Mass Spectrom. 2001;12:1153–62. doi: 10.1016/S1044-0305(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 121.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–14. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 122.Zhang HM, Bou-Assaf GM, Emmett MR, Marshall AG. Fast reversed-phase liquid chromatography to reduce back exchange and increase throughput in H/D exchange monitored by FT-ICR mass spectrometry. J Am Soc Mass Spectrom. 2009;20:520–4. doi: 10.1016/j.jasms.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kazazic S, Zhang HM, Schaub TM, Emmett MR, Hendrickson CL, et al. Automated data reduction for hydrogen/deuterium exchange experiments, enabled by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2010;21:550–8. doi: 10.1016/j.jasms.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hotchko M, Anand GS, Komives EA, Ten Eyck LF. Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 2006;15:583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chik JK, Vande Graaf JL, Schriemer DC. Quantitating the statistical distribution of deuterium incorporation to extend the utility of H/D exchange MS data. Anal Chem. 2006;78:207–14. doi: 10.1021/ac050988l. [DOI] [PubMed] [Google Scholar]
- 126.Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, et al. HDX workbench: software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom. 2012;23:1512–21. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guttman M, Weis DD, Engen JR, Lee KK. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J Am Soc Mass Spectrom. 2013;24:1906–12. doi: 10.1007/s13361-013-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jorgenson JW. Capillary liquid chromatography at ultrahigh pressures. Annu Rev Anal Chem (Palo Alto Calif) 2010;3:129–50. doi: 10.1146/annurev.anchem.1.031207.113014. [DOI] [PubMed] [Google Scholar]
- 129.Wu Y, Engen JR, Hobbins WB. Ultra performance liquid chromatography (UPLC) further improves hydrogen/deuterium exchange mass spectrometry. J Am Soc Mass Spectrom. 2006;17:163–7. doi: 10.1016/j.jasms.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 130.Plumb R, Castro-Perez J, Granger J, Beattie I, Joncour K, Wright A. Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:2331–7. doi: 10.1002/rcm.1627. [DOI] [PubMed] [Google Scholar]
- 131.Chalmers MJ, Busby SA, Pascal BD, Southern MR, Griffin PR. A two-stage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J Biomol Tech. 2007;18:194–204. [PMC free article] [PubMed] [Google Scholar]
- 132.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–20. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jones LM, Zhang H, Vidavsky I, Gross ML. Online, high-pressure digestion system for protein characterization by hydrogen/deuterium exchange and mass spectrometry. Anal Chem. 2010;82:1171–4. doi: 10.1021/ac902477u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ruotolo BT, Gillig KJ, Stone EG, Russell DH. Peak capacity of ion mobility mass spectrometry: separation of peptides in helium buffer gas. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;782:385–92. doi: 10.1016/s1570-0232(02)00566-4. [DOI] [PubMed] [Google Scholar]
- 135.Hilderbrand AE, Myung S, Barnes CA, Clemmer DE. Development of LC-IMS-CID-TOFMS techniques: analysis of a 256 component tetrapeptide combinatorial library. J Am Soc Mass Spectrom. 2003;14:1424–36. doi: 10.1016/j.jasms.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 136.Valentine SJ, Liu X, Plasencia MD, Hilderbrand AE, Kurulugama RT, et al. Developing liquid chromatography ion mobility mass spectometry techniques. Expert Rev Proteomics. 2005;2:553–65. doi: 10.1586/14789450.2.4.553. [DOI] [PubMed] [Google Scholar]
- 137.Iacob RE, Murphy JP, 3rd, Engen JR. Ion mobility adds an additional dimension to mass spectrometric analysis of solution-phase hydrogen/deuterium exchange. Rapid Commun Mass Spectrom. 2008;22:2898–904. doi: 10.1002/rcm.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fadgen KE, Wales TE, Stapels M, Eggertson MJ, Engen JR. Validation of an ion mobility hydrogen/deuterium exchange mass spectrometry system. J Biomol Tech. 2011;22(Suppl):S63. [Google Scholar]
- 139.Wales TE, Eggertson MJ, Engen JR. Considerations in the analysis of hydrogen exchange mass spectrometry data. Methods Mol Biol. 2013;1007:263–88. doi: 10.1007/978-1-62703-392-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–56. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 141.Rand KD, Zehl M, Jorgensen TJ. Measuring the Hydrogen/Deuterium Exchange of Proteins at High Spatial Resolution by Mass Spectrometry: Overcoming Gas-Phase Hydrogen/Deuterium Scrambling. Acc Chem Res. 2014 doi: 10.1021/ar500194w. in press. [DOI] [PubMed] [Google Scholar]
- 142.Pan J, Han J, Borchers CH, Konermann L. Hydrogen/deuterium exchange mass spectrometry with top-down electron capture dissociation for characterizing structural transitions of a 17 kDa protein. J Am Chem Soc. 2009;131:12801–8. doi: 10.1021/ja904379w. [DOI] [PubMed] [Google Scholar]
- 143.Huang RY, Garai K, Frieden C, Gross ML. Hydrogen/deuterium exchange and electron-transfer dissociation mass spectrometry determine the interface and dynamics of apolipoprotein E oligomerization. Biochemistry. 2011;50:9273–82. doi: 10.1021/bi2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Landgraf RR, Chalmers MJ, Griffin PR. Automated hydrogen/deuterium exchange electron transfer dissociation high resolution mass spectrometry measured at single-amide resolution. J Am Soc Mass Spectrom. 2012;23:301–9. doi: 10.1007/s13361-011-0298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bobst CE, Kaltashov IA. Enhancing the quality of H/D exchange measurements with mass spectrometry detection in disulfide-rich proteins using electron capture dissociation. Anal Chem. 2014;86:5225–31. doi: 10.1021/ac500904p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang G, Abzalimov RR, Bobst CE, Kaltashov IA. Conformer-specific characterization of nonnative protein states using hydrogen exchange and top-down mass spectrometry. Proc Natl Acad Sci U S A. 2013;110:20087–92. doi: 10.1073/pnas.1315029110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pan J, Borchers CH. Top-down mass spectrometry and hydrogen/deuterium exchange for comprehensive structural characterization of interferons: implications for biosimilars. Proteomics. 2014;14:1249–58. doi: 10.1002/pmic.201300341. [DOI] [PubMed] [Google Scholar]
- 148.Burkitt W, O’Connor G. Assessment of the repeatability and reproducibility of hydrogen/deuterium exchange mass spectrometry measurements. Rapid Commun Mass Spectrom. 2008;22:3893–901. doi: 10.1002/rcm.3794. [DOI] [PubMed] [Google Scholar]
- 149.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–86. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tiyanont K, Wales TE, Siebel CW, Engen JR, Blacklow SC. Insights into Notch3 Activation and Inhibition Mediated by Antibodies Directed against Its Negative Regulatory Region. J Mol Biol. 2013;425:3192–204. doi: 10.1016/j.jmb.2013.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nakazawa S, Ahn J, Hashii N, Hirose K, Kawasaki N. Analysis of the local dynamics of human insulin and a rapid-acting insulin analog by hydrogen/deuterium exchange mass spectrometry. Biochim Biophys Acta. 2013;1834:1210–4. doi: 10.1016/j.bbapap.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 152.Underbakke ES, Iavarone AT, Chalmers MJ, Pascal BD, Novick S, et al. Nitric oxide-induced conformational changes in soluble guanylate cyclase. Structure. 2014;22:602–11. doi: 10.1016/j.str.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wei H, Ahn J, Yu YQ, Tymiak A, Engen JR, Chen G. Using hydrogen/deuterium exchange mass spectrometry to study conformational changes in granulocyte colony stimulating factor upon PEGylation. J Am Soc Mass Spectrom. 2012;23:498–504. doi: 10.1007/s13361-011-0310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Creighton TE. Pathways and mechanisms of protein folding. Adv Biophys. 1984;18:1–20. doi: 10.1016/0065-227x(84)90004-2. [DOI] [PubMed] [Google Scholar]
- 155.Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–72. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- 156.Engen JR, Smithgall TE, Gmeiner WH, Smith DL. Identification and localization of slow, natural, cooperative unfolding in the hematopoietic cell kinase SH3 domain by amide hydrogen exchange and mass spectrometry. Biochemistry. 1997;36:14384–91. doi: 10.1021/bi971635m. [DOI] [PubMed] [Google Scholar]
- 157.Sivaraman T, Robertson AD. Kinetics of conformational fluctuations by EX1 hydrogen exchange in native proteins. Methods Mol Biol. 2001;168:193–214. doi: 10.1385/1-59259-193-0:193. [DOI] [PubMed] [Google Scholar]
- 158.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006;17:1498–509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 159.Zhang J, Ramachandran P, Kumar R, Gross ML. H/D exchange centroid monitoring is insufficient to show differences in the behavior of protein states. J Am Soc Mass Spectrom. 2013;24:450–3. doi: 10.1007/s13361-012-0555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fang J, Rand KD, Beuning PJ, Engen JR. False EX1 signatures caused by sample carryover during HX MS analyses. Int J Mass Spectrom. 2011;302:19–25. doi: 10.1016/j.ijms.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Majumdar R, Manikwar P, Hickey JM, Arora J, Middaugh CR, et al. Minimizing carry-over in an online pepsin digestion system used for the H/D exchange mass spectrometric analysis of an IgG1 monoclonal antibody. J Am Soc Mass Spectrom. 2012;23:2140–8. doi: 10.1007/s13361-012-0485-9. [DOI] [PubMed] [Google Scholar]
- 162.Yang H, Smith DL. Kinetics of cytochrome c folding examined by hydrogen exchange and mass spectrometry. Biochemistry. 1997;36:14992–99. doi: 10.1021/bi9717183. [DOI] [PubMed] [Google Scholar]
- 163.Chen S, Brier S, Smithgall TE, Engen JR. The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 2007;16:572–81. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Engen JR, Wales TE, Chen S, Marzluff EM, Hassell KM, et al. Partial cooperative unfolding in proteins as observed by hydrogen exchange mass spectrometry. Int Rev Phys Chem. 2013;32:96–127. doi: 10.1080/0144235X.2012.751175. [DOI] [PMC free article] [PubMed] [Google Scholar]