Abstract
Resident gut microbes co-exist with transient bacteria to form the gut microbiota. Despite increasing evidence suggesting a role for transient microbes on gut microbiota function, the interplay between resident and transient members of this microbial community is poorly defined. We aimed to determine the extent to which a host's autochthonous gut microbiota influences niche permissivity to transient bacteria using a fermented milk product (FMP) as a vehicle for five food-borne bacterial strains. Using conventional and gnotobiotic rats and gut microbiome analyses (16S rRNA genes pyrosequencing and reverse transcription qPCR), we demonstrated that the clearance kinetics of one FMP bacterium, Lactococcus lactis CNCM I-1631, were dependent on the structure of the resident gut microbiota. Susceptibility of the resident gut microbiota to modulation by FMP intervention correlated with increased persistence of L. lactis. We also observed gut microbiome configurations that were associated with altered stability upon exposure to transient bacteria. Our study supports the concept that allochthonous bacteria have transient and subject-specific effects on the gut microbiome that can be leveraged to re-engineer the gut microbiome and improve dysbiosis-related diseases.
Introduction
Mammals are holobionts that host microbial communities of astounding density and complexity within their gastrointestinal (GI) tracts. Microorganisms from maternal and environmental microbiomes rapidly colonize the GI tract of the newborn, and a stable microbial ecosystem develops within the first three years of life (Koenig et al., 2011). This microbial stability is then continuously challenged by daily ingestion of environmental bacteria originating from sources such as diet (van Hylckama Vlieg et al., 2011), indoor environments (Lax et al., 2014), human co-inhabitants (Song et al., 2013) and, more recently, by symbionts used to restore a perturbed microbiota (Reeves et al., 2012; Atarashi et al., 2013; Deriu et al., 2013; Laval et al., 2015; Martin et al., 2015). One of the many traits ascribed to the autochthonous (that is, resident) gut microbiota is its ability to prevent colonization by allochthonous (that is, exogenous) bacteria, especially pathogens. This function of the microbial ecosystem is known as ‘colonization resistance' or ‘the barrier effect' (van der Waaij et al., 1971). Colonization resistance has been well-established with respect to Escherichia coli, Clostridium difficile and Salmonella spp. (Que and Hentges 1985; Wilson et al., 1986; Vollaard et al., 1990; Stecher et al., 2005) and has been linked to certain features of the gut microbiota, for example, community complexity as well as the presence of specific taxa (de La Cochetiere et al., 2010; Manges et al., 2010; Stecher et al., 2010; Rousseau et al., 2011).
Bacteria in foodstuffs are a major source of allochthonous bacteria, ranging from 104 to 109 colony-forming units per gram of food with fermented foods having the highest viable bacterial counts (Lang et al., 2014). These food-borne bacteria can temporarily integrate into the gut microbiome and constitute what can be called the transient microbiome (McNulty et al., 2011; David et al., 2014; Veiga et al., 2014; Eloe-Fadrosh et al., 2015). Emerging evidence suggests a significant role of transient food-borne bacteria on the overall gut microbiota community structure and function (McNulty et al., 2011; Veiga et al., 2014; Derrien and van Hylckama Vlieg 2015; Unno et al., 2015).
In the present study, we examined if a host's autochthonous gut microbiota influences niche permissivity (that is, colonization resistance) for transient bacteria administered in a fermented milk product (FMP) containing a consortium of five strains: Bifidobacterium animalis subsp. lactis CNCM I-2494, Lactococcus lactis subsp. lactis CNCM I-1631 Lactobacillus delbrueckii subsp. bulgaricus CNCM I-1632, L. delbrueckii subsp. bulgaricus CNCM I-1519 and Streptococcus thermophilus CNCM I-1630. Following FMP administration to conventional rats, we observed that one subgroup of rats (hereafter called ‘resistant') eliminated L. lactis CNCM I-1631 as fast as a GI transit marker, whereas another subgroup (hereafter called ‘permissive') shed the strain over an additional 24–48 h. Gut microbiota analyses showed that resistant and permissive rats differed in their abundance of Lachnospiraceae and that resistant rats had a microbiota less susceptible to FMP-induced changes compared with the permissive rats. Based on these findings, we re-analyzed the 16S ribosomal RNA (rRNA) amplicon survey data from the McNulty et al.'s, study (2011), which investigated the effects of a similar FMP on human gut microbiota (n=14), and observed similar patterns. We then used fecal transplantation in germ-free rats to demonstrate that the resistant and permissive phenotypes were gut microbiota-dependent.
Materials and methods
Study product
The product was an FMP (Danone Research), which contains the following strains: L. lactis subsp. lactis (strain I-1631 from the French National Collection of Cultures of Microorganisms (CNCM), Paris, France), B. animalis subsp. lactis CNCM I-2494, L. delbrueckii subsp. bulgaricus CNCM I-1632, L. delbrueckii subsp. bulgaricus CNCM I-1519 and S. thermophilus CNCM I-1630. The FMP contains ~108 colony-forming units L. lactis ml−1, B. lactis ml−1, L. bulgaricus ml−1 and 6 × 108 S. thermophilus ml−1. The energy density of the FMP was 6.0–7.2 kcal g−1 and the pH values were 4.35–4.5.
Conventional animal study
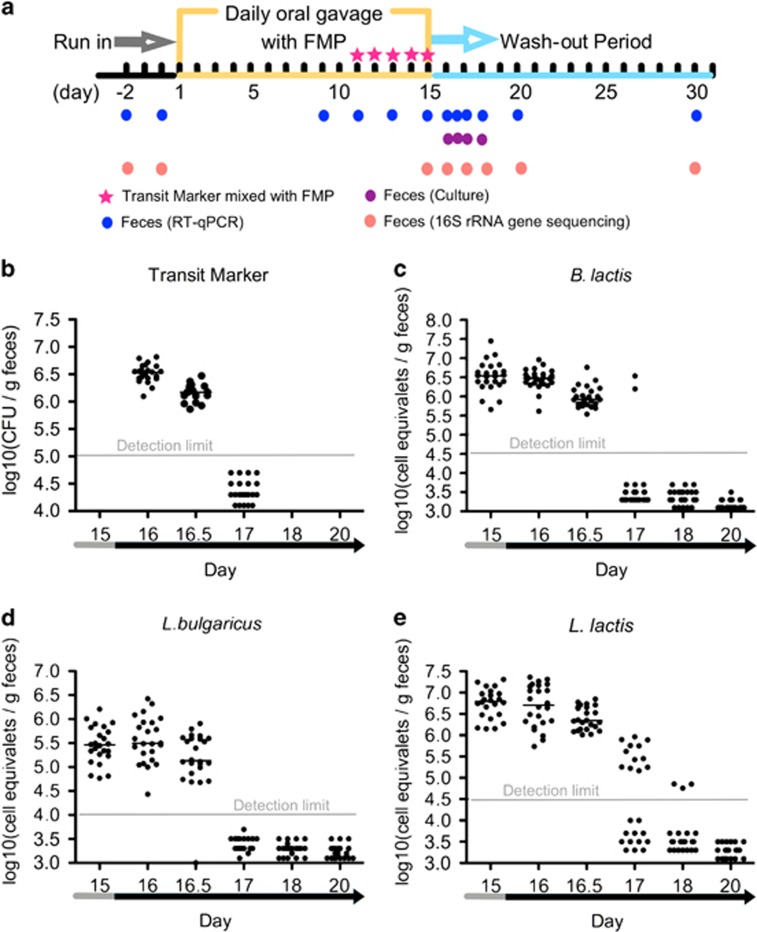
Animal studies and experiments were approved by the National Ethics committee on Animal Experimentation and carried out according to its guidelines (Sous le numéro 45). Eight-week-old male adult Fisher 344 rats (purchased from IcoCrl Charles River Laboratories, L'Arbresle, France, n=24; originating from 14 different litters) were maintained in a specific pathogen-free animal facility, and fed a standard autoclaved chow diet (ref. R03, SAFE, Augy, France) ad libitum. After adaptation and 15 days of run-in, the rats were gavaged with the FMP (0.5 ml per day) for 15 days (Day 1–15). During the last 5 days of the gavage period, Geobacillus stearothermophilus spores (Merck, Darmstadt, Germany) were added to the FMP as a GI transit marker (107 day per rat). Spores collected from fecal samples were germinated at 65 °C in G-spore medium (Drouault et al., 2002). The 15 days after the FMP gavage served as a wash-out period (Day 16–30). The feces of the rats were collected during the experimental period and the collection time points are shown in Figure 1a.
Figure 1.
Experimental design and fecal abundance of L. lactis, B. lactis, L. bulgaricus and G. stearothermophilus spores in conventional rat. (a) Experimental design. (b) Fecal abundance of Geobacillus stearothermophilus spores. Each symbol represents a sample from an individual rat (n=24). RT-qPCR quantification of (c) L. lactis, (d) B. lactis and (e) L. bulgaricus. Each symbol represents a sample from a given individual. Data expressed as log10 (equivalent cells gm−1 feces). Medians are reported.
RNA and DNA extraction
The fecal samples were stored at −80 °C until RNA and DNA extraction. The RNA was extracted by High Pure Isolation Kit (Roche, Branford, CT, USA) with an improved protocol described previously (Tap et al., 2015). A frozen aliquot (200 mg) of each fecal sample was suspended in 250 μl of guanidine thiocyanate, 0.1 m Tris (pH 7.5) and 40 μl of 10% N-lauroyl sarcosine, and DNA was extracted as previously described (Manichanh et al., 2006). RNA and DNA concentration and molecular weight were estimated using a nanodrop instrument (Thermo Scientific, Wilmington, DE, USA) and agarose gel electrophoresis, respectively.
Fecal quantitative reverse transcription PCR
The bacterial culture used for standard curves, the primers and quantitative reverse transcription PCR system and protocol were described previously (Veiga et al., 2010) (Supplementary Table S1). The quantity of each FMP strain was normalized by the number of total bacteria. We converted the number of detected molecules (RNA) into cell equivalents.
Pyrosequencing of the V3–V4 region of 16S rRNA genes
The PCR of the V3–V4 region of 16S rRNA genes and pyrosequencing was performed by Genoscreen (France, www.genoscreen.com) with GS-FLX platform (Roche). The following universal 16S rRNA primers were used for the PCR reactions: V3F (TACGGRAGGCAGCAG, 343–357 E. coli position) and V4R (GGACTACCAGGGTATCTAAT, 787–806 E. coli position).
Bioinformatics and statistical analysis
The quality control of raw sequences, operational taxonomic units (OTUs) classification, alignment of the representative sequence of each OTU, chimera removal, taxonomic assignment and alpha and beta diversity analyses were performed with QIIME (macQIIME 1.7) (Caporaso et al., 2010). The sequences were split based on barcodes (10 nt) and filtered according to the following quality criteria: length between 250–1000 nt, no mismatch allowed in barcodes and primers, and exclusion of homopolymers larger than 6 nt, quality above 25 over a 50 base pairs window). Resulting sequences were used to pick OTUs. The delineation of OTUs was conducted with Uclust using 97% cutoff, the representative sequence of each OTU was aligned using PyNAST and chimeric sequences were removed using ChimeraSlayer. The representative sequence of each OTU was assigned the taxonomic classification with the Ribosomal Database Project Classifier with a minimum bootstrap threshold of 80%. Rarefaction curves were generated with Faith's phylogenetic diversity (PD_Whole tree), Chao 1 and Observed Species method in QIIME. Raw data from 127 samples generated by McNulty et al., (2011) were obtained and processed in QIIME v 1.8 (OTU were picked against the May 2013 Greengenes database) in order to generate taxonomy profiles and beta diversity matrixes. Lactococcus relative abundance was calculated as the average of the Lactococcus-assigned OTUs through the FMP administration period (that is, weeks 5, 6, 8, 11 as per the McNulty et al., study) or wash-out periods (that is, weeks 13, 15 as per the McNulty et al., study) in order to smooth temporal intra-individual variations. LEfSe analyses were performed on the website http://huttenhower.sph.harvard.edu/galaxy (Segata et al., 2011) to identify OTU separating permissive or resistant subjects. The differential features were identified at the OTU (97% similarity), Phylum, Class, Order, Family and genus levels using the following parameters: (1) the alpha value for the factorial Kruskal–Wallis test among classes is <0.05 and (2) the threshold on the logarithmic LDA score for discriminative features is >2.0. Comparison between UniFrac distance kinetics of Lactoc+ and Lactoc− subjects was carried out using linear mixed models with repeated measures and a random effect. Partial-Least Square Discriminant Analyses and receiving operator characteristic analyses were performed with the R packages ‘muma' and ‘pROC'.
Canonical analysis of principal coordinates (CAP) and permutational multivariate analysis of variance were performed by using the method implemented in the R ‘BiodiversityR' and ‘vegan' package, respectively. The Bray–Curtis method was used for calculating ecological distance in CAP and permutational multivariate analysis of variance. Permutation tests using the trace and the greatest root statistics were performed to assess the significance of the discrimination. The permuted P-values were obtained with 999 permutations in CAP and permutational multivariate analysis of variance.
Gnotobiotic rat study
Germ-free male Fischer 344 rats (8-week-old) (Anaxem INRA, Jouy-en-Josas, France) were maintained in two plastic flexible film isolators (n=8 per isolator), and fed a standard autoclaved chow diet (ref. R03, SAFE) ad libitum. Rats in one isolator were conventionalized with two gavages of a fecal suspension originating from one randomly selected permissive conventional rat while rats in the other isolator were conventionalized with two gavages of a fecal suspension originating from one randomly selected resistant conventional rat. After 4 weeks of conventionalization, both groups of conventionalized rats were gavaged with the FMP similar to the experiment performed in conventional rats.
Results
Clearance kinetics of food-borne bacteria varies across rats
Twenty-four conventional rats received an FMP containing five bacterial strains (L. lactis subsp. lactis CNCM I-1631, B. animalis subsp. lactis CNCM I-2494, L. delbrueckii subsp. bulgaricus CNCM I-1632, L. delbrueckii subsp. bulgaricus CNCM I-1519 and S. thermophilus CNCM I-1630) by daily oral gavage for 15 days (Figure 1a). FMP species were not detected in feces before the start of the gavage. During the FMP intervention, the ingested species of L. lactis, B. lactis and L. bulgaricus were detected and quantified (>105 cell equivalents gm−1 feces) in 100% of the stool samples. In contrast, during the FMP intervention, S. thermophilus CNCM I-1630′s DNA was recovered in only 50% of the stool samples at concentrations too low to allow reliable qPCR quantification. Owing to the poor recovery of S. thermophilus DNA, the dynamics of this strain could not be further investigated. We thus focused on the other FMP strains.
We quantified the abundance of the FMP species during the FMP intervention and observed a high intra-individual variability (that is, within a two-log range) when assessed at four time points (Supplementary Figure S1). These data suggest that fecal abundance of the FMP strains during FMP administration could not be a reliable marker of colonization resistance. Subsequently, we focused on the elimination kinetics of the FMP species during the wash-out period.
We verified that the GI transit time was comparable for all animals by measuring the fecal abundance of G. stearothermophilus spores, a commonly used experimental GI transit marker. Its levels dropped below the limit of detection after two days (Day 17) in all the rats (Figure 1b). Unlike B. lactis and L. bulgaricus, L. lactis was still detected in the feces of >50% of rats (n=12) after 2 days (Figures 1c–e). We confirmed the elimination dynamics of L. lactis in two additional independent experiments (n= 52) (Supplementary Figure S2) and again observed no effect on the other FMP strains (data not shown). The differential pattern of L. lactis elimination kinetics suggested that the transient persistence of this strain differed between individual rats. Thus, we separated the rats into two groups, ‘resistant' and ‘permissive', based on their elimination kinetics of L. lactis, with resistant rats showing an elimination of L. lactis similar to the transit marker and permissive rats having a longer persistence of L. lactis compared with resistant rats.
Permissive and resistant phenotypes display distinct autochthonous gut microbiota in rats
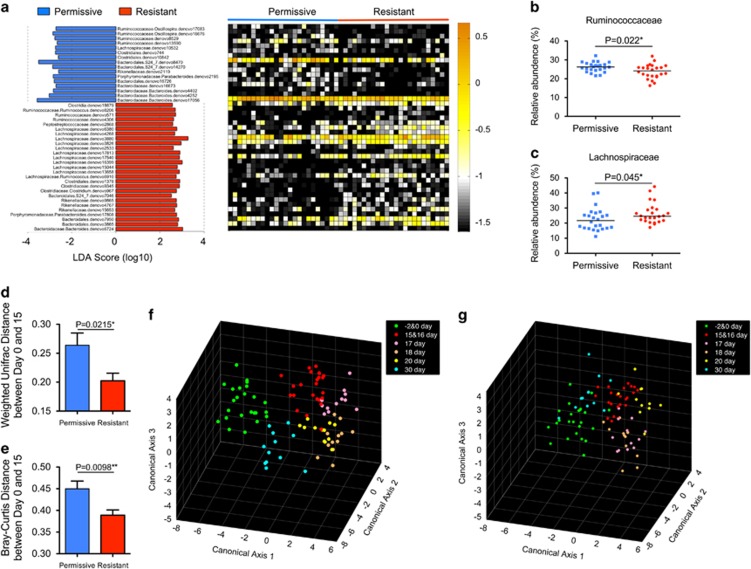
To determine whether permissivity to L. lactis was associated with a specific gut microbiota structure, we analyzed the gut microbiota of the rats before the FMP intervention (Day −2 and 0), at the end of treatment (Day 15), during the wash-out period (Days 16, 17, 18 and 20) and 2 weeks after the wash-out (Day 30) using bar-coded pyrosequencing. After quality-control filtering, we obtained 646 627 reads of the V3–V4 region of 16S rRNA genes from 192 samples with an average of 3368 reads per sample (±303.3 s.d.) and 12 161 OTUs were delineated with a 97% cutoff (Rarefaction curves (Supplementary Figure S3)). ‘Permissive' and ‘resistant' rats showed no significant differences in diversity based on observed OTUs, PD whole-tree and Shannon diversity indexes. Firmicutes (54.5%±9.8%, mean±s.d.) and Bacteroidetes (41.0%±9.1%, mean±s.d.) constituted the two most dominant phyla of the rat fecal bacterial communities, followed by Proteobacteria (2.5%±1.1%, mean±s.d.) and Tenericutes (1.4% ±1.8%, mean ±s.d.) (Supplementary Figure S4A). The relative abundance of the bacteria phylogenetically close to the FMP species was extremely low at baseline (that is, Bifidobacteriaceae<0.04%, Lactobacillaceae <0.26% and Streptococcaceae <0.04%, Supplementary Figure S4B). We used LEfSe, a linear discriminant analysis effect size tool (Segata et al., 2011), to discriminate differential abundance of bacterial phylotypes between permissive (n=12) and resistant rats (n=12) prior to the FMP intervention. Forty-three OTUs were found to discriminate between the gut microbiota of resistant and permissive rats (Figure 2a and Supplementary Table S2). We next used a Partial-Least Square Discriminant Analysis coupled with a receiving operator characteristic analysis and confirmed that LEfSe successfully identified OTUs discriminating between permissive and resistant rats with an Area Under the Curve of 0.90 (Supplementary Figure S5). At the family level, permissive rats had a higher relative abundance of Ruminococcaceae, and a lower relative abundance of Lachnospiraceae, as compared with resistant rats (Figures 2b and c). Similar results were obtained in a second independent experiment (n=12) (Supplementary Figure S6). Conflicting results were observed for Bacteroidetes assigned OTUs between the two experiments. In one experiment (Figure 2a), Bacteroidetes OTUs did not efficiently discriminate between permissive and resistant rats, whereas they appear to be more discriminant in the other experiment (Supplementary Figure S6).
Figure 2.
Permissive and resistant rats differ in their gut microbiota and in the amplitude of ecological changes induced by the FMP. (a) LDA scores computed for taxa differentially abundant between permissive and resistant rats at baseline (Day –2 and 0). The heat map shows the relative abundance (log10 transformation) of OTUs in each sample. Abundance of (b) Ruminococcaceae and (c) Lachnospiraceae. Each symbol represents a sample from an individual rat. Data are expressed as relative abundance (%). The median of the data is shown. **P<0.01 and *P<0.05 by Kruskal–Wallis (KW) sum-rank test. The distance between Day 0 and 15 of each rat was calculated using the (d) UniFrac and (e) Bray–Curtis distances, mean±s.e.m. **P<0.01 and *P<0.05 by Student's t-test. Canonical analysis of principal coordinates (CAP) of the gut microbiota in (f) permissive (n=12) and (g) resistant (n=12) rats prior to (Day −2 and 0), during (Day 15 and 16) and after FMP administration period (Day 17, 18, 20 and 30).
Gut microbiota robustness observed in ‘resistant' rats
To estimate the effect of the FMP intervention on the gut microbiota structure, we calculated the phylogenetic dissimilarities of gut microbial communities between time points at baseline and after the end of the FMP intervention for each rat using weighted UniFrac and Bray–Curtis methods. The results showed that the variation of the gut microbiota induced by the FMP intervention was higher in permissive rats than in resistant ones (Figures 2d and e). In line with this observation, a CAP (Anderson and Willis, 2003) based on Bray–Curtis distance revealed a greater dispersion of the gut microbiota composition in the permissive compared with resistant rats during the FMP intervention (Figures 2f and g). We further used a cross-validated correct classification rates to quantify such dispersion using the first 46 and 37 principal coordinates accounting for 84.9% and 79.2% of the total variations in the permissive and resistant groups, respectively. Consistent with the previous analysis, the correct classification rates of the permissive group was higher (62.9%) compared with the resistant group (47.4%). The CAP model of the rats in the second independent experiments had a similar distribution (Supplementary Figure S7). Using a permutational multivariate analysis of variance, a method that can assess the effects of factors directly based on Bray–Curtis distance matrices (McArdle and Anderson, 2001), we confirmed that the FMP intervention induced limited changes in the gut microbiota structure of the resistant compared to permissive rats (Supplementary Table S3).
To further assess the resilience of the gut microbiota, we focused our analysis on the kinetics of FMP-modulated phylotypes as identified by LEfSe. In the resistant group, the FMP increased Turicibacteraceae and Desulfovibrionaceae, whereas it decreased Porphyromonadaceae, Prevotellaceae and Cyanobacteria (Supplementary Figure S8A and Supplementary Table S4). These taxa recovered to their baseline levels after only 2 days of wash-out. At the lower phylogenetic level, the trend was similar (Supplementary Figure S9). In permissive rats, Lachnospiraceae and Unclassified Bacteroidales were increased upon the FMP administration while unclassified Clostridiales, Ruminococcaceae and Porphyromonadaceae taxa decreased (Supplementary Figure S8B and Supplementary Table S5). In contrast with the resistant group, the majority of these phylotypes (four out of five) did not recover to their baseline levels at Day 17; indicating a lower degree of resilience of the gut microbiota in permissive rats.
Transfer of the permissive and resistant phenotypes by gut microbiota transplantation
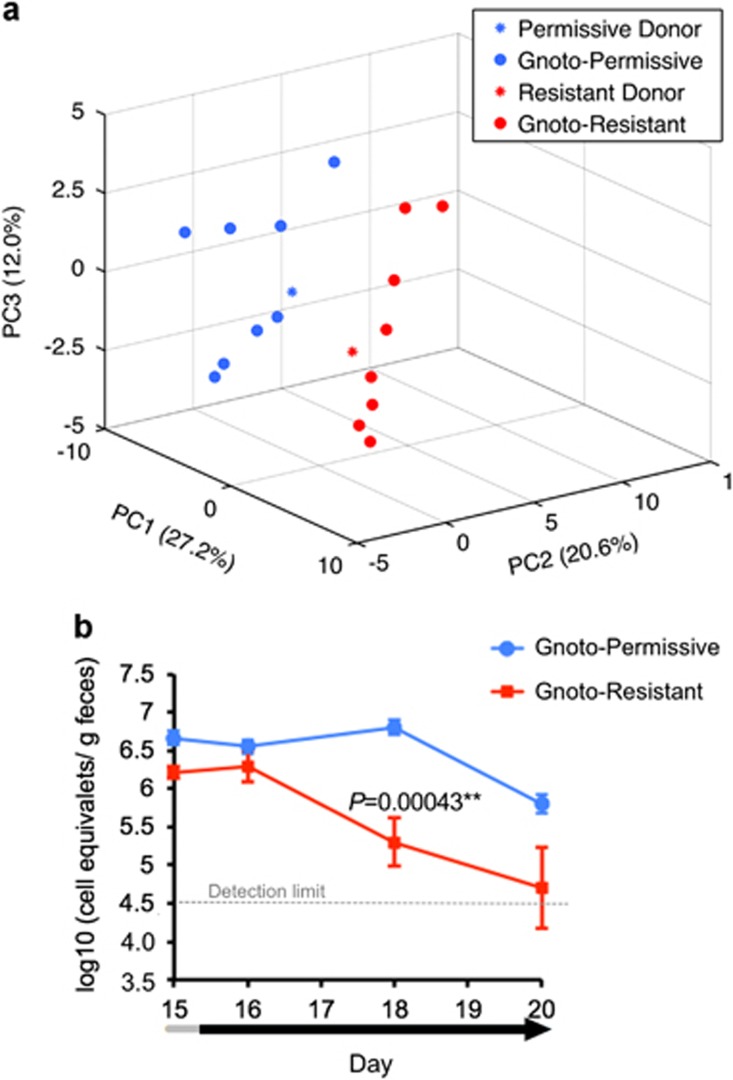
Fecal microbiota samples from selected donors of the permissive and resistant groups were transplanted into separate groups (n=8 × 2) of 8-week-old male Fisher 344 germ-free rats. Four weeks after transplantation, we verified that the gut microbiota of the recipient rats resembled those of the donors (Figure 3a and Supplementary Figure S10). Next, we administered the FMP to the two groups of conventionalized rats over 15 days and assessed the transit time using G. stearothermophilus spores. We confirmed that the transit time of the conventionalized rats were similar across recipient rats (Supplementary Figure S11) and observed that L. lactis CNCM I-1631 persisted longer in permissive recipients compared with resistant recipients (Figure 3b), indicating that permissivity for L. lactis CNCM I-1631 was microbiota-dependent.
Figure 3.
Transplantation of fecal microbiota from permissive and resistant donors into germ-free rats. (a) Principal component analysis (PCA) of the fecal bacterial communities of permissive or resistant donors and their recipients. (b) Abundance of L. lactis in gnotobiotic rats inoculated with a permissive microbiota (Gnoto-permissive; n=8) or a resistant microbiota (Gnoto-resistant; n=8). Fecal samples were collected after 15 days of daily FMP administration and during the wash-out period. Data are expressed as log10 (equivalent cells gm−1 feces) and mean±se.m. for each group. **P<0.01 by Student's t-test.
Evidence of permissive and resistant phenotypes in human
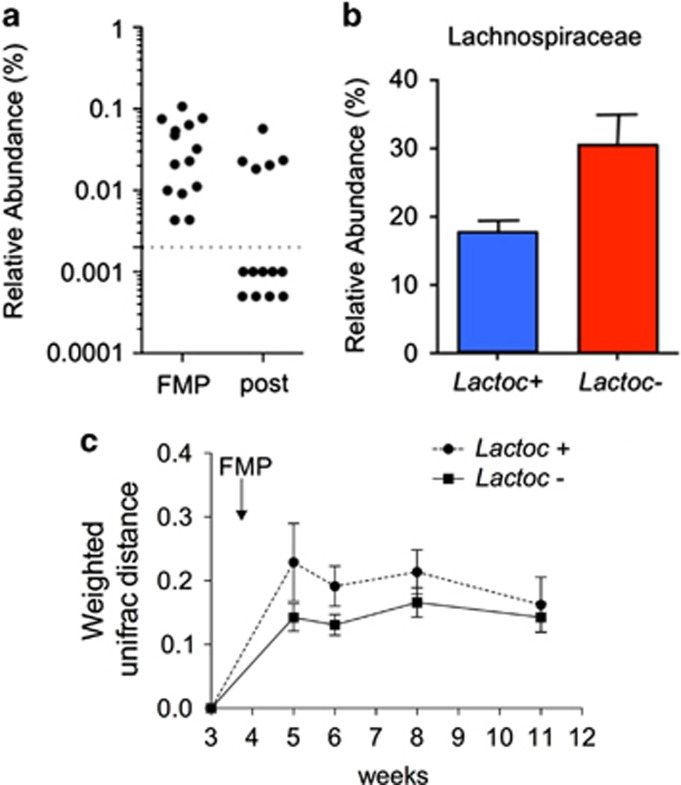
To test whether humans contain gut microbiota signatures similar to permissive and resistant rats, we examined the gut microbiota data from a previous clinical trial, which used the same FMP in 14 healthy females and followed a design resembling the one of our rat study (McNulty et al., 2011). Based on 16S rDNA sequencing data, all individuals were positive for Lactococcus during the FMP consumption period, and Lactococcus prevalence decreased from 100 to 36% during the wash-out period (Figure 4a). This strong correlation between Lactococcus levels and the FMP consumption periods indicated that Lactococcus abundance was a reliable marker of L. lactis I-1631′s shedding. Lactococcus kinetics data enabled us to discriminate between two groups of subjects: the Lactococcus carriers (n=5) and the Lactococcus non-carriers (n=9) during the wash-out period; analogs of the permissive and resistant rats (Figure 4a). Next, we analyzed the baseline gut microbiota of the Lactococcus carriers and non-carriers. At the family level, Lactococcus carriers had a higher relative abundance of Barnesiellaceae (P=0.01), Odoribacteraceae (P=0.03) and Clostridiaceae (P=0.05) and tended to have a higher relative abundance of Streptococcaceae (P=0.08) and Lachnospiraceae (P=0.109) (Mann–Whitney test, Figure 4b and Supplementary S12). Although it did not reach statistical significance, the abundance of Lachnospiraceae was the only common signature between humans and rats discriminating Lactococcus carriers from non-carriers (Figure 4b). We also observed that Lachnospiraceae was the only family with comparable levels in humans and rats (Supplementary Figure S13), whereas the others (Barnesiellaceae, Clostridiaceae, Streptococcaceae and to a lesser extend Odoribacteraceae) were less abundant in rats (Supplementary Figure S13).
Figure 4.
Evidence of permissive and resistant phenotypes in human (a) Distribution of Lactococcus carriers (Lactoc+) and non-carriers (Lactoc–) during and after the FMP administration. (b) Relative abundance of Lachnospiraceae in Lactococcus carriers and non-carriers. (c) Kinetics of weighted UniFrac distances of Lactoc+ and Lactoc– subjects expressed as mean±s.e.m. A linear mixed model showed a difference (P=0.086) between groups across the intervention. x axis label (weeks) were numbered as per the Mc Nulty et al. study.
To assess the effect of the FMP on the structure of the gut microbiota of Lactococcus carriers and non-carriers, we calculated the weighted Unifrac and Bray–Curtis distances between the two groups during baseline and the FMP consumption period. We used a linear mixed model to test the difference between the kinetics of the groups and observed a trend (P=0.086) toward higher inter-individual Weighted UniFrac distances in the Lactococcus carriers compared with the non-carriers (Figure 4c). Interestingly, this difference reached its maximum after the first week (P=0.0502). No significant difference was observed with the Bray–Curtis metrics.
Discussion
Our study sheds light on the inter-individual variability of ecological forces at play in resistance and resilience mechanisms in response to allochthonous bacteria. We showed that individuals with a ‘resistant' phenotype, as assessed by clearance of L. lactis following FMP intervention, have a more robust microbiota leading to limited FMP-induced changes and faster resilience kinetics compared with permissive rats. Of interest, similar trends were observed in humans despite differences between rat and human gut microbiota and limited statistical power (cf. Supplementary Material; Supplementary Figure S11–S13).
There is increasing evidence that food-borne bacteria, principally contained in fermented products, are biologically active in the colon (Oozeer et al., 2005; McNulty et al., 2011; David et al., 2014; Veiga et al., 2014; Eloe-Fadrosh et al., 2015), indicating that they might participate in gut microbial community function. In some cases, such activities might be part of the mechanisms underlying beneficial effects. The FMP used in this study has been shown to ameliorate symptoms associated with irritable bowel syndrome in humans and to decrease intestinal inflammation in mice with variable efficacy across individuals (Veiga et al., 2010; Marteau et al., 2013). Our results suggest that overall ecological robustness of the gut microbial community might be a previously unrecognized cause of inter-individual variation responses to FMP interventions in humans and rodents.
A previous study showed that baseline levels of resident Enterobacteriaceae predicted the susceptibility of mice to be colonized by Salmonella, a pathogen belonging to the Enterobacteriaceae family (Stecher et al., 2010). This observation led the authors to define the ‘like-to-like' rule, which predicts that gut microbiota with high abundance of autochthonous Enterobacteriaceae are more likely to provide favorable conditions for allochthonous Enterobacteriaceae—including pathogens such as Salmonella. The same concept seemed to hold true for the non-pathogenic bacteria Lactobacillus reuteri (Stecher et al., 2010). In our rat data set, the relative abundance of Streptococcaceae (to which L. lactis belongs to) was extremely low (<0.034%) and rare (only detected in two rats), excluding the possibility of a strong association with our phenotypes. In our human cohort, the levels of Streptococcaceae were lower in the permissive individuals suggesting that the ‘like-to-like' rule might not prevail in the case of the persistence of L. lactis. Similarly, detection of Lactobacillus rhamnosus DR20 after a 6-month-consumption period was inversely associated with the baseline abundance of fecal Lactobacillus spp. (Tannock et al., 2000).
Consistent with the known ecological niches of L. lactis (for example, dairy and plant environments (Bachmann et al., 2012)), we and others (Kimoto et al., 2003) have shown that L. lactis does not persist >3 days in the colon of conventional animals. However, in the absence of ecological competition (that is, germ-free animals), L. lactis can colonize and thrive in the mouse intestines (Roy et al., 2008) demonstrating its capacity to utilize gut-derived carbon sources. Proteomic analyses of L. lactis mono-colonized mice indicated a shift of the lactococcal metabolism from lactose catabolism to N-acetylglucosamine and mannose utilization (Roy et al., 2008). Known sources of mannose and N-acetylglucosamine in the intestine are extracellular glycans (that is, mucins) produced by the gut epithelium (Derrien et al., 2010). Inactivation of the E. coli genes involved in mannose or N-acetylglucosamine utilization led to an impairment of E. coli growth in mouse intestine; showing that these two carbohydrates are relevant carbon sources for metabolically active bacteria in the gut (Fabich et al., 2008). A previous study demonstrated that intestinal growth rates and adhesion are two factors influencing the kinetics of elimination of exogenous bacteria (de Jong et al., 2007). In resistant rats, L. lactis CNCM I-1631 was eliminated as fast as the transit marker suggesting a passive clearance with no growth or adhesion of the ingested strain. In permissive rats, L. lactis CNCM I-1631 was retained longer in the colon; raising the possibility that the ingested strain can either grow in vivo, adhere to intestinal wall or both. Growth of L. lactis in the gut requires carbon sources such as N-acetylglucosamine and/or mannose as discussed above. These two carbohydrates are likely to be highly demanded by members of the gut microbiota using such carbon sources. Lachnospiraceae and Ruminococcaceae species are among the known species able to utilize mucin-derived carbohydrates (Tailford et al., 2015), suggesting a possible competition between Ruminococcaceae and/or Lachnospiraceae. As the FMP consumption was associated with a decrease of Ruminococcaceae and an increase of Lachnospiraceae in permissive rats, one can hypothesize that L. lactis competes with Ruminococcaceae. Another hypothesis would be that L. lactis, and possibly the other bacteria of the FMP, might stimulate Lachnospiraceae which in turn compete with Ruminococcaceae. This last hypothesis is supported by prior studies showing that the same FMP increased Lachnospiraceae species in human (Veiga et al., 2014) and in a mouse model of intestinal inflammation (Veiga et al., 2010). No effects of the FMP on the Ruminococcaceae family were reported in these studies (Rooks et al., 2014; Veiga et al., 2010, 2014).
Adhesion is another factor ascribed to probiotic persistence in the gut (Mandlik et al., 2008). In silico analyses of L. lactis CNCM I-1631 genome showed that proteins with host-binding domains were predicted to be anchored to L. lactis' peptidoglycan via a Sortase A (SrtA)-dependent mechanism (Supplementary Information; Supplementary Table S6). To further assess the functionality of these L. lactis cell wall proteins in vivo, we piloted an experiment with a L. lactis ΔsrtA mutant, which was administered to the conventionalized ex-gnotobiotic rats. We observed a shorter persistence of this ΔsrtA mutant in permissive individuals (Supplementary Figure S14E). Although these preliminary data require to be confirmed with a complemented ΔsrtA mutant to unambiguously confirm the role of SrtA in the persistence of L. lactis, the correlation between the inactivation of the SrtA enzyme and the loss of lactococcal persistence in permissive rats already allows us to hypothesize that lactococcal adhesion might be involved in the observed permissive phenotype. In addition, genes encoding surface exposed proteins, or other pathways, might be differentially regulated in permissive and resistant rats.
Our study also sheds light on the inter-individual variability of the gut microbiota of animals housed in the same facility. This observation is consistent with previous studies, which reported intra-facility variations of gut microbiota-related features such as enterotype distribution, antibiotic response or mucus permeability (da SQ-MK et al., 2005; Rooks et al., 2014; Jakobsson et al., 2015).
In humans, 4 out of 14 individuals showed a 2-week persistency of B. lactis CNCM I-2494 after the end of the FMP consumption, indicating that this strain can integrate and transiently persist in some humans in contrast to what we observed in rats. In general, the intestinal abundance of Bifidobacterium in human is higher in average (~4.5%) (Arumugam et al., 2011) compared with laboratory rats, which harbor in average<0.1% Bifidobacterium as observed by us and others (da SQ-MK et al., 2005; Delroisse et al., 2008; Ketabi et al., 2011; Dossou-Yovo et al., 2014). This difference may reflect intestinal conditions and/or diets more favorable to the growth of Bifidobacterium species (including B. lactis CNCM I-2494) in humans compared with laboratory rats.
Collectively, our results suggest that the composition of the autochthonous gut microbial community might account for the amplitude and persistence of effects of supplements containing allochthonous microbes (for example, symbionts or probiotics). Beneficial microbes-based therapies directed at ‘resistant' individuals might require a tailor-made diet able to maximize the transit time of beneficial microbes. Recent studies have shown that diet and dairy matrix might account for the survival and persistence of lactic acid bacteria in the gut (Zhou et al., 2008; Tachon et al., 2014). In our case, further studies using an FMP without probiotics or probiotic species without FMP are warranted. In conclusion, evaluation of gut microbiota directed therapies involving administration of allochthonous microbes should consider the endogenous gut microbiota as a stratifying factor.
Accession numbers
The raw pyrosequencing and Illumina read data for all samples has been deposited in the Sequence Read Archive under the accession number SRP055846.
Acknowledgments
We thank Jean-Christophe Piard for kindly sharing the pDelA plasmid, Chloë Béal for her technical help, Nathan McNulty and Tanya Yatsunenko for generously sharing clinical study data, Timothy Swartz for his critical reading of the manuscript. We thank Thierry Haddad, Sacha van Hijum and Miaomiao Zhou for their kind contribution to the LocateP analysis of our strain (Zhou et al., 2008). We wish to thank T Meylheuc (INRA- MIMA2 platform: www.jouy.inra.fr/mima2) for SEM analysis. The quantitative reverse transcription PCR used was operated under Yakult License (YIFSCAN technology).
Author contributions
CZ and FL conducted animal trial and microbiota analyses; MD and RB analyzed the data from the human clinical trial; M-C D, GQ, PG performed the L. lactis ΔsrtA-related work; SB, JK performed the adhesion assays. PV, JD, CZ, MD, JV, WG participated in the experimental design and the manuscript writing.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson MJ, Willis TJ. (2003). Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84: 511–525. [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al. (2011). Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500: 232–236. [DOI] [PubMed] [Google Scholar]
- Bachmann H, Starrenburg MJ, Molenaar D, Kleerebezem M, van Hylckama Vlieg JE. (2012). Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res 22: 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da SQ-MK, Costa GE, da Silva N, Reis SM, de Oliveira AC. (2005). Bifidogenic effect of dietary fiber and resistant starch from leguminous on the intestinal microbiota of rats. Nutrition 21: 602–608. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong P, Vissers MM, van der Meer R, Bovee-Oudenhoven IM. (2007). In silico model as a tool for interpretation of intestinal infection studies. Appl Environ Microbiol 73: 508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Cochetiere MF, Montassier E, Hardouin JB, Carton T, Le Vacon F, Durand T et al. (2010). Human intestinal microbiota gene risk factors for antibiotic-associated diarrhea: perspectives for prevention. Risk factors for antibiotic-associated diarrhea. Microb Ecol 59: 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delroisse JM, Boulvin AL, Parmentier I, Dauphin RD, Vandenbol M, Portetelle D. (2008). Quantification of Bifidobacterium ssspp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol Res 163: 663–670. [DOI] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H et al. (2013). Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. (2010). Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1: 254–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, van Hylckama Vlieg JE. (2015). Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 23: 354–366. [DOI] [PubMed] [Google Scholar]
- Dossou-Yovo F, Mamadou G, Soudy ID, Limas-Nzouzi N, Miantezila J, Desjeux JF et al. (2014). Metronidazole or Cotrimoxazole therapy is associated with a decrease in intestinal bioavailability of common antiretroviral drugs. PloS One 9: e89943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouault S, Anba J, Corthier G. (2002). Streptococcus thermophilus is able to produce a beta-galactosidase active during its transit in the digestive tract of germ-free mice. Appl Environ Microbiol 68: 938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloe-Fadrosh EA, Brady A, Crabtree J, Drabek EF, Ma B, Mahurkar A et al. (2015). Functional dynamics of the gut microbiome in elderly people during probiotic consumption. mBio 6: e00231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D et al. (2008). Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson HE, Rodriguez-Pineiro AM, Schutte A, Ermund A, Boysen P, Bemark M et al. (2015). The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 16: 164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketabi A, Dieleman LA, Ganzle MG. (2011). Influence of isomalto-oligosaccharides on intestinal microbiota in rats. J Appl Microbiol 110: 1297–1306. [DOI] [PubMed] [Google Scholar]
- Kimoto H, Nomura M, Kobayashi M, Mizumachi K, Okamoto T. (2003). Survival of lactococci during passage through mouse digestive tract. Can J Microbiol 49: 707–711. [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 108: 4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JM, Eisen JA, Zivkovic AM. (2014). The microbes we eat: abundance and taxonomy of microbes consumed in a day's worth of meals for three diet types. PeerJ 2: e659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval L, Martin R, Natividad JN, Chain F, Miquel S, Desclee de Maredsous C et al. (2015). Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes 6: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM et al. (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345: 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. (2008). Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol 16: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges AR, Labbe A, Loo VG, Atherton JK, Behr MA, Masson L et al. (2010). Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridum difficile-associated disease. J Infect Dis 202: 1877–1884. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L et al. (2006). Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteau P, Guyonnet D, Lafaye de Micheaux P, Gelu S. (2013). A randomized, double-blind, controlled study and pooled analysis of two identical trials of fermented milk containing probiotic Bifidobacterium lactis CNCM I-2494 in healthy women reporting minor digestive symptoms. Neurogastroenterol Motil 25: 331–e252. [DOI] [PubMed] [Google Scholar]
- Martin R, Miquel S, Chain F, Natividad JM, Jury J, Lu J et al. (2015). Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol 15: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle BH, Anderson MJ. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82: 290–297. [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL et al. (2011). The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3: 106ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oozeer R, Furet JP, Goupil-Feuillerat N, Anba J, Mengaud J, Corthier G. (2005). Differential activities of four Lactobacillus casei promoters during bacterial transit through the gastrointestinal tracts of human-microbiota-associated mice. Appl Environ Microbiol 71: 1356–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que JU, Hentges DJ. (1985). Effect of streptomycin administration on colonization resistance to Salmonella typhimurium in mice. Infect Immun 48: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. (2012). Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun 80: 3786–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M et al. (2014). Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J 8: 1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Levenez F, Fouqueray C, Dore J, Collignon A, Lepage P. (2011). Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. J Clin Microbiol 49: 858–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Meyrand M, Corthier G, Monnet V, Mistou MY. (2008). Proteomic investigation of the adaptation of Lactococcus lactis to the mouse digestive tract. Proteomics 8: 1661–1676. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D et al. (2013). Cohabiting family members share microbiota with one another and with their dogs. eLife 2: e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Macpherson AJ, Hapfelmeier S, Kremer M, Stallmach T, Hardt WD. (2005). Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect Immun 73: 3228–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC et al. (2010). Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathogens 6: e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachon S, Lee B, Marco ML. (2014). Diet alters probiotic Lactobacillus persistence and function in the intestine. Environ Microbiol 16: 2915–2926. [DOI] [PubMed] [Google Scholar]
- Tailford LE, Crost EH, Kavanaugh D, Juge N. (2015). Mucin glycan foraging in the human gut microbiome. Front Genet 6: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW, Munro K, Harmsen HJ, Welling GW, Smart J, Gopal PK. (2000). Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol 66: 2578–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J, Furet JP, Bensaada M, Philippe C, Roth H, Rabot S et al. (2015). Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol 17: 4954–4964. [DOI] [PubMed] [Google Scholar]
- Unno T, Choi JH, Hur HG, Sadowsky MJ, Ahn YT, Huh CS et al. (2015). Changes in human gut microbiota influenced by probiotic fermented milk ingestion. J Dairy Sci 98: 3568–3576. [DOI] [PubMed] [Google Scholar]
- van der Waaij D, Berghuis-de Vries JM, Lekkerkerk L-v. (1971). Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hygiene 69: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hylckama Vlieg JE, Veiga P, Zhang C, Derrien M, Zhao L. (2011). Impact of microbial transformation of food on health - from fermented foods to fermentation in the gastro-intestinal tract. Curr Opin Biotechnol 22: 211–219. [DOI] [PubMed] [Google Scholar]
- Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, DuBois A et al. (2010). Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci USA 107: 18132–18137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga P, Pons N, Agrawal A, Oozeer R, Guyonnet D, Brazeilles R et al. (2014). Changes of the human gut microbiome induced by a fermented milk product. Sci Rep 4: 6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollaard EJ, Clasener HA, van Saene HK, Muller NF. (1990). Effect on colonization resistance: an important criterion in selecting antibiotics. DICP 24: 60–66. [DOI] [PubMed] [Google Scholar]
- Wilson KH, Sheagren JN, Freter R, Weatherbee L, Lyerly D. (1986). Gnotobiotic models for study of the microbial ecology of Clostridium difficile and Escherichia coli. J Infect Dis 153: 547–551. [DOI] [PubMed] [Google Scholar]
- Zhou M, Boekhorst J, Francke C, Siezen RJ. (2008). LocateP: genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinformatics 9: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.