Abstract
Overgrowth of filamentous bacteria in activated sludge wastewater treatment plants (WWTPs) leads to impaired sludge settleability, a condition known as bulking, which is a common operational problem worldwide. Filaments with the Eikelboom 0092 morphotype are commonly associated with such bulking episodes. Members of the uncultured B45 phylotype, which is embraced within the phylum Chloroflexi, were recently shown to exhibit this morphology. Although these organisms are among the most abundant populations recorded in activated sludge processes, nothing is known about their metabolic characteristics. In this study, a genome sequence, representing the B45 phylotype, was retrieved from a metagenome generated from an activated sludge WWTP. The genome consisted of two chromosomes and one plasmid, which were 4.0, 1.0 and 0.04 Mbps in size, respectively. A metabolic model was constructed for this organism, based on annotation of its genome, showing its ability to generate energy by respiration, utilizing oxygen, nitrite or nitrous oxide as electron acceptors, or by fermentation of sugars. The ability of B45 members to ferment sugars under anaerobic conditions was validated in situ with microautoradiography—fluorescence in situ hybridization. The provisional name of ‘Candidatus Promineofilum breve' is proposed for this species. This study represents the first detailed information on an uncultured genus of filamentous organisms from activated sludge.
Introduction
The filamentous bacteria in wastewater treatment plants (WWTPs) have attracted considerable attention because of their role in the operational problems of bulking and foaming (Eikelboom, 1975; Nielsen et al., 2009). These organisms are thought to have an important role in the formation of flocs with good settling properties by providing the structural matrix around which floc material aggregates. However, their excessive extension from the floc surface into the bulk liquid may lead to inter-floc bridging, which interferes with floc settleability, causing ‘bulking'. Several filamentous phylotypes with hydrophobic cell surfaces also stabilize the formation of thick foam or scum on the surface of reactors (Petrovski et al., 2011). Bulking and foaming incidents continue to be a serious and global problem, probably due to a lack of information about the basic physiology and ecology of the organisms responsible (Seviour and Nielsen, 2010). As such, most control strategies are nonspecific, empirical, and applied only after the abundance of filaments becomes problematic.
These filamentous bacteria were originally separated on the basis of few morphological characters and staining reactions, leading to the recognition of several morphotypes (Eikelboom, 1975; Jenkins et al., 2004). This scheme is still widely used by the industry, but many of these bacteria have now been properly identified phylogenetically, following their isolation into axenic cultures or by fluorescence in situ hybridization (FISH) (see Nielsen et al., 2009). Notably, several of these filament morphotypes, long associated with bulking and foaming episodes, have now been identified as members of the phylum Chloroflexi. These phylotypes (classified with the MiDAS taxonomy 1.20; McIlroy et al., 2015b) include ‘Kouleothrix' spp. (Eikelboom type 1851; Beer et al., 2002; Kohno et al., 2002), B45 (type 0092; Speirs et al., 2009), P2CN44 (type 0803; Kragelund et al., 2011) and ‘Candidatus Sarcinathrix' spp. (type 0914; Speirs et al., 2011).
Large-scale plant surveys reveal that the Chloroflexi genus-level phylotypes B45 and P2CN44, together with the actinobacterial genus ‘Candidatus Microthrix', are the most abundant filaments in full-scale nutrient removal processes. The relatively high observed abundances of these groups indicates that they are likely the most important filamentous organisms in these systems (Mielczarek et al., 2012; McIlroy et al., 2015b) and they simply must make a substantial contribution to the bulk nutrient transformations observed. Although the ecophysiology of the ‘Candidatus Microthrix parvicella' and P2CN44 phylotypes has been described (Kragelund et al., 2011; McIlroy et al., 2013), minimal ecological and physiological information is available for members of the B45 phylotype.
Members of B45 exhibit the Eikelboom 0092 morphotype of Neisser stain positive short blunt-ended trichomes, which extend from the flocs or are freely suspended in the bulk liquid (Speirs et al., 2009). Semiquantitative survey data suggest that these organisms are particularly well suited to flourish under the dynamic electron acceptor conditions, where carbon substrates prevail under the anaerobic conditions, characteristic of plants designed to remove phosphorus (P) and nitrogen (N) (Speirs et al., 2009; Mielczarek et al., 2012). This suggests an ability for anaerobic carbon storage and/or respiration/fermentation. Neither polyphosphate nor lipid storage has been detected in B45 by standard in situ histochemical staining methods (Speirs et al., 2009). Phylum-level in situ investigations of the Chloroflexi in activated sludge biomass suggest they have a role in degradation of complex polymers and display a preference for sugar utilization (Kragelund et al., 2007, 2011). Whether this holds true for B45 phylotype members remains to be determined.
A detailed characterization for most of the abundant activated sludge organisms, including the B45 phylotype, has not been feasible because of our inability to isolate these in pure culture. However, it is now possible to assemble complete genome sequences from metagenome data sets, allowing unprecedented access to detailed information about the metabolic potentials of these organisms (Albertsen et al., 2013). Additional in situ based validation is also essential to understand their ecology as organisms are known to be more specialized than their genome annotation data or behavior in axenic culture would suggest (for example, Kindaichi et al., 2013; McIlroy et al., 2015a). Combining genomic-based investigations and in situ analyses proved to be powerful tools in determining the ecology of ‘Candidatus M. parvicella' (Andreasen and Nielsen, 2000; McIlroy et al., 2013; Muller et al., 2014).
In this study, a complete genome for a representative of the B45 phylotype was assembled from a metagenome prepared from a full-scale activated sludge WWTP. A genome-based metabolic model is proposed and partially validated in situ with microautoradiography (MAR)-FISH. The data presented here give the first detailed insight into the ecology of this biotechnologically important filament and extends the comparatively poor genome sequence coverage of the Chloroflexi phylum.
Materials and methods
Sampling
All biomass samples for this study were taken from activated sludge WWTPs in Slovenia and Denmark. Sample details are given in Supplementary Table S1.
Metagenomics and genome assembly
Biomass samples for metagenomic analyses were taken from Kočevje WWTP in Slovenia (Supplementary Table S1). DNA was extracted using the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA), essentially as described by the manufacturer, from two 0.5 ml aliquots of activated sludge (designated as K1 and K2). Bead beating was performed at 6 m s–1 with a Fast Prep FP120 (MP Biomedicals). Beating times were varied for the two aliquots, with 4 × 40 s and 1 × 40 s applied for K1 and K2, respectively.
Two paired-end sequencing libraries, K1.PE and K2.PE, were prepared using the Nextera DNA Library Preparation Kit (Illumina, San Diego, CA, USA) and the TruSeq DNA PCR-Free Library Preparation Kit (Illumina), respectively. A mate-pair sequencing library (K2.MP) was prepared using the Nextera DNA Library Preparation Kit (Illumina). Sequencing libraries were validated with gel electrophoresis using Tapestation2200 and High Sensitivity D1K screentapes (Agilent Technologies, Santa Clara, CA, USA). The K1.PE and K2.MP libraries were sequenced (2 × 150 bp) on an Illumina HiSeq2000 using the TruSeq PE Cluster Kit v3-cBot-HS and TruSeq SBS kit v.3-HS sequencing kit (Illumina). The K2.PE library was sequenced (2 × 300 bp) on an Illumina MiSeq instrument using MiSeq reagent kit v3 (Illumina).
NextClip (Leggett et al., 2014) was applied to trim (min length 100) the K2.MP mate-pair FASTQ data and remove poor quality pairs (category D). Pre-processed mate-pair reads and paired-end FASTQ data were imported into CLC Genomics Workbench v. 7.0.3 (CLC Bio, Aarhus, Denmark) and processed. The K1.PE and K2.PE paired-end data were trimmed using the Trim Sequences tool (phred ⩾20, min length 64 bp, no ambiguous bases and removal of Truseq/Nextera adaptor sequences). De novo metagenome assembly was performed using the De Novo Assembly tool (word size 64, automatic bubble size, min scaffold length 1000 bp, auto-detect paired distances, perform scaffolding) with K2.PE and K2.MP libraries as input. Assembly read coverage in K1.PE, K2.PE and K2.MP was estimated individually by mapping each library to the metagenome assembly using the Map Reads to Reference tool (length: 0.95, similarity: 0.95).
The mmgenome workflow (M Albertsen and others, unpublished: http://madsalbertsen.github.io/mmgenome/) was used to extract the genome by utilizing the differential coverage in samples K1 and K2 introduced by the different bead-beating time during DNA extraction. In short, open reading frames were predicted in the metagenome assembly using Prodigal (Hyatt et al., 2010). The translated open reading frames were screened for 107 essential single copy genes using HMM models (Dupont et al., 2012) and HMMER3(-cut_tc) (http://hmmer.janelia.org/). The positive hits were taxonomically classified using BLASTP (e-value: 1e-5) against the RefSeq protein database (v. 52), and MEGAN (Huson et al., 2011). Putative scaffold connections were extracted with the network.pl script on the aforementioned read mappings exported in SAM file format. Metagenome sequences, essential gene taxonomic information and coverage estimates were imported into R (v. 3.1.3, R Foundation for Statistical Computing, Vienna, Austria) using RStudio (v. 0.98.953, RStudio, Inc., Boston, MA, USA), and the genome bin was extracted using the mmgenome R-package.
Based on the initial genome bin, the genome was re-assembled and curated manually. The bin was imported into CLC workbench v. 7.0.3 (CLC Bio) and each of the three libraries was mapped to the genome bin using the Map Reads to Reference tool (length: 0.7, similarity: 1.00). The mapped reads were extracted and links between broken paired reads were re-established with a custom bash script. The extracted paired reads were used for reassembly using SPAdes 3.1.1 (—only-assembler, —careful, -k 21,33,55,77,99,127) (Bankevich et al., 2012). The assembly was curated manually by iterative cycles of (A) read mapping in CLC genomics workbench (see above); (B) tracking broken paired reads in the read mappings using the Find Broken Pair Mates tool in CLC genomics workbench as well as Circos (Krzywinski et al., 2009) and Cytoscape (Cline et al., 2007), following the workflow from Albertsen et al. (2013); (C) breaking up scaffolds and/or manually scaffolding in R using the packages biostrings (Pages et al., 2014) and rtracklayer (Lawrence et al., 2009). (D) Gaps between scaffold contigs were filled using GapFiller v.1.11 (-m 30 –o 10 –t 50 –d 2000 –n 20 –i 1) (Boetzer and Pirovano, 2012). Single-nucleotide polymorphisms were detected by read mapping and resolved manually.
The raw metagenome reads and the annotated genome sequence data have been submitted to the European Nucleotide Archive (ENA) under the study accession number PRJEB11413.
Genome annotation and metabolic model reconstruction
The assembled genome was uploaded to the automated Microscope platform (Vallenet et al., 2009). Automatic annotations were manually curated for the genes involved in selected metabolic pathways of interest based on the analyses of several integrated bioinformatics database tools that include UniProt (The UniProt Consortium, 2012), COG (Tatusov et al., 2003), InterPro (Hunter et al., 2009) and PRIAM (Claudel-Renard, 2003). Assessment of pathways was also assisted by the integrated MicroCyc (Caspi et al., 2014) and KEGG (Kyoto Encyclopedia of Genes and Genomes; Kanehisa et al., 2014) databases.
Fluorescence in situ hybridization
FISH with the CFX197 probe, designed to cover the B45 phylotype (Speirs et al., 2009), was performed essentially as described previously (Nielsen, 2009), at a hybridization buffer formamide concentration of 40% [v/v]. Both the 5′ and 3′ ends of oligonucleotide probe were labeled with the sulfoindocyanine dye Cy3 (DOPE-FISH; Stoecker et al., 2010). The EUBmix probes, targeting most bacteria (Amann et al., 1990; Daims et al., 1999), were labeled with either 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (FLUOS) or Cy5. The NON-EUB nonsense probe was used as a negative hybridization control (Wallner et al., 1993). Microscopic analysis was performed with either an Axioskop epifluorescence microscope (Carl Zeiss, Oberkochen, Germany) or a LSM510 Meta laser scanning confocal microscope (Carl Zeiss).
Microautoradiography
Activated sludge was collected from the aeration tank of the Randers and Ejby Mølle WWTPs (Supplementary Table S1). Biomass from the Kočevje WWTP in Slovenia, from which the metagenome was prepared, was not used because of the need for fresh biomass for the MAR-FISH procedure. This assumes that the B45 members in these plants have a similar physiology; noting that the MAR-FISH and genome-based analyses was consistent (see later). The MAR-FISH procedures were carried out within 24 h of sampling essentially as described previously (Nielsen and Nielsen, 2005). Before MAR incubations, the activated sludge was aerated for 1 h to remove any residual organic substrates present. An additional 1-h anaerobic pre-incubation was performed for all anoxic incubations to remove traces of electron acceptors present. Pre-incubated mixed liquor was diluted to 1 g SS l–1 with sludge supernatant, giving a total volume of 2 ml, in 10 ml serum bottles sealed with thick rubber stoppers. Unlabeled and labeled substrates were added to give final concentrations of 2 mM and 10 μCi ml–1, respectively. Radiolabeled compounds used included: [14C]-propionate, [14C]-butyrate, [3H]-amino acid mix, [3H]-ethanol, [3H]-glycerol, [3H]-N-acetyl glucosamine (American Radiolabeled Chemicals Inc., St Louis, MO, USA); [3H]-acetate, [14C]-pyruvate, [3H]-glucose (PerkinElmer Inc., Waltham, MA, USA); [3H]-oleic acid (Amersham Bioscience, Hillerød, Denmark). Uptake of all substrates was assessed under aerobic conditions and those taken up were additionally assessed under anoxic conditions. To achieve anoxic conditions, oxygen was removed before substrate addition by repeated evacuation of headspace and injection of oxygen-free nitrogen. After 3-h incubation, the cells were fixed immediately with cold 4% [w/v] paraformaldehyde (final concentration) and washed three times with sterile filtered tap water. Aliquots of 20 μl were homogenized gently between glass coverslips. Following FISH, slides were coated with Ilford K5D emulsion (Polysciences Inc., Warrington, PA, USA), exposed in the dark for 8 days at 4 °C and developed with Kodak D-19 developer (Artcraft Camera and Digital, Kingston, NY, USA).
In situ staining protocols
Activated sludge was sampled from the aeration tank of the Aalborg West WWTP (Supplementary Table S1) and aerated for 1 h before staining. The presence of exoenzymatic activity was determined using enzyme-labeled fluorescence substrates (ELF-97, Molecular Probes, Eugene, OR, USA) for chitinase, esterase, β-d-galactosidase, β-d-glucuronidase and phosphatase activities, as described in Kragelund et al. (2005). Exoenzymatic activity of α-amylase was tested using Enz-Chek Ultra Amylase Assay Kit (Molecular Probes) following the manufacturer's instructions. Metabolic inhibitors were added to exclude false-positive signals from cells feeding on the fluorescent product of hydrolyzed substrate as detailed by Xia et al. (2008). After staining, the samples were applied to microscopic slides, air-dried and subjected to the FISH procedure and microscopic analysis as described previously. Cell surface properties were determined using the microsphere adhesion to cells assay as described previously (Zita and Hermansson, 1997). After staining, microscopic images were recorded, FISH was performed as described above and surface hydrophobicity of the recorded B45 filaments was examined after relocation.
Results and Discussion
This study applied metagenomics to obtain genome sequence information from a member of the B45 phylotype, which is among the most abundant bacterial genera of many full-scale activated sludge communities (McIlroy et al., 2015b). The selected sample for metagenomic analyses was highly enriched in filaments with the 0092 morphotype, which were confirmed by FISH as members of the B45 phylotype (see Figure 1a).
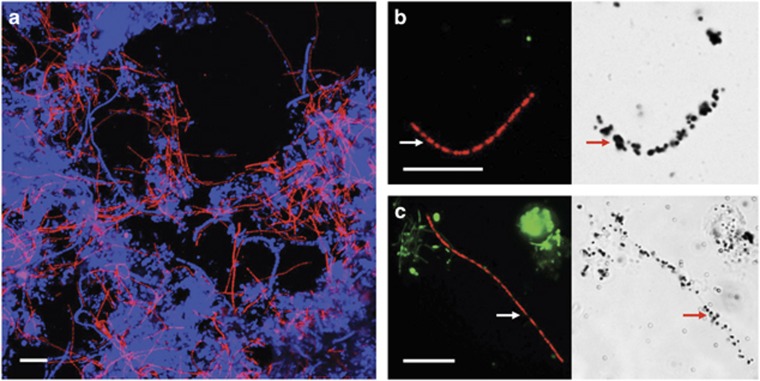
Figure 1.
Micrographs of B45 taxon members in full-scale sludge. (a) FISH image of biomass from the KČN Kŏcevje WWTP (CFX197 (Cy3=red)+EUBmix (Cy5=blue)). (b, c) Representative FISH (CFX197 (Cy3=red)+EUBmix (FLUOS=green)) and corresponding brightfield MAR images after incubation with radiolabeled 3H-glucose under (b) aerobic (Randers WWTP biomass) and (c) anaerobic conditions (Ejby Mølle WWTP biomass). CFX197-positive filaments are indicated with an arrow. Formation of black silver granules on the filament surface in MAR images indicated positive uptake. Scale bars represent 10 μm.
Genome assembly and characteristics of the Chloroflexi genome
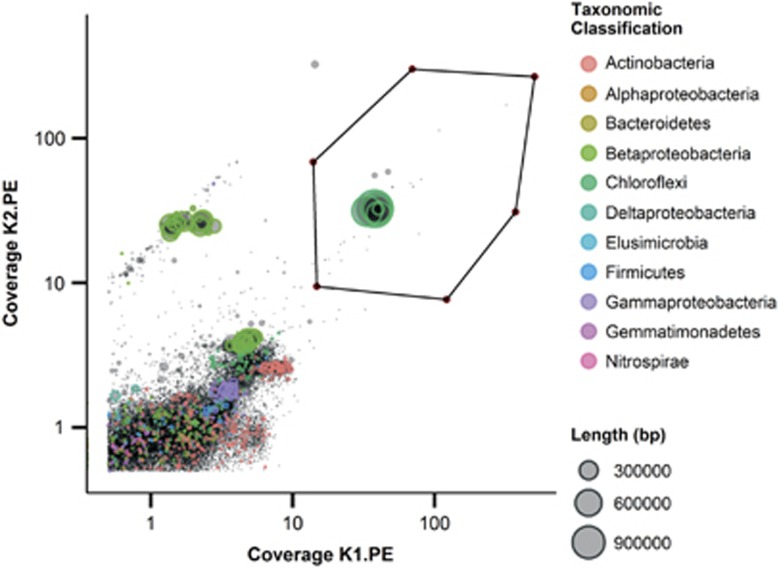
Metagenome sequencing of DNA from the full-scale plant sample resulted in two paired-end libraries (K1.PE, 9.7 Gbp, insert size 190 bp and K2.PE, 5.4 Gbp, insert size 543 bp) and one mate-pair (K2.MP, 2.1 Gbp, insert size 3650 bp). The K1 sample was more enriched, relative to the K2 sample, in the target Chloroflexi, suggesting that use of different bead-beating intensities during DNA extraction can be a powerful aid for differential coverage binning of metagenomes. The Chloroflexi genome bin was extracted using primarily differential coverage, taxonomic color overlay and paired-end information (see Figure 2). The genome reassembly and polishing resulted in three complete circular replicons with a total size of 5 033 966 bp. The statistics for the metagenome assembly, genome bin and final assembly can be found in Table 1 and Supplementary Tables S2 and S3. Annotated essential genes were split between the two larger replicons, indicating that the B45 phylotype genome (designated Cfx-K) consists of two chromosomes and one plasmid. Duplicates of essential single copy genes were observed in the genome, which is usually an indicator of contamination. However, they are believed to be genuine duplicates as no evidence of misassembly, such as broken paired reads or sudden changes in read coverage, was found in the region around the genes. The plasmid was linked to the genome based on read coverage and the presence of an identical nucleotide region of approximately 3200 bp in both the plasmid and chromosome 1. No phage-related genes were found in the repeat region, and based on paired read data no connection between the repeat region and other contigs in the metagenome was observed, which supports the link between the plasmid and the genome. Based on read coverage in sample K1.PE, it is estimated that the relative abundance of the Cfx-K genome in the microbial community was 18.8%.
Figure 2.
Extraction of the Chloroflexi genome bin from the metagenome assembly using differential read coverage and taxonomic classification (phylum) color overlay. The read coverage of the samples has been normalized using the mean sample read coverage to better visualize enrichment difference in the two samples. The axis are log scaled. The genome bin is enclosed by the polygon.
Table 1. Summary of Cfx-K genome properties.
| Property | Replicon 1 | Replicon 2 | Replicon 3 |
|---|---|---|---|
| Size (Mbp) | 4.0 | 1.0 | 0.04 |
| GC content (%) | 64 | 65 | 62 |
| Protein coding density (%) | 92 | 89 | 93 |
| CDS | 3656 | 866 | 61 |
| CDS assigned function (%)a | 23 | 17 | 2 |
| Essential genes | 97b | 9 | — |
| rRNA operons | 1 | — | — |
Abbreviations: CDS, coding sequence; rRNA, ribosomal RNA; GC, guanine-cytosine.
MaGe prediction classes 1–3.
The value does not count duplicates. Further details can be found in Supplementary Table S2.
The arrangement of the Cfx-K genome shares similar properties to those of other bacteria with multiple chromosomes. The difference in G+C mol % content of the two chromosomes is <1%, and the distribution of the ‘essential' single copy genes is skewed toward the dominant larger chromosome, whereas the proportion of unclassified genes is higher for the smaller chromosome (see Table 1). Most of the genes involved in energy metabolism and central carbon transformations (discussed later) are also located on the larger chromosome. Notably, some genes essential for the glycolysis, pentose phosphate, and Calvin–Benson–Bassham pathways, and for the fermentation of pyruvate to acetate (see later), are found only on the smaller chromosome (Supplementary Figure S1). The term ‘chromid' has been suggested as a term to describe additional bacterial chromosomes, given that they share characteristics with both chromosomes and plasmids (Harrison et al., 2010). Chromids are often seen in bacteria, being estimated to constitute approximately 1 in 10 of the available genomes from multiple phyla, including Chloroflexi members (Harrison et al., 2010; Jha et al., 2012). What advantage the split genome has for the Cfx-K species is unclear, with the role of such an arrangement in bacteria being generally poorly understood (Egan et al., 2005; Jha et al., 2012). One prevalent suggestion is that a split genome allows faster DNA and hence cell replication, although an elevated growth rate is not a universal feature of bacteria with divided genomes (Egan et al., 2005).
Phylogeny of the Cfx-K organism
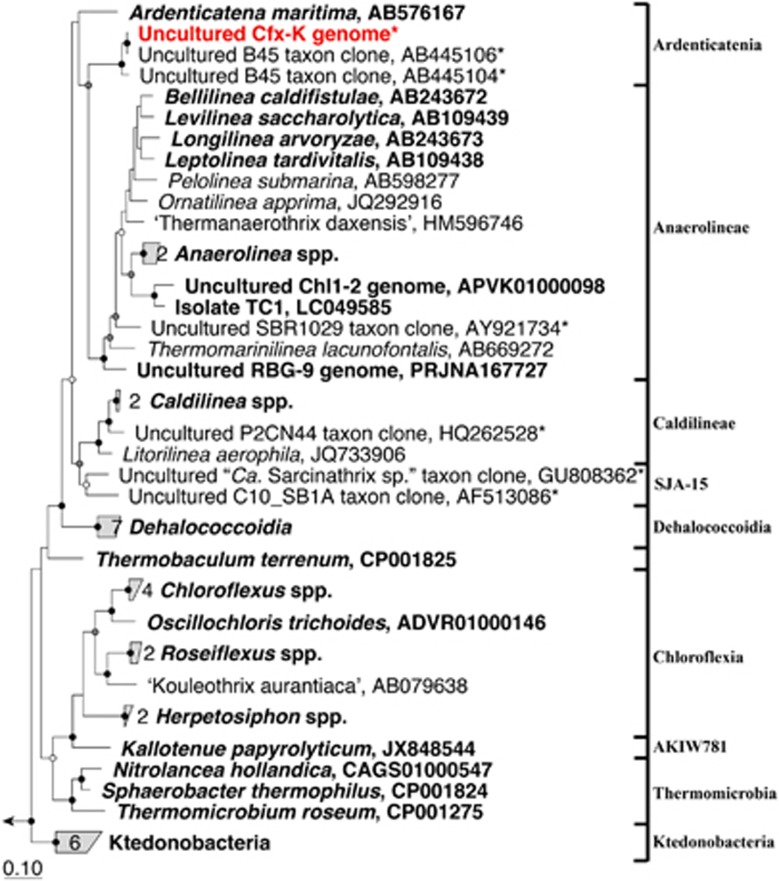
Evolutionary relationships of the Cfx-K genome to those of other Chloroflexi were analyzed and phylogenetic trees, based on both 16 S ribosomal RNA (rRNA) gene sequences and those of the concatenated alignment of 43 conserved marker genes, are shown in Figure 3 and Supplementary Figure S2, respectively. Using 16 S rRNA gene sequence data, the Cfx-K genome is confirmed to be a member of the B45 phylotype, being most closely related to isolated members of the classes Anaerolineae (82–84%), Caldilineae (83%) and Ardenticatenia (83%) (Figure 3). Phylogenetic placement based on the genome sequence is congruent with the 16 S rRNA gene-based phylogeny (Supplementary Figure S2). Given the lack of close relatives, the Cfx-K genome represents the only genome of what is at least a novel order in the phylum (based on the taxonomic sequence identity threshold recommendations of Yarza et al., 2014).
Figure 3.
Maximum-likelihood (PhyML) 16 S rRNA gene phylogenetic tree for members of the phylum Chloroflexi constructed using the ARB software package (Ludwig et al., 2004). The 16 S rRNA gene sequence representing the Cfx-K genome was imported and aligned into the MiDAS database (version 1.20), which is a version of the SILVA database (Release 119 NR99; Quast et al., 2013) curated for activated sludge organisms (McIlroy et al., 2015b). Selected sequences include: isolates, metagenome-derived-genomes and selected representatives of characterized activated sludge uncultured phylotypes. *Indicates sequences from genera that are abundant (median ⩾0.5% of amplicon reads; MiDAS survey; see McIlroy et al., 2015b) in full-scale activated sludge WWTPs in Denmark. Class level classifications are given to the right. Organisms with available genomes (see Supplementary Figure S2) are given in bold typeface. Bootstrap values from 100 re-samplings are indicated for branches with >50% (white circle), 50–70% (gray) and >90% (black) support. The scale bar represents substitutions per nucleotide base.
With the MiDAS taxonomy (McIlroy et al., 2015b), the Cfx-K 16 S rRNA gene falls into the class Ardenticatenia, for which there is only one other described and characterized isolate (Kawaichi et al., 2013), although our analyses supports its possible inclusion into the class Anaerolineae (see Figure 3 and Supplementary Figure S2). The current composition of Chloroflexi classes is based on 16 S rRNA gene sequence phylogeny and includes few isolates and genomes (see Figure 3 and Supplementary Figure S2). Given the weak support for the clustering of the six classes into a single phylum, it has been suggested that membership of the Chloroflexi be confined to members of classes Chloroflexi and Thermomicrobium (Gupta et al., 2013). The generation of further genome sequences is required to resolve the taxonomy of the remaining classes, along with the precise phylogenetic position of members of the B45 genus. Metagenomic approaches, as applied here and in other Chloroflexi studies (Hug et al., 2013; Campbell et al., 2014), will be important in providing coverage of members of the phylogenetic tree presently recalcitrant to isolation into pure culture.
Surface properties of the B45 phylotype
Although members of the Chloroflexi were once proposed to be diderms (organisms with two membranes) because of their Gram-negative stain reaction and presence of multi-layered cell envelopes by electron microscopy, subsequent genome analyses of small numbers of Chloroflexi have reinforced the view that they are indeed monoderms (their cell envelope contains one membrane) (Sutcliffe, 2011). Analysis of the Cfx-K genome was consistent with that of a bacterium with a Gram-positive cell wall architecture (Supplementary Figure S3). Lipopolysaccharides are present in the outer membrane of almost all diderm bacteria (Sutcliffe, 2010). However, no genes associated with the biogenesis of lipopolysaccharides could be identified in the Cfx-K genome (Supplementary Figure S3). Outer membrane proteins with conserved antigenic Pfam domains (PF07244 and PF01103) are also often found in diderm bacteria (Albertsen et al., 2013), but the encoding genes were also absent in the Cfx-K genome. The same conclusion was reached for all examined outer membrane transporter genes, including those associated with the secretin-based type II and III secretion systems (Supplementary Figure S3; Desvaux et al., 2009). Genes encoding membrane-associated proteins with Pfam domains, commonly seen in monoderm bacteria (PF02645, PF03816, PF09269 and PF01424; Albertsen et al., 2013), could all be identified in the Cfx-K genome (Supplementary Figure S3). Sortases are used by almost all monoderm bacteria to anchor covalently surface proteins to the cell wall envelope (Ton-That et al., 2004; Schneewind and Missiakas, 2014). Accordingly, genes encoding a sortase (CFX0092_v1_a1813) and a sortase-like acyltransferase (CFX0092_v1_a1454) were identified in the Cfx-K genome.
Based on the microsphere adhesion to cells fluorescent staining procedure performed in situ, it was concluded that the cell surface of B45 filaments are not hydrophobic in nature, consistent with the observation that members of the 0092 morphotype are not associated with problematic foaming episodes.
Members of the Chloroflexi in activated sludge have been suggested to have an important role in the hydrolysis of complex carbohydrates, a suggestion reinforced by the in situ detection of extracellular surface-associated catalytic enzymes for some species (Kragelund et al., 2007). Annotation of the Cfx-K genome revealed putatively secreted enzymes (based on the detection of a signal peptide sequence (Bendtsen et al., 2004), including an alpha-galactosidase (CFX0092_a0397) and several glycoside hydrolase, esterase and protease family proteins. Filaments targeted by CFX197 FISH probe showed negative in situ results for the limited number of available surface-associated enzymes tested in this study (data not shown). Therefore, involvement of B45 members in the hydrolysis of complex carbon sources remains to be demonstrated.
The potential for aerobic respiration
Cfx-K genome annotation confirmed the potential for aerobic respiration (Supplementary Table S5; Figure 4). Although only few of the described Cfx-K-related species (of the classes Anaerolineae, Caldilineae and Ardenticatenia) are able to utilize oxygen as an electron acceptor (see Supplementary Table S4), aerobic uptake of substrates is a common feature of members of this phylum in full-scale activated sludge systems (Kragelund et al., 2007, 2011) and was also confirmed in situ for the B45 phylotype in this study (see later).
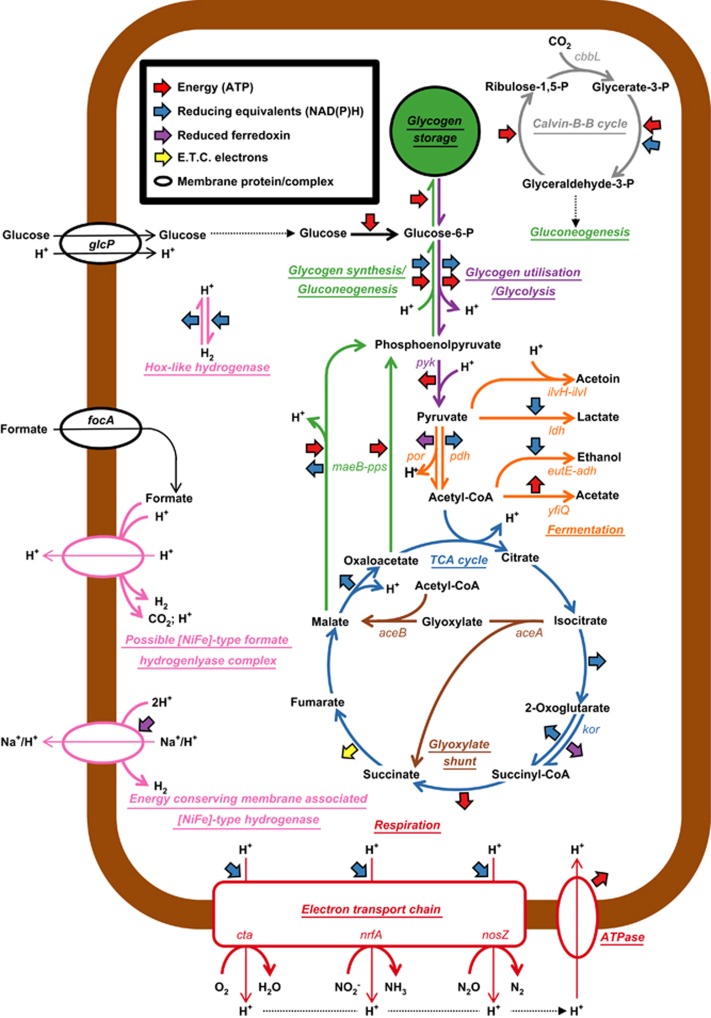
Figure 4.
Metabolic model for members of the B45 phylotype in activated sludge. Selected pathways are color coded and are related to energy and central carbon metabolism. For more detailed information, see Supplementary Table S5 and Supplementary Figure S1.
Potential contribution of the B45 phylotype to denitrification
No putative nitrate reductase gene was annotated in the Cfx-K genome, although the presence of a putative cytochrome c nitrite reductase (NrfA) suggests an ability for dissimilatory nitrite reduction to ammonia (Simon et al., 2000). No nitric oxide reductase (NorBC) encoding gene was detected either, although nitrite reductase (NrfA) has been shown to reduce nitric oxide to ammonia, a reaction that was suggested to be associated with protection from nitrosative stress (Poock et al., 2002). Putative nitrite reductase genes (nrfA) have been found in the genomes of Caldilinea aerophila (65% protein sequence identity to the NrfA of Cfx-K; accession no. WP_044276490) and Anaerolinea thermophila (46% protein identity; accession no. WP_013561276). However, the ability of these isolates to utilize nitrite as an electron acceptor to support anaerobic growth is yet to be assessed (Sekiguchi et al., 2003). The related Ardenticatena maritima genome reveals the potential for reduction of nitrate to either dinitrogen gas or ammonia (Kawaichi et al., 2015), although only the latter product was detected in pure culture studies (Kawaichi et al., 2013).
Interestingly, the Cfx-K genome contains a periplasmic nitrous oxide reductase (NosZ), indicating that nitrous oxide may also act as a terminal electron acceptor under anoxic conditions. A homolog of this gene is also possessed by several other Chloroflexi members; including A. maritima (71% protein sequence identity; GAP62557) and C. aerophila (63% protein sequence identity; WP_014431995).
Central carbon pathways
Annotations of the Cfx-K genome provide data consistent with it being a chemoorganoheterotroph. Candidate genes for a complete tricarboxylic acid cycle, pentose phosphate pathway and Embden–Meyerhof–Parnas glycolysis pathway were annotated, whereas key genes encoding enzymatic steps diagnostic for the Entner–Doudoroff (ED) version of the latter pathway were absent (see Figure 4 and Supplementary Figure S1; Supplementary Table S5).
In situ substrate uptake profile for the B45 phylotype
Substrate specificity is particularly difficult to deduce from genome annotation data, as bacteria are much more specialized in situ than their complete metabolic potential suggests (Kindaichi et al., 2013). Equally, the specificity of transporters is notoriously difficult to infer conclusively from gene and protein sequence homology alone (Gelfand and Rodionov, 2008). As we are interested ultimately in describing the in situ activities of these filaments, characterization with MAR-FISH was applied to assess which substrates were utilized in activated sludge. Positive substrate uptake was observed only with radiolabeled glucose, with no observed uptake of pyruvate, acetate, propionate, butyrate, oleate, glycerol, N-acetyl glucosamine, ethanol or amino acids (see Figure 1 and Supplementary Figure S4). Such a specialized uptake profile is consistent with previous investigations with a wide diversity of the Chloroflexi in activated sludge, which reported a clear preference for sugars (Kragelund et al., 2007).
Ability to generate energy by fermentation
In situ MAR-FISH analyses revealed that B45 members could assimilate glucose under both aerobic and anaerobic conditions (see Figures 1b and c). This is in agreement with the results showing most of the B45-related isolates in the classes Anaerolineae and Caldilineae can grow by fermentation of carbohydrates or amino acids (see Supplementary Table S4) as well as with the genome annotation data of related uncultured Chloroflexi species (Hug et al., 2013; Campbell et al., 2014). Other Chloroflexi members in activated sludge have also been reported as being able to ferment sugars. These include ‘Kouleothrix aurantiaca' and P2CN44 (Kohno et al., 2002; Kragelund et al., 2007; Miura et al., 2007). The annotated Cfx-K genome supports the potential for fermentation of sugars, with acetate, lactate, acetoin and ethanol as possible end products (see Figure 4).
Hydrogen may also be produced during fermentative growth, because of the annotated presence of group 3d NAD-linked and group 4 membrane-bound energy-conserving (NiFe) hydrogenases (Vignais and Billoud, 2007; see Supplementary Figure S5). The putative multi-subunit membrane-bound (NiFe) hydrogenase theoretically couples the oxidation of excess reduced ferredoxin, produced during fermentation, to the reduction of H+ to H2 and the export of a proton, contributing to a proton gradient (Hedderich and Forzi, 2005). A putative formate dehydrogenase H (FdhH) encoding gene suggests that H+ reduction may alternatively be coupled to formate oxidation (Andrews et al., 1997). In the absence of any candidate gene for a pyruvate-formate lyase (EC 2.3.1.54), it is not clear if formate is produced during sugar fermentations. The putative fdhH gene is co-located with that for a probable formate transporter (FocA) (CFX0092_v1_a0448), supporting possible utilization of exogenous sources of formate, which are produced by other fermentative activated sludge bacteria under anaerobic conditions (Kong et al., 2008). The additional putative cytoplasmic Hox-like bi-directional hydrogenase (Schmitz et al., 2002) was detected in the Cfx-K genome, which may act as an electron valve; either oxidizing excess NADH that is generated during fermentation, or by providing reducing power for anabolic pathways when operating in reverse.
Potential for lithotrophy
Interestingly, the Cfx-K genome sequence data indicate an ability for carbon fixation utilizing the Calvin–Benson–Bassham cycle. However, no genes associated with the oxidation of inorganic electron donors, such as nitrite, ammonia or iron, were found in the Cfx-K genome. It is therefore generally unclear as to what role the Calvin–Benson–Bassham pathway has for this organism, noting that it is unlikely to have a major role in the activated sludge environment where it would be an expensive alternative to the use of the available organic carbon.
Potential storage polymers
Pathways for the synthesis and degradation of glycogen were identified in the Cfx-K genome, suggesting that this polysaccharide may serve as a possible storage compound under unbalanced growth conditions. The absence of an annotated polyhydroxyalkanoate synthase (PhaC) indicates an inability for polyhydroxyalkanoate storage, which is supported by in situ Nile blue A staining results (Speirs et al., 2009). However, annotation of a putative acyl-CoA:DAG acyltransferases (AtfA), catalyzing the final step in the synthesis of triacylglycerols, indicates the potential for lipid storage (Kalscheuer and Steinbüchel, 2003). However, triacylglycerols are unlikely to accumulate in large amounts, given that such inclusions would probably stain positive with Nile Blue A (Serafim et al., 2002) and none were detected in the B45 phylotype (Speirs et al., 2009).
A metabolic model for members of the B45 phylotype in activated sludge
Based on the annotation of the Cfx-K genome and in situ analysis, we are able to suggest a metabolic model for the behavior of the B45 phylotype in nutrient removal WWTPs where anaerobic and aerobic conditions are imposed on the microbial community. It seems that these operational conditions favor the growth of this phylotype, as anecdotal evidence suggests they are relatively less abundant in continuous aerated plants (Jenkins et al., 2004).
The key metabolic features of the B45 phylotype include the following: under anaerobic conditions sugars are assimilated and fermented, and presumably these filaments grow. Potential end products of fermentation include acetate, lactate, acetoin, ethanol and hydrogen. Under the subsequent carbon-deficient aerobic conditions that prevail in nutrient removal WWTPs, carbon and energy sources supporting further growth of B45 members may originate from sugars released from the hydrolyzes of cellular detritus and exopolysaccharides. Whether or not B45 members directly contribute to the hydrolysis of complex carbon sources remains to be demonstrated. The aerobic utilization of complex organic material has been demonstrated for other members of the Chloroflexi in nutrient removal systems (Miura et al., 2007; Kragelund et al., 2011). Nitrite and nitrous oxide may both serve as alternate electron acceptors for the strain with the Cfx-K genome. Other organisms may provide nitrous oxide, with some known to produce this compound as the end product of denitrification. Such groups in activated sludge include some Tetrasphaera spp. (Kristiansen et al., 2013), which is the most abundant genus in Danish full-scale treatment plants (McIlroy et al., 2015b).
Concluding remarks
Members of the B45 phylotype represent one of the most abundant filamentous bacteria in activated sludge treatment plants and thus likely have a substantial impact on both nutrient transformations and sludge settleability of nutrient removal WWTPs. In this study, we have used metagenomics and in situ FISH-based characterization to successfully elucidate their basic metabolic activities in these systems. Members of the group appear to be facultative anaerobes, being potentially metabolically active under both aerobic and anoxic conditions, which may explain their presence usually in higher abundance in plants of this configuration than in continuously aerated processes. They are fermentative organisms with a preference for sugars as substrates. This attribute should be kept in mind if B45-linked bulking episodes follow supplementation of carbon with dosing of sugar-based products, such as molasses, or changes in the composition of wastewater from industries. The retrieval of the genome provides the foundation for more detailed investigation of these process important bacteria. In addition, this genome importantly extends the relatively poor coverage of the Chloroflexi phylum, being the first of what is at least a novel phylogenetic order.
Taxonomic proposal for ‘Candidatus Promineofilum breve'
In the absence of a pure culture, in accordance with the recommendations of Murray and Stackebrandt (1995), we propose the provisional taxonomic assignment ‘Candidatus Promineofilum breve'.
Pro.'mi.nee.oh (Latin verb, combining form of prominere, to project, to jut out); fi.lum (latin neutral noun, filament); bre'veh (Latin adject, neutral form to agree with filum). Promineofilum breve denoting the appearance of short filaments protruding from activated sludge flocs into the bulk liquid.
Acknowledgments
This work was supported by the Danish Council for Independent Research (grant no. 4093-00127 A), Innovation Fund Denmark (EcoDesign and NomiGas), the Villum Foundation and Aalborg University. We acknowledge the LABGeM team at Genoscope for providing access to their automated annotation pipeline as well as for database maintenance and technical assistance. We would also like to thank Dr Tom Burton for his assistance with the naming etymology, and Valter Tandoi and Irena Čarman for supplying the B45-enriched sample used for the metagenome.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisolm SW, Devereux R, Stahl DA. (1990). Combination of 16 S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Env Microbiol 56: 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen K, Nielsen PH. (2000). Growth of Microthrix parvicella in nutrient removal activated sludge plants: studies of in situ physiology. Water Res 34: 1559–1569. [Google Scholar]
- Andrews SC, Berks BC, McClay J, Ambler A, Quail MA, Golby P et al. (1997). A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology 143: 3633–3647. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer M, Seviour EM, Kong Y, Cunningham M, Blackall LL, Seviour RJ. (2002). Phylogeny of the filamentous bacterium Eikelboom Type 1851, and design and application of a 16 S rRNA targeted oligonucleotide probe for its fluorescence in situ identification in activated sludge. FEMS Microbiol Lett 207: 179–183. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. (2012). Toward almost closed genomes with GapFiller. Genome Biol 13: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AG, Schwientek P, Vishnivetskaya T, Woyke T, Levy S, Beall CJ et al. (2014). Diversity and genomic insights into the uncultured Chloroflexi from the human microbiota. Environ Microbiol 16: 2635–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42: D459–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudel-Renard C. (2003). Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res 31: 6633–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C et al. (2007). Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22: 434–444. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Hébraud M, Talon R, Henderson IR. (2009). Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol 17: 139–145. [DOI] [PubMed] [Google Scholar]
- Dupont CL, Rusch DB, Yooseph S, Lombardo MJ, Richter RA, Valas R et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6: 1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ES, Fogel MA, Waldor MK. (2005). Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol 56: 1129–1138. [DOI] [PubMed] [Google Scholar]
- Eikelboom DH. (1975). Filamentous organisms observed in activated sludge. Water Res 9: 365–388. [Google Scholar]
- Gelfand MS, Rodionov DA. (2008). Comparative genomics and functional annotation of bacterial transporters. Phys Life Rev 5: 22–49. [Google Scholar]
- Gupta RS, Chander P, George S. (2013). Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexi class. nov. into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochloridaceae and the family Chloroflexaceae fam. nov., and the suborder Roseiflexineae subord. nov., containing the family Roseiflexaceae fam. nov. Antonie Van Leeuwenhoek 103: 99–119. [DOI] [PubMed] [Google Scholar]
- Harrison PW, Lower RPJ, Kim NKD, Young JPW. (2010). Introducing the bacterial ‘chromid': not a chromosome, not a plasmid. Trends Microbiol 18: 141–148. [DOI] [PubMed] [Google Scholar]
- Hedderich R, Forzi L. (2005). Energy-converting [NiFe] hydrogenases: more than just H2 activation. J Mol Microbiol Biotechnol 10: 92–104. [DOI] [PubMed] [Google Scholar]
- Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR et al. (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D, Richard MG, Daigger GT. (2004) Manual on the causes and control of activated sludge bulking, foaming and other solids separation problems 3rd. CRC Press LLC: London, England. [Google Scholar]
- Jha JK, Baek JH, Venkova-Canova T, Chattoraj DK. (2012). Chromosome dynamics in multichromosome bacteria. Biochim Biophys Acta 1819: 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer R, Steinbüchel A. (2003). A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem 278: 8075–8082. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42: D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaichi S, Ito N, Kamikawa R, Sugawara T, Yoshida T, Sako Y. (2013). Ardenticatena maritima gen. nov., sp. nov., a ferric iron- and nitrate-reducing bacterium of the phylum ‘Chloroflexi' isolated from an iron-rich coastal hydrothermal field, and description of Ardenticatenia classis nov. Int J Syst Evol Microbiol 63: 2992–3002. [DOI] [PubMed] [Google Scholar]
- Kawaichi S, Yoshida T, Sako Y, Nakamura R. (2015). Draft genome sequence of a heterotrophic facultative anaerobic thermophilic bacterium, Ardenticatena maritima strain 110 S T. Genome Announc 3: e01145–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindaichi T, Nierychlo M, Kragelund C, Nielsen JL, Nielsen PH. (2013). High and stable substrate specificities of microorganisms in enhanced biological phosphorus removal plants. Environ Microbiol 15: 1821–1831. [DOI] [PubMed] [Google Scholar]
- Kohno T, Sei K, Mori K. (2002). Characterization of type 1851 organism isolated from activated sludge samples. Water Sci Technol 46: 111–114. [PubMed] [Google Scholar]
- Kong Y, Xia Y, Nielsen PH. (2008). Activity and identity of fermenting microorganisms in full-scale biological nutrient removing wastewater treatment plants. Env Microbiol 10: 2008–2019. [DOI] [PubMed] [Google Scholar]
- Kragelund C, Levantesi C, Borger A, Thelen K, Eikelboom D, Tandoi V et al. (2007). Identity, abundance and ecophysiology of filamentous Chloroflexi species present in activated sludge treatment plants. FEMS Microbiol Ecol 59: 671–682. [DOI] [PubMed] [Google Scholar]
- Kragelund C, Nielsen JL, Thomsen TR, Nielsen PH. (2005). Ecophysiology of the filamentous alphaproteobacterium Meganema perideroedes in activated sludge. FEMS Microbiol Ecol 54: 111–122. [DOI] [PubMed] [Google Scholar]
- Kragelund C, Thomsen TR, Mielczarek AT, Nielsen PH. (2011). Eikelboom's morphotype 0803 in activated sludge belongs to the genus Caldilinea in the phylum Chloroflexi. FEMS Microbiol Ecol 76: 451–462. [DOI] [PubMed] [Google Scholar]
- Kristiansen R, Nguyen HTT, Saunders AM, Nielsen JL, Wimmer R, Le VQ et al. (2013). A metabolic model for members of the genus Tetrasphaera involved in enhanced biological phosphorus removal. ISME J 7: 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Gentleman R, Carey V. (2009). rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25: 1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett RM, Clavijo BJ, Clissold L, Clark MD, Caccamo M. (2014). NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics 30: 566–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Lapidus A, Thomsen T, Han J, Haynes M, Lobos E et al. (2015. a). High quality draft genome sequence of Meganema perideroedes str. GR1T and a proposal for its reclassification to the family Meganemaceae fam. nov. Stand Genomic Sci 10: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Saunders A, Albertsen M, Nierychlo M, McIlroy B, Hansen A et al. (2015. b). MiDAS: the field guide to the microbes of activated sludge. Database 2015: bav062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Kristiansen R, Albertsen M, Karst SM, Rossetti S, Nielsen JL et al. (2013). Metabolic model for the filamentous ‘Candidatus Microthrix parvicella' based on genomic and metagenomic analyses. ISME J 7: 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielczarek AT, Kragelund C, Eriksen PS, Nielsen PH. (2012). Population dynamics of filamentous bacteria in Danish wastewater treatment plants with nutrient removal. Water Res 46: 3781–3795. [DOI] [PubMed] [Google Scholar]
- Miura Y, Watanabe Y, Okabe S. (2007). Significance of Chloroflexi in performance of submerged membrane bioreactors (MBR) treating municipal wastewater. Environ Sci Technol 41: 7787–7794. [DOI] [PubMed] [Google Scholar]
- Muller EEL, Pinel N, Laczny CC, Hoopmann MR, Narayanasamy S, Lebrun LA et al. (2014). Community-integrated omics links dominance of a microbial generalist to fine-tuned resource usage. Nat Commun 5: 5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RG, Stackebrandt E. (1995). Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol 45: 186–187. [DOI] [PubMed] [Google Scholar]
- Nielsen JL. (2009) Protocol for fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotides. In: Nielsen PH, Daims H, Lemmer H (eds) FISH Handbook for Biological Wastewater Treatment. IWA Publishing: London, pp 73–84. [Google Scholar]
- Nielsen JL, Nielsen PH. (2005). Advances in microscopy: microautoradiography of single cells. Methods Enzymol 397: 237–256. [DOI] [PubMed] [Google Scholar]
- Nielsen PH, Kragelund C, Seviour RJ, Nielsen JL. (2009). Identity and ecophysiology of filamentous bacteria in activated sludge. FEMS Microbiol Rev 33: 969–998. [DOI] [PubMed] [Google Scholar]
- Pages H, Aboyoun P, Gentleman R, DebRoy S. (2014) Biostrings: String Objects Representing Biological Sequences, and Matching Algorithms R package version 2.36.1 Available from http://bioconductor.org/packages/Biostrings/.
- Petrovski S, Dyson ZA, Quill ES, McIlroy SJ, Tillett D, Seviour RJ. (2011). An examination of the mechanisms for stable foam formation in activated sludge systems. Water Res 45: 2146–2154. [DOI] [PubMed] [Google Scholar]
- Poock SR, Leach ER, Moir JWB, Cole JA, Richardson DJ. (2002). Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J Biol Chem 277: 23664–23669. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz O, Boison G, Salzmann H, Bothe H, Schütz K, Wang S et al. (2002). HoxE—a subunit specific for the pentameric bidirectional hydrogenase complex (HoxEFUYH) of cyanobacteria. Biochim Biophys Acta 1554: 66–74. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Missiakas D. (2014). Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochim Biophys Acta - Mol Cell Res 1843: 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi Y, Yamada T, Hanada S, Ohashi A, Harada H, Kamagata Y. (2003). Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int J Syst Evol Microbiol 53: 1843–1851. [DOI] [PubMed] [Google Scholar]
- Serafim LS, Lemos PC, Levantesi C, Tandoi V, Santos H, Reis MAM. (2002). Methods for detection and visualization of intracellular polymers stored by polyphosphate-accumulating microorganisms. J Microbiol Methods 51: 1–18. [DOI] [PubMed] [Google Scholar]
- Seviour RJ, Nielsen PH. (2010) Microbial Ecology of Activated Sludge. IWA Publishing: London. [Google Scholar]
- Simon J, Gross R, Einsle O, Kroneck PM, Kröger A, Klimmek O. (2000). A NapC/NirT-type cytochrome c (NrfH) is the mediator between the quinone pool and the cytochrome c nitrite reductase of Wolinella succinogenes. Mol Microbiol 35: 686–696. [DOI] [PubMed] [Google Scholar]
- Speirs L, Nittami T, McIlroy S, Schroeder S, Seviour RJ. (2009). Filamentous bacterium Eikelboom type 0092 in activated sludge plants in Australia is a member of the phylum Chloroflexi. Appl Env Microbiol 75: 2446–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs LBM, McIlroy SJ, Petrovski S, Seviour RJ. (2011). The activated sludge bulking filament Eikelboom morphotype 0914 is a member of the Chloroflexi. Env Microbiol Rep 3: 159–165. [DOI] [PubMed] [Google Scholar]
- Stoecker K, Dorninger C, Daims H, Wagner M. (2010). Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl Environ Microbiol 76: 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe IC. (2010). A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol 18: 464–470. [DOI] [PubMed] [Google Scholar]
- Sutcliffe IC. (2011). Cell envelope architecture in the Chloroflexi: a shifting frontline in a phylogenetic turf war. Environ Microbiol 13: 279–282. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinform 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. (2012). Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71–D75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. (2004). Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta - Mol Cell Res 1694: 269–278. [DOI] [PubMed] [Google Scholar]
- Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A et al. (2009). MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009: bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais PM, Billoud B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev 107: 4206–4272. [DOI] [PubMed] [Google Scholar]
- Wallner G, Amann R, Beisker W. (1993). Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14: 136–143. [DOI] [PubMed] [Google Scholar]
- Xia Y, Kong Y, Nielsen PH. (2008). In situ detection of starch-hydrolyzing microorganisms in activated sludge. FEMS Microbiol Ecol 66: 462–471. [DOI] [PubMed] [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16 S rRNA gene sequences. Nat Rev Microbiol 12: 635–645. [DOI] [PubMed] [Google Scholar]
- Zita A, Hermansson M. (1997). Determination of bacterial cell surface hydrophobicity of single cells in cultures and in wastewater in situ. FEMS Microbiol Lett 152: 299–306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.