Abstract
Groundwater ecosystems are conventionally thought to be fueled by surface-derived allochthonous organic matter and dominated by heterotrophic microbes living under often-oligotrophic conditions. However, in a 2-month study of nitrate amendment to a perennially suboxic aquifer in Rifle (CO), strain-resolved metatranscriptomic analysis revealed pervasive and diverse chemolithoautotrophic bacterial activity relevant to C, S, N and Fe cycling. Before nitrate injection, anaerobic ammonia-oxidizing (anammox) bacteria accounted for 16% of overall microbial community gene expression, whereas during the nitrate injection, two other groups of chemolithoautotrophic bacteria collectively accounted for 80% of the metatranscriptome: (1) members of the Fe(II)-oxidizing Gallionellaceae family and (2) strains of the S-oxidizing species, Sulfurimonas denitrificans. Notably, the proportion of the metatranscriptome accounted for by these three groups was considerably greater than the proportion of the metagenome coverage that they represented. Transcriptional analysis revealed some unexpected metabolic couplings, in particular, putative nitrate-dependent Fe(II) and S oxidation among nominally microaerophilic Gallionellaceae strains, including expression of periplasmic (NapAB) and membrane-bound (NarGHI) nitrate reductases. The three most active groups of chemolithoautotrophic bacteria in this study had overlapping metabolisms that allowed them to occupy different yet related metabolic niches throughout the study. Overall, these results highlight the important role that chemolithoautotrophy can have in aquifer biogeochemical cycling, a finding that has broad implications for understanding terrestrial carbon cycling and is supported by recent studies of geochemically diverse aquifers.
Introduction
Microbial communities in nature are seldom exposed to a static environment, and their responses to changes in their physicochemical environment can help us to better understand and predict what organisms and processes will drive biogeochemical cycling under a range of conditions (e.g, Allison and Martiny, 2008; Hamilton et al., 2014; Faust et al., 2015). In this study, we characterized the response of an aquifer microbial community to an increase in the flux of nitrate, a naturally occurring electron acceptor, at a field site in Rifle, Colorado (USA). This nitrate perturbation study was motivated in part by observations of dynamic fluctuations in nitrate concentrations within the capillary fringe of this aquifer (ranging from ~50 μm to more than 6000 μm over time periods of 2 months and vertical distances of 0.5 m; unpublished data). This study allowed us to observe, in a controlled manner, how the aquifer microbial community could respond to nitrate being transported into the saturated zone from the vadose zone.
The response to an influx of a thermodynamically favorable electron acceptor like nitrate could be expected to enhance microbial oxidation of organic electron donors (e.g., via heterotrophic denitrification; Korom, 1992; Starr and Gillham, 1993; Bengtsson and Bergwall, 1995), consistent with current heterotroph-dominant paradigms of groundwater ecology (Baker et al., 2000; Foulquier et al., 2010), as well as inorganic electron donors (e.g., via chemolithoautotrophic denitrification; Pauwels et al., 1998; Beller et al., 2004; Herrmann et al., 2015). A wide range of factors could modulate the response to a nitrate perturbation, but a predictive and mechanistic understanding of these factors requires highly resolved empirical studies, ideally at genome-scale resolution such that strain-level behavior could be correctly interpreted (Allen et al., 2007; Sharon et al., 2013; Luo et al., 2015).
Here, we used strain-resolved metagenomic reconstructions combined with metatranscriptomic analysis to derive transcriptional evidence of metabolic activity and highlight important biogeochemical processes. Although an increasing number of metatranscriptomic analyses of environmental systems have been reported over the past decade (Poretsky et al., 2005; Frias-Lopez et al., 2008; Gilbert et al., 2008; Urich et al., 2008; Poretsky et al., 2009; Shrestha et al., 2009; Lesniewski et al., 2012; Quaiser et al., 2014; Kopf et al., 2015), most have focused on marine, freshwater and surface soil systems, and rarely on aquifer environments, where studies have typically targeted selected, diagnostic transcripts rather than the entire metatranscriptome (e.g., Chandler et al., 2006; O'Neil et al., 2008; Alfreider and Vogt, 2012; Herrmann et al., 2015).
Materials and methods
Field site
The field experiment was conducted at the US Department of Energy (DOE) Rifle field site near Rifle, Colorado (USA), which hosts an alluvial aquifer with groundwater characterized by low dissolved oxygen (DO) concentrations, typically <20 μm, elevated concentrations of dissolved iron and sulfate, nitrate concentrations generally below 200 μm and ammonium concentrations ranging from 0.6 to 70 μm near the study area (Zachara et al., 2013; unpublished data). Microbial Fe(III) and sulfate reduction at the site have contributed to the presence of reduced Fe- and S-containing compounds in the aquifer (Williams et al., 2011); for example, after previous acetate amendment studies at the site, total reduced inorganic sulfur concentrations in aquifer sediment ranged from 0.2 to 11 mmol kg−1 (Williams et al., 2011). Further details of the Rifle site geology, hydrogeology and history are presented elsewhere (Williams et al., 2011).
Nitrate injection, analytical and biomass collection procedures
The experimental plot in which this 2013 nitrate injection occurred (Supplementary Figure S1) had previously been used for successive injection experiments in the preceding 3 years (Long et al., 2015; Luef et al., 2015; D Pan, KH Williams, MJ Robbins, and KA Webber, personal communication). Groundwater from an unamended background well (CU01) was pumped into a storage tank and amended with sodium nitrate to achieve a tank concentration of ca. 2.9 mm. The conservative tracer deuterium (as D2O; average tank composition δD=+210.8‰) was added to assess the downgradient migration of the injectate to account for the non-conservative behavior of nitrate. The tank contents were injected via peristaltic pump into five wells (CG01–CG05) over a period of 34 days. An injection rate of 36 ml min−1 per well was used over the first 23 days to achieve an aquifer nitrate concentration of 0.5 mm, with the rate increasing to 120 ml min−1 per well from days 23 to 34 in order to increase the nitrate loading and resultant aquifer concentration (2 mm). To improve the advective delivery of the injectate to downgradient wells, cross-well mixing pumps were used to circulate injection well contents, as described previously (Williams et al., 2011, 2013). Over the course of the experiment, groundwater samples for geochemical (cations, anions, pH, Fe(II), δD) and biological samples were collected from a well (CD01) located 2.5 m downgradient from the region of nitrate/deuterium injection. Sampling and analytical protocols have been described elsewhere (Williams et al., 2011; Long et al., 2015). Before injection and at regular intervals thereafter, groundwater was pumped from well CD01 and serially filtered through 293-mm diameter polyethersulfone filters having pore sizes of 1.2 and 0.2 μm; filtered volumes were ca. 100 L collected within 2 h. Following filtration, both filters were placed into gas-impermeable Mylar bags, flash frozen in liquid nitrogen and stored at −80 °C until nucleic acid extraction. As biomass samples in this study were captured on groundwater filters, they represent microbes that were planktonic or associated with colloidal particles. As such, the degree to which they represent microbes in aquifer sediment is uncertain.
Conservative vs non-conservative tracer breakthrough
Methods used to predict the concentrations of nitrate in groundwater that would have been present if injected nitrate had behaved conservatively (i.e., was not biologically active) are presented in the Supplementary Information.
Nucleic acid extractions, library construction and library sequencing
The methods used for DNA and RNA extraction, metagenomic and metatranscriptomic library construction and library sequencing can be found in the Supplementary Information.
Analysis of metagenomics and metatranscriptomic short reads
Details of the following steps in the bioinformatic analysis of the metagenomic and metatranscriptomic libraries can be found in the Supplementary Information: (1) metagenome sequencing quality control, trimming and filtering, (2) metagenome assembly, (3) binning, bin evaluation and bin curation, (4) functional annotation, (5) metatranscriptomic sequence data analysis, (6) estimation of transcript abundances and (7) additional bioinformatic analyses.
Results
Groundwater geochemistry
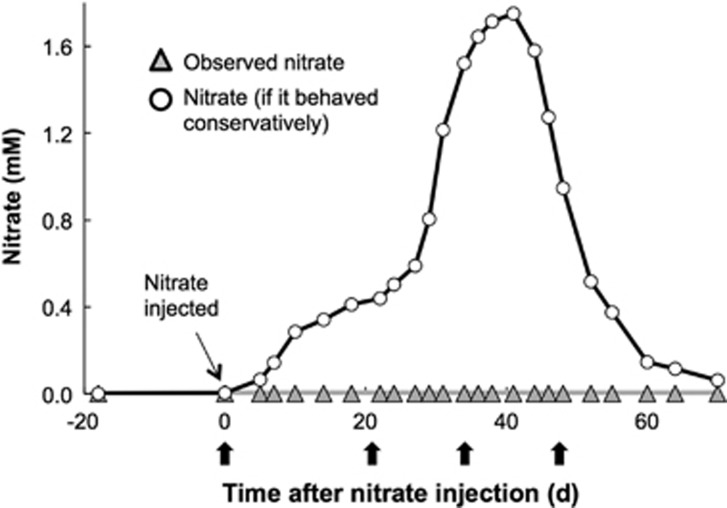
Geochemical data were generated for groundwater samples collected throughout the 2-month study from a monitoring well-located 2.5 m downgradient of the nitrate injection gallery (Supplementary Table S1). Nitrate was consumed very rapidly throughout the study (Figure 1). Whereas injected nitrate concentrations should have reached 1.75 mm at the monitoring well (if nitrate were not consumed, i.e., behaved conservatively), observed nitrate concentrations were rarely above the 5 μm detection limit (Figure 1 and Supplementary Table S1). While pH remained relatively constant throughout the study (7.1±0.1), sulfate concentrations increased after time zero and reached maximum values after 18 days (Supplementary Figure S2).
Figure 1.
Nitrate concentrations in groundwater (well CD01) collected 2.5 m downgradient of injection wells. Observed nitrate concentrations are shown along with calculated concentrations that would be expected if injected nitrate had behaved conservatively, that is, was not consumed by biological activity (see text). The black arrows represent the time of collection for metagenomic/metatranscriptomic samples.
Overview of metatranscriptomic data
For each of the four 0.2-μm groundwater filter samples whose metagenomes and metatranscriptomes were analyzed for this study (day 0, 21, 35 and 47), 2.8 to 3.4 Gb of cDNA were sequenced and passed quality filters (Supplementary Table S2B). The data are available in the Supplementary Information. The open reading frames with the highest 100 RPKM (reads per kilobase per million mapped reads) values corresponded to the 99.95th percentile of the RPKM distribution in each of the four samples and are listed in Supplementary Table S3; a full listing of all detected transcripts with their corresponding RPKM values can be found in the Supplementary Information. The majority of the most highly expressed genes encoded ribosomal, cold-shock and housekeeping proteins, or were uncharacterized. However, many of the top 100 transcripts encoded proteins linked to chemolithoautotrophic bacterial metabolism.
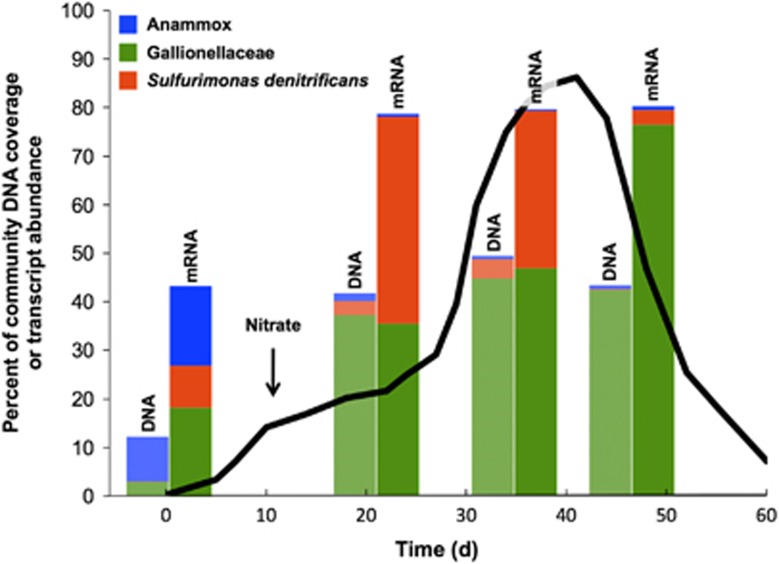
Perhaps the most striking trend in the metatranscriptome data is the prevalent activity of a small but diverse group of chemolithoautotrophic bacteria that accounted for a very large portion of overall microbial community gene expression. More specifically, chemolithoautotrophic bacteria collectively accounting for ca. 40–80% of the total mapped metatranscriptome in all samples (including the prerelease sample) belonged to only three groups: (1) members of the Fe(II)-oxidizing Gallionellaceae family, (2) anammox bacteria, that is, planctomycetes capable of anaerobic ammonia oxidation (Jetten et al., 2009; Zhou et al., 2012; Kartal et al., 2013) and (3) strains of the S-oxidizing species, Sulfurimonas denitrificans (Figure 2). At the maximum point of their activities, Gallionellaceae family members collectively accounted for 75% of the total metatranscriptome (day 47), S. denitrificans accounted for 42% (day 21) and anammox bacteria accounted for 16% (day 0). Notably, the proportion of the metatranscriptome accounted for by these three groups was considerably greater than the proportion of the metagenome coverage that they represented (Figure 2); in other words, overall gene expression for these chemolithoautotrophic taxa was disproportionately large in relation to overall gene abundance. No single group of known heterotrophic bacteria or archaea represented >2% of the total metatranscriptome after day 0 (Supplementary Table S4). The following sections present transcriptional trends observed for the three chemolithoautotrophic groups that were most highly represented in the community gene expression data.
Figure 2.
Percent of total community mapped DNA (as depth of coverage) and total community mapped mRNA (as RPKM) for the three most active chemolithoautotrophic groups: anammox bacteria (bin 16), Gallionellaceae (bins 22.1–22.9) and Sulfurimonas denitrificans (bins 62 and 93). For each pair of bars, the bar on the left represents metagenome data and the bar on the right represents metatranscriptome data. The profile of nitrate concentration if it had behaved conservatively (as in Figure 1) is displayed to provide more context on the collection times for the four samples represented. The data at time zero were collected before the nitrate release commenced.
Anammox bacteria highly active before nitrate injection
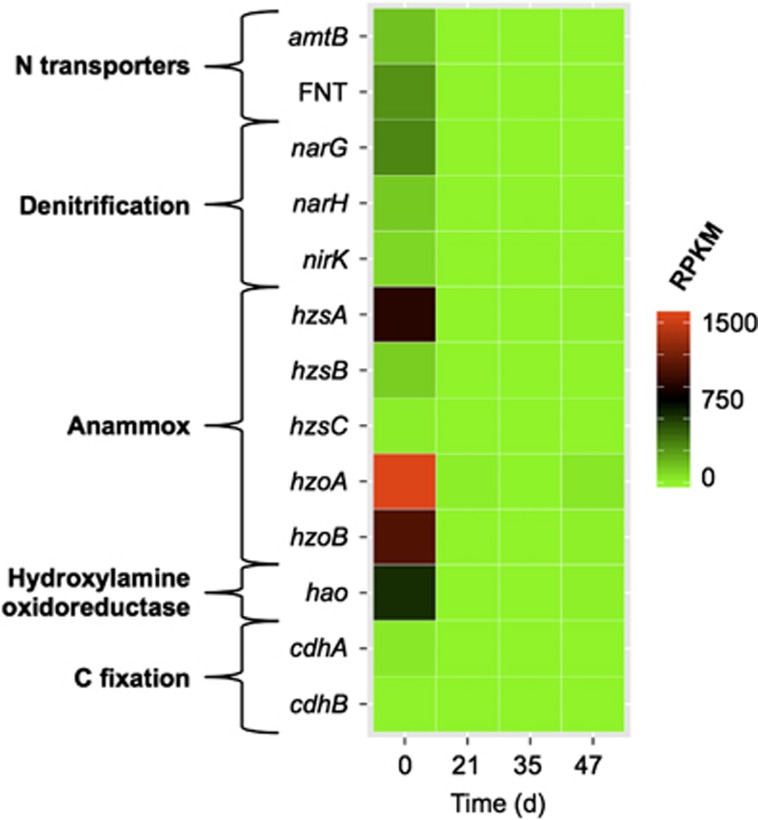
Anammox bacteria closely related to Candidatus Brocadia sinica (Oshiki et al., 2015) and Candidatus Jettenia caeni (Strous et al., 2006; Ali et al., 2015), largely represented by metagenome bin 16, constituted 16% of the community metatranscriptome at day 0 but declined in relative expression after the nitrate injection (Figure 2). Among the more highly expressed genes in bin 16 were those associated with unique anammox biochemistry, nitrate and nitrite reduction, CO2 fixation and transport of ammonium and other solutes (Figure 3). Approximately one-third of the 50 most highly expressed transcripts from this bin were related to nitrogen metabolism, whereas one-third had gene products of unknown function (Supplementary Table S5). Figure 3 highlights highly expressed genes that are diagnostic of key metabolic processes in anammox bacteria that mediate N and C cycling, including hydrazine synthase (hzsA, hzsB, hzsC) and hydrazine oxidoreductase (hzoA, hzoB) from the anammox process; dissimilatory, membrane-bound nitrate reductase (narG, narH); copper-containing nitrite reductase (nirK); carbon monoxide dehydrogenase/acetyl-CoA synthase from the Wood–Ljungdahl pathway (cdhA, cdhB); and transporters for nitrite and ammonium (FNT, amtB). Genes encoding two essential anammox enzymes (hydrazine synthase and particularly hydrazine oxidoreductase; Figure 3) were among the most highly expressed genes in the time-zero sample (hzoA, hzoB were in the 99.99th percentile of transcripts at day 0). Transcripts represented in Figure 3 were all at their maxima at day 0; RPKM values for these genes at the later sampling dates never exceeded 4% of the time-zero values. The translated sequence identity relative to the corresponding genes in anammox species Ca. J. caeni and Ca. B. sinica JPN1 ranged from 88% to 96%, except for nirK, which had 37% identity to Nitrospina sp. SCGC_AAA799_C22 (Supplementary Table S6). In addition to these transcripts, there were also six putative copies of hydroxylamine oxidoreductase (hao), which is often found in multiple copies in the genomes of anammox bacteria (Strous et al., 2006; Shimamura et al., 2008) and whose gene product has recently been found to interconvert NO and hydroxylamine in in vitro studies (Irisa et al., 2014; Maalcke et al., 2014), but its physiological role in anammox is uncertain.
Figure 3.
Temporal expression profiles for selected genes associated with N and C metabolism in the most active anammox bacterial strain (bin 16). A scale (RPKM) is shown on the right. Genes: amtB, ammonium transporter; FNT, formate/nitrite transporter; narG and narH, α and β subunits of membrane-bound nitrate reductase; nirK, copper-containing nitrite reductase; hzsA, hzsB, hzsC, hydrazine synthase A, B and C subunits; hzoA and hzoB, hydrazine oxidoreductase genes; hao, hydroxylamine oxidoreductase; cdhA and cdhB, α and β subunits of CO dehydrogenase/acetyl-CoA synthase.
The emerging activity of Gallionellaceae family members following nitrate injection
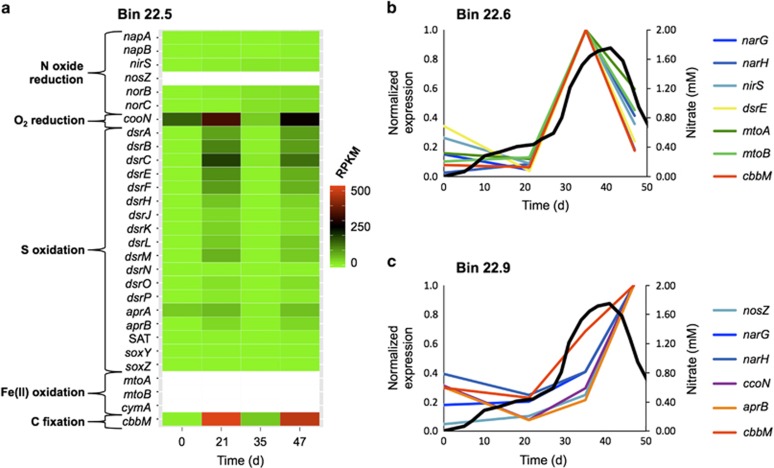
Collectively, transcripts from members of the Gallionellaceae increased from 18% of the total metatranscriptome at time zero to 75% at day 47 (Figure 2). Most genes belonging to members of the Gallionellaceae family, which is best known for Fe(II)-oxidizing activity, were assigned to nine reconstructed genome bins (bins 22.1–22.9). Three of the bins, 22.5, 22.6 and 22.9, displayed unique transcriptional profiles for metabolism relevant to Fe, N, S and C cycling (Figure 4 and Supplementary Table S7), as described below.
Figure 4.
Temporal expression profiles for selected genes associated with C, S, N and Fe metabolism in three of the most active Gallionellaceae strains (bins 22.5, 22.6 and 22.9). (a) Heat map of relative expression of selected genes in bin 22.5; a scale is shown on the right. White rows represent genes that were found in other Gallionellaceae bins but not in bin 22.5. (b) Relative expression of selected genes over time for Gallionellaceae bin 22.6. For each gene, RPKM values have been normalized to their maximum value on the four collection dates. The profile of nitrate concentration if it had behaved conservatively (as in Figure 1) is displayed in panels b and c to provide context for gene expression. (c) Relative expression of selected genes over time for Gallionellaceae bin 22.9. Genes: napA and napB, α and β subunits of periplasmic nitrate reductase; narG and narH, α and β subunits of membrane-bound nitrate reductase; nirS, cytochrome cd1-nitrite reductase; nosZ, nitrous oxide reductase; norBC, subunits I and II of nitric oxide reductase; ccoN, subunit I of cbb3-type cytochrome c oxidase; dsrABCEFHJKLMNOP, reverse dissimilatory sulfite reductase genes; mtoA, mtoB and cymA, Fe(II) oxidation genes; cbbM, large subunit of form II RubisCO.
Gene expression data indicated that bin 22.5 was the most active Gallionellaceae strain, with highest transcript abundances on days 21 and 47, time points that flank the maximum consumption of nitrate. On those 2 days, bin 22.5 transcripts represented 29% and 33% of the total metatranscriptome, respectively, while representing only 14% and 4% of metagenome coverage (Supplementary Table S4). Transcriptional data for bin 22.5 indicated metabolic activity related to denitrification, S-compound oxidation, oxygen reduction (via cytochrome cbb3 oxidase) and CO2 fixation (Figure 4a). No genes typically associated with neutrophilic Fe(II) oxidation, mtoA, mtoB and cymA (Ilbert and Bonnefoy, 2013), were assigned to this bin, thus their transcripts were not detected (Figure 4a). In this case, and in all such cases in which an expected key gene was missing, we performed multiple BLASTP searches against a custom metagenomic database constructed for this study using closely related sequences as queries.
The unexpected emergence during the nitrate perturbation of Gallionellaceae, whose isolated members are microaerophiles and not nitrate reducers (Emerson et al., 2013), is consistent with our transcriptional data indicating expression of structural genes involved in denitrification. Specifically, for bin 22.5 (Figure 4a), transcripts were observed for a periplasmic, Mo-containing nitrate reductase (napAB; 78% sequence identity to NapA of Thioalkalivibrio nitratireducens; Supplementary Table S7), cytochrome cd1-nitrite reductase (nirS; 80% sequence identity to NirS of Sulfuricella denitrificans) and nitric oxide reductase (norBC; 85–95% sequence identity to NorB and NorC of Gallionella capsiferriformans). There were no copies of nitrous oxide reductase (nosZ) assigned to bin 22.5, thus there could be no nosZ transcripts mapping to this bin. The gene cluster in bin 22.5 including napA and napB is presented in Figure 5.
Figure 5.
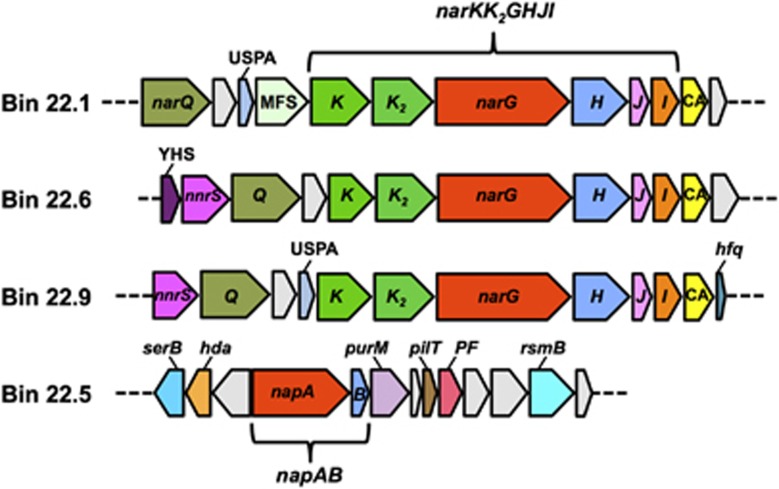
Portions of Gallionellaceae scaffolds containing putative copies of nap genes (bin 22.5) or nar genes (bins 22.1, 22.6 and 22.9), encoding nitrate reductases. Genes plotted include: narQ, nitrate/nitrite sensor protein; uspA, universal stress protein A; narGHI, membrane-bound nitrate reductase; narJ, nitrate reductase molybdenum cofactor assembly chaperone 1; narK and narK2, nitrate/nitrite transporter, narQ, nitrate/nitrite-specific signal-transduction histidine kinase; MFS, MFS transporter; CA, carbonic anhydrase; YHS, YHS protein; nnrS, NnrS family protein; hfq, RNA-binding protein Hfq; napA and napB, α and β subunits of periplasmic nitrate reductase; serB, phosphoserine phosphatase; hfq, DnaA regulatory inactivator Hfq; purM, phosphoribosylaminoimidazole synthetase; pilT, PilT protein domain protein; PF, phosphoribosylglycinamide formyltransferase; rsmB, SAM-dependent methyltransferase; light gray, hypothetical or uncharacterized proteins. The regions shown extend from scaffold-2213_2214_33 to scaffold-2213_2214_44 for bin 22.1, scaffold-6558_6559_58 to scaffold-6558_6559_69 for bin 22.6, scaffold-958_959_56 to scaffold-958_959_43 for bin 22.9 and scaffold-1168_1169_107 to scaffold-1168_1169_119 for bin 22.5.
Transcriptional data for bin 22.5 also indicate S-compound oxidation during the nitrate perturbation, particularly on days 21 and 47 (Figure 4a). Sulfur oxidation transcripts were observed for (reverse) dissimilatory sulfite reductase genes dsrABEFHCMKLJOPN (Supplementary Table S7) (this cluster includes dsr genes specific to S oxidation rather than sulfite reduction, such as dsrEFH and dsrL; Ghosh and Dam, 2009), ATP sulfurylase and soxYZ. These gene clusters were in close proximity to each other on a single scaffold, and shared a high degree of synteny to homologs encoded in the genome of Sideroxydans ES-1 (Supplementary Table S7; Emerson et al., 2013). Other sox genes normally found in a cluster with soxYZ in Sideroxydans ES-1, namely soxX, soxA and soxB, were not identified in bin 22.5. The most highly expressed gene associated with S-compound oxidation was dsrC, whose role in S oxidation has been discussed (Venceslau et al., 2014).
Among the more highly expressed genes in bin 22.5 (Figure 4a) were subunits of a cbb3-type cytochrome c oxidase (ccoN, ccoO, ccoQ and ccoP) (Supplementary Table S7), a high-oxygen-affinity cytochrome oxidase (Pitcher and Watmough, 2004) consistent with oxygen reduction in a suboxic environment. The most highly expressed gene represented in Figure 4a is cbbM, which encodes form II ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO). Form II RubisCO is consistent with CO2 fixation via the Calvin–Benson–Bassham cycle under suboxic or anoxic conditions (Beller et al., 2006a, b; Hernandez et al., 1996). Also very highly expressed was a putative homolog of the Fe(II)-oxidizing protein, Cyc2, a high-potential, outer-membrane c-type cytochrome (scaffold-1168_1169_219). Alignment of the translated sequence of this bin 22.5 gene against Cyc2PV-1, a putative Fe(II) oxidase from the neutrophilic Fe(II) oxidizer Mariprofundus ferrooxydans PV-1 (Barco et al., 2015) and Cyc2, the well-characterized Fe(II) oxidase from Acidithiobacillus ferrooxidans (Yarzábal et al., 2002), revealed sequence similarity in the N-terminal portion of the protein, including a CXXCH heme-binding motif and a predicted signal peptide. A homologous cytochrome in bin 22.6 (scaffold-40806_40807_9; 56.9% sequence identity to the version in bin 22.5) was also very highly expressed (as high as the 100th percentile in one sample).
Gallionellaceae bins 22.6 and 22.9 had different temporal expression trends than bin 22.5 for many of the functional genes involved in the above-mentioned pathways. A striking trend for transcripts in bin 22.6 in particular is an extremely strong correspondence between expression profiles and nitrate input and consumption (Figure 4b), indicating a strong response of this strain to nitrate. As shown in Figure 4b, expression levels for key genes related to nitrate reduction (narG, narH), nitrite reduction (nirS), S-compound oxidation (dsrE), Fe(II) oxidation (mtoA, mtoB) and CO2 fixation (cbbM) all maximized at day 35, suggesting coupling between these metabolic processes (such as nitrate-dependent Fe(II) and S oxidation). A related but different expression trend was observed for bin 22.9 (Figure 4c). For this organism, expression levels peaked at day 47, not day 35, but the data still suggested a response to nitrate. Genes plotted in Figure 4c relate to nitrate reduction (narG, narH), nitrous oxide reduction (nosZ), reduction of low levels of dissolved O2 (ccoN), S-compound oxidation (aprB) and CO2 fixation (cbbM) (Supplementary Table S7). Notably, in bins 22.6 and 22.9, a different nitrate reductase was encoded and expressed than in bin 22.5. Instead of the periplasmic napAB, which was included in bin 22.5, the strains represented by bins 22.6 and 22.9 expressed the membrane-bound dissimilatory nitrate reductase narGHI. The nar gene clusters in bins 22.1, 22.6 and 22.9 and shared a high degree of synteny (e.g., the canonical narKK2GHJI order (Philippot, 2002); Figure 5) and sequence similarity (e.g., translated narG copies in the three bins shared 86–100% sequence identity). An interesting feature shared by the nar clusters in Gallionellaceae bins was a carbonic anhydrase gene immediately downstream of narI, a feature that also occurs in Methylophaga nitratireducenticrescens.
Beyond their diagnostic value for indicating key, active biogeochemical processes, several of the genes described above were among the most highly expressed in the postrelease samples. Bin 22.5 included carbon fixation and S-compound oxidation genes, cbbM and dsrC, in the 99.95th and 99.91st percentile of the metatranscriptome, respectively, and a putative Fe(II) oxidase (Cyc2 homolog) in the 99.99th–100th percentile in all postinjection samples.
The emergence of activity of S. denitrificans strains following nitrate injection
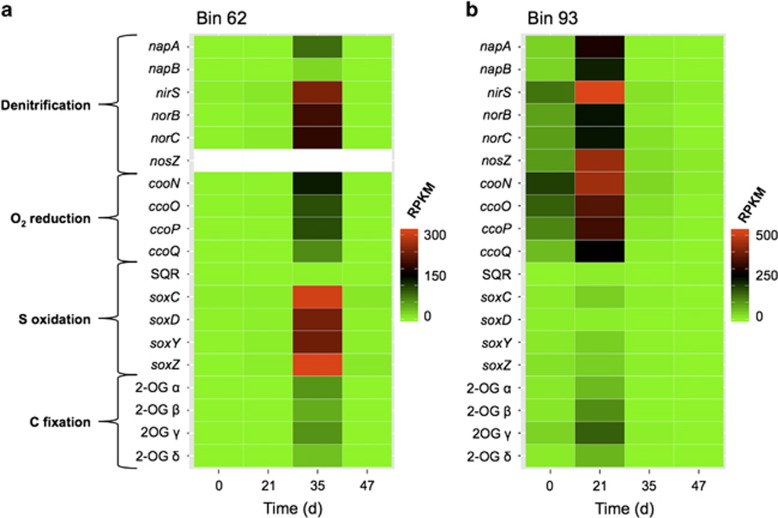
Two strains of S. denitrificans, represented by bins 62 and 93 (Supplementary Table S8), were prominent in the metranscriptomes of samples collected after the nitrate release commenced. These two bins share 880 putative orthologs (>60% sequence identity) out of 2142 open reading frames in bin 62 and 1491 open reading frames in bin 93. Reconstructed genomes with similar apparent metabolic potential and phylogeny were reported in another study at the Rifle site (Handley et al., 2012). Although the two strains represented by bins 62 and 93 had different expression profiles (Figure 6), both strains expressed genes associated with denitrification (e.g., napA, napB, nirS, norB, norC), S-compound oxidation (e.g., soxC, soxD, soxY, soxZ, sulfide:quinone oxidoreductase), carbon fixation via the reverse tricarboxylic acid cycle (e.g., the diagnostic gene ferredoxin-dependent 2-oxoglutarate synthase; Berg, 2011) and oxygen reduction via cbb3-type cytochrome c oxidase (ccoNOPQ). The most evident difference between the expression patterns for bins 62 and 93 is that they had maxima on days 35 and 21, respectively (Figure 6). Another clear difference is in the relative expression of sox genes; these S-compound oxidation genes were much more highly expressed in bin 62 than in bin 93. In light of the less powerful evidence of S-compound oxidation for bin 93, we looked for evidence of use of alternative electron donors documented for S. denitrificans (Sievert et al., 2008), such as H2 or formate. However, we found no evidence of the expression of either a hydrogenase or formate dehydrogenase in either bin. There were some common features in the expression profiles for bins 62 and 93: both bins displayed relatively high expression of certain denitrification genes (particularly nirS) and genes for cbb3-type cytochrome c oxidase.
Figure 6.
Temporal expression profiles for selected genes associated with C, S and N metabolism in the two most active S. denitrificans strains. (a) Heat map of relative expression of selected genes in bin 62. (b) Heat map of relative expression of selected genes in bin 93. Note that the scales differ in panels a and b. Genes: napA and napB, α and β subunits of periplasmic nitrate reductase; nirS, cytochrome cd1 nitrite reductase; nosZ, nitrous oxide reductase; norBC, subunits I and II of nitric oxide reductase; ccoNOPQ, subunits I, II, III and IV of cbb3-type cytochrome c oxidase; sqr, sulfide:quinone oxidoreductase; soxYZBCD, sulfur-oxidizing Sox enzyme system; 2-OG αβγδ, 2-oxoglutarate ferredoxin oxidoreductase α, β, γ, δ subunits.
Discussion
In this study, we characterized the response of an aquifer microbial community to an increase in the flux of nitrate, a naturally occurring electron acceptor at the Rifle field site. This nitrate perturbation study was motivated, in part, by observations of dynamic fluctuations in nitrate concentrations in the Rifle aquifer. We took advantage of synoptically collected metagenomic, metatranscriptomic and geochemical data during the 2-month study to characterize community response, with a focus on transcriptional evidence of metabolic activity for bacteria represented by strain-resolved metagenomic reconstructions.
Although influx of a thermodynamically favorable electron acceptor like nitrate could be expected to enhance microbial oxidation of organic electron donors as well as inorganic ones, the metatranscriptomic data unequivocally indicated a strong chemolithoautotrophic response, with coupled reduction of nitrate and other nitrogen oxides, oxidation of Fe(II) and reduced S compounds, and CO2 fixation. Thus, pervasive and diverse chemolithoautotrophic activities mediated C, S, N and Fe cycling in the aquifer both before and during the nitrate release. Notably, the very high level of chemolithoautotrophic activity indicated by the metatranscriptomic data would generally not have been inferred based on metagenomic data alone (Figure 2).
The three most active groups of chemolithoautotrophic bacteria in this study, Gallionellaceae, S. denitrificans and anammox bacteria, have overlapping metabolisms that allowed them to occupy different yet related metabolic niches throughout the study. Although members of all three groups respired nitrate, they had markedly different temporal expression profiles and activity maxima, even among closely related species/strains within the three groups (Figures 2, 3, 4 and 6). Temporal and spatial variations in geochemistry, such as nitrate influx and composition of minerals containing Fe(II) and reduced S, and in community structure, likely established the foundations for temporal and spatial niche variation. Anammox bacterial activity was relatively high only on day 0 (Figure 3), whereas Gallionellaceae metabolic activity was highest on days 21 and 47 (bin 22.5) or day 35 (bin 22.6) or day 47 (bin 22.9) (Figure 4), and S. denitrificans activity was greatest on day 21 (bin 93) or day 35 (bin 62) (Figure 6). An example of apparent niche differentiation between and within the major chemolithoautotrophic groups can be illustrated with S-compound oxidation. Different expression patterns were observed for S-compound oxidation pathways, such as sox and reverse dsr, among two S. denitrificans strains (Figure 6) and several of the putative Sideroxydans spp. within the Gallionellaceae group (Figure 4). The observation of S-compound oxidation in Gallionellaceae strains was unusual, as these bacteria are typically characterized by Fe(II) oxidation. Although growth of Sideroxydans ES-1 with a reduced S compound (thiosulfate) has been observed in the laboratory (Emerson et al., 2013), this is one of the few reports of S oxidation by Sideroxydans spp. in the field (Purcell et al., 2014). It appears that both nitrate-dependent S oxidation and nitrate-dependent Fe(II) oxidation were metabolic strategies used in situ by members of the Gallionellaceae group, including both putative Gallionella and Sideroxydans species. Neither of these nitrate-dependent activities has been documented previously for isolated members of the Gallionellaceae, although nitrate-dependent Fe(II) oxidation has been inferred for a Sideroxydans member of an enrichment culture (Blöthe and Roden, 2009).
The observation that the genomes of various Gallionellaceae strains encoded two different nitrate reductases, the periplasmic NapAB and the membrane-bound NarGHI, is particularly noteworthy because the reported genomes of Gallionellaceae isolates do not contain any genes encoding dissimilatory nitrate reductases. These annotations of nitrate reductases entail a high level of confidence; for example, alignment of the translated napA from bin 22.5 vs structurally characterized versions of NapA reveals conservation of all six active-site residues (Moura et al., 2004) and a high degree of sequence identity (Supplementary Figure S3). The scaffold containing napA had ribosomal proteins with high sequence identity (89–95%) to the corresponding proteins in Gallionella capsiferriformans. A putative Gallionella genome from a different Rifle aquifer study also included napAB genes (JF Banfield and A Shelton, personal communication), although the gene organization was different than that observed in bin 22.5 (Figure 5). In metagenomic data from the CO2-rich Crystal Geyser aquifer (Green River, UT, USA) (Emerson et al., 2015), nitrate reductase (narG, narH) and sox genes were putatively placed in a Gallionellales bin. The genome of the isolate Sideroxydans ES-1 encodes a nitrite reductase, but nitrite-dependent Fe(II) or S oxidation have not been reported in this strain. Overall, the presence and expression of nitrate reductases in Gallionellaceae strains in this study compels us to expand our view of the metabolic diversity of these chemolithoautotrophic bacteria.
Anammox metabolic activity appears to be a favorable niche at the study site under background conditions, as anammox gene expression was greatest before the nitrate release and accounted for a substantial portion of the metatranscriptome on day 0. Although a number of aquifer studies have focused on 16S rRNA and functional gene surveys of anammox bacteria, and of their activity using isotopic methods (López-Archilla et al., 2007; Clark et al., 2008; Smits et al., 2009; Hirsch et al., 2011), this is the first study to document the activity of an anammox population in an aquifer using metatranscriptomic data. The presence of a population with high relative activity under background conditions suggests that the relevance of anammox in such systems may be underrepresented, particularly in studies that focus solely on metagenomic analysis or marker genes. The precipitous decrease in anammox activity after the initiation of the nitrate release is confounding, particularly because nitrite is the electron acceptor for this process. We explored two possibilities that may have contributed to the marked decrease in anammox gene expression after the addition of nitrate: the production of sulfide by sulfate-reducing bacteria, and the introduction of small amounts of oxygen during the nitrate injection. The active sulfate-reducing bacterial population only constituted ca. 1% of the metatranscriptome at day 0, and decreased to ca. 0.3% by day 47, and diagnostic sulfate reduction pathway genes were not highly expressed in any sample (<35 RPKM). Thus, while low levels of sulfide irreversibly inhibit anammox bacteria (Jin et al., 2013), the amount of sulfide produced under field conditions was probably too low to have markedly inhibited the anammox bacterial population. A more likely cause for the decreased anammox activity is the introduction of low levels of oxygen during the nitrate injection. Anammox bacteria are obligate anaerobes that are reversibly inhibited by molecular oxygen; a wide range of inhibitory O2 concentrations has been reported (Strous et al., 1997; Carvajal-Arroyo et al., 2013). Although we did not collect DO data for this study, we did observe transcriptional evidence for low concentrations of DO. Relatively high expression of genes encoding cbb3-type cytochrome c oxidase, a high-oxygen-affinity cytochrome oxidase consistent with oxygen reduction in a suboxic environment, was observed both for Gallionellaceae strains (Figure 4) and S. denitrificans strains (Figure 6). This constitutes evidence of DO within the aquifer test zone at concentrations sufficient to elicit a biological response from two different groups of bacteria. Note that DO concentrations may have been sufficient to inhibit anammox bacteria but may not have supported respiration by other prominent chemolithoautotrophs. S. denitrificans DSM 1251 has been characterized as an obligate denitrifier that is sensitive to oxygen, and its function for cbb3-type cytochrome c oxidase could be to prevent oxygen poisoning, not to respire O2 (Sievert et al., 2008). Further, background DO concentrations are typically very low (<20 μm) at the study site (D Pan, KH Williams, MJ Robbins, and KA Webber, personal communication).
The preceding discussion raises an important point about the role of transcriptional data in this study: it informed interpretation of biogeochemical processes beyond what could have been inferred based on metagenomic and geochemical data. Transcriptional data clearly highlighted highly active organisms (Figure 2) and metabolic pathways (Figures 3, 4 and 6) that would not have been highlighted by metagenomic data alone. Further, transcriptional data revealed considerable information on S-compound oxidation in situ, even though no data were collected on any solid-phase S-containing compounds in the aquifer, which were likely the electron donors for S oxidation and potentially also included some of the oxidized products. Such solid-phase compounds would have posed significant analytical challenges if we had attempted to measure them routinely. The only S-containing compound that was routinely measured in this study was sulfate, and it indeed increased markedly in concentration after the nitrate release started (Supplementary Figure S2), consistent with S-compound oxidation, but the transcriptional data is a powerful complementary indicator of this biogeochemical activity.
Overall, these results demonstrating pervasive and diverse chemolithoautotrophy at the Rifle site have implications beyond this site and these study conditions. Although groundwater ecosystems were conventionally thought to be fueled by surface-derived allochthonous organic matter and dominated by heterotrophic microbes living under often-oligotrophic conditions, recent studies at a range of field sites have indicated the importance of chemolithoautotrophy in subsurface carbon cycling (Alfreider et al., 2003, 2009; Alfreider and Vogt, 2012; Herrmann et al., 2015). For example, in a study of two adjacent limestone aquifers, quantitative PCR surveys of form I and II RubisCO genes revealed that up to 17% of the microbial population had the capability to fix CO2 by the Calvin cycle, and that S-oxidizing Sulfuricella strains and ammonia-oxidizing Nitrosomonas strains accounted for substantial portions of the cbbM and cbbL transcript pools (Herrmann et al., 2015). Further, PCR surveys of RubisCO genes and transcripts in groundwater samples from a range of pristine and polluted, shallow and deep aquifers indicated that autotrophs were widespread inhabitants of these groundwater systems (Alfreider et al., 2003, 2009; Alfreider and Vogt, 2012). Finally, in a study of the Rifle aquifer under background (unperturbed) conditions, Handley et al. (2014) documented that an S-oxidizing chemolithoautotrophic bacterium, Candidatus Sulfuricurvum sp. RIFRC-1, composed ~47% of the sediment bacterial community based on metagenomic data. The present work adds to the collective evidence of the importance of chemolithoautotrophy in aquifers while providing a high degree of resolution of the metabolic pathways being expressed and elucidating metabolic lifestyles that expand our appreciation of the metabolic diversity of known chemolithoautotrophic taxa. Ongoing and future studies include attempts to isolate novel Gallionellaceae strains capable of nitrate-dependent Fe(II) and S oxidation to better understand their underlying biochemistry and efforts to quantify the role of chemolithoautotrophy in the subsurface carbon budget via quantitative environmental proteomics.
Acknowledgments
We thank Jill Banfield (University of California, Berkeley, CA, USA) for valuable discussions and David M Silberman for technical assistance. This paper is dedicated to the memory of Richard (Dick) Dayvault, whose commitment to supporting scientific research at DOE's Rifle field site was instrumental to the success of this experiment and all those preceding it. This work was supported as part of the Subsurface Biogeochemical Research Scientific Focus Area funded by the US Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-AC02-05CH11231. This work used the Vincent J Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alfreider A, Vogt C. (2012). Genetic evidence for bacterial chemolithoautotrophy based on the reductive tricarboxylic acid cycle in groundwater systems. Microbes Environ 27: 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfreider A, Vogt C, Geiger-Kaiser M, Psenner R. (2009). Distribution and diversity of autotrophic bacteria in groundwater systems based on the analysis of RubisCO genotypes. System Appl Microbiol 32: 140–150. [DOI] [PubMed] [Google Scholar]
- Alfreider A, Vogt C, Hoffmann D, Babel W. (2003). Diversity of ribulose-1, 5-bisphosphate carboxylase/oxygenase large-subunit genes from groundwater and aquifer microorganisms. Microb Ecol 45: 317–328. [DOI] [PubMed] [Google Scholar]
- Ali M, Oshiki M, Awata T, Isobe K, Kimura Z, Yoshikawa H et al. (2015). Physiological characterization of anaerobic ammonium oxidizing bacterium ‘Candidatus Jettenia caeni'. Environ Microbiol 17: 2172–2189. [DOI] [PubMed] [Google Scholar]
- Allen EE, Tyson GW, Whitaker RJ, Detter JC, Richardson PM, Banfield JF. (2007). Genome dynamics in a natural archaeal population. Proc Natl Acad Sci 104: 1883–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SD, Martiny JB. (2008). Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA 105: 11512–11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MA, Valett HM, Dahm CN. (2000). Organic carbon supply and metabolism in a shallow groundwater ecosystem. Ecology 81: 3133–3148. [Google Scholar]
- Barco RA, Emerson D, Sylvan JB, Orcutt BN, Meyers MEJ, Ramírez GA et al. (2015). New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol 81: 5927–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller HR, Chain PS, Letain TE, Chakicherla A, Larimer FW, Richardson PM et al. (2006. a). The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J Bacteriol 188: 1473–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller HR, Letain TE, Chakicherla A, Kane SR, Legler TC, Coleman MA. (2006. b). Whole-genome transcriptional analysis of chemolithoautotrophic thiosulfate oxidation by Thiobacillus denitrificans under aerobic versus denitrifying conditions. J Bacteriol 188: 7005–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller HR, Madrid V, Hudson GB, McNab WW, Carlsen T. (2004). Biogeochemistry and natural attenuation of nitrate in groundwater at an explosives test facility. Appl Geochem 19: 1483–1494. [Google Scholar]
- Bengtsson G, Bergwall C. (1995). Heterotrophic denitrification potential as an adaptive response in groundwater bacteria. FEMS Microbiol Ecol 16: 307–317. [Google Scholar]
- Berg IA. (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blöthe M, Roden EE. (2009). Composition and activity of an autotrophic Fe (II)-oxidizing, nitrate-reducing enrichment culture. Appl Environ Microbiol 75: 6937–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Arroyo JM, Sun W, Sierra-Alvarez R, Field JA. (2013). Inhibition of anaerobic ammonium oxidizing (anammox) enrichment cultures by substrates, metabolites and common wastewater constituents. Chemosphere 91: 22–27. [DOI] [PubMed] [Google Scholar]
- Chandler DP, Jarrell AE, Roden ER, Golova J, Chernov B, Schipma MJ et al. (2006). Suspension array analysis of 16S rRNA from Fe-and SO42--reducing bacteria in uranium-contaminated sediments undergoing bioremediation. Appl Environ Microbiol 72: 4672–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I, Timlin R, Bourbonnais A, Jones K, Lafleur D, Wickens K. (2008). Origin and fate of industrial ammonium in anoxic ground water—15N evidence for anaerobic oxidation (anammox). Groundwater Monit Remed 28: 73–82. [Google Scholar]
- Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C et al. (2013). Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JB, Thomas BC, Alvarez W, Banfield JF. (2015). Metagenomic analysis of a high carbon dioxide subsurface microbial community populated by chemolithoautotrophs and bacteria and archaea from candidate phyla. Environ Microbiol e-pub ahead of print 28 February 2015 doi:10.1111/1462-2920.12817. [DOI] [PubMed]
- Faust K, Lahti L, Gonze D, de Vos WM, Raes J. (2015). Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr Opin Microbiol 25: 56–66. [DOI] [PubMed] [Google Scholar]
- Foulquier A, Simon L, Gilbert F, Fourel F, Malard F, Mermillod-Blondin F. (2010). Relative influences of DOC flux and subterranean fauna on microbial abundance and activity in aquifer sediments: new insights from 13C-tracer experiments. Freshwater Biol 55: 1560–1576. [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW et al. (2008). Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105: 3805–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh W, Dam B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol Rev 33: 999–1043. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P et al. (2008). Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3: e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TL, Koonce E, Howells A, Havig JR, Jewell T, José R et al. (2014). Competition for ammonia influences the structure of chemotrophic communities in geothermal springs. Appl Environ Microbiol 80: 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley KM, Bartels D, O'Loughlin EJ, Williams KH, Trimble WL, Skinner K et al. (2014). The complete genome sequence for putative H2- and S-oxidizer Candidatus Sulfuricurvum sp., assembled de novo from an aquifer-derived metagenome. Environ Microbiol 16: 3443–3462. [DOI] [PubMed] [Google Scholar]
- Handley KM, Wrighton KC, Piceno YM, Andersen GL, DeSantis TZ, Williams KH et al. (2012). High-density PhyloChip profiling of stimulated aquifer microbial communities reveals a complex response to acetate amendment. FEMS Microbiol Ecol 81: 188–204. [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Baker SH, Lorbach SC, Shively JM, Tabita FR. (1996). Deduced amino acid sequence, functional expression, and unique enzymatic properties of the form I and form II ribulose bisphosphate carboxylase/oxygenase from the chemoautotrophic bacterium Thiobacillus denitrificans. J Bacteriol 178: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M, Rusznyák A, Akob DM, Schulze I, Opitz S, Totsche KU et al. (2015). Large fractions of CO2-fixing microorganisms in pristine limestone aquifers appear to be involved in the oxidation of reduced sulfur and nitrogen compounds. Appl Environ Microbiol 81: 2384–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch MD, Long ZT, Song B. (2011). Anammox bacterial diversity in various aquatic ecosystems based on the detection of hydrazine oxidase genes (hzoA/hzoB. Microb Ecol 61: 264–276. [DOI] [PubMed] [Google Scholar]
- Ilbert M, Bonnefoy V. (2013). Insight into the evolution of the iron oxidation pathways. Biochim Biophys Acta 1827: 161–175. [DOI] [PubMed] [Google Scholar]
- Irisa T, Hira D, Furukawa K, Fujii T. (2014). Reduction of nitric oxide catalyzed by hydroxylamine oxidoreductase from an anammox bacterium. J Biosci Bioeng 118: 616–621. [DOI] [PubMed] [Google Scholar]
- Jetten MS, Niftrik LV, Strous M, Kartal B, Keltjens JT, Op den Camp HJ. (2009). Biochemistry and molecular biology of anammox bacteria. Crit Rev Biochem Mol Biol 44: 65–84. [DOI] [PubMed] [Google Scholar]
- Jin R-C, Yang G-F, Zhang Q-Q, Ma C, Yu J-J, Xing B-S. (2013). The effect of sulfide inhibition on the ANAMMOX process. Water Res 47: 1459–1469. [DOI] [PubMed] [Google Scholar]
- Kartal B, de Almeida NM, Maalcke WJ, den Camp HJO, Jetten MS, Keltjens JT. (2013). How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev 37: 428–461. [DOI] [PubMed] [Google Scholar]
- Kopf A, Kostadinov I, Wichels A, Quast C, Glöckner FO. (2015). Metatranscriptome of marine bacterioplankton during winter time in the North Sea assessed by total RNA sequencing. Marine Genom 19: 45–46. [DOI] [PubMed] [Google Scholar]
- Korom SF. (1992). Natural denitrification in the saturated zone: a review. Water Resour Res 28: 1657–1668. [Google Scholar]
- Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ. (2012). The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J 6: 2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long PE, Williams KH, Davis JA, Fox PM, Wilkins MJ, Yabusaki SB et al. (2015). Bicarbonate impact on U (VI) bioreduction in a shallow alluvial aquifer. Geochim Cosmochim Acta 150: 106–124. [Google Scholar]
- Luef B, Frischkorn KR, Wrighton KC, Holman H-YN, Birarda G, Thomas BC et al. (2015). Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6: article number 6372 doi:10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D. (2015). ConStrains identifies microbial strains in metagenomic datasets. Nature Biotechnol 33: 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Archilla AI, Moreira D, Velasco S, López-García P. (2007). Archaeal and bacterial community composition of a pristine coastal aquifer in Donana National Park, Spain. Aquat Microb Ecol 47: 123–139. [Google Scholar]
- Maalcke WJ, Dietl A, Marritt SJ, Butt JN, Jetten MS, Keltjens JT et al. (2014). Structural basis of biological NO generation by octaheme oxidoreductases. J Biol Chem 289: 1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura JJ, Brondino CD, Trincão J, Romão MJ. (2004). Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases. J Biol Inorg Chem 9: 791–799. [DOI] [PubMed] [Google Scholar]
- O'Neil RA, Holmes DE, Coppi MV, Adams LA, Larrahondo MJ, Ward JE et al. (2008). Gene transcript analysis of assimilatory iron limitation in Geobacteraceae during groundwater bioremediation. Environ Microbiol 10: 1218–1230. [DOI] [PubMed] [Google Scholar]
- Oshiki M, Shinyako-Hata K, Satoh H, Okabe S. (2015). Draft genome sequence of an anaerobic ammonium-oxidizing bacterium, 'Candidatus Brocadia sinica'. Genome Announc 3: e00267–00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels H, Kloppmann W, Foucher J-C, Martelat A, Fritsche V. (1998). Field tracer test for denitrification in a pyrite-bearing schist aquifer. Appl Geochem 13: 767–778. [Google Scholar]
- Philippot L. (2002). Denitrifying genes in bacterial and archaeal genomes. Biochim Biophys Acta 1577: 355–376. [DOI] [PubMed] [Google Scholar]
- Pitcher RS, Watmough NJ. (2004). The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta 1655: 388–399. [DOI] [PubMed] [Google Scholar]
- Poretsky RS, Bano N, Buchan A, LeCleir G, Kleikemper J, Pickering M et al. (2005). Analysis of microbial gene transcripts in environmental samples. Appl Environ Microbiol 71: 4121–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. (2009). Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol 11: 1358–1375. [DOI] [PubMed] [Google Scholar]
- Purcell AM, Mikucki JA, Achberger AM, Alekhina IA, Barbante C, Christner BC et al. (2014). Microbial sulfur transformations in sediments from Subglacial Lake Whillans. Front Microbiol 5: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A, Bodi X, Dufresne A, Naquin D, Francez A-J, Dheilly A et al. (2014). Unraveling the stratification of an iron-oxidizing microbial mat by metatranscriptomics. PLoS One 9: e102561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. (2013). Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res 23: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura M, Nishiyama T, Shinya K, Kawahara Y, Furukawa K, Fujii T. (2008). Another multiheme protein, hydroxylamine oxidoreductase, abundantly produced in an anammox bacterium besides the hydrazine-oxidizing enzyme. J Biosci Bioeng 105: 243–248. [DOI] [PubMed] [Google Scholar]
- Shrestha PM, Kube M, Reinhardt R, Liesack W. (2009). Transcriptional activity of paddy soil bacterial communities. Environ Microbiol 11: 960–970. [DOI] [PubMed] [Google Scholar]
- Sievert SM, Scott KM, Klotz MG, Chain PS, Hauser LJ, Hemp J et al. (2008). Genome of the epilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl Environ Microbiol 74: 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits TH, Hüttmann A, Lerner DN, Holliger C. (2009). Detection and quantification of bacteria involved in aerobic and anaerobic ammonium oxidation in an ammonium-contaminated aquifer. Bioremed J 13: 41–51. [Google Scholar]
- Starr RC, Gillham RW. (1993). Denitrification and organic carbon availability in two aquifers. Groundwater 31: 934–947. [Google Scholar]
- Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440: 790–794. [DOI] [PubMed] [Google Scholar]
- Strous M, Van Gerven E, Kuenen JG, Jetten M. (1997). Effects of aerobic and microaerobic conditions on anaerobic ammonium-oxidizing (anammox) sludge. Appl Environ Microbiol 63: 2446–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC. (2008). Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3: e2527–e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venceslau S, Stockdreher Y, Dahl C, Pereira I. (2014). The 'bacterial heterodisulfide' DsrC is a key protein in dissimilatory sulfur metabolism. Biochim Biophys Acta 1837: 1148–1164. [DOI] [PubMed] [Google Scholar]
- Williams KH, Long PE, Davis JA, Wilkins MJ, N'Guessan AL, Steefel CI et al. (2011). Acetate availability and its influence on sustainable bioremediation of uranium-contaminated groundwater. Geomicrobiol J 28: 519–539. [Google Scholar]
- Williams KH, Wilkins MJ, N'Guessan A, Arey B, Dodova E, Dohnalkova A et al. (2013). Field evidence of selenium bioreduction in a uranium-contaminated aquifer. Environ Microbiol Rep 5: 444–452. [DOI] [PubMed] [Google Scholar]
- Yarzábal A, Brasseur G, Ratouchniak J, Lund K, Lemesle-Meunier D, DeMoss JA et al. (2002). The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J Bacteriol 184: 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachara JM, Long PE, Bargar J, Davis JA, Fox P, Fredrickson JK et al. (2013). Persistence of uranium groundwater plumes: contrasting mechanisms at two DOE sites in the groundwater–river interaction zone. J Contam Hydrol 147: 45–72. [DOI] [PubMed] [Google Scholar]
- Zhou Y-j, Wang S-m, Zhang Z-j, Chen S-h. (2012). Metabolic pathways of anammox bacteria and related key enzymes: a review. Chin J Ecol 3: 036. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.