SUMMARY
Tetherin encodes an interferon-inducible antiviral protein that traps a broad spectrum of enveloped viruses at infected cell surfaces. Despite the absence of any clearly related gene or activity, we describe possible scenarios by which tetherin arose that exemplify how protein modularity, evolvability and robustness can create and preserve new functions. We find that tetherin genes in various organisms exhibit no sequence similarity and share only a common architecture and location in modern genomes. Moreover, tetherin is part of a cluster of three potential sister genes encoding proteins of similar architecture, some variants of which exhibit antiviral activity while others can be endowed with antiviral activity by a simple modification. Only in slowly evolving species (e.g. coelacanths) does tetherin exhibit sequence similarity to one potential sister gene. Neofunctionalization, drift and genetic conflict appear to have driven a near complete loss of sequence similarity among modern tetherin genes and their sister genes.
Graphical abstract
INTRODUCTION
Cells have evolved a variety of mechanisms to inhibit the replication of viruses. In animal cells, the detection of viruses can lead to the expression of interferons (IFNs), which in turn induce hundreds of IFN-stimulated genes (Schoggins and Rice, 2011; Iwasaki, 2012; Yan and Chen, 2012). Some of these genes function autonomously and directly to inhibit viral replication, through a variety of mechanisms. Tetherin (also known as BST-2, CD317, or HM1.24) is one such antiviral protein that causes entrapment of a broad spectrum of enveloped viruses at infected cell surfaces (Neil et al., 2008; Van Damme et al., 2008). Tetherin is a membrane glycoprotein comprised of a short N-terminal cytoplasmic tail (CT), a single pass transmembrane (TM) helix, a helical coiled-coil (CC) ectodomain that drives parallel homodimer formation, and a C-terminal glycosylphosphatidylinositol (GPI) membrane anchor. This unusual architecture, not primary sequence, is critical for Tetherin function (Perez-Caballero et al., 2009). Axially configured Tetherin dimers directly trap virions, primarily through the insertion of their GPI anchors into the lipid envelopes of budding virions (Venkatesh and Bieniasz, 2013).
The uniqueness of this antiviral mechanism, the absence of any known analogous process in the normal functioning of cells and the general problem in biology of tracing the origins of orphan genes, prompted us to ask the question of how this gene and antiviral mechanism arose. New functions could be generated through minor adaptation of cellular genes whose products have an intrinsic capacity to perform a function that is closely related to their antiviral activity. Alternatively, innovations or trace side activities can become the selected function of a gene (Bergthorsson et al., 2007). In either case, gene duplication is an important driver of the acquisition of new functions because it provides genetic “raw material” (i.e. a duplicated paralog) that can be released from evolutionary constraints imposed on a parental gene (Ohno, 1970). The fate of any gene can depend on its ability to preserve and adapt its function while accumulating mutations in a changing environment, i.e. its robustness and evolvability. The interplay between robustness and evolvability might allow evolving genes to sample variants while resisting potentially deleterious mutations (Masel and Trotter, 2010; Wagner, 2011). For certain antiviral proteins such as the APOBEC3 proteins, their ability to inhibit HIV-1 replication has obviously arisen via duplication and adaptation of homologous genes with related (cytidine deamination) functions (Refsland and Harris, 2013). Conversely, the antiviral activity of TRIM5 likely initially arose via the amplification of a trace side activity exhibited by other TRIM proteins (Zhang et al., 2006).
Every sequenced genome harbors a significant fraction of “orphan” genes whose origins are obscure due to the absence of homologs (Khalturin et al., 2009; Tautz and Domazet-Loso, 2011; Palmieri et al., 2014). Unlike other antiviral restriction factors, tetherin is an orphan gene in eutherian mammals whose origins are unknown. Sequence similarity searches for possible ancestors of tetherin yield no candidate genes. Moreover, tetherin is a non-essential gene in mice (Liberatore and Bieniasz, 2011) and is only expressed in response to interferon in most cells. Thus, it is difficult to envisage a scenario in which a gene of similar and necessary function gave rise to tetherin.
Here, we trace the ancestry of this orphan gene and propose models by which tetherin arose in ancient jawed vertebrates. Specifically, tetherin is part of a cluster of three genes that encode proteins with related protein architecture, but lack sequence similarity. Proteins encoded by the putative sister genes of tetherin in mammals have a TM-CC configuration but lack a C-terminal GPI anchor that is critical for antiviral function. However, antiviral function can be endowed by the addition of a GPI anchor, and in some divergent vertebrates, variants of one of these sister genes have the potential to encode a GPI anchor and exhibit antiviral activity.
RESULTS
Candidate ancestors of tetherin
Sequence similarity searches have hitherto revealed no reasonable candidates for genes that might share a common ancestor with tetherin. However, orphan genes that are unusually divergent might preserve structural but not sequence similarity with their parents (Domazet-Loso and Tautz, 2003). Therefore, we searched for candidate sister genes that encode proteins of similar predicted architecture, irrespective of sequence similarity. We initially focused on the human and mouse genomes because they are well curated and updated, and searched for annotated genes whose products are predicted to have the following features: (i) a single TM domain, (ii) a CC domain C-terminal to the TM domain, and, (iii) a GPI modified C-terminus (see Supplemental Information). Among the 22,691 and 22,709 annotated human and mouse protein coding genes (Ensembl Release 71) (Flicek et al., 2013), Tetherin was the only protein predicted to have a TM-CC-GPI architecture.
We reasoned that the genesis of tetherin might have involved the acquisition of a third domain by genes encoding 2 out of the 3 contiguous Tetherin features, i.e. proteins with either TM-CC or CC-GPI domains. A search of the Ensembl database found 66 to 211 human and 47 to 175 mouse TM-CC proteins, depending on the length requirement imposed to form a bona fide coiled-coil (Dataset S1). Analysis of a previously predicted set of GPI-modified human proteins revealed that Tetherin and 12 other proteins had a CC-GPI configuration (Pierleoni et al., 2008) (Dataset S1). Overall, ~1% of all annotated human or mouse genes encode proteins that have TM-CC or CC-GPI architectures from which Tetherin could plausibly have arisen through acquisition of its third defining feature.
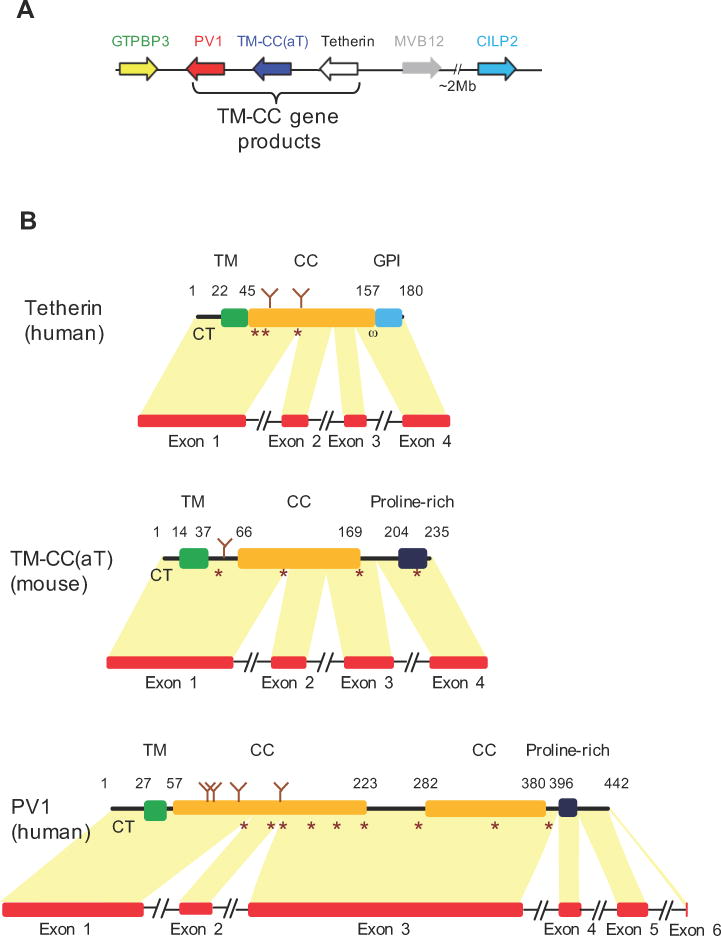
One likely mechanism by which tetherin arose is the duplication and neofunctionalization of another gene (Ohno, 1970). Because duplicated genes are often positioned adjacent to each other in genomes (Pan and Zhang, 2008), we inspected the organization of genes proximal to tetherin in human and mouse genomes. Strikingly, two genes whose protein product had a TM-CC configuration were positioned proximally to tetherin in human and mouse genomes (Figure 1A). One such gene is pv1 that encodes an essential component of the stomatal diaphragms of caveolae and the diaphragms of fenestrae and transendothelial channels (Stan et al., 1999b; Stan et al., 2012). Another gene encoding a TM-CC protein, that we designated tm-cc, adjacent to tetherin, tm-cc(at), is of unknown function and is located between pv1 and tetherin (Figure 1A). tm-cc(at) was not identified in our initial searches of mouse and human genomes because it is not annotated in the Ensembl database and is the product of a Gnomon-predicted transcript (GenBank: XM_011242347.1) whose existence in mammals is supported only by a single cDNA sequence (GenBank: AK141396.1) from an early mouse embryo. Thus, it was initially uncertain whether tm-cc(at) is a bona fide gene. However, PCR analysis of day 7 mouse embryo mRNA confirmed that a transcript encoding tm-cc(at) was present therein, and spliced at the predicted intron-exon junctions (Figure S1A). Additionally, tm-cc(at) exons appear to be better conserved compared to flanking genome sequence across mammals (Figure S1B), supporting its existence as a bona fide gene. Moreover, a cDNA encoding a homologous protein has been found in the chicken transcriptome (GenBank: BU332041.1). Even though there is no mRNA evidence for tm-cc(at) expression in humans, a homologous sequence is present in the syntenic genomic location of humans (GenBank: XM_011528476.1). RNA-seq data suggest that it is expressed at low abundance and is spliced in a similar way to mouse TM-CC(aT), but includes a fifth exon (Figure S1C). Thus, tm-cc(at) appears to be a gene that is poorly annotated because it is rarely expressed.
Figure 1. Location and architecture of TM-CC gene products proximal to tetherin in human and mouse genomes.
(A) Diagram of genes surrounding human and mouse tetherin.
(B) Organization of TM-CC genes and their protein products for human Tetherin (GenBank: NP_004326.1), mouse TM-CC(aT) (GenBank: XP_003945491.1), and human PV1 (GenBank: NP_112600.1) proteins. Glycosylation and cysteine residues are indicated as brown Y symbols and stars respectively. Numbers indicate amino acid positions. Structural features of TM-CC(aT) and PV1 are based on predictions using TMHMM, COILS, Pred-GPI and GlycoEP.
See also Figure S1.
Tetherin, PV1 and TM-CC(aT) proteins have a short N-terminal CT, a single TM domain, and a predominantly CC extracellular domain with multiple cysteine residues that, in the case of Tetherin and PV1, are known to stabilize parallel homodimer formation via the formation of disulfide bonds (Stan et al., 1999a; Andrew et al., 2009; Perez-Caballero et al., 2009) (Figure 1B). Analysis of cDNA sequence architecture reveals that tetherin has 4 exons, pv1 has 5–7 exons (depending on the species) and tm-cc(at) has 4 (mouse) or 5 (chicken) exons. The organization of exons and protein coding domains in tetherin, pv1 and tm-cc(at) is similar, particularly when tetherin and tm-cc(at) are compared, with the first exon of all three genes encoding the CT, TM and the N-terminal portion of their CC domains. Notably, human and mouse pv1 and tm-cc(at) both encode proline-rich sequences at their C-termini, rather than the GPI anchor encoded by Tetherin (Figure 1B). The overt and unusual structural similarity of these genes and proteins, along with their adjacent genomic location, suggested that pv1, tm-cc( at) and tetherin might share a common ancestor, despite the absence of sequence similarity.
To determine how unexpected this apparent clustering of TM-CC encoding genes should be, we mined all annotated human and mouse genes for adjacently positioned pairs that encode TM-CC proteins. Among the 211 and 175 TM-CC-encoding genes in the human and mouse genomes respectively, 10 (human) and 6 (mouse) gene pairs were found adjacent to each other and in the same orientation. Most duplicated genes (72–94%) remain adjacent to each other and in the same orientation (Pan and Zhang, 2008). We observed that most of the few adjacent TM-CC gene pairs (7 of 10 human and 5 of 6 mouse) share clear sequence similarity, usually belong to obvious gene families (e.g. CLEG and CTEG proteins) and therefore obviously arose via duplication of the neighboring gene (Dataset S1). Thus, the probability that tetherin originated de novo in a distal chromosomal location, and was then inserted adjacent to two genes that encode proteins of similar TM-CC architecture appears to be low (< ~211 human TM-CC-encoding genes / 22691 total human genes, or <0.01; lower if these genes are in the same orientation).
TM-CC(aT) and PV1 can be endowed with antiviral activity by the addition of a GPI anchor
Both the TM and GPI anchor domains present in Tetherin are essential for virion entrapment at the cell surface (Perez-Caballero et al., 2009). A clear difference in the overall architectures of human and mouse PV1, TM-CC(aT) and Tetherin is the presence of the GPI anchor in Tetherin. If the three proteins indeed share a common ancestor, then a model for the genesis of Tetherin would include acquisition of a GPI anchor by an ancestral paralog. Indeed, given the apparent plasticity of Tetherin protein sequences, this simple modification might be sufficient to endow PV1 and/or TM-CC(aT) with antiviral activity.
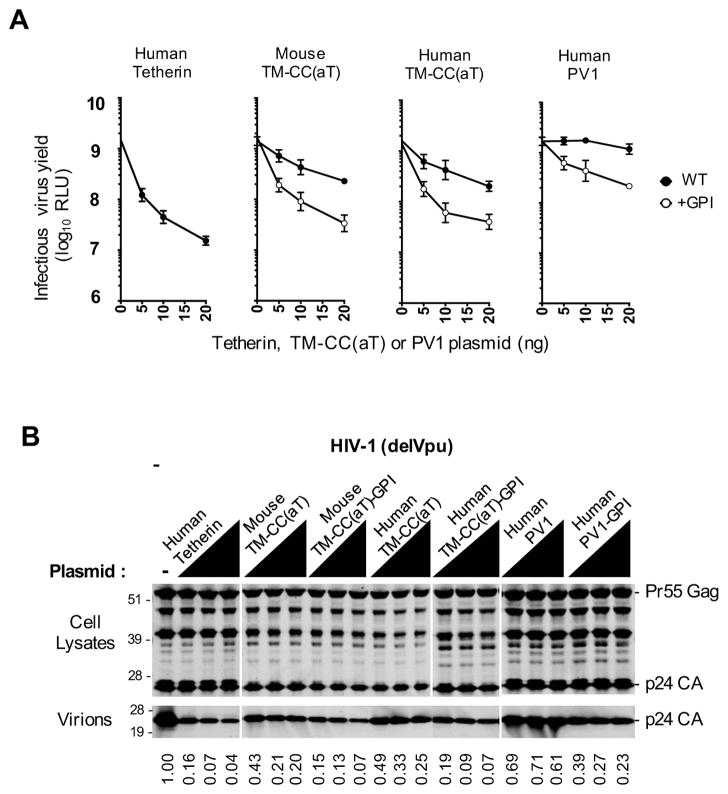
We measured the yield of HIV-1 particles from transfected 293T cells expressing unmodified PV1 and TM-CC(aT) proteins, or derivatives of PV1 and TM-CC(aT) proteins that were appended with a GPI modification signal from human Tetherin. This analysis revealed that the human and mouse TM-CC(aT) proteins engineered with GPI modified C-termini inhibited HIV-1 particle release nearly as potently as human Tetherin (Figures 2A, 2B). Interestingly, the unmodified TM-CC(aT) proteins also had some propensity to inhibit virion release. Conversely, while the unmodified PV1 protein had little, if any, antiviral activity (Figures 2A, 2B) the addition of a GPI anchor endowed PV1 (PV1-GPI) with virion entrapment activity that was less potent than that exhibited by the GPI modified TM-CC(aT) proteins. Western blot analysis of N-terminally HA-tagged derivatives of these proteins confirmed that active and inactive proteins were approximately equivalently expressed (Figure S2A).
Figure 2. Antiviral activity of GPI-modified TM-CC(aT) and PV1 proteins.
(A) Infectious virion yield measured using HeLa TZM-bl indicator cells following transfection with a Vpu-deficient HIV-1 proviral plasmid along with increasing amounts of the indicated unmodified (WT) or GPI-modified (+GPI) Tetherin, TM-CC(aT) or PV1 proteins (RLU= relative light units, mean ± SD, n=3).
(B) Western blot analyses (anti-CA) of cell lysates and virions corresponding to (A). Numbers at the bottom represent virion CA protein levels relative to those obtained in the absence of an inhibitor.
See also Figure S2.
We assessed the antiviral activity of two unrelated TM-CC proteins, CD72 and CLEC1A, which have been demonstrated to form dimers and to reside at the cell surface (Von Hoegen et al., 1990; Sattler et al., 2012). The unmodified CD72 protein had a minor propensity to inhibit HIV-1 virion release (Figure S2B and S2C), but the addition of a C-terminal GPI anchor markedly impaired protein expression (Figure S2C). The addition of a C-terminal GPI anchor to CLEC1A protein did not confer antiviral activity (Figure S2B and S2C), although GPI modified and unmodified proteins were expressed at similar levels, suggesting that this feature is not generalizable to all TM-CC proteins.
Sequence and structural homologs of Tetherin in diverse vertebrates
Typically, sister genes exhibit sequence similarity, but this was not evident in human or mouse PV1, Tetherin and TM-CC(aT) proteins or genes. Therefore, we next sought to delineate the evolutionary history of tetherin by identifying orthologs in diverse species. BLAST searching revealed that orthologs of human Tetherin are present in therian mammals (marsupials and eutherian mammals) (Figure 3A) suggesting an origin predating the emergence of mammals. Proteins that shared sequence similarity with Tetherin could not be found in any other vertebrate species by BLAST searching. However, the marginal sequence similarity among some mammalian Tetherin proteins suggested that sequence divergence over >150 million years may have eroded sequence similarity to a point that Tetherin orthologs might not be detected in more diverse species by sequence similarity searches (e-value ≤ 1e-5).
Figure 3. Antiviral activity of divergent Tetherin/TM-CC-GPI proteins.
(A) Tetherin/TM-CC-GPI Protein sequences from human (GenBank: NP_004326.1), mouse (GenBank: NP_932763.1), opossum (GenBank: XP_007489270.1 and XP_007489271.1), Tasmanian devil (GenBank: XP_012399618.1), turtle (GenBank: XP_008169758.1, XP_005279001.1 and XP_005279003.1), turkey (inferred from GenBank: XP_010723307.1), falcon (inferred from GenBank: XP_005444407.1 and Gnomon prediction: 2189215010.p), alligator (GenBank: XP_006017475.1 and XP_006017476.1), coelacanth (Gnomon prediction: 16424589.p), elephant shark (GenBank: XP_007897024.1) Tetherin/TM-CC-GPI proteins (see also Supplemental Information). The TM, CC domains and GPI anchor are indicated. Conserved residues are highlighted and predicted omega sites (GPI modification) are indicated in grey.
(B) Infectious virion yield measured using HeLa TZM-bl indicator cells following transfection of Vpu-deficient HIV-1 proviral plasmids along with plasmids expressing Tetherin/TM-CC-GPI proteins. (RLU= relative light units, Mean ± SD, n=3).
(C) Western blot analyses (anti-CA) of cell lysates and virions corresponding to (B). Numbers at the bottom represent virion CA protein levels relative to those obtained in the absence of an inhibitor.
See also Figure S3.
Therefore, we next collected all annotated and predicted transcript sequences (Gnomon (Souvorov et al., 2010)) from representative diverse species of monotremes (platypus), birds (chicken, saker falcon, turkey), reptiles (Chinese alligator and painted turtle), amphibians (frog), lobe-finned fish (coelacanth), ray-finned fish (medaka and zebrafish) and cartilaginous fish (elephant shark). We also obtained transcriptomic data for a jawless vertebrate, the sea lamprey (Smith et al., 2013). We searched these actual and putative transcripts for sequences that were predicted to encode TM-CC-GPI proteins. By this approach we were able to identify putative Tetherin-like proteins (i.e. proteins exhibiting a TM-CC-GPI architecture, but lacking sequence similarity to Tetherin) in most of the aforementioned species except platypus, chicken, turkey and frog. Thus, if the genes encoding these proteins share a common ancestor with Tetherin, they diverged from all previously analyzed eutherian mammal tetherin genes ~150 to 500 MYA (Janvier, 2006; Inoue et al., 2010; Venkatesh et al., 2014). There was no statistically significant sequence similarity between these TM-CC-GPI proteins and known or putative Tetherin proteins from mammals.
Although an inspection of other bird genomes did not yield any Tetherin homologs, we found homologs of the saker falcon TM-CC-GPI protein in eagles (Canadian eagle and the bald eagle), ibises (crested ibis), penguins (emperor penguin), hummingbirds (Anna’s hummingbird), cuckoos (common cuckoo) and other falcons (peregrine falcon); some of these are supported by transcriptomic evidence (Canadian eagle, bald eagle, emperor penguin and saker falcon). Blast searches also revealed sequences in the turkey genome that were similar to the falcon TM-CC-GPI protein in the predicted terminal four exons of a proximal gene (cilp2). However, the inclusion of these four exons in turkey cilp2 is unsupported by RNA-seq data and likely represents an annotation error. We also found sequences adjacent to the chicken tm-cc(at) gene that are predicted to encode a TM-CC-GPI protein. These observations suggest that the genomes of both Neoaves (i.e. falcons, eagles, ibises, etc.) and Galloanseres (i.e. chicken and turkey) have the potential to code for Tetherin/TM-CC-GPI proteins.
Antiviral activity of divergent Tetherin and TM-CC-GPI proteins
Previously, only Tetherin proteins encoded by eutherian mammals and by one reptile have been identified and demonstrated to exhibit strong antiviral activity (Arnaud et al., 2010; Takeda et al., 2012; Heusinger et al., 2015). We next determined whether widely divergent Tetherin orthologs encoded by marsupials (opossum and Tasmanian devil), as well as TM-CC-GPI proteins lacking sequence similarity to Tetherin that are encoded by a bird (saker falcon), a reptile (Chinese alligator), a lobe-finned fish (coelacanth), a cartilaginous fish (elephant shark), and a jawless fish (lamprey), possessed antiviral activity.
We transfected 293T cells with a panel of plasmids encoding Tetherin or /TM-CC-GPI proteins, along with a Vpu-deficient HIV-1 (ΔVpu) proviral plasmid. (Figures 3B, 3C, S3A). The putative Tetherin/TM-CC-GPI proteins encoded by opossum, Tasmanian devil, alligator, falcon, coelacanth and elephant shark, all inhibited HIV-1 virion release, in most cases with an apparent potency that was similar to human Tetherin (Figures 3B, 3C, S3A). The addition of a N-terminal HA-tag and western blotting verified that each of these TM-CC-GPI proteins was well expressed. In contrast, the TM-CC-GPI protein encoded in the lamprey genome was poorly expressed and inactive (Figures 3B, 3C, S3A). Two divergent Tetherin proteins (Tasmanian devil and Chinese alligator) were tested for susceptibility to antagonism by HIV-1 Vpu and, predictably, were found to be resistant (Figure S3B, S3C). These findings suggest that virion entrapment is a nearly universal feature of TM-CC-GPI proteins that have arisen in jawed vertebrates within the past ~450 million years.
Genomic loci harboring tetherin/TM-CC-GPI genes
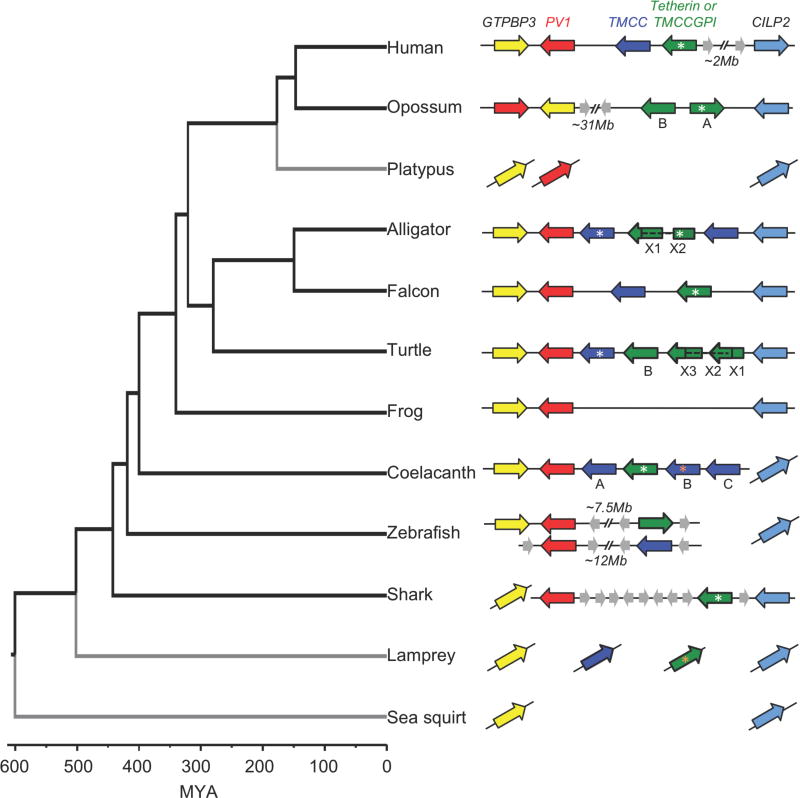
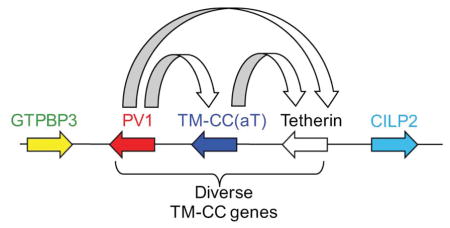
In eutherian mammals, tetherin and its neighbors form a syntenic block of genes: gtpbp3–pv1–tm-cc(at)–tetherin–mvb12–[2Mb]–cilp2 (Figure 1A). While most mammals possess a single tetherin gene, recent duplications have led to the presence of multiple homologous tetherin genes in some species, such as cows, sheep, and bats (Arnaud et al., 2010; Takeda et al., 2012). Inspection of this genomic locus in diverse mammals revealed that in the opossum and the wallaby, tetherin has been recently duplicated, tm-cc(at) is absent, and pv1 is separated from the two tetherin genes by ~31 MB. Moreover, the tetherin genes are adjacent to a gene, cilp2 that is located ~2MB distal to tetherin in eutherian mammals (Figure 4). We could not reconstruct an analogous locus in a monotreme (platypus) because its genome is incompletely assembled.
Figure 4. Organization of pv1-proximal genes in vertebrates.
Diagrams were generated using NCBI, UCSC, Ensembl Genome Browsers and sequence similarity approaches. Branches in gray indicate incomplete genome assemblies. Inclined figures indicate genes in incompletely assembled scaffolds. White and orange asterisks indicate genes that were active or inactive respectively in virion release-inhibition assays. Potential alternatively spliced versions of alligator and turtle tetherin are indicated by dotted lines. The duplicated loci in zebrafish are shown. Phylogeny and speciation dates were based on (Inoue et al., 2010; Janvier, 2006; Venkatesh et al., 2014).
See also Figure S4.
Remarkably, we found that all of the aforementioned TM-CC-GPI encoding genes in birds, reptiles, coelacanth, ray-finned fish and sharks were present in nearly the same location in their respective genomes, i.e. between pv1 and cilp2 (Figure 4). By manual curation of genomic sequence in this interval in some species, including searches of translated genomic sequences, and inspection of RNAseq data, we also found that there likely have been several duplication and deletion events involving tm-cc(at) and/or tm-cc-gpi genes in the gtpbp3–pv1–cilp2 interval during the course of vertebrate evolution (Figure 4). For example, single tm-cc-gpi and tm-cc(at) genes are present in most eutherian mammals and avian species, but tm-cc(at) is not present in a marsupial (opossum). In reptiles, there is a single tm-cc(at) gene and multiple tm-cc-gpi genes including some that appear to generate multiple TM-CC-GPI protein isoforms via alternative splicing of duplicated exons (Figure S4). In an amphibian (frog) the locus lacks a tetherin homolog or a putative tetherin gene. In the coelacanth, a single tm-cc-gpi and three tm-cc(at) genes are present. Consistent with the fact that a whole genome duplication occurred in the ancestor of ray-finned fishes (Glasauer and Neuhauss, 2014), a tm-cc-gpi gene is linked to one of the pv1 copies in zebrafish while a similar tm-cc gene (lacking a predicted GPI modification) is linked to a second pv1 gene (Figure 4). These genes are outside the gtpbp3–cilp2 interval, arranged in a manner that suggests that a segmental inversion occurred. In another ray-finned fish (medaka), a tm-cc-gpi gene is similarly found linked to one of the pv1 copies, but the tm-cc gene is absent. In the elephant shark genome, a tm-cc-gpi gene is present in the gtpbp3–cilp2 interval proximal to cilp2 (separated from it by an intron-less gene) and separated from pv1 by ~223 KB and tm-cc(at) is absent (Figure 4). It was not possible to reconstruct the configuration of this locus in the lamprey due to the fragmented nature of the genome sequence. Nevertheless, these findings suggest that one or more tm-cc(at) and/or tm-cc-gpi genes, including tetherin, appeared in vertebrates at the genomic locus containing gtpbp3–pv1–cilp2 (Figure 4).
Relationship between PV1, TM-CC(aT) and tetherin/TM-CC-GPI proteins and genes
Although these findings suggest that tetherin arose from duplication of an ancestral tm-cc gene (pv1 or tm-cc(at)), a key question is whether tetherin, pv1 and tm-cc(at) should be expected to share sequence similarity, given the time at which they diverged, and the types of selection pressure that have acted on them. Notably, introns share little similarity across pv1 in divergent amniotes, indicating that neutral evolution over ~300 MY is sufficient to diminish sequence similarity to nearly undetectable levels at presumed neutrally evolving sites (Figure S5A).
To potentially facilitate the detection of homology between distantly related sequences, we used maximum likelihood methods to reconstruct ancestral amniote PV1 and TM-CC(aT) protein sequences. Due to the high sequence divergence, it was not feasible to reconstruct reliable ancestral Tetherin sequences. Thus, we searched for sequence similarity among extant and ancient PV1, TM-CC(aT) and Tetherin/TM-CC-GPI protein sequences using BLAST. PV1 sequences were clearly similar across all jawed vertebrates and their phylogenetic relationships resembled vertebrate phylogeny (Figure S5B), perhaps reflecting its essential role in the formation of diaphragms. In contrast, TM-CC(aT) and especially Tetherin/TM-CC-GPI were more variable. For most Tetherin/TM-CC-GPI sequences, significant sequence similarity was found only between closely related species (e.g. among mammals or among ray-finned fish) (Figures 5A, 5B, 5C). The extreme Tetherin/TM-CC-GPI sequence divergence may have been driven, in part, by positive selection. Indeed, codon-based tests of selection have previously demonstrated that primate tetherin has evolved under positive selection, likely driven by viral antagonists (McNatt et al., 2009). The high level of sequence divergence confounded the extension of codon-based tests to other vertebrate lineages, because homologous sites could not be reliably assigned and a high number of synonymous substitutions would undermine the apparent impact of non-synonymous substitutions.
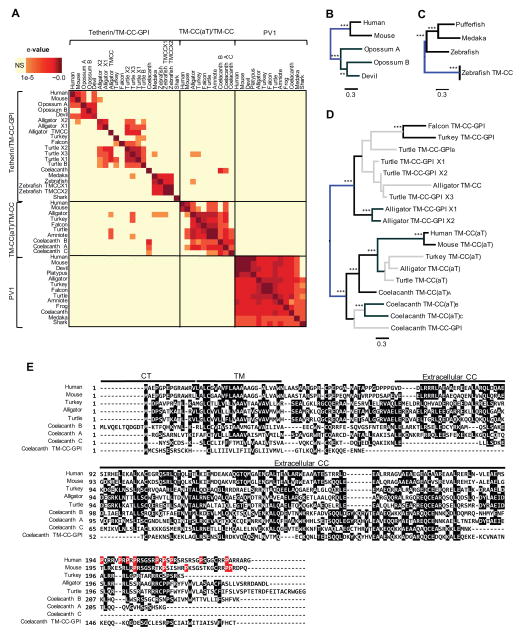
Figure 5. Sequence similarity between PV1, TM-CC(aT) and Tetherin/TM-CC-GPI proteins.
(A) Heat map showing e-values of all combinations of reciprocal BLASTp analyses using the PV1, TM-CC(aT) and Tetherin/TM-CC-GPI proteins in this study.
(B, C and D) Phylogenetic trees of divergent mammalian Tetherin (B) Fish Tetherin/TM-CC-GPI
(C) and other vertebrate Tetherin/TM-CC-GPI and TM-CC(aT) (D) protein sequences. Sequences with significant BLAST hits in (A) were used to construct each tree. Maximum likelihood tree was constructed using RAxML with 1000 bootstrap replicates. Nodes and branches in grey were supported by <80% of the bootstrap replicates. Asterisks indicate bootstrap support for each node. (*) ≥ 80%, (**) ≥ 90%, (***) ≥ 95%. Trees were midpoint rooted (indicated in blue).
(E) Alignment of divergent TM-CC(aT) and Tetherin/TM-CC-GPI protein sequences from human (adapted from GenBank: XP_011526778.1), mouse (GenBank: XP_003945491.1), turkey (GenBank: XP_010723297.1), alligator (GenBank: KQL90195.1), turtle (adapted from GenBank: XP_008169839.1) and coelacanth (GenBank: XP_006001674.1 and XP_014347293.1, Gnomon prediction: 16424589.p and TM-CC(aT)A adapted from RNAseq reads of NW_005819727.1). Sequences spanning the TM, CC domains and GPI anchor are indicated. Residues that comprise the proline-rich domain in human and mouse TM-CC(aT) proteins are indicated in red.
See also Figure S5.
Attempts to detect sequence similarity between PV1 and TM-CC(aT) or Tetherin were statistically inconclusive, even when amniote ancestral sequences were used. However, the TM-CC(aT) proteins from mammals, reptiles, avian, and coelacanth exhibited sequence similarity to each other, indicating an unambiguous common origin for vertebrate tm-cc(at) genes (Figures 5A, 5D and 5E). As noted above, the coelacanth has three tm-cc(at) genes and one tetherin/tm-cc-gpi gene in the pv1–cilp2 interval. Strikingly, the coelacanth Tetherin/TM-CC-GPI protein exhibited clear sequence similarity to the TM-CC(aT) proteins, particularly the TM-CC(aT)B protein that is encoded by the neighboring gene (Figures 5A, 5D and 5E). Additionally, some reptilian Tetherin/TM-CC-GPI genes (e.g. Turtle X3) show local sequence similarity with both reptilian Tetherin/TM-CC-GPI and TM-CC(aT) proteins (Figure 5A). Alignment of Tetherin/TM-CC-GPI and TM-CC(aT) protein sequences (Figure 5E) and construction of a phylogenetic tree (Figure 5D) revealed that the coelacanth Tetherin/TM-CC-GPI protein, which is antiviral (Figures 3B and 3C), shares clear sequence similarity and likely, therefore, a common ancestor with vertebrate TM-CC(aT) proteins.
Potential for tm-cc(at) to encode a GPI anchor in some species
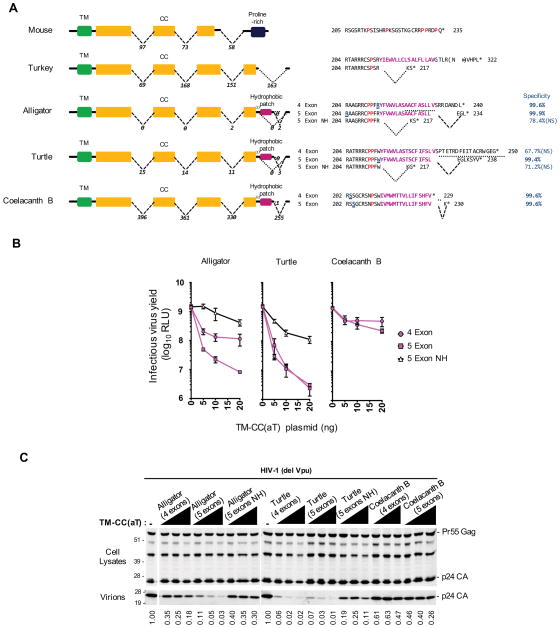
Although tm-cc(at) and tetherin/tm-cc-gpi genes share no sequence similarity in most vertebrates, the finding that tm-cc(at) and tetherin/tm-cc-gpi are clear homologs in the coelacanth prompted us to ask how one might have arisen from the other. Because a key difference between these two proteins in mammals is the presence of a proline rich C-terminus in TM-CC(aT) versus a C-terminal GPI anchor in Tetherin, we inspected the annotated 3′ exons of tm-cc(at) in non-mammalian species to determine how a GPI modification might have arisen. Notably, bona fide, near full length tm-cc(at) cDNA sequences from mouse and chicken differ in the number of exons that contribute to sequence encoding the tm-cc(at) C-terminus. Specifically, mouse tm-cc(at) has four exons, with the fourth exon encoding a proline-rich C-terminal sequence (Figure 6A). Conversely the chicken tm-cc(at) fourth exon is truncated by splicing to a fifth exon encoding only two C-terminal amino acids (Figure 6A). Human tm-cc(at) is predicted to include a fifth exon that appends seven C-terminal amino acids that are absent in mouse tm-cc(at). RNAseq data indicates that coelacanth tm-cc(at)A and tm-cc(at)B includes a fifth exon that encodes only two or one amino acid respectively, while the 3′ end of the fourth exon in tm-cc(at)B encodes C-terminal sequences that have a high probability conferring GPI modification, followed by a stop codon (Figure 6A). Thus, unlike mammalian and avian TM-CC(aT) proteins, it is highly likely that coelacanth TM-CC(aT)B is GPI modified at its C-terminus, whether or not the fifth exon is used.
Figure 6. Antiviral activity of non-mammalian TM-CC(aT) variants.
(A) Transcript structure and C-terminal protein sequences of potential alternatively spliced isoforms of tm-cc(at) in non-mammalian species. The TM, CC and proline-rich domains and hydrophobic patch are indicated in color. The omega site (underlined in blue) and specificity (1 – false positive rate) were predicted using PredGPI. The number of RNAseq reads supporting the occurrence or absence of splicing events are indicated between the exons.
(B) Infectious virion yield measured using HeLa TZM-bl indicator cells following transfection of Vpu-deficient HIV-1 proviral plasmids along with plasmids expressing alternatively spliced isoforms of TM-CC(aT) proteins. NH= no hydrophobic (isoforms lacking the hydrophobic patch). (RLU= relative light units, Mean ± SD, n=3).
(C) Western blot analyses (anti-CA) of cell lysates and virions corresponding to (B). Numbers at the bottom represent virion CA protein levels relative to those obtained in the absence of an inhibitor.
See also Figure S6.
The tm-cc(at) gene in two reptile species (painted turtle and Chinese alligator) has sequences that potentially encode a hydrophobic amino acid-rich sequence immediately 3′ to the fourth exon, while a potential fifth exon codes for 3–7 amino acids and a termination codon. We reasoned that, similar to the coelacanth variant, the hydrophobic amino acid-rich sequence in reptilian TM-CC(aT) proteins might confer GPI modification (Figure 6A). Due to a paucity of RNAseq data, it is unknown whether modern reptile tm-cc(at) genes encode four- or five-exon proteins or if they are GPI-modified at their C-termini. However, unlike their mammalian counterparts, reptile TM-CC(aT) genes have the potential to encode C-terminally GPI-modified proteins.
We constructed cDNAs expressing the coelacanth TM-CC(aT)B protein and the potential alternatively spliced versions of the alligator and turtle TM-CC(aT) proteins. The coelacanth TM-CC(aT)B proteins were poorly active, despite abundant expression (Figures 6B, 6C, and S6). However, the inclusion of the hydrophobic amino acid-rich sequence (i.e. GPI-modified) in the alligator and turtle TM-CC(aT) proteins conferred antiviral activity that was enhanced by the presence of the fifth exon (Figures 6B and 6C). Strikingly, both the four and five-exon versions of the turtle TM-CC(aT) proteins that contained the hydrophobic sequence potently inhibited the release of infectious HIV-1 particles by ~100-fold at the highest dose tested (Figures 6B and 6C). These data suggest that tm-cc(at) genes in reptiles have the potential to encode GPI-modified proteins, and some isoforms exhibit potent antiviral activity.
DISCUSSION
Elucidating the ancient evolutionary origins of tetherin is challenging given the absence of genes with sequence similarity. Our findings strongly suggest that tetherin arose by duplication and neofunctionalization of an ancestor of a neighboring gene encoding a TM-CC protein, i.e. pv1 and/or tm-cc(at). This conclusion is based on the findings that: (i) genes encoding TM-CC proteins constitute ~1% of all genes and most of those that are arranged contiguously obviously share a common ancestor; (ii) pv1, tm-cc(at) and tetherin/tm-cc-gpi genes are proximal in many modern species and share a similar exon-intron structure; (iii) PV1 and especially TM-CC(aT) are able to trap virions following a simple manipulation that could have plausibly been acquired to enable GPI modification; (iv) In the coelacanth, TM-CC(aT) and Tetherin/TM-CC-GPI are obvious homologs; (v) some modern tm-cc(at) genes have the potential to encode GPI-modified proteins and exhibit antiviral activity.
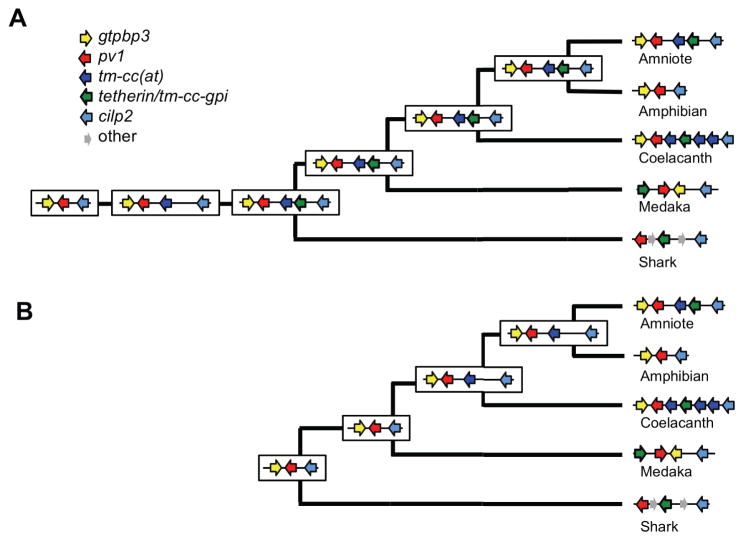
In considering models for the genesis of tetherin/tm-cc-gpi genes, the conservation and essential function of pv1 strongly suggests that it was present in the ancestor of vertebrates, with one or two duplications of it giving rise to tm-cc(at) and/or tetherin/tm-cc-gpi in the various vertebrate lineages. The high antiviral potency of some GPI-modified TM-CC(aT) proteins, the sequence similarity between coelacanth TM-CC(aT) and functional Tetherin/TM-CC-GPI proteins, and shared gene structure suggest that most modern Tetherin proteins arose from TM-CC(aT)-like proteins. In one plausible model, pv1 first duplicated to give pv1–tm-cc(at) and then tm-cc(at) duplication yielded pv1–tm-cc(at)–tetherin/tm-cc-gpi to give a single common tetherin/tm-cc-gpi ancestor that was derived from tm-cc(at) prior to division of sharks from other jawed vertebrate lineages (Figure 7A). In this scenario, gene loss and rearrangement events in sharks, ray-finned fish and amphibians would give the modern genome configurations, with sequence similarity between tm-cc(at) and the ancestral tetherin/tm-cc-gpi surviving only in the coelacanth lineage (and to some extent in reptiles), although gene conversion events might contribute to preservation of similarity. In this model, our finding that all vertebrate Tetherin/TM-CC-GPI proteins tested exhibit potent antiviral activity, would suggest that tetherin/tm-cc-gpi neofunctionalization occurred early during vertebrate evolution, prior to the division of shark from other vertebrate lineages.
Figure 7. Possible evolutionary scenarios for the emergence of tetherin/tm-cc-gpi gene(s) in the pv1–cilp2 locus.
(A) Tetherin/TM-CC-GPI originated once, prior to the division of sharks from other jawed vertebrate lineages via sequential duplications of pv1 and tm-cc(at).
(B) Tetherin/TM-CC-GPI originated independently in multiple vertebrate lineages via duplications of pv1 and tm-cc(at).
See also Figure S7.
However, it is not necessarily the case that a single ancestral tetherin/tm-cc-gpi gene arose in vertebrates on one occasion, with all modern Tetherin/TM-CC-GPI proteins being descended from this ancestor. In another possible model, pv1 duplicated to give pv1–tm-cc(at) in the ancestor of lobe-finned fish, amphibians and amniotes (i.e. after the separation of sharks and ray-finned fish from remaining vertebrates). Thereafter, separate pv1 duplication events in sharks and ray-finned fish, and tm-cc(at) duplication events in coelacanths and the various amniote lineages yielded tetherin/tm-cc-gpi genes in their modern configurations (Figure 7B). In this model, it is implausible that tm-cc-gpi genes could have arisen on multiple occasions in nearly the same genomic location, unless they are indeed duplications of proximal TM-CC genes (i.e. pv1 or tm-cc(at)).
Although it is intuitive to expect sequence similarity between proteins that have a common ancestor, our findings suggest that the lack of sequence similarity between tetherin and its sister genes in most species should be expected, due to their contrasting evolutionary histories. PV1 is essential for mouse viability and has a key role in the generation of diaphragms, thus selective pressures to preserve this function have resulted in sequence similarity across vertebrates in its coding but not intronic sequence. The function(s) of TM-CC(aT) proteins is unknown, but the apparent absence of tm-cc(at) in opossums, as well as in fish and shark lineages suggests that it does not play an essential role in the life of cells, and RNA-Seq data suggests that it is poorly or rarely expressed in humans and is detected only in d7 mouse embryos. Notably, some versions of tm-cc(at), like pv1, encode a proline rich-C terminus, others are predicted to encode a GPI anchor, and still others have no defining C-terminal feature. It is thus conceivable that tm-cc(at) has different functions in different species. It is even possible that tm-cc(at) is/was a functional tetherin in some species, particularly since some reptile tm-cc(at) genes have the potential to encode a protein with potent antiviral activity.
Our results suggest the loss or acquisition of splicing signals played a key role in the genesis of Tetherin proteins, enabling the acquisition of the critical C-terminal GPI anchor. Presumably, nascent tetherin/tm-cc-gpi sequences were optimized thereafter in various species through positive selection pressures imposed by pathogenic viruses that enhanced potency or enabled the avoidance of viral antagonists. In hominids, an antiviral role may have been further expanded by acquisition of signaling capability (Cocka and Bates, 2012; Galao et al., 2012; Tokarev et al., 2013). Overall, neutral evolution and positive selection for some fraction of ~450 MYA, is expected to have erased sequence similarity within tetherin orthologs in other species to nearly the same degree as its sister genes, ultimately resulting in extreme diversity of Tetherin/TM-CC-GPI proteins and its orphaned status in modern vertebrates. Nevertheless, probabilistic models used to detect distant homologs (HMMER3 (Finn et al., 2011)) found a significant hit (e-value = 2E-5) between coelacanth PV1 and a HMM profile of TM-CC(aT) (Figures 5C and S7). This finding, coupled with our observation that the coelacanth Tetherin/TM-CC-GPI protein bears significant sequence similarity with a coelacanth TM-CC(aT) protein, might be a reflection of the slow neutral substitution rate in the coelacanth (Amemiya et al., 2013).
While this manuscript was in preparation, a different and less complete account of early tetherin evolution was published (Heusinger et al., 2015). Heusinger et al. inspected annotated genes in the pv1–cilp2 interval that were presumed to be tetherin orthologs but did not recognize that an ancient duplication leads to distinguishable tm-cc(at) and tetherin/tm-cc-gpi genes, only some of which are annotated in genome databases. Thus the “tetherin” genes whose sequences were inspected in Heusinger et al. are actually a mixture of tm-cc(at) and tetherin/tm-cc-gpi genes. Heusinger et al. also functionally tested two non-mammalian “Tetherin” proteins, from coelacanth and alligator. However, the coelacanth “Tetherin” protein tested in Heusinger et al, is actually equivalent to the TM-CC(aT)B protein tested herein, and in agreement with Heusinger et al. we find that it has little antiviral activity. Conversely we find that the neighboring, not previously annotated, coelacanth gene encodes a bona fide Tetherin/TM-CC-GPI protein that has potent antiviral activity.
To our knowledge, the pv1–tm-cc(at)–tetherin cluster is the only known gene triplet whose members share structural, but not sequence similarity with their sister genes, and have unrelated functions. Tetherin is unusual in that its biological function can be attributed largely to its unique TM-CC-GPI dual-anchored topology, rather than to its specific amino acid sequence (Perez-Caballero et al., 2009). As such, the modular structure and rather simple function of Tetherin appears to have resulted in an unusual combination of extreme robustness and evolvability that first led to its genesis from a protein of unrelated function, and then enabled it to adapt to evade viral antagonists while maintaining antiviral activity. These findings highlight the remarkable capacity of genomes to innovate novel functions following gene duplication. Moreover, predicted structure comparisons, synteny and functional analyses might serve as valuable approaches for revealing the evolutionary history of other orphan genes.
EXPERIMENTAL PROCEDURES
Bioinformatics and sequence comparisons
Sequences for mammalian Tetherin, PV1 and TM-CC(aT) gene and protein sequences were retrieved from Ensembl (Release 71) and GenBank. Tetherin, TM-CC(aT) and PV1 genes that have not being annotated were identified using BLAT and tblastn searches guided by published RNA-seq data. Protein sequences were used to construct multiple alignments and phylogenetic trees. To determine the genomic organization proximal to the tetherin gene, sequences from various vertebrate species were retrieved using UCSC, NCBI and Ensembl genome browsers and synteny was assessed using the Genomicus Browser.
To identify human and mouse TM-CC proteins, a Perl script was written to automate a pipeline to find protein sequences that contain one TM domain followed by coiled-coil domains. Sequence similarity between TM-CC proteins that are adjacent to each other was determined using BLASTp searches. Analysis of introns and neutrally evolving sequences were carried out using the mVISTA tool for glocal sequence alignments. BLASTp sequence similarity searches were performed using either extant or ancestral PV1, TM-CC(aT) or Tetherin sequences as queries of a database of PV1, Tetherin, and other TM-CC proteins.
Plasmid construction
All Tetherin, PV1 and TM-CC(aT) proteins were transiently expressed using pCR3.1 (Invitrogen) based plasmids. The human PV1 cDNA was constructed using PCR-based gene synthesis methods. All other Tetherin and TM-CC(aT) cDNAs were codon optimized (human) and custom synthesized (Genewiz).
Virion yield assays
Cells (293T) were co-transfected with wild-type or Vpu-deficient proviral plasmids along with varying amounts of Tetherin, PV1, TM-CC(aT) or TM-CC expression plasmids and a plasmid expressing YFP, to monitor transfection efficiency. The culture medium was replaced the following day. At 48 hours post transfection, the culture supernatants were harvested, clarified by centrifugation at 3000 rpm, and filtered through a 0.2 μm PVDF membrane (Millipore). Infectious virus yield was determined by inoculating sub-confluent monolayers of HeLa TZM-bl cells that were seeded in 96-well plates at 10,000 cells/well with 100 μl of serially diluted supernatants. At 48 hours post infection, β-galactosidase activity was determined using GalactoStar reagent, in accordance with the manufacturer’s instructions (Applied Biosystems). Physical particle yield was determined by layering virion containing supernatant onto 1 ml of 20% sucrose in PBS followed by centrifugation at 20,000xg for 90 minutes at 4°C. Virion pellets were then analyzed by Western blotting.
Western blot assays
Pelleted virions and cell lysates were resuspended in SDS-PAGE loading buffer, with the addition of β-mercaptoethanol, and resolved on NuPAGE Novex 4–12% Bis-Tris Mini Gels (Invitrogen) in MOPS running buffer. Proteins were blotted onto nitrocellulose membranes (HyBond, GE-Healthcare) in transfer buffer (25 mM Tris, 192 mM glycine). The blots were then blocked with Odyssey blocking buffer and probed with mouse monoclonal anti-HIV-1 capsid (NIH), mouse monoclonal anti-HA (Covance), and mouse monoclonal anti-PV1 (Abcam) primary antibodies. The bound primary antibodies were detected using a fluorescently labeled secondary antibody (IRDye 800CW Goat Anti-Mouse Secondary Antibody, LI-COR Biosciences). Fluorescent signals were detected using a LI-COR Odyssey scanner and quantitated with Odyssey software (LI-COR Biosciences).
Further details can be found in Supplemental information.
Supplementary Material
Acknowledgments
We thank Theodora Hatziioannou and members of the Bieniasz Lab for helpful discussions. This work was supported by a grant from the NIH (R01AI50111).
Footnotes
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: D.B-M., S.V., P.D.B. Computational analyses: D.BM. Performed the experiments: D.B-M., S.V. Analyzed the data: D.B-M., S.V., P.D.B. Wrote the paper: D.B-M., S.V., P.D.B.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AJ, Miyagi E, Kao S, Strebel K. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology. 2009;6:80. doi: 10.1186/1742-4690-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud F, Black SG, Murphy L, Griffiths DJ, Neil SJ, Spencer TE, Palmarini M. Interplay between ovine bone marrow stromal cell antigen 2/tetherin and endogenous retroviruses. J Virol. 2010;84:4415–4425. doi: 10.1128/JVI.00029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Andersson DI, Roth JR. Ohno’s dilemma: evolution of new genes under continuous selection. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocka LJ, Bates P. Identification of alternatively translated Tetherin isoforms with differing antiviral and signaling activities. PLoS Pathog. 2012;8:e1002931. doi: 10.1371/journal.ppat.1002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 2003;13:2213–2219. doi: 10.1101/gr.1311003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galao RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell host & microbe. 2012;12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasauer SM, Neuhauss SC. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 2014;289:1045–1060. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- Heusinger E, Kluge SF, Kirchhoff F, Sauter D. Early Vertebrate Evolution of the Host Restriction Factor Tetherin. J Virol. 2015;89:12154–12165. doi: 10.1128/JVI.02149-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Lam K, Tay BH, Danks JA, Bell J, Walker TI, Venkatesh B. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol. 2010;27:2576–2586. doi: 10.1093/molbev/msq147. [DOI] [PubMed] [Google Scholar]
- Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier P. Palaeontology: modern look for ancient lamprey. Nature. 2006;443:921–924. doi: 10.1038/443921a. [DOI] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Liberatore RA, Bieniasz PD. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18097–18101. doi: 10.1073/pnas.1113694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Trotter MV. Robustness and evolvability. Trends Genet. 2010;26:406–414. doi: 10.1016/j.tig.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Palmieri N, Kosiol C, Schlotterer C. The life cycle of Drosophila orphan genes. Elife. 2014;3:e01311. doi: 10.7554/eLife.01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Zhang L. Tandemly arrayed genes in vertebrate genomes. Comp Funct Genomics. 2008:545269. doi: 10.1155/2008/545269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler S, Reiche D, Sturtzel C, Karas I, Richter S, Kalb ML, Gregor W, Hofer E. The human C-type lectin-like receptor CLEC-1 is upregulated by TGF-beta and primarily localized in the endoplasmic membrane compartment. Scand J Immunol. 2012;75:282–292. doi: 10.1111/j.1365-3083.2011.02665.x. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer HB, Minx P, Warren DE, Shedlock AM, Thomson RC, Valenzuela N, Abramyan J, Amemiya CT, Badenhorst D, Biggar KK, et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013;14:R28. doi: 10.1186/gb-2013-14-3-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–421. 421e411–412. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souvorov A, Kapustin Y, Kiryutin B, Chetvernin V, Tatusova T, Lipman D. Gnomon - NCBI eukaryotic gene prediction tool. 2010 ( http://www.ncbi.nlm.nih.gov/genome/guide/gnomon.shtml.)
- Stan RV, Ghitescu L, Jacobson BS, Palade GE. Isolation, cloning, and localization of rat PV-1, a novel endothelial caveolar protein. The Journal of cell biology. 1999a;145:1189–1198. doi: 10.1083/jcb.145.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan RV, Kubitza M, Palade GE. PV-1 is a component of the fenestral and stomatal diaphragms in fenestrated endothelia. Proceedings of the National Academy of Sciences of the United States of America. 1999b;96:13203–13207. doi: 10.1073/pnas.96.23.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan RV, Tse D, Deharvengt SJ, Smits NC, Xu Y, Luciano MR, McGarry CL, Buitendijk M, Nemani KV, Elgueta R, et al. The diaphragms of fenestrated endothelia: gatekeepers of vascular permeability and blood composition. Dev Cell. 2012;23:1203–1218. doi: 10.1016/j.devcel.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda E, Nakagawa S, Nakaya Y, Tanaka A, Miyazawa T, Yasuda J. Identification and functional analysis of three isoforms of bovine BST-2. PLoS One. 2012;7:e41483. doi: 10.1371/journal.pone.0041483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- Tokarev A, Suarez M, Kwan W, Fitzpatrick K, Singh R, Guatelli J. Stimulation of NFkappaB activity by the HIV restriction factor BST2. J Virol. 2013;87:2046–2057. doi: 10.1128/JVI.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell host & microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Bieniasz PD. Mechanism of HIV-1 virion entrapment by tetherin. PLoS Pathog. 2013;9:e1003483. doi: 10.1371/journal.ppat.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoegen I, Nakayama E, Parnes JR. Identification of a human protein homologous to the mouse Lyb-2 B cell differentiation antigen and sequence of the corresponding cDNA. J Immunol. 1990;144:4870–4877. [PubMed] [Google Scholar]
- Wagner A. The molecular origins of evolutionary innovations. Trends Genet. 2011;27:397–410. doi: 10.1016/j.tig.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353:396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.