Abstract
Bacteria of the genus Burkholderia establish an obligate symbiosis with plant species of the Rubiaceae and Primulaceae families. The bacteria, housed within the leaves, are transmitted hereditarily and have not yet been cultured. We have sequenced and compared the genomes of eight bacterial leaf nodule symbionts of the Rubiaceae plant family. All of the genomes exhibit features consistent with genome erosion. Genes potentially involved in the biosynthesis of kirkamide, an insecticidal C7N aminocyclitol, are conserved in most Rubiaceae symbionts. However, some have partially lost the kirkamide pathway due to genome erosion and are unable to synthesize the compound. Kirkamide synthesis is therefore not responsible for the obligate nature of the symbiosis. More importantly, we find evidence of intra-clade horizontal gene transfer (HGT) events affecting genes of the secondary metabolism. This indicates that substantial gene flow can occur at the early stages following host restriction in leaf nodule symbioses. We propose that host-switching events and plasmid conjugative transfers could have promoted these HGTs. This genomic analysis of leaf nodule symbionts gives, for the first time, new insights in the genome evolution of obligate symbionts in their early stages of the association with plants.
Introduction
Many microbes can establish a wide range of beneficial interactions with plants, often contributing to plant nutrition, for example, mineral acquisition or nitrogen fixation, or plant defense through the synthesis of secondary metabolites (Sachs and Simms, 2006). Most of these mutualistic associations are facultative and have been widely studied (Philippot et al., 2013).
In contrast to animal symbioses where the life cycle of the symbiont is often tied to the host, symbioses with vertical transmission are extremely rare in higher plants (Leigh, 2010). There is only one case known where the bacterial symbiont is vertically transmitted and its presence is critical for the development of the host. This unique symbiosis is established between bacteria from the genus Burkholderia and some species from the Rubiaceae and Primulaceae family.
Leaf nodule symbiosis is characterized by the presence of specialized structures within the leaves called leaf galls or nodules (Miller, 1990). This association has been described in three genera of Rubiaceae: Psychotria, Pavetta and Sericanthe (Lemaire et al., 2012). Nodulated species are endemic to tropical and sub-tropical Africa, with most nodulated Psychotria found in savannah habitats (Lachenaud, 2013). Morphological and ontological studies early in the 20th century revealed the presence of extracellular bacteria within the leaf nodules of Psychotria (Zimmermann, 1902; Von Faber, 1912). These bacteria were further detected in all life stages of the plant suggesting a closed symbiotic cycle, in which the bacterial symbionts could be transmitted hereditarily via colonization of the seeds. Bacteria are essential to the development of the plants as seeds treated with heat germinate into aposymbiotic seedlings that fail to reach maturity and die after several months (Miller, 1990; Van Oevelen et al., 2001). Conversely, bacteria have lost their autonomy and have not yet been cultured outside of the host. Only culture-independent molecular techniques assigned the symbiotic bacteria to the genus Burkholderia (Van Oevelen et al., 2002).
The genome of Candidatus Burkholderia kirkii, the leaf nodule symbiont of Psychotria kirkii, has provided new insights into the genome evolution and molecular nature of this symbiosis (Carlier and Eberl, 2012). The genome of Ca. B. kirkii is half the size of other closely related free-living plant-associated Burkholderia and contains a massive amount of pseudogenes and transposable elements (Carlier and Eberl, 2012). These properties are common in recently evolved and vertically transmitted symbionts such as the obligate cyanobiont of the water-fern Azolla filiculoides (Ran et al., 2010), the facultative tsetse fly symbiont Sodalis glossinidius (Belda et al., 2010) or the aphid Cinara tujafilina co-obligate symbiont Serratia symbiotica (Manzano-Marín and Latorre, 2014), suggesting that Ca. B. kirkii recently switched to a host-restricted lifestyle. The process of genome reduction is well documented in intracellular, obligate symbionts of animals where host restriction and cellular isolation of bacteria are thought to cause the accumulation of deleterious mutations, promote gene loss and prevent horizontal gene transfer (HGT) (McCutcheon and Moran, 2012).
The molecular nature of leaf nodule symbiosis is still unknown. Previous speculations, including hormone production and nitrogen fixation (Centifanto and Silver, 1964; Edwards and Lamotte, 1975), have been ruled out with the genomic and proteomic information obtained from Ca. B. kirkii (Carlier and Eberl, 2012; Carlier et al., 2013). Genomic and chemical evidence instead suggests a plant protective role of the leaf nodule bacteria. The functional analysis of Ca. B. kirkii's genome revealed the presence of a biosynthetic gene cluster for 2-epi-5-epi-valiolone-derived aminocyclitols from the C7N aminocyclitol family. These genes, located on the 140 kb plasmid pKIR01, are most likely involved in the synthesis of kirkamide, a C7N aminocyclitol found in the leaves of nodulated P. kirkii but absent in stunted, aposymbiotic specimens (Sieber et al., 2015). Interestingly, no homologs of the putative C7N aminocyclitol biosynthesis cluster have been found in other Burkholderia species, indicating that these genes were probably acquired relatively recently by HGT (Carlier and Eberl, 2012). Pure kirkamide is cytotoxic and insecticidal and may therefore increase the fitness of the plant via protection against herbivory (Sieber et al., 2015).
The aim of this study was to (i) characterize the genome reduction process in leaf nodule obligate symbionts and compare our findings with the classical models inferred from the study of vertically transmitted, intracellular animal symbionts (ii) investigate the importance of secondary metabolism for the symbiosis and (iii) identify genetic determinants potentially involved in the obligate nature of the symbiosis. Our results show that production of C7N aminocyclitol secondary metabolites is conserved in most leaf nodule symbionts, highlighting the importance of secondary metabolism for the symbiosis. We further demonstrate that cross-infection events may explain the lack of strict co-evolution. We also provide evidence of recent events of HGT of C7N aminocyclitol biosynthetic genes. Together, our data suggest that in leaf nodule symbionts, host restriction and early stages of genome reduction do not preclude gene flow.
Materials and methods
Sample collection
Seven plant species of the Rubiaceae family were selected for this study and material was obtained from the National Botanic Garden of Belgium (the collection acquisition numbers are given in parenthesis when available), except for Psychotria punctata for which material was obtained from an authenticated specimen kept in the greenhouse of the Department of Plant and Microbial Biology of the University of Zurich. The species used in this study are: Psychotria humilis collection acquisition number: BR2009135940, Psychotria pumila (BR2004143571), Psychotria verschuerenii (BR19750204), Psychotria umbellata (BR2007130262), Psychotria brachyanthoides (BR2009844596), Psychotria punctata and Pavetta schumanniana (BR2000194257). Silica-dried leaves from single specimens were obtained from the Botanic Garden Meise for all species except P. punctata, for which fresh material was obtained (see below). Around 1000 nodules per specimen containing symbiotic bacteria were dissected using a Harris Uni-core 2.0-mm core-punching tool (Whatman Inc., Kent, UK) and rehydrated in 5 ml of TNE buffer (100 mM ethylenediaminetetraacetic acid, 200 mm NaCl, 10 mm Tris.Cl pH8) on ice for 1 h. The rehydrated nodules were ground with a mortar and pestle. Plant debris was removed by filtering through a 10 μm Millipore Nylon filter. The flow-through was collected and bacteria were pelleted by centrifugation at 6000 rpm. High molecular weight DNA was extracted according to Wilson (2001). The quality of the DNA was assessed by agarose gel electrophoresis and PCR amplification of the bacterial 16S ribosomal RNA gene. Levels of plant DNA contamination in genomic DNA preparations were assessed by semi-quantitative PCR using primers designed on the P. kirkii chloroplast rbcL gene (5′-CCGGTACTGTAGTCGGGAAA and 5′-CCAAAGATCTCCGTCAGAGC) and standards consisting of bacteria-free DNA prepared from non-nodulated parts of the plants under investigation. The proportion of bacterial genomic DNA in our preparations from nodules was consistently higher than 80% (w/w).
Leaf nodules containing Ca. B. punctata were obtained from a P. punctata specimen kept at the botanical garden of the University of Zurich. Bacteria were separated from plant material by density-gradient centrifugation according to a previously published protocol (Carlier and Eberl, 2012). Genomic DNA was prepared as described above.
Genome sequencing, bacterial reads selection and assembly
Genomic DNA was sequenced at the Functional Genomics Center of Zurich (FGCZ). Paired-end 250 bp libraries were prepared using the Nextera XT kit (Illumina Inc., San Diego, CA, USA) and sequenced using Illumina Miseq sequencing platform. The reads were prepared for assembly using the adapter trimming function of the Scythe software (https://github.com/vsbuffalo/scythe) and sequencing reads with a Phred score below 30 were removed using the FastQC package (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). The average coverage for the seven sequenced samples was 50 × and the reads length were 230 bp on average. In order to assemble sequencing reads of bacterial origin from samples containing contaminating plant genomes, a binning approach was used (Albertsen et al., 2013). Preliminary assembly of the reads was done using CLC Genomics Workbench v7.0 (http://www.clcbio.com) (CLC Bio-Qiagen, Aarhus, Denmark) with low stringency parameters. Guanine and Cytosine (GC) content and average read coverage of the resulting contigs were plotted using the ggplot package implemented in R (Wickham, 2009) (Supplementary Figure S1). Because the genomes of the plants are several orders of magnitude larger than that of the symbionts and have a low GC%, clusters of contigs with high coverage (ca. above 70 ×) and high GC% were selected as bacterial contigs and the rest were considered plant contaminants. In order to avoid false positives all the contigs were searched against National Center for Biotechnology Information (NCBI)-nt database using the Blast software (Altschul et al., 1990). Sequencing reads with one mate-pair mapped were extracted from the contigs assigned to the bacterial genomes using SAMtools (Li et al., 2009) and ad-hoc Python scripts. Extracted reads were used for de novo assembly using SPAdes v3.0 assembler with k-mer lengths of 55, 77 and 100 (Bankevich et al., 2012). The QUAST program was used to generate the summary statistics of the assembly (N50, maximum contig length, GC%) (Gurevich et al., 2013). The total length of the contigs was used to estimate the genome size.
Partial chloroplast genome assembly was done by mapping the sequencing reads using the CLC Genomics Workbench v7.0 (http://www.clcbio.com; CLC Bio-Qiagen) onto the P. kirkii chloroplast genome, obtained as part of the Ca. B. kirkii sequencing project (Carlier and Eberl, 2012).
Genomic DNA of Ca. B. punctata was used for single molecule real-time (SMRT) library preparation and sequencing using the P2 chemistry on the Pacific Biosciences (PacBio) RSII instrument at the FGCZ. One 0.5 kb-insert library was sequenced using 13 SMRT cells to generate high-quality circular consensus sequence reads. A second 6 kb-insert library was sequenced using four SMRT cells to generate continuous long reads. Raw read data was filtered and analyzed using the PacBio SMRTportal v1.4 pipeline. The high-quality long reads were generated by mapping the circular consensus sequence reads into continuous long reads reads using the PacBiotoCA utility of the Celera assembler package (Koren et al., 2013). Assembly of the corrected reads was done using the MIRA v3.4 software (Chevreux et al., 1999). Forty-nine contigs with an average coverage of 42 × were obtained and edited in GAP5 (Bonfield and Whitwham, 2010).
Annotation
Open reading frames were detected with the Prodigal software (Hyatt et al., 2010), transfer RNA genes using tRNAscan (Schattner et al., 2005) and ribosomal RNA genes with RNAmmer (Lagesen et al., 2007). Coding DNA sequences were annotated with the rapid annotation using subsystem technology (RAST) online annotation service and metabolic information was obtained from the kyoto encyclopedia of genes and genomes annotation tool implemented in RAST (Aziz et al., 2008). Gene ontology information was collected using the Blast2GO program (Conesa et al., 2005) and subcellular localization prediction was carried out using the InterProScan (Zdobnov and Apweiler, 2001). Stretches of sequence over 1 kb lacking annotations, possibly because of divergent codon usage or GC bias, were manually annotated with Artemis and the NCBI Blast software suite (Altschul et al., 1990; Rutherford et al., 2000). Functional genes and pseudogenes were predicted according to the pipeline developed by Carlier et al., 2013. Further, pseudogenes were annotated by homology search of intergenic regions using the NCBI blastx program against a custom database of protein sequences predicted from Burkholderia genomes. Only hits over 50% identity and with an e-value <10−6 were considered. Hit regions were aligned with the best-scoring subject protein sequence using the tfasty program of the FASTA software suite v3.6 in order to find the boundaries of the pseudogenes (Pearson, 2000). Assignment of pseudogenes to COG functional categories was done using the sequence of the best blast hit protein (see below).
Repetitive sequences were detected with the program RepeatScout v1.0.5 (Price et al., 2005). To characterize the diversity of insertion sequences (IS), the whole genome was analyzed with ISsaga (Varani et al., 2011). To refine the annotation, repeats longer than 500 bp were searched against the ISfinder database using the NCBI blastx program. The annotated genomes were deposited at Genbank under the accession numbers LFMX00000000, LFLF00000000, LFKV00000000, LFJJ00000000, LFJI00000000, LELG00000000 and LFJH00000000.
Phylogenetic analysis
In order to reveal the phylogenetic relationships between Burkholderia leaf nodule symbionts and other plant-associated Burkholderia species we reconstructed a maximum likelihood phylogenetic tree of a concatenated alignment of 534 single-copy orthologs. Amino-acid sequences were aligned with MUSCLE (Edgar, 2004) and back translated into nucleotides with T-Coffee (Notredame et al., 2000). Positions of the alignment with gaps in more than 50% of the sequences were removed with TrimAl (Capella-Gutiérrez et al., 2009). Maximum likelihood reconstruction was done using RAxML v8.0 (Stamatakis, 2014) with GTRGAMMA nucleotide substitution model and 1000 rapid bootstrap analyses. One-to-one ortholog genes were obtained comparing a genome database comprising six Burkholderia species with a beneficial association with plants (B. phymatum STM815, B. phytofirmans PsJN, B. xenovorans LB400, B. sp. CCGE1001, B. sp. CCGE1002, B. sp. CCGE1003), three commonly found in soil (B. sp. YI23, B. sp. SJ98, B. glathei), one bean bug symbiont (B. sp. RPE64) and eight Burkholderia leaf nodule symbionts (Ca. B. kirkii, Ca. B. punctata, Ca. B. humilis, Ca. B. umbellata, Ca. B. schumanniana, Ca. B. verschuerenii, Ca. B. brachyanthoides, Ca. B. pumila). To root the tree we used two species (B. cenocepacia J2315 and B. ambifaria AMMD) belonging to the Burkholderia cepacia complex (Suárez-Moreno et al., 2012).
The maximum likelihood tree for the seven plant species was obtained from a concatenated alignment of a conserved 30 kb segment of chloroplast genome sequence. The conserved region was identified in the genomes using the Mauve software (Darling et al., 2010) and aligned using the MUSCLE algorithm with standard settings. The Pa. schumanniana chloroplast sequence was used as outgroup to root the tree (Denoeud et al., 2014).
Evolutionary rates and natural selection analyses
To determine the evolutionary rates of leaf nodule symbionts, rates of synonymous (dS) and nonsynonymous (dN) substitutions were estimated for the gene families with strictly one ortholog in each of the eight species. Protein sequences for each ortholog cluster were aligned with MUSCLE (Edgar, 2004) and subsequent nucleotide codon alignments were generated and trimmed using Pal2nal perl script (Suyama et al., 2006).
Pairwise dN/dS values were estimated with the Codeml module of the PAML v4.4 package using the following settings (model=−2 and codon frequency=2) (Yang, 2007). Genes with dS values near saturation (dS>2) were excluded from the analyses.
In order to increase sensitivity, we analyzed sites under positive selection with a site model test (Yang and dos Reis, 2011) implemented in Codeml. We contrasted the null model M7 (with beta distribution for ω) to the alternative model M8 (beta distribution for ω and an extra class of sites under positive selection with ω>1). The likelihood ratio test was performed for each model and the results were compared with a χ2-distribution with two degrees of freedom and alpha value of 0.01.
Functional characterization and genome comparisons
Clusters of Orthologous Groups (COG) functional categories were assigned to each open reading frame. This was achieved with a reversed position specific blast (RPS-BLAST) against the NCBI conserved domain database (e-value cutoff of 1 × 10−3). Genes belonging to each COG category were counted and the distributions were compared using a Pearson's χ2-test run with the R software package (R Development Core Team., 2011).
In order to compare the genomes of leaf nodule symbionts, predicted protein sequences were clustered using OrthoMCL v1.4 software with reciprocal blastp (e-value cutoff of 1e10−6 and 50% identity and 50% match length cutoff) (Li et al., 2003). Core and accessory genes of leaf nodule symbionts were defined based on gene family clustering as previously described (Carlier and Eberl, 2012).
Detection of C7N aminocyclitols by gas chromatography-mass spectrometry (GC-MS)
Whole leaves were obtained for each plant specimen and dried on silica gel. Samples were ground to a powder, macerated in methanol for 24 h, filtered and dried. Part of the extract was dissolved in water (500 μl), filtered, transferred in a high-performance liquid chromatography vial and dried by lyophilization. MSTFA (N-Methyl-N-(trimethylsilyl) trifluoroacetamide) (Sigma Aldrich, St Louis, MO, USA) was added, the mixture was stirred for 30 min at 70 °C, pyridine (dry, same volume as MSTFA) was added, and the reaction was filtered. Samples of 5 μl were analyzed on a Thermo Scientific (Waltham, MA, USA) TRACE 1300 series gas chromatograph equipped with a Thermo Scientific ISQ Series Single Quadrupole mass spectrometer using a HP-Ultra-1 column (Agilent Technologies (Santa Clara, CA, USA), 25 m × 0.200 mm, 0.35 μm). The GC-MS runs were performed using an initial temperature of 40 °C (kept for 1 min), linear ramping of 7 °C min−1 and a final temperature of 330 °C (kept for 10 min). As a control, derivatization and GC-MS analysis was also carried out with synthetic kirkamide and the data were used as analytical standard (see Supplementary Information Methods).
Quantification by proton nuclear magnetic resonance (1H-NMR) spectroscopy
A solution of maleic acid as internal standard (Sigma Aldrich, TraceCERT) was prepared at a concentration of 0.52 μmol ml−1 in D2O (99.9% deuterated) and the NMR experiments were performed on a Bruker Avance III NMR spectrometer operating at 500 MHz proton frequency.
The protons chosen for maleic acid displayed a chemical shift between 6.30 and 6.20 ppm, for streptol glucoside between 5.966 and 5.941 ppm and for kirkamide between 5.887 and 5.874 ppm (Supplementary Figure S2A). The delay time were determined for these protons and the values of 7.5 s for maleic acid, 1.7 s for streptol glucoside and 2.2 s for kirkamide were found. The quantitative 1H-NMR measurements were operating with a relaxation delay (D1) of 40 s, a number of scan (ns) of 128 or 256, a spectral width (sw) of 1.2 ppm and a transmitter offset (O1) of 6 ppm. The water peak at 4.79 ppm was used as the reference for the calibration of the maleic acid peak based on a second 1H-NMR spectrum recorded for every measurement. The concentration of streptol glucoside and kirkamide were calculated by comparing their peak integration with the one measured for maleic acid. For the determination of kirkamide in the P. kirkii leaves extract a mixing experiment was carried out by spiking with 40 μg of synthetic kirkamide. The leaf extracts, prepared as described for the GC-MS measurement, were weighted using an analytical balance (Mettler-Toledo XA105DU).
Results and discussion
Genome properties
The assembly of the Illumina Miseq paired-end reads resulted in several hundreds of scaffolds for most of the genomes sequenced (except for Ca. B. punctata whose genome was assembled using long read data) and over 50 × average coverage (detailed information in Table 1). Fragmentation of the assemblies was mostly caused by repetitive sequences derived from mobile elements.
Table 1. Genome properties of leaf nodule symbionts.
| Ca. B. kirkii | Ca. B. punctata | Ca. B. pumila | Ca. B. schumanniana | Ca. B. umbellata | Ca. B. brachyanthoides | Ca. B humilis | Ca. B. verschuerenii | |
|---|---|---|---|---|---|---|---|---|
| Number of contigs | 305 | 48 | 519 | 283 | 307 | 685 | 354 | 446 |
| Genome size (Mb) | 4 | 3.9 | 3.7 | 2.4 | 4.2 | 3.5 | 5.1 | 6.2 |
| GC content (%) | 63 | 64 | 59 | 63 | 62 | 61 | 61 | 62 |
| Total number of ORFs | 6260 | 4802 | 5145 | 2262 | 5913 | 3508 | 5256 | 5939 |
| Number of intact ORFs | 2069 | 2323 | 1737 | 1459 | 2002 | 2137 | 3121 | 4699 |
| Mobile elements in the genome (%) | 10 | 11 | 5.6 | 5 | 3.5 | 2.4 | 1.4 | 2.9 |
| rRNA | 4 | 7 | 3 | 3 | 6 | 3 | 3 | 3 |
| tRNA | 52 | 47 | 51 | 48 | 52 | 47 | 48 | 53 |
Abbreviations: GC, guanine and cytosine; ORFs, open reading frames; rRNA, ribosomal RNA; tRNA, transfer RNA.
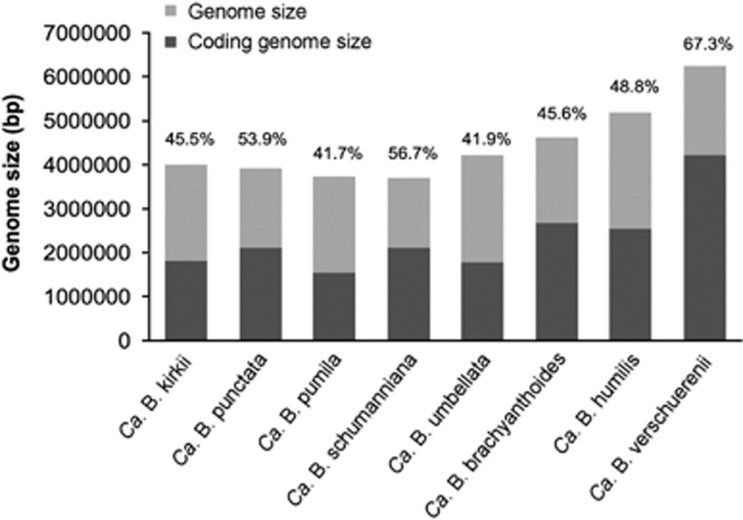
Analysis of the contigs showed the presence of a single species of symbiotic bacteria per plant sample. The genome sizes of the leaf nodule symbionts sequenced in this study varied between 2.4 Mb for Ca. B. schumanniana and 6.1 Mb for Ca. B. verschuerenii (Table 1). The genomes also showed different coding capacities ranging from 41.7% for Ca. B. pumila to 67.3% for Ca. B. verschuerenii (Figure 1).
Figure 1.
Genome sizes and coding proportions of leaf nodule Burkholderia. Genome size estimates for all the leaf nodule bacteria are shown in gray. The proportion of the coding genome and their corresponding and values in percentage are shown in black.
Reductive genome evolution in leaf nodule symbionts
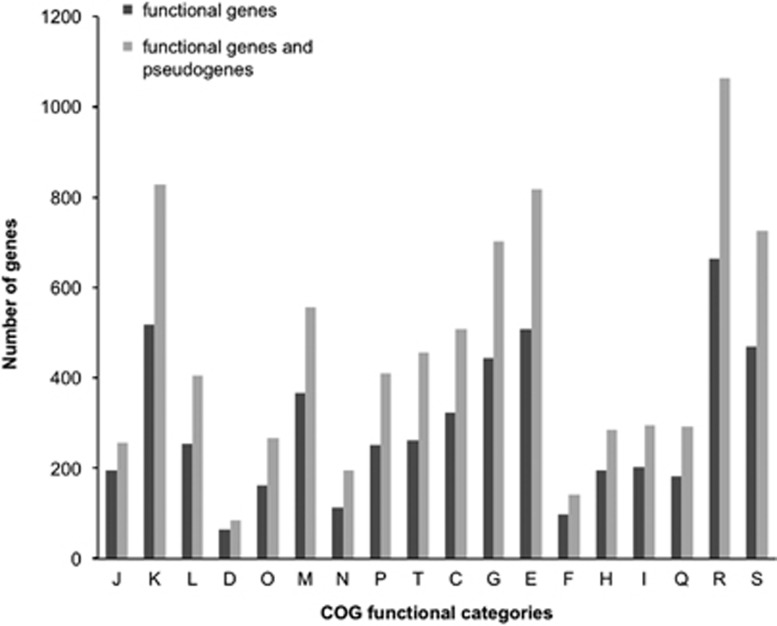
The transition from free-living to an obligate symbiosis often results in marked changes in genome architecture (Moya et al., 2008; Toft and Andersson, 2010; McCutcheon and Moran, 2012). Apart from a reduced genome size and coding capacity, leaf nodule symbionts share other traits with obligate symbionts in the early stages of the symbiosis such as the accumulation of pseudogenes and transposable elements (Toh et al., 2006; Burke and Moran, 2011; Oakeson et al., 2014). Around 60% of the DNA-coding regions are interrupted by frameshift or null mutations and no significant bias toward the loss of any particular functional gene category was observed (Figure 2). This indicates that genetic drift is the main evolutionary force driving the erosion of gene functions in the leaf nodule symbiont genomes, similar to what has been observed for other obligate symbionts (Mira and Moran, 2002).
Figure 2.
Distribution of the genes conserved in all the leaf nodule symbionts in functional categories of COGs. For each COG category, the number of functional genes (gray bars) and the total number of genes including pseudogenes (black bars) were compared. (J)=translation, ribosomal structure and biogenesis; (K)=transcription; (L)=DNA replication, recombination and repair; (D)=cell division and chromosome partitioning; (O)=posttranslational modification, protein turnover, chaperones; (M)=cell envelope biogenesis, outer membrane; (N)=cell motility and secretion/Intracellular trafficking and secretion; (P)=inorganic ion transport and metabolism; (T)=signal transduction mechanisms; (C)=energy production and conversion; (G)=carbohydrate transport and metabolism; (E)=amino-acid transport and metabolism; (F)=nucleotide transport and metabolism; (H)=coenzyme metabolism; (I)=lipid metabolism; (Q)=secondary metabolites biosynthesis, transport and catabolism; (R)=general function prediction only; (S)=function unknown.
Transposable elements are abundant in all the leaf nodule symbionts and may have an important role in the first steps of genome reduction. At least nine different IS families were represented in leaf nodule symbionts. The distribution of these families is patchy and most of the identified mobile elements lack a functional copy in the genome (Supplementary Table S1). We could only trace full copies corresponding with the families IS3, IS4, IS5, IS66 and IS200 across several genomes.
The variability in copy number and localization of IS elements from the same family across leaf nodule symbiont genomes suggest that: (i) proliferation of an IS family in a given genome is triggered randomly and (ii) the recent transition to a host-specific lifestyle may have stimulated the rapid proliferation of IS elements. These observations are in accordance with the proposed neutral punctuated model for IS elements proliferation (Iranzo et al., 2014). This model predicts instability of IS copy number following a change in the environment, such as that following the establishment of a host-specific lifestyle. The fact that we find many inactivated or truncated IS in the leaf nodule symbiont genomes may indicate either an overall strong deletion bias or a return to a more stable condition where the multiplication and elimination of active IS copies return to equilibrium.
All the leaf nodule symbiont genomes show extremely low synteny, which is consistent with extensive genome shuffling caused by transposable elements. We compared the genome of Ca. B. punctata, which we could assemble in large contigs, with that of the related stinkbug symbiont Burkholderia sp. RPE64. The alignment shows mostly small syntenic blocks greater than 100 kb. By contrast, the more distantly related, free-living soil-isolated B. sp. YI23 shows a relatively high synteny with Burkholderia symbiont sp. RPE64 with stretches of syntenic DNA spanning entire replicons (Supplementary Figure S3). Many rearrangements are observed within scaffolds and in many cases syntenic blocks are flanked by intact or inactivated transposases. This suggests that mobile elements, possibly in combination with homologous recombination, facilitated the massive DNA rearrangements observed with the leaf nodule genomes.
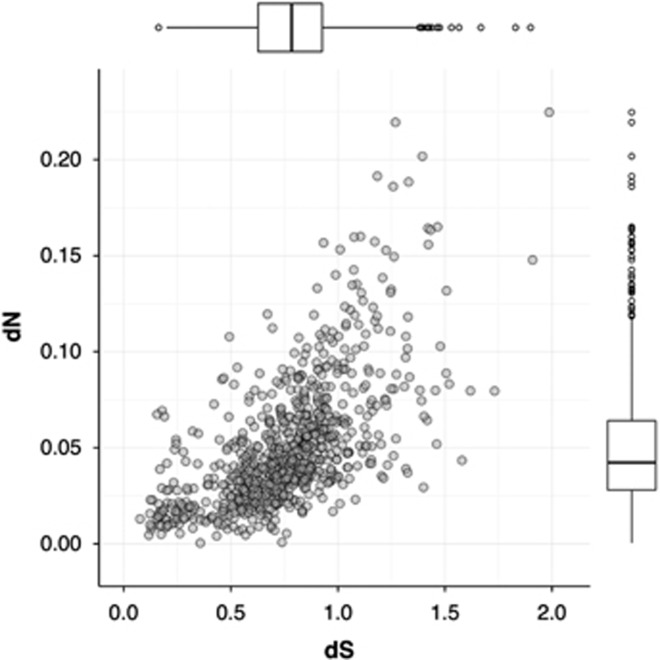
Genome rearrangement, IS proliferation and large numbers of pseudogenes indicate that genetic drift is a major driving force in shaping the genomes of the leaf nodule symbionts. In other obligate and vertically transmitted symbionts, genetic drift is known to be the major evolutionary force shaping the genome architecture, whereas purifying selection is less effective (Kuo et al., 2009). This often results in the accumulation of deleterious mutations (a process known as Muller's ratchet) (Muller, 1964; Felsenstein and Yokoyama, 1976; Moran, 1996; McCutcheon and Moran, 2012). Because ongoing genome erosion often translates into high rates of nonsynonymous (dN) to synonymous (dS) substitutions across the genome, we calculated the dN/dS rates of substitutions for the 798 true orthologs in all the leaf nodule symbiont genomes. The average dN/dS ratio was 0.07 (Figure 3), whereas free-living bacteria or facultative symbionts have an average dN/dS ratio between 0.02 and 0.06 (Kuo et al., 2009).
Figure 3.
Synonymous (dS) and nonsynonymous (dN) substitution rates inferred from pairwise comparisons of the 798 true orthologous genes (gray dots) in all leaf nodule symbiont genomes. The boxplots in the adjacent axes represent the distribution of dS and dN values and the outliers.
Our observations are so far consistent with what has been described for other obligate symbionts, including endosymbionts of insects (McCutcheon and Moran, 2012). In these, relaxed selection usually results in enrichment of the genome in adenine and thymine (A+T) and a lower GC% in obligate symbionts compared with free-living relatives (McCutcheon and Moran, 2012). In contrast, average GC% of leaf nodule symbionts's genomes (60–63%) is within the range of related free-living Burkholderia (62–65%). One reason for the high GC% of the genomes may be attributed to the relatively young age of the symbiosis and to the fact that DNA repair mechanisms are still intact in most of the leaf nodule symbionts. However, the average GC% of pseudogenes, which could be assumed to be under relaxed selection, is not significantly different from that of functional genes (Supplementary Table S2). In other organisms, pseudogenes have a lower GC% than their functional counterparts (Lerat and Ochman, 2004). This lack of deviation from average GC% suggests instead that substitutions are not biased toward A+T in Burkholderia leaf nodule symbionts. This interpretation is supported by experimental data demonstrating an unusual mutational bias toward G+C in other Burkholderia species (Dillon et al., 2015).
Core genome of obligate endophytes
We computed the core genome of leaf nodule symbionts and compared it with the core genome of 52 free-living Burkholderiaceae species, which serve as an approximation of the essential gene set of Burkholderia species (Juhas et al., 2012). The core genome of leaf nodule symbionts contains 798 genes and only seven Burkholderiaceae core genes were missing in all leaf nodule symbiont genomes. However, none of them appears to belong to any essential metabolic pathway (Supplementary Table S3), suggesting that despite rampant genome reduction, leaf nodule symbionts may not be dependent on the host for essential housekeeping functions. This indirectly confirms the earlier assumption that the inability to grow leaf nodule bacteria axenically may be due to their lack of adaptability to changing environmental conditions (Carlier and Eberl, 2012).
We also explored the set of the non-essential genes (accessory genes of free-living Burkholderia) conserved in all leaf nodule symbiont genomes in order to find specific pathways that could be important for the symbiosis or for life as an endophyte. We identified 314 non-essential genes, mostly involved in coenzyme metabolism and cell envelope and outer membrane biogenesis that were conserved in all leaf nodule symbionts of Rubiaceae (Supplementary Figure S4, Supplementary Table S4).
Among the genes involved in coenzyme metabolism, we could identify complete pathways for the biosynthesis of thiamin, cobalamin and glutathione. These vitamins are known to promote plant–microbe interactions and could therefore be potentially important for the obligate nature of leaf nodule symbiosis. There is only one case of cobalamin-dependent symbiosis described, namely between Volvox carteri algae and its cyanobacteria symbiont (Helliwell et al., 2011). Contrary to algae, higher plants do not require cobalamin and they are not able to produce it. Cobalamin biosynthesis is confined only to bacteria where the vitamin acts as a cofactor for multiple enzymes such as the methionine synthase (Banerjee and Ragsdale, 2003). Leaf nodule symbionts retain the cobalamin-dependent version of methionine synthase and a complete cobalamin biosynthetic pathway. The amounts of free Met in ultra performance liquid chromatography analyses of extracts of aposymbiotic P. kirkii were indistinguishable from nodulated plants, suggesting that leaf nodule symbionts synthesize Met to fulfill their own metabolic needs and not those of the host (data not shown).
Bacterial antioxidants can act as a defense mechanism against plant reactive oxygen species (Nanda et al., 2010). The relevance of enzymatic and non-enzymatic reactive oxygen species scavengers such as the superoxide dismutase enzyme and glutathione in plant–microbe interactions has been widely demonstrated to be important in legume-rhizobium symbioses (Santos et al., 2000; Rubio et al., 2004; Pauly et al., 2006). These scavenging functions (for example, superoxide dismutase are conserved in all leaf nodule symbionts, indicating that they may also have an important role for the symbiosis and maybe nodule formation (Supplementary Table S4). Alternatively, mechanisms for coping with reactive oxygen species might be necessary to survive in the harsh, photo-oxidative environment of the leaf (Triantaphylidès et al., 2008).
Cell wall and host–symbiont interaction
As previously mentioned, leaf nodule symbionts share non-essential genes responsible for the lipopolisaccharides (LPS) biosynthesis, which may be important for host–symbiont recognition (Raetz and Whitfield, 2002). Lipopolisaccharides are ‘microbe-associated molecular patterns' that constitute the major component of the cell surface (Boller and Felix, 2009). They are considered the first barrier against plant defenses and mediate the communication with the host plant (Soto et al., 2009). As the leaf nodule symbionts colonize plant species from the same family, similar structural composition of the O-antigen was expected. However, the cluster potentially involved in assembly, modification and export (Bkir_c27_1818–1838) is not fully conserved in all leaf nodules and some of them even lack a glycosyltranferase enzyme (Bkir_c27_1818), presumably involved in the assembly of the O-antigen. The absence of a conserved cluster for the synthesis of O-antigen in leaf nodule symbionts does not entirely rule out the possibility that lipopolisaccharide is involved in symbiotic communication. However, lipopolisaccharide does not seem to be the principal mediator and other microbe-associated molecular patterns (for example, GroEL, elongation factor, peptidoglycan and so on) could be involved in the recognition of the symbiont (Newman et al., 2013; Chaudhary et al., 2014).
Evidence of host switching
Vertical transmission of bacterial symbionts through the germ line of the host is one of the most prevalent strategies for the maintenance of obligate symbiosis and often results in phylogenetic congruence or co-speciation between partners (Moran et al., 1993; Russell and Moran, 2005; Sloan and Moran, 2012). Despite host specificity and vertical transmission of the leaf nodule symbionts, the phylogenies of the Rubiaceae species and their Burkholderia symbionts are incongruent. A vertical transmission mode with occasional horizontal transmission has been proposed to explain this lack of strict co-evolution (Lemaire et al., 2011). Moreover, the interlaced presence of Burkholderia soil isolates and insect symbionts within the phylogenetic clade of leaf nodule symbionts suggest multiple sources of transmission (plants, insects and soil).
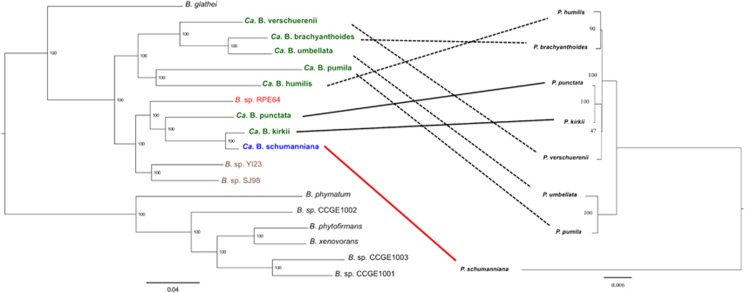
Incongruence between hosts and symbionts phylogenies suggest that the mode of transmission of the bacteria is mixed rather than strictly vertical (Figure 4). Most intriguingly, we detected a conspicuous host switching or re-infection event between the P. kirkii/Ca. B. kirkii and the Pa. schumanniana/Ca. B. schumanniana species pairs. The bacterial symbionts are closely related and share similar degrees of genome reduction despite having diverged <300 000 years ago (Supplementary Figure S5). However, P. kirkii and Pa. schumanniana belong to distinct genera that are thought to have diverged >60 million years ago (Lemaire et al., 2011). The simplest explanation is that the symbiont of Pa. schumanniana was recently replaced with the leaf nodule symbiont of P. kirkii. The possibility of a re-infection by bacteria living in a soil reservoir as previously hypothesized (Lemaire et al., 2012) seems unlikely. The high degree of genome reduction, together with the very high sequence homology observed between the genomes of the symbionts of Pa. schumanniana and P. kirkii, rather suggest that both species have been restricted to a host for millions of years. The likelihood of host-switching events may be increased by the overlap of the geographic ranges of P. punctata, P. kirkii and Pa. schumanniana (Bremekamp, 1934; Lachenaud, 2013).
Figure 4.
Maximum likelihood phylogenetic reconstruction of Burkholderia species (left) and Psychotria species (right). The Burkholderia tree was reconstructed based on the analysis of a concatenated alignment of 534 true ortholog genes. The reconstruction of the Psychotria plant species was based on a 30 kb chloroplast-conserved fragment. In green are highlighted the leaf nodule bacterial symbionts of Psychotria plant species. The leaf nodule bacterial symbiont of Pavetta plant species is highlighted in blue. Highlighted in red is a symbiont of Riptortus (stinkbug) and in brown, soil isolates. Burkholderia ambifaria AMMD was used as outgroup (not shown). Bootstrap values are shown.
A hint about the mechanism of symbiont transmission between plants may come from the nested clade of soil isolates (B. sp. YI23 and B. sp. SJ98) and the stinkbug Riptortus pedestris symbiont (B. sp. RPE64), which form a sister group with Ca. B. punctata, Ca. B. kirkii and Ca. B. schumanniana (Figure 4). Burkholderia isolates, B. sp. YI23 and B. sp. SJ98, have been isolated from soil and are capable of degrading the pesticides 2-chloro-4-nitrophenol (in B. sp. SJ98) (Min et al., 2014) and fenitrothion (in B. sp. YI23) (Lim et al., 2012). B. sp. RPE64 has been isolated from stinkbugs (Kikuchi et al., 2012). This group of Burkholderia species, to which the leaf nodule symbionts of Rubiaceae belong, live freely in soil or are associated with plants and insects. Hence, one can envision that the ancestor of the leaf nodule symbionts was a Burkholderia species with a remarkably broad host range. It is therefore conceivable that some of the leaf nodule species have retained the ability for transient association with insects, which could act as potential vectors (whereas non-vector insects may be targets of the insecticidal activity of kirkamide).
Secondary metabolism is important but possibly not essential for leaf nodule symbiosis
In order to get better insight into the determinants of leaf symbiosis, we focused on the genes uniquely present in leaf nodule symbionts and absent in other closely related Burkholderia species. The genome comparison of the bacterial leaf nodule symbionts with 19 closely related Burkholderia species revealed only one gene that is unique to the leaf nodule lineage. This gene codes for a putative 2-epi-5-epi-valiolone synthase, involved in the first step of the 2-epi-5-epi-valiolone-derived aminocyclitols from the C7N aminocyclitol family. Closer inspection revealed that the gene is truncated in Ca. B. brachyanthoides, and biosynthesis of C7N aminocyclitols seems to have been lost in this species (Table 2). However, the core genome of the seven other genomes contains a set of six genes (2-epi-5-epi-valiolone synthase, N-acetylmannosamine kinase, sugar hydrolase, glycosidase, sugar dehydrogenase, Gcn5-related N-acetylransferase family) potentially involved in the synthesis of the kirkamide molecule (Table 2). Almost all the genes are conserved and are in the same gene order and only the acetyltransferase is in a different location in Ca. B. humilis. Interestingly, this cluster of genes is in some cases flanked by the remnants of transposable elements (Supplementary Figure S6).
Table 2. Genes of the secondary metabolism in leaf nodule symbionts.
| Putative product | Ca. B. kirkii | Ca. B. punctata | Ca. B. pumila | Ca. B. humilis | Ca. B. schumanniana | Ca. B. verschuerenii | Ca. B. umbellata | Ca. B. brachyanthoides |
|---|---|---|---|---|---|---|---|---|
| 2-epi-5-epi-valiolone synthase (Bkir_c149_4879) | Ψ | |||||||
| N-acetylmannosamine kinase (Bkir_c149_4878) | ||||||||
| Aminosugar transaminase (Bkir_c149_4877) | Ψ | |||||||
| Glycoside hydrolase (Bkir_c149_4876) | ||||||||
| Sugar dehydrogenase (Bkir_c149_4875) | ||||||||
| HAD family hydrolase (Bkir_c149_4874) | ||||||||
| Sugar phosphate isomerase (Bkir_c149_4873) | Ψ | |||||||
| Acetyltransferase, GNAT family (Bkir_c149_4873) | ||||||||
| Dioxygenase (Bkir_c48_3583) | ||||||||
| Methyltransferase (Bkir_c48_3584) | ||||||||
| C7-cyclitol-7-kinase (Bkir_c48_3591) | ||||||||
| 3-dehydroquinate synthase (Bkir_c48_3593) | ||||||||
| dTDP-4-dehydrorhamnose reductase (Bkir_c48_3594) | ||||||||
| NUDIX family hydrolase (Bkir_c48_3606) | ||||||||
| Kirkamide | + | + | + | + | − | + | − | − |
Genes of Ca. B. kirkii are used as a reference. Functional homologues are highlighted in grey and pseudogenes indicated by Ψ. Conserved genes are shown in bold. Detection of kirkamide in crude leaf extracts is indicated by a + symbol. A − symbol indicates kirkamide concentrations below detection levels (see Materials and Methods).
Because the common ancestor of all modern Rubiaceae leaf nodule symbionts likely possessed C7N aminocyclitol secondary metabolism, we speculate that acquisition of the ability to synthesize this protective compound enabled the shift from perhaps a commensal to a mutualistic lifestyle. However, secondary metabolism and the associated fitness advantage alone does not explain the strict dependence of the host plant on the presence of bacteria, which may have evolved independently (De Mazancourt et al., 2001; Leigh, 2010; Sachs et al., 2011).
To test this, we assayed leaf extracts for kirkamide by GC-MS and 1H-NMR (see Supplementary Information Methods). We could detect kirkamide in leaves extract from P. kirkii, P. punctata, P. verschuerenii, P. humilis and P. pumila (Table 2, Supplementary Figures S2D and E) and concentrations ranging from 0.2 to 0.4% of the dry weight were determined in leaves of P. kirkii and P. punctata. All of these species harbor the putative kirkamide biosynthetic pathway. In contrast, we could not detect kirkamide in extracts of P. brachyanthoides, whose symbionts have lost most of the kirkamide biosynthesis cluster. We were also unable to detect kirkamide in extracts of Pa. schumanniana and P. umbellata, both species harboring the enzymes of the kirkamide pathway (Table 2, Supplementary Figures S2D and E). Interestingly, a putative major facilitator superfamily transporter gene attached to the kirkamide operon is a pseudogene in Ca. B. umbellata. The orthologous gene in Ca. B. schumanniana is detached from the main kirkamide operon after IS-mediated rearrangement. It is therefore possible that transport of kirkamide or of precursors is compromised in both of these organisms. An alternative hypothesis is that another undetected C7N aminocyclitol is produced in P. umbellata and Pa. schumanniana. We could, for instance, isolate a new C7 cyclitol compound, streptol glucoside (Supplementary Figure S2A) from leaves of P. kirkii. The structure is epimeric at the anomeric center when compared to A-79197–2, which was isolated from an epiphytic Actinomyces species (Kizuka et al., 2002) (Supplementary Figure S2F). Because streptol glucoside displays a strong inhibition of lettuce seeds germination (confirmed with the purified compound from P. kirkii), it might confer an allelopathic advantage to P. kirkii (see Supplementary Information Methods). The host plant enzymatic repertoire might also contribute to the diversity of secondary metabolites produced by the association.
Evidence for HGT of secondary metabolites genes
The lack of complete synteny in the conserved secondary metabolite genes and the presence of flanking transposable elements suggest that HGT events might have occurred. This is also supported by the incongruence of the gene phylogenies, indicating multiple events of HGT within leaf nodule symbionts' clade (Supplementary Figure S7). Although C7N aminocyclitol biosynthesis was likely present in the ancestor of the modern leaf nodule symbionts, HGT seems to have happened at least twice (Supplementary Figure S8). Because the putative kirkamide biosynthetic genes are located on plasmid pKIR01 in Ca. B. kirkii, conjugative transfer could explain the mixed phylogenetic trees of the secondary metabolism genes of the leaf nodule symbionts. We previously identified the repA gene of pKIR01, located in the immediate vicinity of a parA/parB partition gene pair and possibly coding for a replication initiation protein (Carlier and Eberl, 2012; Carlier et al., 2013). We could retrieve complete or partial sequences of repA homologs from all the genomes of leaf nodule symbionts (Supplementary Figure S9). Remarkably, we could detect two copies of repA in the genome of Ca. B. kirkii, one of which being a pseudogene, belonging to a distinct branch of the repA tree. This is clear evidence that acquisition of the pKIR01 replicon happened at least twice in Ca. B. kirkii. Moreover, we find little evidence of co-evolution between the repA genes and the rest of the genomes, except for Ca. B. punctata, Ca. B. kirkii and Ca. B. schumanniana. Together, these data suggest that transfer of the pKIR01 replicon is common in leaf nodule symbionts and may explain the suspected HGT of C7N aminocyclitol biosynthetic genes in leaf nodule symbionts.
Acquisition of secondary metabolite production by HGT has been documented in other symbioses, notably the insect-defensive symbiosis that occurs in the Asian citrus psyllid Diaphorina citri (Nakabachi et al., 2013b). Further, HGT events have been hypothesized between the obligate intracellular symbiont Candidatus Profftella armatura and the phloem-restricted pathogen ‘Candidatus Liberibacter asiaticus' (Nakabachi et al., 2013a). However, the timing of the HGT events is unclear in those examples. Our data strongly suggest that HGT of kirkamide synthetic genes still occurred after leaf nodule symbiosis first evolved. Intra-clade HGT are rare in obligate symbiotic bacteria but HGT of bacteriophage-related genes have been documented in facultative Wolbachia insect symbionts, which are also known to co-infect their hosts (Bordenstein and Wernegreen, 2004). We are documenting here the first case of intra-clade horizontal transfer of genes, which appear to have a central role in obligate leaf nodule symbiosis.
Conclusions
Although the genomes of all leaf nodule symbionts are in a similar state of erosion, we did observe some variability in their coding capacity, indicating dynamic and ongoing genome reduction. Our data also show that limited intra-clade HGT can still occur at the early stages of genome reduction. Transient co-habitation of symbionts and plasmid conjugative transfer may explain the presence of HGT in leaf nodule symbionts. Indeed, our data provide evidence in favor of a mode of transmission of the symbionts that is not strictly vertical. We propose insects as possible vectors for the horizontal transmission of leaf nodule bacteria to novel hosts. Host-switching events could result in transient co-habitation of distinct bacterial species within a leaf nodule, thereby promoting HGT among leaf nodule symbionts.
Our data suggest that plasmid-mediated HGT is a common occurrence among leaf nodule symbionts. We hypothesize, however, that as leaf nodule bacteria species diverge, exchange of genetic material will become limited because of narrowing of plasmid host range or the loss of the ability to colonize an insect vector. In fact, our data show that plasmids of the pKIR01 group can become integrated in the chromosome, precluding further genetic exchange. In this regard, the genomes of leaf nodule symbionts offer a unique possibility to investigate the early stages of genome reduction following the transition to an obligate symbiotic lifestyle.
In this study, we observed that not all the leaf nodule symbionts are capable of producing kirkamide. These results suggest that kirkamide has an important defensive role but is not responsible for the obligate nature of the symbiosis. Interestingly, a recent phylogenetic study on Psychotria leaf nodulated species revealed the presence of two non-nodulated plant species (P. tetragonopus and P. limba) within the nodulated clade, suggesting that symbiotic traits, have been lost in those species (Lachenaud, 2013).
Our genomic study is fully consistent with other studies that have dated the origin of the leaf nodule symbiosis back to the Miocene (in a range of 5.5–13.82 Mya) (Supplementary Figure S5) (see Supplementary Information Methods) (Lemaire et al., 2011; Barrabé et al., 2014). The obligate interaction between leaf nodule symbionts and their host plant could have evolved from an initial facultative and commensal association (Figure 5). The acquisition of secondary metabolites synthesis by the symbiotic bacteria may likely have been essential for the transition to a mutualistic interaction and the establishment of the leaf nodule symbiosis. Subsequently, the association became permanent and the bacterial genomes started to erode. Isolating intermediate stages of the symbiosis could give valuable information into this process. It would thus be interesting to explore the genome properties of the recently described Burkholderia endophytes of non-nodulated Rubiaceae plants (Verstraete et al., 2013).
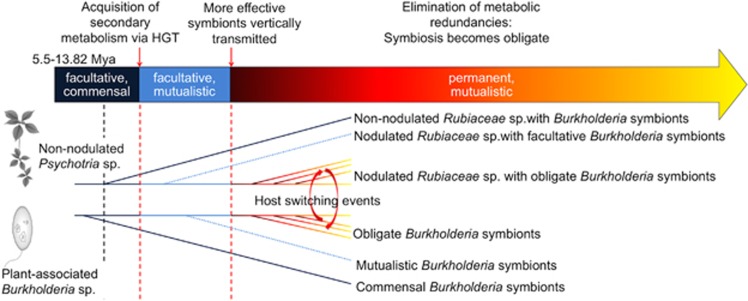
Figure 5.
Evolutionary history hypothesis for the leaf nodule symbiosis. Ancestors of modern Psychotria sp. were probably in transient association with endophytic, or possibly epiphytic commensal bacteria. About 5.5–13.8 Mya, the bacteria acquired genes for the production of protective secondary metabolites, thereby giving a net fitness benefit to the association. As more effective symbionts were selected, the symbiotic bacteria became vertically transmitted, making the symbiosis permanent. A consequence of this permanent association is the increased dependency of the host on the leaf nodule bacteria, possibly through the loss of redundant metabolic pathways. All described modern nodulated Psychotria species are locked in the relationship with the leaf nodule bacteria, regardless of the production of protective metabolites. Intermediate stages shown in dashed lines (mutualistic Burkholderia symbionts, nodulated Rubiaceae with facultative symbionts) are hypothetical.
Acknowledgments
We are grateful to Rolf Kümmerli and Kentaro Shimizu for their help and comments on the manuscript. MPC acknowledges support from the University of Zurich URPP Evolution in Action. This work was supported by the Swiss National Foundation under grant CRSII3_154430.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 3: 403–410. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, Ragsdale SW. (2003). The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem 72: 209–247. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich Aa, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrabé L, Maggia L, Pillon Y, Rigault F, Mouly A, Davis AP et al. (2014). New Caledonian lineages of PsychotriaRubiaceae reveal different evolutionary histories and the largest documented plant radiation for the archipelago. Mol Phylogenet Evol 71: 15–35. [DOI] [PubMed] [Google Scholar]
- Belda E, Moya A, Bentley S, Silva FJ. (2010). Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 11: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Whitwham A. (2010). Gap5—editing the billion fragment sequence assembly. Bioinformatics 26: 1699–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Wernegreen JJ. (2004). Bacteriophage flux in endosymbionts (Wolbachia: infection frequency, lateral transfer, and recombination rates. Mol Biol Evol 21: 1981–1991. [DOI] [PubMed] [Google Scholar]
- Bremekamp. (1934). A monograph of the genus Pavetta L. In: Repert specierum Nov regni Veg, pp 171–172.
- Burke GR, Moran NA. (2011). Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol 3: 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier AL, Eberl L. (2012). The eroded genome of a Psychotria leaf symbiont: hypotheses about lifestyle and interactions with its plant host. Environ Microbiol 14: 2757–2769. [DOI] [PubMed] [Google Scholar]
- Carlier AL, Omasits U, Ahrens CH, Eberl L. (2013). Proteomics analysis of Psychotria leaf nodule symbiosis: improved genome annotation and metabolic predictions. Mol Plant Microbe Interact 26: 1325–1333. [DOI] [PubMed] [Google Scholar]
- Centifanto YM, Silver WS. (1964). Leaf-nodule symbiosis I endophyte of Psychotria bacteriophila. J Bacteriol 88: 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary R, Atamian HS, Shen Z, Briggs SP, Kaloshian I. (2014). GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc Natl Acad Sci 111: 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux, Wetter, Suhai. (1999). Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. In: Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB), 99: 45–56.
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed]
- Darling AE, Mau B, Perna NT. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mazancourt C, Loreau M, Dieckmann U. (2001). Can the evolution of plant defense lead to plant-herbivore mutualism? Am Nat 158: 109–123. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M et al. (2014). The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345: 1181–1184. [DOI] [PubMed] [Google Scholar]
- Dillon MM, Sung W, Lynch M, Cooper VS. (2015). The rate and molecular spectrum of spontaneous mutations in the GC-rich multichromosome genome of Burkholderia cenocepacia. Genetics 200: 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards WJ, Lamotte CE. (1975). Evidence for cytokinin in bacterial leaf nodules of Psychotria punctata (Rubiaceae). Plant Physiol 56: 425–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, Yokoyama S. (1976). The evolutionary advantage of recombination. II. Individual selection for recombination. Genetics 83: 845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. (2011). Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol 28: 2921–2933. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo J, Gómez MJ, López de Saro FJ, Manrubia S. (2014). Large-scale genomic analysis suggests a neutral punctuated dynamics of transposable elements in bacterial genomes. PLoS Comput Biol 10: e1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Eberl L, Church GM. (2012). Essential genes as antimicrobial targets and cornerstones of synthetic biology. Trends Biotechnol 30: 601–607. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. (2012). Symbiont-mediated insecticide resistance. Proc Natl Acad Sci USA 109: 8618–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizuka M, Enokita R, Shibata K, Okamoto Y, Inoue Y, Okazaki T. (2002). Studies on actinomycetes isolated from plant leaves —new plant growth inhibitors A-79197-2 and -3 from Dacthylosporangium aurantiacum SANK 61299. Actinomycetology 16: 14–16. [Google Scholar]
- Koren S, Harhay GP, Smith TPL, Bono JL, Harhay DM, Mcvey SD et al. (2013). Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol 14: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-H, Moran NA, Ochman H. (2009). The consequences of genetic drift for bacterial genome complexity. Genome Res 19: 1450–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenaud O. (2013). Le genre Psychotria (Rubiaceae) en Afrique occidentale et centrale: taxonomie, phylogénie et biogéographie Université Libre de Bruxelles. [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh EG. (2010). The evolution of mutualism. J Evol Biol 23: 2507–2528. [DOI] [PubMed] [Google Scholar]
- Lemaire B, Lachenaud O, Persson C, Smets E, Dessein S. (2012). Screening for leaf-associated endophytes in the genus Psychotria (Rubiaceae). FEMS Microbiol Ecol 81: 364–372. [DOI] [PubMed] [Google Scholar]
- Lemaire B, Vandamme P, Merckx V, Smets E, Dessein S. (2011). Bacterial leaf symbiosis in angiosperms: host specificity without co-speciation. PLoS One 6: e24430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Ochman H. (2004). Psi-Phi: exploring the outer limits of bacterial pseudogenes. Genome Res 14: 2273–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JS, Choi BS, Choi AY, Kim KD, Kim DI, Choi IY et al. (2012). Complete genome sequence of the fenitrothion-degrading Burkholderia sp. strain YI23. J Bacteriol 194: 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Latorre A. (2014). Settling down: the genome of Serratia symbiotica from the aphid Cinara tujafilina zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol Evol 6: 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. (2012). Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10: 13–26. [DOI] [PubMed] [Google Scholar]
- Miller IM. (1990) Bacterial leaf nodule symbiosis. Wright State University, Dayton, OH, USA. [Google Scholar]
- Min J, Zhang J-J, Zhou N-Y. (2014). The gene cluster for para-nitrophenol catabolism is responsible for 2-chloro-4-nitrophenol degradation in Burkholderia sp. strain SJ98. Appl Environ Microbiol 80: 6212–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Moran NA. (2002). Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb Ecol 44: 137–143. [DOI] [PubMed] [Google Scholar]
- Moran NA. (1996). Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci 93: 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. (1993). A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc B Biol Sci 253: 167–171. [Google Scholar]
- Moya A, Peretó J, Gil R, Latorre A. (2008). Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet 9: 218–229. [DOI] [PubMed] [Google Scholar]
- Muller HJ. (1964). The relation of recombination to mutational advance. Mutat Res Mol Mech Mutagen 1: 2–9. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Nikoh N, Oshima K, Inoue H, Ohkuma M, Hongoh Y et al. (2013. a). Horizontal gene acquisition of liberibacter plant pathogens from a bacteriome-confined endosymbiont of their psyllid vector. PLoS One 8: e82612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, Gurgui M et al. (2013. b). Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol 23: 1478–1484. [DOI] [PubMed] [Google Scholar]
- Nanda AK, Andrio E, Marino D, Pauly N, Dunand C. (2010). Reactive oxygen species during plant-microorganism early interactions. J Integr Plant Biol 52: 195–204. [DOI] [PubMed] [Google Scholar]
- Newman M-A, Sundelin T, Nielsen JT, Erbs G. (2013). MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci 4: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. (2000). T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Oakeson KF, Gil R, Clayton AL, Dunn DM, von Niederhausern AC, Hamil C et al. (2014). Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol 6: 76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly N, Pucciariello C, Mandon K, Innocenti G, Jamet A, Baudouin E et al. (2006). Reactive oxygen and nitrogen species and glutathione: key players in the legume–Rhizobium symbiosis. J Exp Bot 57: 1769–1776. [DOI] [PubMed] [Google Scholar]
- Pearson WR. (2000). Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol 132: 185–219. [DOI] [PubMed] [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11: 789–799. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. (2005) De novo identification of repeat families in large genomes. In: Proceedings of the 13 Annual International conference on Intelligent Systems for Molecular Biology (ISMB-05) Detroit, Michigan.
- R Development Core Team. (2011) R: A Language and Environment for Statistical Computing. The R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Raetz CRH, Whitfield C. (2002). Lipopolysaccharide endotoxins. Annu Rev Biochem 71: 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran L, Larsson J, Vigil-Stenman T, Nylander JAA, Ininbergs K, Zheng W-W et al. (2010). Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One 5: e11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MC, James EK, Clemente MR, Bucciarelli B, Fedorova M, Vance CP et al. (2004). Localization of superoxide dismutases and hydrogen peroxide in legume root nodules. Mol Plant Microbe Interact 17: 1294–1305. [DOI] [PubMed] [Google Scholar]
- Russell JA, Moran NA. (2005). Horizontal transfer of bacterial symbionts: heritability and fitness effects in a novel aphid host. Appl Environ Microbiol 71: 7987–7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream Ma et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Simms EL. (2006). Pathways to mutualism breakdown. Trends Ecol Evol 21: 585–592. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Skophammer RG, Regus JU. (2011). Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA 108(Suppl): 10800–10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Herouart D, Puppo A, Touati D. (2000). Critical protective role of bacterial superoxide dismutase in Rhizobium-legume symbiosis. Mol Microbiol 38: 750–759. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber S, Carlier A, Neuburger M, Grabenweger G, Eberl L, Gademann K. (2015). Isolation and total synthesis of kirkamide, an aminocyclitol from an obligate leaf nodule symbiont. Angew Chem Int Ed Engl 54: 7968–7970. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. (2012). Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol 29: 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto MJ, Domínguez-Ferreras A, Pérez-Mendoza D, Sanjuán J, Olivares J. (2009). Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol 11: 381–388. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 1–2. [DOI] [PMC free article] [PubMed]
- Suyama M, Torrents D, Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonça-Previato L, James EK, Venturi V. (2012). Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb Ecol 63: 249–266. [DOI] [PubMed] [Google Scholar]
- Toft C, Andersson SGE. (2010). Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet 11: 465–475. [DOI] [PubMed] [Google Scholar]
- Toh H, Weiss BL, Perkin SAH, Yamashita A, Oshima K, Hattori M et al. (2006). Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res 16: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M et al. (2008). Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol 148: 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oevelen S, De Wachter R, Vandamme P, Robbrecht E, Prinsen E. (2002). Identification of the bacterial endosymbionts in leaf galls of PsychotriaRubiaceae, angiosperms) and proposal of ‘Candidatus Burkholderia kirkii' sp. nov. Int J Syst Evol Microbiol 52: 2023–2027. [DOI] [PubMed] [Google Scholar]
- Van Oevelen S, Prinsen E, De Wachter R, Robbrecht E. (2001). The taxonomic value of bacterial symbiont identification in African PsychotriaRubiaceae. Syst Geogr 71: 557–563. [Google Scholar]
- Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. (2011). ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol 12: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraete B, Janssens S, Smets E, Dessein S. (2013). Symbiotic ß-proteobacteria beyond legumes: Burkholderia in Rubiaceae. PLoS One 8: e55260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Faber FC. (1912). Das erbliche Zusammenleben von Bakterien und tropischen Pflanzen. Jb wiss Bot 51: 285–375. [Google Scholar]
- Wickham H. (2009) ggplot2: elegant graphics for data analysis. Springer: New York, NY, USA. [Google Scholar]
- Wilson K. (2001) Preparation of genomic DNA from bacteria. In: Current Protocols in Molecular Biology. John Wiley & Sons, Inc: New York, NY, USA. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, dos Reis M. (2011). Statistical properties of the branch-site test of positive selection. Mol Biol Evol 28: 1217–1228. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. (2001). InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848. [DOI] [PubMed] [Google Scholar]
- Zimmermann A. (1902). Uber Bakterienknoten in den Blattern einiger Rubiaceen. Jb wiss Bot 37: 1–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.