Abstract
Like other obligate intracellular bacteria, the Chlamydiae feature a compact regulatory genome that remains uncharted owing to poor genetic tractability. Exploiting the reduced number of transcription factors (TFs) encoded in the chlamydial (pan-)genome as a model for TF control supporting the intracellular lifestyle, we determined the conserved landscape of TF specificities by ChIP-Seq (chromatin immunoprecipitation-sequencing) in the chlamydial pathogen Waddlia chondrophila. Among 10 conserved TFs, Euo emerged as a master TF targeting >100 promoters through conserved residues in a DNA excisionase-like winged helix-turn-helix-like (wHTH) fold. Minimal target (Euo) boxes were found in conserved developmentally-regulated genes governing vertical genome transmission (cytokinesis and DNA replication) and genome plasticity (transposases). Our ChIP-Seq analysis with intracellular bacteria not only reveals that global TF regulation is maintained in the reduced regulatory genomes of Chlamydiae, but also predicts that master TFs interpret genomic information in the obligate intracellular α-proteobacteria, including the rickettsiae, from which modern day mitochondria evolved.
Introduction
Regulation at the level of transcription initiation represents the most commonly used strategy for cellular reprogramming and is generally orchestrated by one or several master (global) transcriptional factors (TFs) that directly coordinate the activity of hundreds of genes by binding to their promoters (Laub et al., 2007; Cole and Young, 2008; Losick and Desplan 2008; Whyte et al., 2013; Wolanski et al., 2014; Panis et al., 2015). Despite the apparent simplicity of bacteria and their compact genome size compared with their eukaryotic counterparts, free-living bacteria typically encode hundreds of TFs in their genomes. Obligate intracellular bacteria such as the Chlamydiae encode far fewer TFs in their reduced genomes, making them ideal models for determining the ‘regulatory pan-genome' during intracellular growth. We defined here the regulatory pan-genome as the genomic regulatory sites that are targeted by TFs common to all members of the phylum Chlamydiae. In other bacteria, the regulatory program implements stochastic and/or deterministic cell fate and transcriptional switches that promote productive acute or chronic infections of host cells (Arnoldini et al., 2014; Deghelt et al., 2014; Diard et al., 2014; Manina et al., 2015; Panis et al., 2015). However, defining the landscape of TF specificities within the host is challenging in the case of facultative bacterial pathogens that also grow outside cells. Obligate intracellular bacteria may thus present an optimal system for this goal.
Chlamydiaceae family, the most described in the Chlamydiae phylum, includes members able to infect a wide host range (Coulon et al., 2012; Kebbi-Beghdadi et al., 2011, 2015) and that are well-characterized human and animal pathogens with important zoonotic implications (Wheelhouse and Longbottom, 2012). Although the estimated incidence of sexually-transmitted infections by Chlamydia trachomatis is >100 million per year (Baud et al., 2008), other members of the Chlamydiaceae family, such as Chlamydia pneumoniae, are also pathogenic towards humans, being implicated in lung infections (Grayston, 2000; Baud et al., 2008; Senn et al., 2011; Taylor et al., 2014). In addition to the Chlamydiaceae, several new Chlamydia-related bacteria, including Waddlia chondrophila, were recently discovered in diverse environments (Dilbeck et al., 1990; Fritsche et al., 1993; Kahane et al., 1995; Amann et al., 1997; Rurangirwa et al., 1999; Collingro et al., 2005; Thomas et al., 2006; Lienard et al., 2011). Some of these are harmless symbionts of ameobae, while others are implicated as emerging pathogens for humans and animals (Greub and Raoult, 2004; Lamoth and Greub, 2010). The ecological niche as well as the potential reservoirs and vectors of these bacteria are not well described yet, despite some hints suggesting the role of ticks as potential vectors (Croxatto et al., 2014; Pilloux et al., 2015), as well as reports supporting a role of free-living amoebae as widespread reservoir of these Chlamydia-related bacteria in water environment (Thomas et al., 2006; Corsaro et al., 2009). Moreover, the complexity and diversity of the Chlamydiae phylum is largely underestimated as suggested recently by metagenomic and phylogenetic analyses revealing 181 putative families present mainly in marine environments (Lagkouvardos et al., 2014). Members of the Chlamydiae phylum are related to free-living aquatic bacteria that belong to the Verrucomicrobia and Planctomycetes (PVC superphylum) (Jackson and Weeks, 2008; Satinsky et al., 2015) and are mostly (currently) genetically intractable. Interestingly, at least one member of the Verrucomicrobia has recently been identified as an important constituent of the human microbiota controlling obesity, brown fat tissue and cold tolerance (Chevalier et al., 2015).
The members of the Verrucomicrobia, Planctomycetes and of most chlamydial families (for example, W. chondrophila) have a larger genome and therefore presumably an expanded metabolic capacity (Greub et al., 2009; Bertelli et al., 2010) compared with the 1–1.2 Mbp genome of C. trachomatis and C. pneumoniae (Stephens et al., 1998; Shirai et al., 2000; Collingro et al., 2011). Nevertheless, all chlamydial genomes encode a limited number of TFs (Stephens et al., 1998; Shirai et al., 2000; Collingro et al., 2011; Siegl and Horn, 2012) (see below and Figure 1) to control the interactions with different eukaryotic hosts and an infectious cycle involving two morphotypes (Rockey and Matsumoto, 2000; Siegl and Horn, 2012; Tan, 2012). The cycle can be divided into three stages. In the first, an infectious non-dividing elementary body (EB) enters the host cells and differentiates into the non-infectious and replicative reticulate body (RB). The RB features a de-condensed genome and expresses cytokinetic proteins, to permit rapid proliferation and division of RBs within a vacuole (called the inclusion) during the mid-stage. In the late stage, the RBs differentiate back into EBs and are released by extrusion or cell lysis (Moulder, 1991). Changes in the chlamydial transcriptome are thought to underlie these developmental stages, reflected by three corresponding temporal transcript classes (early, mid and late) (Shaw et al., 2000; Belland et al., 2003; Nicholson et al., 2003). Thus, by analogy to other developmental systems, we speculate that one or several conserved master TFs target promoters of developmentally-regulated genes.
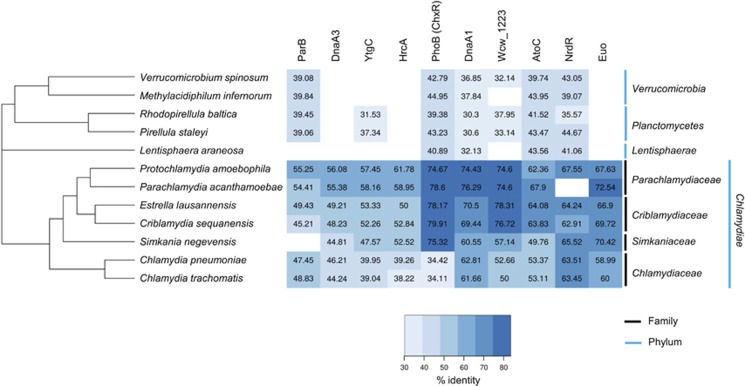
Figure 1.
The 10 conserved TFs in Waddlia chondrophila and their orthologues in the PVC superphylum. Presence (blue) or absence (white) of the 10 TFs in the PVC superphylum. The % of identity, compared with W. chondrophila, is indicated in each square. The proteins are indicated on the top, organisms on the left and the family (black) and the phylum (blue) on the right. Organisms are ordered on the basis of the phylogenetic tree performed with the maximum likelihood method using the 158 core genes. The topology of the Chlamydiae phylum derived from the concatenation of the 158 core genes is similar to what is already known (Pillonel et al, 2015). Euo, HrcA and DnaA3 are only present in the Chlamydiae. PhoB, AtoC and NrdR are conserved in the PVC superphylum. ParB is not present in Simkania negevensis and NrdR is not present in Parachlamydia acanthamoebae.
At least 10 conserved TFs (Figure 1, note that TFs are defined here as proteins that are not predicted to be structural components of RNA polymerase holoenzyme, RNAP, or regulators of RNAP enzymatic activity) are predicted within the Chlamydiae (Greub et al., 2009; Bertelli et al., 2010). As the chlamydial TF regulatory network remains experimentally untested, we unmasked the regulatory pan-genome defined by these 10 TFs by chromatin-immunoprecipitation followed by deep-sequencing (ChIP-Seq) hoping to learn whether these 10 conserved TFs and/or a possible master TF might control the developmental and obligate intracellular proliferation cycle of chlamydia. The immunochemistry of ChIP-Seq has the advantage of minimizing the contaminating host nucleic acids compared with chlamydial transcriptome studies (Albrecht et al., 2010, 2011) and, within one stroke, records the TF landscape of an obligate intracellular bacterium within its host cell.
Materials and methods
Cell culture and bacterial strains
Vero cells (ATCC CCL-81) were cultivated in Dulbecco's modified minimal essential medium (DMEM; GE Healthcare, Pasching, Austria) supplemented with 10% fetal bovine serum (GE Healthcare), at 37 °C in the presence of 5% CO2.
Waddlia chondrophila strain ATCC VR-1470 was co-cultivated at 32 °C with Acanthamoeba castellani strain ATCC 30010 in 75 cm2 flask containing 30 ml of peptone-yeast-extract-glucose broth. After 7 days of culture, the suspension was filtered on 5 μm filter to eliminate trophozoites and cysts and to isolate bacteria in the filtrate. The filtrate was then diluted at the appropriate dilution in DMEM to proceed to infection of Vero cells.
Infection procedure
One day before the infection, 5 × 105 Vero cells were seeded per well in 24-well plates. A 1/2000 dilution of W. chondrophila was used to infect Vero cells. This corresponded to a multiplicity of infection 2–3, as estimated by qPCR. This infectious dose led to 50% of infected cells with 2–3 bacteria, as determined by confocal microscopy. The infected Vero cell suspension was centrifuged at 1790 g for 10 min at room temperature and then incubated for 15 min at 37 °C with 5% CO2. To remove non-internalized bacteria to obtain a synchronous infection, cells were washed with the DMEM. Then, infected cells were incubated for different time periods.
Quantitative PCR
The number of bacteria at different time post-infection was determined using real-time quantitative PCR (qRT-PCR). Infected cells were recovered after cell scraping. Genomic DNA extraction was performed on 50 μl of cell suspension using the WizardSV Genomic DNA purification kit (Promega, Madison, WI, USA) and eluted in 100 μl. The quantitative PCR was performed as described previously using iTaq supermix with ROX (Bio-Rad, Reinach, Switzerland) (Goy et al., 2009). The reaction mixture contained 10 μl of iTaq supermix, 200 nm of primers WadF4 and WadR4, 100 nm of the probe WadS2 and 5 μl of DNA. The qPCRs were performed on the Step-One system (Applied Biosystems, Zug, Switzerland) using the following cycling conditions: 3 min at 95 °C, 40 cycles of 15 s at 95 °C and 1 min at 60 °C.
Crosslinking experiments
Five μm of His6-Euo was incubated, during 15 min at 25 °C, in the presence or not of 1 mm of bis(maleimido)hexane (Pierce, Rockford, IL, USA) a crosslinking agent. When indicated, 80 ng of PCR amplified DNA fragment was added to the reaction mix. Four μl of 6 × denaturing Laemmli loading dye was added to each sample. The samples were then separated by SDS-PAGE on a 12% polyacrylamide gel (Bio-Rad) and transferred onto nitrocellulose. The His6-Euo was detected by immunodetection using mouse monoclonal anti-His6 (Sigma-Aldrich, St Louis, MO, USA) (see Immunoblotting section).
Data access, supplementary data sets and supplementary information
All ChIP-Seq data files have been deposited at the GEO database under accession number GSE68059. Supplementary tables, figures and additional experimental procedures are available for download from the ISME Journal website.
Results
Conservation of TFs within the PVC
To investigate the conservation of chlamydial TFs, we first identified homologues of 10 putative W. chondrophila TFs in all known chlamydial genera and representatives of two sister clades of the Chlamydiae phylum, the Planctomycetes and Verrucomicrobia. We then conducted pairwise sequence comparisons between the 12 members of the PVC superphylum. This revealed that several of these 10 TFs are conserved within the entire PVC superphylum (PhoB, AtoC, DnaA1, NrdR and to a lesser extent ParB), while others are not (DnaA3, HrcA and Euo). Euo may have a key role in Chlamydiae as it is the only conserved TF unique to the Chlamydiae and shows a remarkable degree of sequence conservation (>70% identity in some cases, Figure 1). Wcw_1223 is present within the PVC and in several representatives of the Bacteroidetes phylum, albeit with a significantly lower (ca. 35%) sequence identity level. By contrast, the paralogues of the bifunctional replication initiator/TF DnaA and orthologues of the heat-sensitive repressor HrcA are also found outside the PVC lineage. The phosphate response regulator PhoB is also found in other lineages, but it is unusual because it features a high degree of conservation (near 80% identity) across many chlamydial families (especially those that replicate in amoebae), except for the Chlamydiaceae family (Figure 1). As the sequence identity between W. chondrophila PhoB and the orthologues encoded in Planctomycetes, Verrucomicrobia, Lentisphaerae and the other Chlamydiaceae genomes is considerably lower, we hypothesize that this reflects a functional specialization of PhoB within the chlamydia to control a distinct regulon. This notion and that Euo is a master chlamydial TF gained further supported by the ChIP-Seq experiments described below.
Developmental regulation of TFs in W. chondrophila
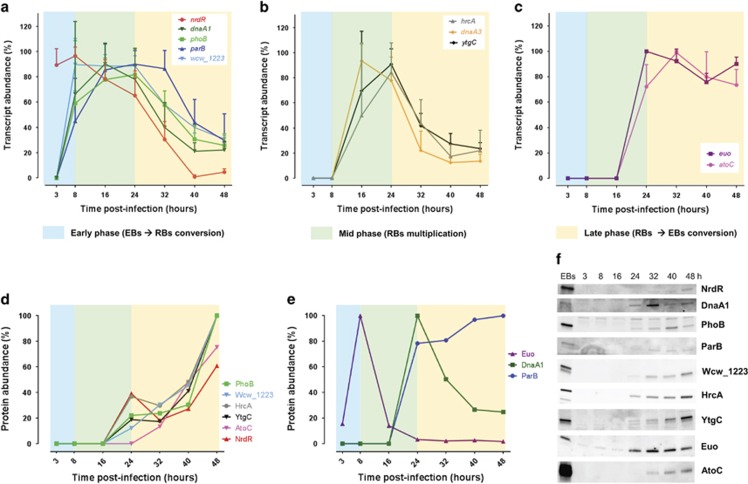
To determine whether these conserved TFs are temporally regulated during the developmental cycle of W. chondrophila, we measured their transcript levels by qRT-PCR at various time points during intracellular growth, that is, post infection (p.i.) of Vero cells with W. chondrophila. We observed a peak in the steady-state levels of the dnaA1, phoB, parB, wcw_1223, hrcA, dnaA3 and ytgC transcripts in mid-phase (16–24 h p.i., Figures 2a and b). By contrast, the nrdR transcript levels are high at 3 h and 8 h p.i. (Figure 2a), followed by a progressive drop until 40 h p.i. The two remaining transcripts, euo and atoC, were only detected from 24 h p.i. onwards and remained high henceforth (Figure 2c), suggesting that these TFs are important late in the current developmental cycle or may be needed for an early event in the ensuing cycle.
Figure 2.
Temporal expression of the 10 TFs during the developmental cycle of W. chondrophila. Samples (equal volume) of infected Vero cells were harvested at different times p.i. and analysed by qRT-PCR (a–c) or by immunoblotting using specific polyclonal antibodies to the TFs (d–f). The transcript abundance (%) during the developmental cycle (early (blue shading), mid (green) and late (yellow)) was determined for each TF (a–c). The data are shown as mean values±standard deviation of three independent experiments. Most of the genes show a mid-phase transcript profile (a, b) except for euo and atoC, which exhibit a late-phase transcript profile (c). The TF abundance (%, d and e) was quantified by normalization of the signal detected on the blot (f) according to the number of bacteria/well defined by qPCR. Most of the TFs accumulated steadily along the progression of the developmental cycle (d). Euo and DnaA1 exhibited a peak in abundance at 8 h and 24 h, respectively (e). Note that we could not unambiguously detect DnaA3 by immunoblotting and thus omitted it from this analysis.
To determine the relative TF protein abundance during the developmental cycle, we conducted immunoblotting (see Materials and methods) using polyclonal antibodies to each of these TFs (Figures 2d–f). We observed a relative increase in abundance of NrdR, AtoC, YtgC, HrcA, Wcw_1223 and PhoB along the developmental cycle (Figures 2d and f). By contrast, ParB levels surged at 24 h p.i. and then plateaued (Figures 2e and f), Euo and DnaA1 peaking at 8 h and 24 h p.i., respectively. The abundance of the Euo protein at an early time point is surprising considering that its transcript abundance is highest from 24 h onwards. One possibility is that the euo transcript is synthesized during the conversion of RBs into EBs (late phase) and stored in EBs, thus allowing immediate translation during the early stage of the infection in the next cycle. A similar peak in euo transcripts has been observed for C. psittaci euo at 15 h p.i. (Zhang et al., 1998), suggesting that the developmental control of euo in different chlamydial families is conserved. It is also possible that the translation and/or stability of Euo is differentially regulated.
Landscape of TF specificities in W. chondrophila
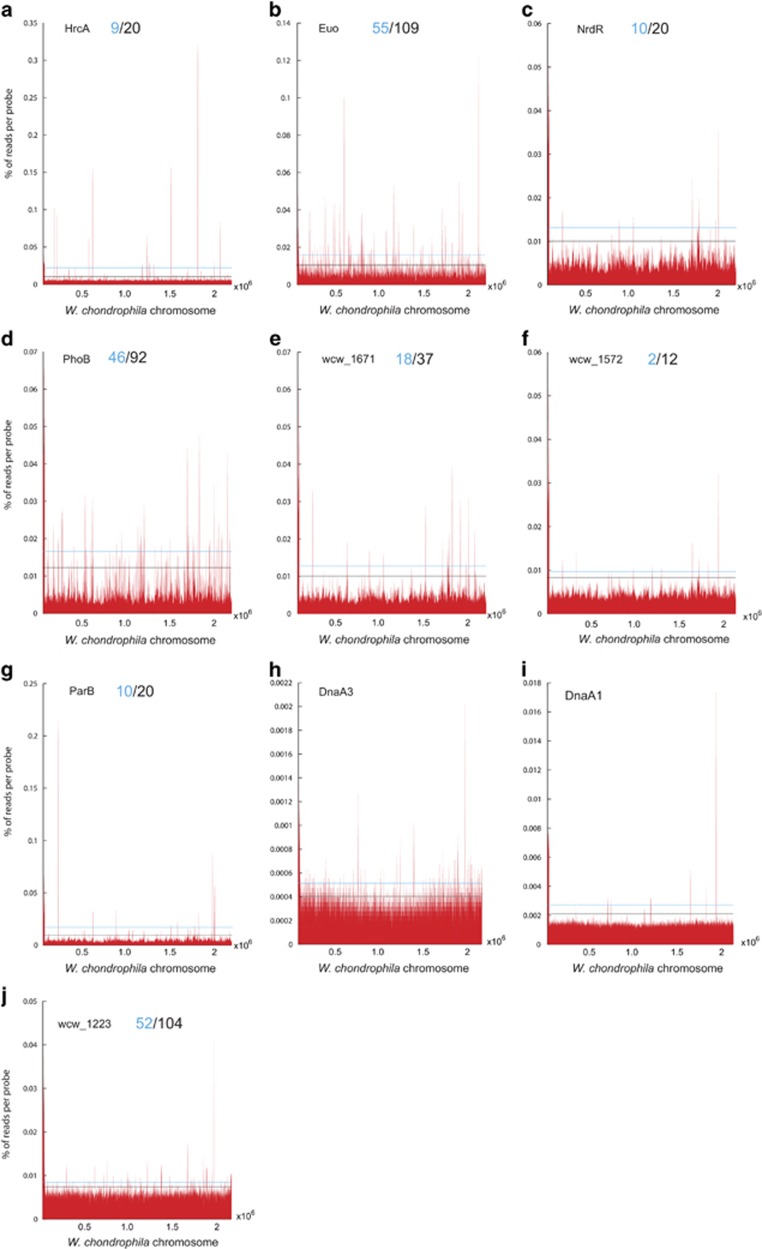
To chart the landscape of TF specificities, we conducted ChIP-Seq experiments in W. chondrophila with the antibodies to the TFs (Supplementary Figure 2A–C, Supplementary Table 1). Using our peak-finding strategy (see Supplementary Information), we observed a range of total TF target sites from >100 to <10 in some cases at different levels of enrichment (that is, enrichment of 0.5–1 times above the 2 s.d. cutoff, Figures 3a–f). We defined a subset of high confidence targets within in each set using the median value of relative abundance as a reference point compared with the standard 2 s.d. cutoff value (Figures 3a–f), with the predicted targets for Euo, for example, lying primarily in putative promoters or intergenic regions (Supplementary Figure 2D–F). Our analysis predicted 9/18 (HrcA), 55/109 (Euo), 46/92 (PhoB) high confidence/total sites within promoter regions (400 bp upstream to 100 bp downstream of the start codon of a predicted coding sequence, Figures 3a, b and d; Supplementary Table 2). The high confidence target lists for HrcA contains genes such as the groES and groEL chaperone genes, which are targets of the C. trachomatis HrcA orthologue in vivo (Chen et al., 2011; Rosario and Tan, 2012; Domman and Horn, 2015; Hanson and Tan, 2015). By contrast, no putative target sites are known for PhoB in other Chlamydiae. Fewer conserved PhoB targets were found for the Chlamydiaceae family than for other families (Supplementary Table 1). Importantly, we successfully derived distinct consensus motifs by Multiple Em for Motif Elicitation (MEME)-based analyses (see Supplementary Information, Supplementary Figure 2A–C) from the target lists of these three TFs (Euo, HrcA and PhoB) and validated them as described below.
Figure 3.
Occupancy of the 10 conserved TFs on the W. chondrophila genome. (a–j) ChIP-Seq profiles of the 10 TFs. Black line on the graphs depicts the cutoff used to identify the total predicted targets of each TF, while the blue line denotes the median score used to select the predicted high confidence targets (blue) from the total predicted target sites (black). The coordinates below the graph (x axis) indicate the nucleotide (nt) position along the W. chondrophila genome and the y axis shows the relative abundance of the corresponding nt position in the precipitated sample. Owing to the poor quality of the DnaA1 and DnaA3 precipitates we elected not to predict targets.
Validation of HrcA and PhoB target sites
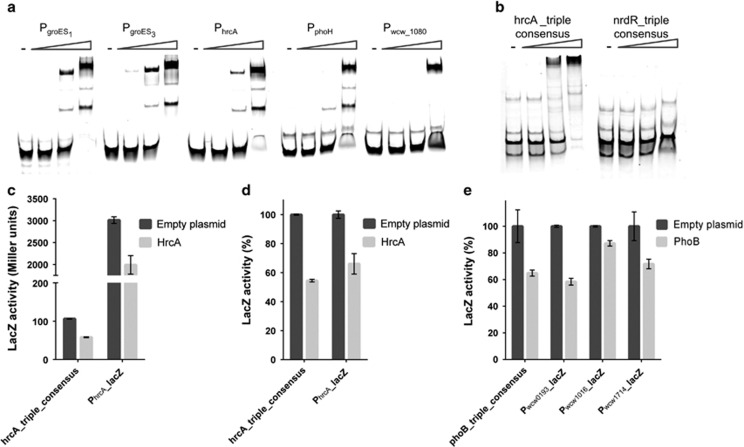
Five out of the nine predicted high confidence in vivo targets for HrcA (Figure 3a) were selected for testing by in vitro electrophoretic mobility shift assay (EMSA) with a purified recombinant His6-tagged version of W. chondrophila HrcA (His6-HrcA) (see Materials and methods) and fluorescently-labelled target promoter fragments (PhrcA, PgroES1, PgroES3, PphoH and Pwcw_1080) as probes. All five probes were band-shifted by His6-HrcA (Figure 4a). Since the consensus motif for HrcA deduced above (Supplementary Figure 2C; Supplementary Table 2, 5′TAGCA-(N)15-TGCTAA-3′) matches the CIRCE element-containing inverted repeat that is bound by HrcA in other bacteria and that predicted for C. trachomatis (Hecker et al., 1996; Narberhaus, 1999; Minder et al., 2000; Wilson and Tan, 2004; Hanson and Tan, 2015), we tested whether His6-HrcA also band-shifts a synthetic fragment harbouring a triple repeat of our HrcA consensus motif (Figure 4b). This was the case, but not for an analogous synthetic fragment harbouring a triple repeat of the unrelated NrdR consensus motif (see Supplementary Information, Figure 4b).
Figure 4.
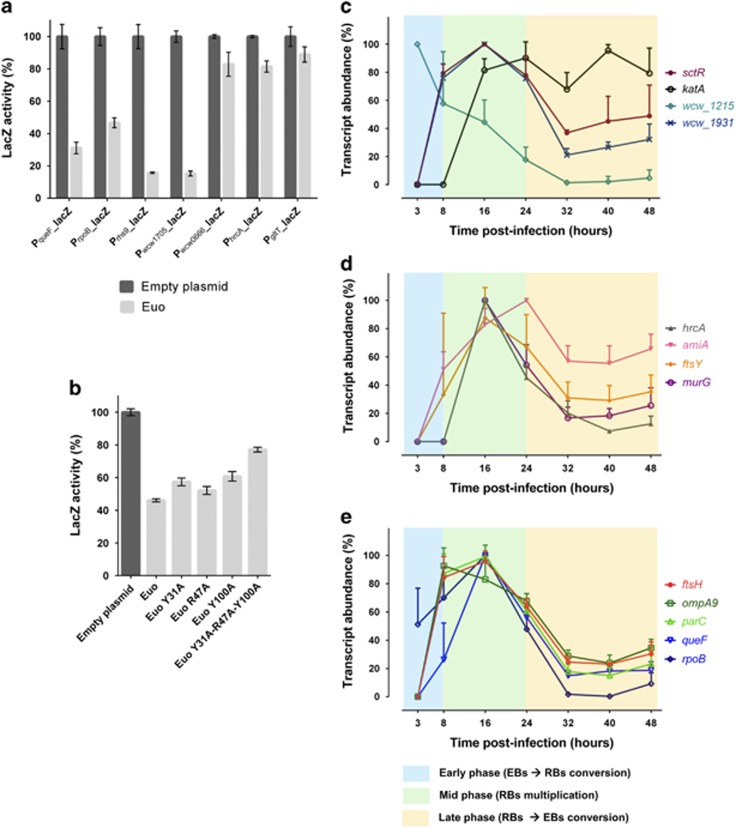
ChIP-Seq validation for HrcA and PhoB. (a) Binding of His6-HrcA on promoters identified by ChIP-Seq (PgroES1, PgroES3, PhrcA, PphoH and Pwcw_1080). The promoters were amplified by PCR using specific primers coupled to Cy5. DNA fragments were incubated in the absence (−) or in presence of an increasing concentration of His6-HrcA (50, 250 and 1250 nm) and analysed by EMSA. (b) EMSA analysis showing the binding of His6-HrcA to a synthetic fragment harbouring the HrcA-triple consensus. As a negative control, we used an analogous triple-repeat fragment harbouring the predicted NrdR consensus motif. (c, d) In vivo binding of HrcA on the triple consensus and on its own promoter (PhrcA) using lacZ reporter gene in E. coli. HrcA was expressed from an arabinose-inducible promoter on pBAD22 (Guzman et al., 1995). As a negative control, the expression empty vector was used. Data are means±standard deviation of three biological triplicates. Panel (c) absolute values, panels (d) and (e) normalized value according to the empty plasmid (set as 100%). When HrcA was expressed, a decrease of 40% of the LacZ activity was observed, suggesting that HrcA is able to bind and to repress the expression of lacZ. (e) In vivo binding of PhoB (expressed from pBAD22) on three promoters (Pwcw_0193, Pwcw_1016 and Pwcw_1714) identified by ChIP-Seq and on the PhoB-triple consensus using lacZ reporter gene in E. coli. A decrease of the LacZ activity was observed for the four lacZ promoter-probe plasmids.
Having validated the HrcA target motif in vitro, we next tested if HrcA binds the consensus motif in vivo in a surrogate host. To this end, we used an E. coli β-galactosidase (LacZ)-based transcription interference assay in which a synthetic promoter containing the triple repeat of the HrcA consensus motif directs the expression of a promoterless lacZ on a low-copy plasmid in E. coli. We used this plasmid to test whether W. chondrophila HrcA expressed heterologously in E. coli can downregulate promoter activity (measured as LacZ activity). We observed that LacZ activity dropped by 40% when HrcA was expressed compared with E. coli cells harbouring the empty vector (Figures 4c and d). Since ChIP-Seq suggested autoregulation of HrcA, we conducted a similar LacZ-based interference assay in E. coli with native W. chondrophila PhrcA promoter using the PhrcA-lacZ reporter plasmid (Figures 4c and d). A commensurate decrease of ±35% of the LacZ activity compared with the empty vector was also seen for PhrcA, establishing that HrcA also binds this site and consensus in vivo.
Next, we used a BBH (bidirectional best blast hit) approach to compile a list of orthologous target genes of HrcA encoded in 13 Chlamydiales (Supplementary Figure 3A). This list clustered into two main groups. The first cluster contains genes conserved across the Chlamydiales order (>40% identity) and includes genes targeted by C. trachomatis HrcA in vitro for example groES1(ct111/wcw_1342), groEL1(ct110/wcw_1343), groES3(wcw_1848), groEL3(wcw_1849) and hrcA(ct394/wcw_1636) (Tan et al., 1996; Wilson and Tan, 2004; Hanson and Tan, 2015). The second larger cluster harbours targets mostly restricted to W. chondrophila, including 12 genes that seem to define the accessory (specific) W. chondrophila HrcA target regulon (see Discussion).
Next, we validated the predicted target sites of PhoB (Supplementary Table 3). The deduced 21-bp consensus motif computed by MEME (5′-(T/A)NTNAA(G/A)AAANTGN(T/A)AAATTT-3′, Supplementary Figure 2B; Supplementary Table 3) includes a sequence (in bold) resembling the predicted half site for the distantly related PhoB orthologue ChxR of C. trachomatis (Hickey et al., 2011). As described above for HrcA, we used an E. coli transcriptional interference assay with the native target promoters (identified by ChIP-Seq, Supplementary Figure 2B) or a synthetic promoter harbouring a triple repeat of the PhoB consensus (PhoB-triple consensus) upstream of the promoterless lacZ gene and expressed PhoB from a second plasmid (arabinose-inducible promoter). We observed a reduction in LacZ activity by ±40% comparing the Pwcw_0193 and PphoB-triple-consensus and by ±15% and 30% for Pwcw_1016 versus Pwcw_1714 promoter-probe plasmids, respectively, compared with the empty vector (Figure 4e). Thus, W. chondrophila PhoB binds these sites in vivo. BBH-based conservation analysis (Supplementary Figure 3B) revealed a small group of PhoB-regulated genes highly conserved across the phylum Chlamydiae (for example, the sctE[wcw_1612] predicted to encode a needle chaperone involved in inclusion modification and in chlamydial pathogenesis), specifically within the Chlamydia-related bacteria. The other group of PhoB-regulated genes includes W. chondrophila orthologues present also in other members of the Chlamydiae phylum, but with a lower degree of conservation and of unknown function.
Euo has properties of a master TF
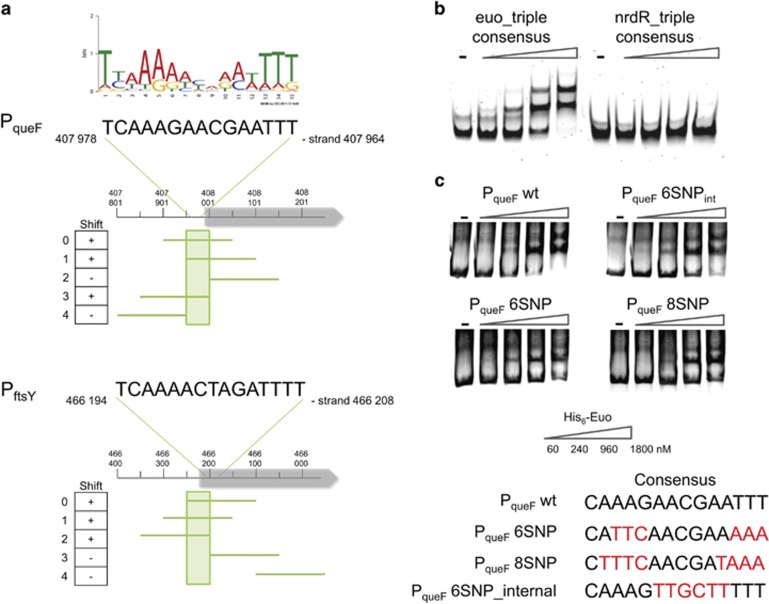
Of the >100 predicted Euo target sites, 19 (of 21 tested) were band-shifted by W. chondrophila His6-Euo in EMSAs (Table 1, Supplementary Figure 4A). Moreover, probing a blotted EMSA gel with antibodies to Euo confirmed that the shifted bands were indeed His6-Euo-DNA nucleoprotein complexes (Supplementary Figure 4B). The Euo consensus motif predicted by MEME (5′-TTAAAAACAAATTTT-3′, Supplementary Figure 2A; Supplementary Table 4), resembles part of the proposed extended Euo binding sites (5′-AGTAGGTAACAACCAAGTACTTGGGTTTT-3′ and 5′-TTTTAAAAAACAATTGATATAATTTTTATT-3′) deduced for C. trachomatis and C. psittaci based on targets identified in vitro (Zhang et al., 2000; Rosario and Tan, 2012). To explore whether our W. chondrophila Euo consensus motif is necessary for binding by His6-Euo, we designed five specific EMSA probes for PqueF and PftsY with 50 bp shifts including (or not) the predicted Euo DNA-binding motif (Figure 5a and Supplementary Table 6). EMSAs revealed that His6-Euo bound only PqueF, probes that include the predicted 15-bp Euo consensus motif, but not those lacking it. Interestingly, His6-Euo also shifted PftsY probes containing only a part of the Euo consensus motif (Figure 5a).
Table 1. Summary of the validation of Euo target sites from ChIP-Seq by EMSA.
| Gene | Predicted function | Synonym | Shift |
|---|---|---|---|
| wcw_0093 | Flagellar biosynthesis pathway, component FliP | sctR | ++++ |
| wcw_0203 | Histone H1-like protein Hc1 | hctA | – |
| wcw_0354 | N-acetylmuramoyl-l-alanine amidase | amiA | +++ |
| wcw_0365 | Enzyme related to GTP cyclohydrolase I | queF | ++++ |
| wcw_0390 | Rhs family protein | rhs9 | ++++ |
| wcw_0423 | Signal recognition particle GTPase | ftsY | +++ |
| wcw_0430 | NADH dehydrogenase, FAD-containing subunit | ndh | ++++ |
| wcw_0544 | Integrase | wcw_0544 | – |
| wcw_0547 | Transposase | wcw_0547 | + |
| wcw_0592 | DNA-directed RNA polymerase, beta subunit/140 kD subunit | rpoB | +++ |
| wcw_0655 | Catalase | katA | ++++ |
| wcw_0685 | ATP-dependent Zn proteases | ftsH | + |
| wcw_1215 | Outer membrane protein | + | |
| wcw_1315 | Hypothetical protein | OmpA9 | ++ |
| wcw_1386 | UDP-N-acetylglucosamine:LPS N-acetylglucosamine transferase | murG | + |
| wcw_1561 | Type IIA topoisomerase (DNA gyrase/topo II, topoisomerase IV), A subunit | parC | ++ |
| wcw_1636 | Transcriptional regulator of heat shock gene | hrcA | ++++ |
| wcw_1705 | Predicted ATPase or kinase | wcw_1705 | +++ |
| wcw_1731 | Na+/H+-dicarboxylate symporters | gltT | ++++ |
| wcw_1924 | Secreted protein | ++++ | |
| wcw_1931 | Cell division protein FtsI/penicillin-binding protein 2 | ftsI | +++ |
Abbreviations: ChIP-Seq, chromatin-immunoprecipitation-sequencing; EMSA, electrophoretic mobility shift assay. ++++ for completely shifted at 1800 nm; +++ for shifted band bigger than the PCR band at 1800 nm; ++ for shifted band appears at 240 nm; +for shifted band appears at 960 nm; − for no shifted band observed.
List of target promoters used as probes to test for binding by His6-Euo by EMSA. The number of plus symbols indicates the efficiency of binding.
Figure 5.
Euo consensus binding site analysis. (a) Identification of 50-bp region which includes the consensus and is necessary for the binding of His6-Euo. PCR probes shifted by 50 bp were designed for the PqueF and PftsY promoters and all fragments were used for in vitro binding assay (EMSA) with His6-Euo. A total of 80 ng of DNA fragments were incubated in the absence or in the presence of an increasing concentration of His6-Euo, protein–DNA complexes were detected using GelRed. Results are presented in the left column. + or − indicates whether or not shifted bands were observed. (b) EMSA analysis of His6-Euo binding to the Euo-triple consensus amplified with a specific primer coupled to Cy5. As a negative control, we used the triple repeat of the predicted NrdR consensus binding site (see Figure 4b). (c) Mutations in the consensus poorly affect the binding of His6-Euo tested by EMSA (GelRed detection, see Supplementary Information). His6-Euo concentrations used in the gel shift assay are indicated in (c).
Next, we designed synthetic EMSA probes in which the Euo consensus was repeated three times (Euo-triple consensus) and showed that His6-Euo bound this probe, whereas no shift was observed with the NrdR-triple repeat consensus (Figure 5b). Transcriptional interference assays in E. coli revealed that expression of Euo decreased by 50% the LacZ activity for PqueF, PrpoB, Prhs9 and Pwcw_1705 and only decreased by 15–20% for Pwcw_0666, PhrcA and PgltT (see Figure 7a). These results show that Euo binds these promoters in vivo in E. coli.
Determinants directing Euo to its targets
Next, we used site-specific mutagenesis to identify the critical positions necessary for Euo to bind its targets. We designed mutant PqueF EMSA probes (Figure 5c, Supplementary Figure 5) carrying specific nucleotide substitutions (Figure 5c), either near the end or in the middle of the predicted Euo target motif. As His6-Euo still band-shifted these probes we concluded that additional determinants must exist in the context of a native W. chondrophila promoter (Figure 5a) to recruit His6-Euo in vitro.
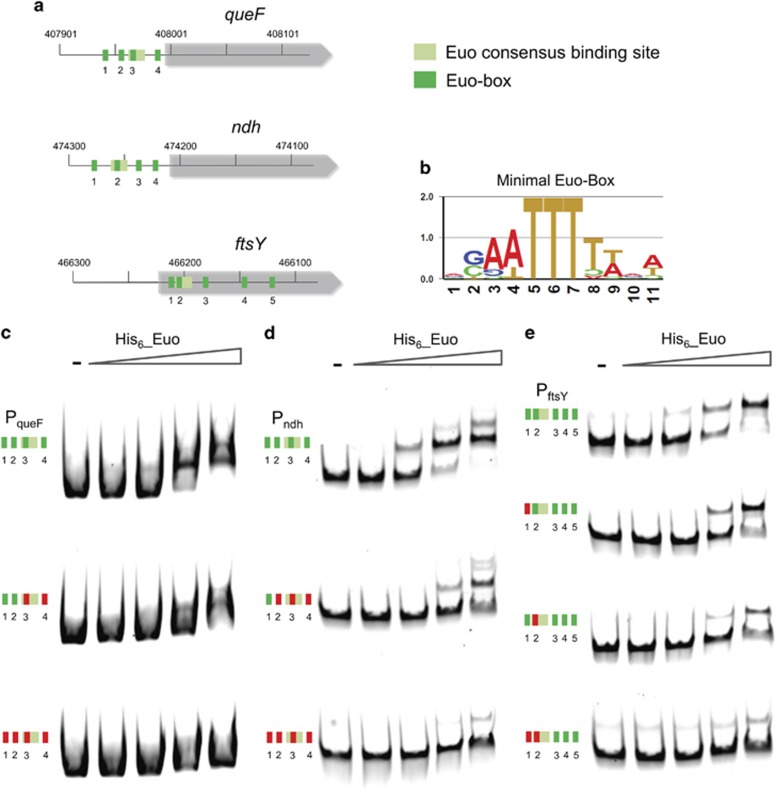
To identify these unknown determinants, we re-inspected the sequence of PqueF, Pndh and PftsY and observed the presence of a repeated, short and conserved box (3′-(A/G)(A/C)(A/T)TTT-5′, henceforth minimal Euo box). Interestingly, one minimal Euo box was always embedded in the long consensus motif predicted by MEME above (Figures 6a and b and Supplementary Figure 6). To explore the role of the minimal box in recruiting His6-Euo, we substituted the TTT by GGG signature one at a time and found for PqueF and Pndh that it was necessary to mutate all four minimal Euo boxes to impede binding of His6-Euo (Figures 6c and d). This was not the case for PftsY where mutations in the first two boxes were sufficient to prevent the binding of His6-Euo to its target sites (Figure 6e). Thus, minimal boxes direct Euo to its targets in vitro.
Figure 6.
Discovery of a minimal Euo target box. (a) Several minimal Euo boxes (green) are present in PqueF, PftsY and Pndh. Euo consensus binding site is also represented in light green. (b) Consensus based on these Euo boxes showing the conservation of the TTT. The TTT was replaced by GGG (shown in red) and the His6-Euo binding was tested by EMSA (c–e). The synthetic DNAs carrying the mutations were amplified using a specific primer coupled to Cy5 and used for EMSA. Mutation (red boxes) of the four Euo boxes (green) in PqueF and Pndh completely abolished the binding of His6-Euo as no band-shift was observed (c, d). Mutations in the two first Euo boxes in PftsY completely abolished the binding of His6-Euo (e).
Structural predictions using HHpred (Soding et al., 2005) revealed a resemblance of Euo to the DNA-binding domain of DNA excisionases such as Xis encoded in Enterococcus faecalis Tn916 or TorI from E. coli controlling recombination of the KlpE1 prophage (Elantak et al., 2005). TorI folds into a winged helix-turn-helix (wHTH) (Elantak et al., 2005), a DNA-binding motif comprising three alpha helices followed by 3–4 beta sheets (the wing), and assembles into stable dimers and multimers in the presence of target DNA. We also observed that the presence of DNA favours dimerization of His6-Euo in vitro (Supplementary Figure 7). Interestingly, the full-length Euo protein appears to harbour TorI-like wHTHs arranged in tandem, the first from residues 6–62 and the second from residues 78–137, suggesting that an Euo dimer can bind multiple minimal Euo boxes and that this tandem arrangement of putative wHTH promotes the formation of a stable nucleoprotein complex at chlamydial promoters, potentially accounting for the binding to the multiple minimal Euo boxes described above. We mutated the conserved arginine and tyrosine residues (individually and in combination) in the Euo wHTH that are required for DNA binding of TorI (Panis et al., 2012) and found that the triple mutation (Y31A/R47A/Y100A) impaired binding of Euo on the PqueF promoter fused to the lacZ gene, in the E. coli transcription interference assay (Figure 7b). Thus, binding of Euo resembles that of TorI-like excisionases.
Figure 7.
Developmental control by Euo. (a) LacZ-based promoter-probe plasmids were co-transformed into E. coli with a plasmid carrying euo under the control of an IPTG-inducible promoter (pSRK-Gm) (Khan et al., 2008). As a control, we used an empty expression vector. The LacZ activity was determined and the data represent mean values±standard deviation of three biological triplicates. When Euo expression is induced, the LacZ activity strongly decreased for the PqueF, PrpoB, Prhs9 and Pwcw_1705 and slightly decreased for the Pwcw_0066, PhrcA and PgltT. (b) Transcriptional interference assays as in (a) with plasmids expressing the point mutant versions of Euo as indicated in the panel. Mutations are located in the two TorI-like wHTH arranged in tandem (individually and in combination) and cloned in a pBAD22 vector under the control of an arabinose-inducible promoter. These plasmids were co-transformed into E. coli with the plasmid carrying the PqueF-lacZ fusion. Euo strongly decreased (by 60%) the LacZ activity for the PqueFpromoter, while the triple mutant only decreased the activity by 30%. (c–e) Temporal expression of Euo target genes was assessed by qRT-PCR at different time p.i. The transcript abundance (%) during the developmental cycle was determined for each Euo target gene. Data are mean values±standard deviation of three independent experiments. Most of the genes exhibited a peak of expression during the mid-phase of the developmental cycle and were considered as mid-phase genes. Wcw_1215 is an early gene since the peak of transcript abundance was at 3 h p.i. whereas katA is a late gene since the expression remained stable during the late phase.
Euo targets are developmentally regulated
As Euo was proposed to regulate late gene expression in C. trachomatis (Rosario and Tan, 2012; Rosario et al., 2014), we explored whether this is also the case for W. chondrophila Euo, by determining the transcript profiles of selected Euo target genes during the W. chondrophila developmental cycle by qRT-PCR. The selected genes were from diverse functional categories, including stress adaptation (hrcA[wcw_1636], ftsH[wcw_0685], dps[wcw_0932]), cell division and morphogenesis (parC[wcw_1561], sctR, amiA[wcw_0354], murG[wcw_1386]), general homeostasis/metabolism (rpoB[wcw_0592], ftsY[wcw_0423], gspE[wcw_1931], queF[wcw_0365], engA[wcw_1705]) and unknown functions (ompA9[wcw_1315], wcw_1215). Almost all transcripts showed a mid-phase profile with a peak in abundance between 16 and 24 h p.i. (Figures 7c–e). Only the wcw_1215 showed an early gene expression profile and the katA a late gene expression profile. Thus, the transcripts of Euo targets peak in mid-phase of the W. chondrophila developmental cycle.
Interestingly, W. chondrophila does not appear to target any genes orthologous to those identified as in vitro targets of Euo in Chlamydia (Zhang et al., 1998, 2000; Rosario and Tan, 2012; Rosario et al., 2014). We therefore investigated the differences of the W. chondrophila Euo regulon across the whole Chlamydiales order by BBH comparison. Clustering of similar sequences revealed the presence of two main groups: a small cluster including highly conserved (>50% identity) proteins across the Chlamydiales order, and a second large cluster of less conserved proteins (<40% identity) (Supplementary Figure 3). Even though this list does not include any genes targeted by C. trachomatis Euo in vitro, our list of W. chondrophila in vivo targets includes several conserved genes whose transcripts are developmentally regulated in C. trachomatis (for example, amiA (ct2687/wcw_0354), hrcA (ct394/ wcw_1636), ftsI (ct682/wcw_1931) and sctR (ct562/wcw_0093)).
Discussion
The reduced regulatory genomes of obligate intracellular Chlamydiae offer a unique opportunity to explore which regulatory systems are dispensable for developmental control. C-di-GMP- and histidine kinase-based regulatory systems that are most commonly used for post-translational regulation in free-living bacteria (West and Stock, 2001; Hengge, 2009) are sparsely (if at all) encoded in chlamydial genomes. Additionally, as only 10 conserved (annotated) TFs (Figure 1) could control transcript abundance over the developmental cycle (Belland et al., 2003; Albrecht et al., 2010), we aimed to define the regulatory genome directing intracellular growth and/or differentiation in a system with low TF multiplicity offered by chlamydiae. Using ChIP-Seq, we unearthed the landscape of conserved chlamydial TF specificities and provided strong evidence that the chlamydial signature protein Euo acts as a master TF that controls developmental transcription. Thus, transcriptional reprogramming during the chlamydial developmental cycle is governed at least in part at the level of transcription initiation by TFs that selectively bind promoters of developmentally-regulated genes and are themselves under temporal control. The pervasive binding of Euo to 109 predicted target sites represents more than 5% of the potential transcriptome that could be influenced directly by this TF and possibly further magnified by (direct or indirect) transcriptional or post-transcriptional control systems (for example, by anti-termination or mRNA degradation, respectively).
The predicted number of direct Euo targets is well within the range for master TFs known from other developmental systems. For example, the α-proteobacterial master TF CtrA (Quon et al., 1996), an extended member of the OmpR superfamily of DNA-binding response regulators, is also predicted to target >100 promoters in the free-living α-proteobacterium Caulobacter crescentus that regulates at least 20% of its transcriptome as a function of its developmental cycle and has a genome twice the size of that of W. chondrophila (Laub et al., 2000, 2002; Nierman et al., 2001; Fiebig et al., 2014; Fumeaux et al., 2014). Interestingly, CtrA acts as a master TF not only in free-living α-proteobacteria (Brilli et al., 2010; De Nisco et al., 2014; Fumeaux et al., 2014; Panis et al., 2015), but has recently been implicated in targeting several developmentally-regulated promoters in the α-proteobacterial intracellular pathogen Ehrlichia chaffeensis (Cheng et al., 2011), the aetiological agent of tick-borne human ehrlichiosis. E. chaffeensis branches with the rickettsial lineage of obligate intracellular α-proteobacteria and exhibits a developmental cycle bearing remarkable similarity to that of chlamydia, with the dense-cored cells acting as infectious forms that enter the host, differentiate into reticulate cells that eventually morph into dense-cored cells (Zhang et al., 2007), released from host cells. Thus, Euo may act as chlamydial counterpart of the α-proteobacterial master (developmental) regulator CtrA. Based on the distinct primary structures of the two proteins, mechanistic differences may clearly underlie the conceptual resemblance. Moreover, it was recently reported that Euo has been vertically inherited during chlamydial evolution strengthening the role of Euo as a master regulator (Domman and Horn, 2015). The overwhelming majority of Euo regulated genes encode mid-phase transcripts (Figure 7) that are required for the rapid proliferative (RB) phase in chlamydial development following the peak of Euo abundance in W. chondrophila (at 8 h p.i., Figure 2).
Speculating on the mode of binding of Euo to DNA via TorI-like wHTHs, it is noteworthy that multiple TorI binding sites are thought to have a crucial role in (i) bending DNA (nucleoprotein filament) and (ii) positioning recombination integrase proteins for proper formation of the excisive nucleoprotein complex (Panis et al., 2010a, b, 2012). Thus, these Euo boxes could also serve in proper positioning of RNAP for open complex formation in transcription initiation. The association of Euo with transposase genes also resembles the repression of the integrase gene by TorI (Panis et al., 2010b). Typically, the formation of higher order nucleoprotein complexes leads to repression while a lower order complex can promote transcriptional activation. Distinct promoter architectures of such minimal Euo boxes may dictate whether Euo acts as activator or repressor, of the many divergent Euo-targeted promoters.
It is certainly possible, or even likely, that the reprogramming of the chlamydial transcriptome is reinforced by other TFs acting (sequentially) in isolation or as modules (Niehus et al., 2008; Vijayan et al., 2009; Case et al., 2010; Tan, 2012; Domman and Horn, 2015). Such strategies are known for α-proteobacteria (Panis et al., 2015) directly at the transcriptional or indirectly at the post-transcriptional level for fine-tuning developmental regulation. Intriguingly, our ChIP-Seq data indicate that the Wcw_1223 TF that is encoded in the genomes of Chlamydiae and Planctomycetes, but has not yet been studied, functions as a master TF across the chlamydial and planctomycetes phyla. Wcw_1223 also targets >100 putative sites in W. chondrophila and like Euo controls genes of different functional categories. Moreover, using the Phyre2 prediction server (Kelley and Sternberg, 2009) we noted a structural similarity to the predicted transcriptional regulator VC0467 from Vibrio cholerae.
Akin to Euo, HrcA is not present in the Lentisphaerae, Verrucomicrobia and Planctomycetes (which seems to encode RpoH/σ32) (Wecker et al., 2009), but is present throughout the phylum Chlamydiae (Figure 1) (Hecker et al., 1996; Tan et al., 1996; Narberhaus, 1999; Wilson and Tan, 2004; Hanson and Tan, 2015). This distribution begs the question whether HrcA has been appropriated for chlamydial development and/or pathogenesis. The W. chondrophila HrcA regulon is quite small (<5%) compared with that of Euo and includes hrcA as well as genes involved in the adaptive response to temperature upshift, such as the (co-) chaperonin encoding genes groEL/ES, dnaK and grpE (Domman and Horn, 2015). In Chlamydia, the regulation of dnaK and groE operons was of great interest because of their role in pathogenesis and because of their induction upon temperature upshift due to a decrease in HrcA binding on these promoters (Hanson and Tan, 2015). In this context, we note that a widely used concept to modify gene expression in pathogenic bacteria upon host cell entry is that of protein thermometers, changing the activity upon temperature upshift in the host compared with the lower temperature in the environment (Kamp and Higgins, 2011; Loh et al., 2013) and HrcA is a thermosensor in the human pathogen Helicobacter pylori (Roncarati et al., 2014). Moreover, some Chlamydia-related bacteria have been shown to be stable endosymbionts of amoebae when present in the environment at a temperature of ⩽30 °C but then exhibit a lytic phenotype towards amoebae when the temperature increased above 32 °C (Greub et al., 2003). Thus, the pathogenic potential of Parachlamydia towards amoebae and higher eukaryotes including humans and bovines (Borel et al., 2007; Greub, 2009) might be due to an activation of temperature-regulated genes. Interestingly, we also note that HrcA targets the promoter of a gene (wcw_0325) encoding a protein featuring a von Willebrand factor type A domain known to act as mechanosensors (Siryaporn et al., 2014), possibly signalling the interaction of the bacterium with its target host cell upon temperature upshift in the host.
By contrast to such phylum-specific regulation, PhoB of the Chlamydiaceae (where it is called ChxR, Figure 1) is not very similar to that encoded in the other chlamydial families. Moreover, unlike PhoB, ChxR does not possess the conserved Asp residue and likely regulates genes expression independently of the activation by phosphorylation. With the putative PhoR-like histidine kinase Wcw_1870 following the same evolutionary pattern (that is, absence from the Chlamydiaceae), we suspect that PhoB may have become functionally specialized as the Chlamydiaceae branched from the ancestral lineage. In support of this, many of the promoters targeted by W. chondrophila PhoB appear to direct expression of hypothetical proteins, and none of the five known members of the C. trachomatis ChxR regulon (Koo et al., 2006; Hickey et al., 2011) were found to be regulated by PhoB in vivo. From an evolutionary perspective, it will be very interesting to define the PhoB regulon in other members of the PVC superphylum to determine whether orthologous genes are targeted by this TF. Indeed, PhoB of the widespread and highly diverse Chlamydia-related bacteria is more similar to PhoB of Planctomycetes, Verrucomicrobia and Lentisphaerae than to the ChxR homologue in Chlamydiaceae, suggesting that this TF may regulate genes implicated in the survival and in the colonization of a large variety of environmental ecosystems. This will be particularly interesting given the different ecology of the free-living versus obligate intracellular PVC superphylum members. Moreover, members of the Verrucomicrobia are important determinants of the human microbiome, and thus also have the capacity to interact (directly or indirectly) with eukaryotic host cells (Chevalier et al., 2015).
Acknowledgments
Funding support was from SNSF, Sinergia grant n° CRSII3-141837 (to PHV and GG), the de Reuter, Vontobel and Boninchi Foundations (PHV).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. (2010). Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res 38: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Sharma CM, Dittrich MT, Muller T, Reinhardt R, Vogel J et al. (2011). The transcriptional landscape of Chlamydia pneumoniae. Genome Biol 12: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R, Springer N, Schonhuber W, Ludwig W, Schmid EN, Muller KD et al. (1997). Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl Environ Microbiol 63: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoldini M, Vizcarra IA, Pena-Miller R, Stocker N, Diard M, Vogel V et al. (2014). Bistable expression of virulence genes in salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS Biol 12: e1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D, Regan L, Greub G. (2008). Emerging role of Chlamydia and Chlamydia-like organisms in adverse pregnancy outcomes. Curr Opin Infect Dis 21: 70–76. [DOI] [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J et al. (2003). Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA 100: 8478–8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, Collyn F, Croxatto A, Ruckert C, Polkinghorne A, Kebbi-Beghdadi C et al. (2010). The Waddlia genome: a window into chlamydial biology. PLoS One 5: e10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel N, Ruhl S, Casson N, Kaiser C, Pospischil A, Greub G. (2007). Parachlamydia spp. and related Chlamydia-like organisms and bovine abortion. Emerg Infect Dis 13: 1904–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M et al. (2010). The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case ED, Peterson EM, Tan M. (2010). Promoters for Chlamydia type III secretion genes show a differential response to DNA supercoiling that correlates with temporal expression pattern. J Bacteriol 192: 2569–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AL, Wilson AC, Tan M. (2011). A Chlamydia-specific C-terminal region of the stress response regulator HrcA modulates its repressor activity. J Bacteriol 193: 6733–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Miura K, Popov VL, Kumagai Y, Rikihisa Y. (2011). Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol Microbiol 82: 1217–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Stojanovic O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C et al. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cell 163: 1360–1374. [DOI] [PubMed] [Google Scholar]
- Cole MF, Young RA. (2008). Mapping key features of transcriptional regulatory circuitry in embryonic stem cells. Cold Spring Harb Symp Quant Biol 73: 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, Toenshoff ER, Taylor MW, Fritsche TR, Wagner M, Horn M. (2005). 'Candidatus Protochlamydia amoebophila', an endosymbiont of Acanthamoeba spp. Int J Syst Evol Microbiol 55: 1863–1866. [DOI] [PubMed] [Google Scholar]
- Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC et al. (2011). Unity in variety—the pan-genome of the Chlamydiae. Mol Biol Evol 28: 3253–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsaro D, Feroldi V, Saucedo G, Ribas F, Loret JF, Greub G. (2009). Novel Chlamydiales strains isolated from a water treatment plant. Environ Microbiol 11: 188–200. [DOI] [PubMed] [Google Scholar]
- Coulon C, Eterpi M, Greub G, Collignon A, McDonnell G, Thomas V. (2012). Amoebal host range, host-free survival and disinfection susceptibility of environmental Chlamydiae as compared to Chlamydia trachomatis. FEMS Immunol Med Microbiol 64: 364–373. [DOI] [PubMed] [Google Scholar]
- Croxatto A, Rieille N, Kernif T, Bitam I, Aeby S, Peter O et al. (2014). Presence of Chlamydiales DNA in ticks and fleas suggests that ticks are carriers of Chlamydiae. Ticks Tick Borne Dis 5: 359–365. [DOI] [PubMed] [Google Scholar]
- De Nisco NJ, Abo RP, Wu CM, Penterman J, Walker GC. (2014). Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc Natl Acad Sci USA 111: 3217–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deghelt M, Mullier C, Sternon JF, Francis N, Laloux G, Dotreppe D et al. (2014). G1-arrested newborn cells are the predominant infectious form of the pathogen Brucella abortus. Nat Commun 5: 4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diard M, Sellin ME, Dolowschiak T, Arnoldini M, Ackermann M, Hardt WD. (2014). Antibiotic treatment selects for cooperative virulence of Salmonella typhimurium. Curr Biol 24: 2000–2005. [DOI] [PubMed] [Google Scholar]
- Dilbeck PM, Evermann JF, Crawford TB, Ward AC, Leathers CW, Holland CJ et al. (1990). Isolation of a previously undescribed Rickettsia from an aborted bovine fetus. J Clin Microbiol 28: 814–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domman D, Horn M. (2015). Following the footsteps of Chlamydial gene regulation. Mol Biol Evol 32: 3035–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elantak L, Ansaldi M, Guerlesquin F, Mejean V, Morelli X. (2005). Structural and genetic analyses reveal a key role in prophage excision for the TorI response regulator inhibitor. J Biol Chem 280: 36802–36808. [DOI] [PubMed] [Google Scholar]
- Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. (2014). A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet 10: e1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche TR, Gautom RK, Seyedirashti S, Bergeron DL, Lindquist TD. (1993). Occurrence of bacterial endosymbionts in Acanthamoeba spp. isolated from corneal and environmental specimens and contact lenses. J Clin Microbiol 31: 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumeaux C, Radhakrishnan SK, Ardissone S, Theraulaz L, Frandi A, Martins D et al. (2014). Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat Commun 5: 4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayston JT. (2000). Background and current knowledge of Chlamydia pneumoniae and atherosclerosis. J Infect Dis 181(Suppl 3): S402–S410. [DOI] [PubMed] [Google Scholar]
- Greub G, La Scola B, Raoult D. (2003). Parachlamydia acanthamoeba is endosymbiotic or lytic for Acanthamoeba polyphaga depending on the incubation temperature. Ann NY Acad Sci 990: 628–634. [DOI] [PubMed] [Google Scholar]
- Greub G, Raoult D. (2004). Microorganisms resistant to free-living amoebae. Clin Microbiol Rev 17: 413–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G. (2009). Parachlamydia acanthamoebae, an emerging agent of pneumonia. Clin Microbiol Infect 15: 18–28. [DOI] [PubMed] [Google Scholar]
- Greub G, Kebbi-Beghdadi C, Bertelli C, Collyn F, Riederer BM, Yersin C et al. (2009). High throughput sequencing and proteomics to identify immunogenic proteins of a new pathogen: the dirty genome approach. PLoS One 4: e8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goy G, Croxatto A, Posfay-Barbe KM, Gervaix A, Greub G. (2009). Development of a real-time PCR for the specific detection of Waddlia chondrophila in clinical samples. Eur J Clin Microbiol Infect Dis 28: 1483–1486. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson BR, Tan M. (2015). Transcriptional regulation of the Chlamydia heat shock stress response in an intracellular infection. Mol Microbiol 97: 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Schumann W, Volker U. (1996). Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol 19: 417–428. [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7: 263–273. [DOI] [PubMed] [Google Scholar]
- Hickey JM, Weldon L, Hefty PS. (2011). The atypical OmpR/PhoB response regulator ChxR from Chlamydia trachomatis forms homodimers in vivo and binds a direct repeat of nucleotide sequences. J Bacteriol 193: 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CR, Weeks AQ. (2008). Influence of particle size on bacterial community structure in aquatic sediments as revealed by 16S rRNA gene sequence analysis. Appl Environ Microbiol 74: 5237–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahane S, Metzer E, Friedman MG. (1995). Evidence that the novel microorganism 'Z' may belong to a new genus in the family Chlamydiaceae. FEMS Microbiol Lett 126: 203–207. [DOI] [PubMed] [Google Scholar]
- Kamp HD, Higgins DE. (2011). A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog 7: e1002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebbi-Beghdadi C, Batista C, Greub G. (2011). Permissivity of fish cell lines to three Chlamydia-related bacteria: Waddlia chondrophila, Estrella lausannensis and Parachlamydia acanthamoebae. FEMS Immunol Med Microbiol 63: 339–345. [DOI] [PubMed] [Google Scholar]
- Kebbi-Beghdadi C, Fatton M, Greub G. (2015). Permissivity of insect cells to Waddlia chondrophila, Estrella lausannensis and Parachlamydia acanthamoebae. Microbes Infect. 17: 749–54. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Khan SR, Gaines J, Roop RM 2nd, Farrand SK. (2008). Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74: 5053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo IC, Walthers D, Hefty PS, Kenney LJ, Stephens RS. (2006). ChxR is a transcriptional activator in Chlamydia. Proc Natl Acad Sci USA 103: 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Weinmaier T, Lauro FM, Cavicchioli R, Rattei T, Horn M. (2014). Integrating metagenomic and amplicon databases to resolve the phylogenetic and ecological diversity of the Chlamydiae. ISME J 8: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoth F, Greub G. (2010). Amoebal pathogens as emerging causal agents of pneumonia. FEMS Microbiol Rev 34: 260–280. [DOI] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. (2000). Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148. [DOI] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH. (2002). Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci USA 99: 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Shapiro L, McAdams HH. (2007). Systems biology of Caulobacter. Annu Rev Genet 41: 429–441. [DOI] [PubMed] [Google Scholar]
- Lienard J, Croxatto A, Prod'hom G, Greub G. (2011). Estrella lausannensis, a new star in the Chlamydiales order. Microbes Infect 13: 1232–1241. [DOI] [PubMed] [Google Scholar]
- Loh E, Kugelberg E, Tracy A, Zhang Q, Gollan B, Ewles H et al. (2013). Temperature triggers immune evasion by Neisseria meningitidis. Nature 502: 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R, Desplan C. (2008). Stochasticity and cell fate. Science 320: 65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manina G, Dhar N, McKinney JD. (2015). Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17: 32–46. [DOI] [PubMed] [Google Scholar]
- Minder AC, Fischer HM, Hennecke H, Narberhaus F. (2000). Role of HrcA and CIRCE in the heat shock regulatory network of Bradyrhizobium japonicum. J Bacteriol 182: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW. (1991). Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55: 143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F. (1999). Negative regulation of bacterial heat shock genes. Mol Microbiol 31: 1–8. [DOI] [PubMed] [Google Scholar]
- Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. (2003). Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol 185: 3179–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus E, Cheng E, Tan M. (2008). DNA supercoiling-dependent gene regulation in Chlamydia. J Bacteriol 190: 6419–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen J et al. (2001). Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA 98: 4136–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panis G, Duverger Y, Champ S, Ansaldi M. (2010. a). Protein binding sites involved in the assembly of the KplE1 prophage intasome. Virology 404: 41–50. [DOI] [PubMed] [Google Scholar]
- Panis G, Duverger Y, Courvoisier-Dezord E, Champ S, Talla E, Ansaldi M. (2010. b). Tight regulation of the intS gene of the KplE1 prophage: a new paradigm for integrase gene regulation. PLoS Genet 6: e1001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panis G, Franche N, Mejean V, Ansaldi M. (2012). Insights into the functions of a prophage recombination directionality factor. Viruses 4: 2417–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panis G, Murray SR, Viollier PH. (2015). Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol Rev 39: 120–133. [DOI] [PubMed] [Google Scholar]
- Pillonel T, Bertelli C, Salamin N, Greub G. (2015). Taxogenomics of the Chlamydiales. Int J Syst Evol Microbiol 65: 1381–1393. [DOI] [PubMed] [Google Scholar]
- Pilloux L, Aeby S, Gaumann R, Burri C, Beuret C, Greub G. (2015). The high prevalence and diversity of Chlamydiales DNA within Ixodes ricinus ticks suggest a role for ticks as reservoirs and vectors of Chlamydia-related bacteria. Appl Environ Microbiol 81: 8177–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. (1996). Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84: 83–93. [DOI] [PubMed] [Google Scholar]
- Rockey DD, Matsumoto A. (2000). The Chlamydial developmental cycle. In: Brun YV, Shimkets LJ (eds), Prokaryotic Development. American Society for Microbiology: Washington, DC, USA, pp 403–425. [Google Scholar]
- Roncarati D, Danielli A, Scarlato V. (2014). The HrcA repressor is the thermosensor of the heat-shock regulatory circuit in the human pathogen Helicobacter pylori. Mol Microbiol 92: 910–920. [DOI] [PubMed] [Google Scholar]
- Rosario CJ, Tan M. (2012). The early gene product EUO is a transcriptional repressor that selectively regulates promoters of Chlamydia late genes. Mol Microbiol 84: 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario CJ, Hanson BR, Tan M. (2014). The transcriptional repressor EUO regulates both subsets of Chlamydia late genes. Mol Microbiol 94: 888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rurangirwa FR, Dilbeck PM, Crawford TB, McGuire TC, McElwain TF. (1999). Analysis of the 16S rRNA gene of micro-organism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int J Syst Bacteriol 49(Pt 2): 577–581. [DOI] [PubMed] [Google Scholar]
- Satinsky BM, Fortunato CS, Doherty M, Smith CB, Sharma S, Ward ND et al. (2015). Metagenomic and metatranscriptomic inventories of the lower Amazon River, May 2011. Microbiome 3: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn L, Jaton K, Fitting JW, Greub G. (2011). Does respiratory infection due to Chlamydia pneumoniae still exist? Clin Infect Dis 53: 847–848. [DOI] [PubMed] [Google Scholar]
- Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. (2000). Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 37: 913–925. [DOI] [PubMed] [Google Scholar]
- Shirai M, Hirakawa H, Kimoto M, Tabuchi M, Kishi F, Ouchi K et al. (2000). Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res 28: 2311–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl A, Horn M. (2012) Lessons from environmental Chlamydiae. In: Tan M, Bavoil PM (eds), Intracellular Pathogens I: Chlamydiales. American Society for Microbiology: Washington, DC, USA, pp 51–73. [Google Scholar]
- Siryaporn A, Kuchma SL, O'Toole GA, Gitai Z. (2014). Surface attachment induces Pseudomonas aeruginosa virulence. Proc Natl Acad Sci USA 111: 16860–16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L et al. (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282: 754–759. [DOI] [PubMed] [Google Scholar]
- Tan M, Wong B, Engel JN. (1996). Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J Bacteriol 178: 6983–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M. (2012). Temporal gene regulation during the Chlamydial developmental cycle. In: Tan M, Bavoil PM (eds), Intracellular Pathogens I: Chlamydiales. American Society for Microbiology: Washington, DC, USA, pp 149–169. [Google Scholar]
- Taylor HR, Burton MJ, Haddad D, West S, Wright H. (2014). Trachoma. Lancet 384: 2142–2152. [DOI] [PubMed] [Google Scholar]
- Thomas V, Casson N, Greub G. (2006). Criblamydia sequanensis, a new intracellular Chlamydiales isolated from Seine river water using amoebal co-culture. Environ Microbiol 8: 2125–2135. [DOI] [PubMed] [Google Scholar]
- Vijayan V, Zuzow R, O'Shea EK. (2009). Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA 106: 22564–22568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecker P, Klockow C, Ellrott A, Quast C, Langhammer P, Harder J et al. (2009). Transcriptional response of the model planctomycete Rhodopirellula baltica SH1(T) to changing environmental conditions. BMC Genomics 10: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AH, Stock AM. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26: 369–376. [DOI] [PubMed] [Google Scholar]
- Wheelhouse N, Longbottom D. (2012). Endemic and emerging chlamydial infections of animals and their zoonotic implications. Transbound Emerg Dis 59: 283–291. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH et al. (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Tan M. (2004). Stress response gene regulation in Chlamydia is dependent on HrcA-CIRCE interactions. J Bacteriol 186: 3384–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolanski M, Jakimowicz D, Zakrzewska-Czerwinska J. (2014). Fifty years after the replicon hypothesis: cell-specific master regulators as new players in chromosome replication control. J Bacteriol 196: 2901–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Popov VL, Gao S, Walker DH, Yu XJ. (2007). The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell Microbiol 9: 610–618. [DOI] [PubMed] [Google Scholar]
- Zhang L, Douglas AL, Hatch TP. (1998). Characterization of a Chlamydia psittaci DNA binding protein (EUO) synthesized during the early and middle phases of the developmental cycle. Infect Immun 66: 1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Howe MM, Hatch TP. (2000). Characterization of in vitro DNA binding sites of the EUO protein of Chlamydia psittaci. Infect Immun 68: 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.