Abstract
Ralstonia solanacearum is a globally distributed soil-borne plant pathogenic bacterium, which shares a broad ecological range with many plant- and soil-associated fungi. We sought to determine if R. solanacearum chemical communication directs symbiotic development of polymicrobial consortia. R. solanacearum produced a diffusible metabolite that induced conserved morphological differentiation in 34 species of fungi across three diverse taxa (Ascomycetes, Basidiomycetes and Zygomycetes). Fungi exposed to this metabolite formed chlamydospores, survival structures with thickened cell walls. Some chlamydospores internally harbored R. solanacearum, indicating a newly described endofungal lifestyle for this important plant pathogen. Using imaging mass spectrometry and peptidogenomics, we identified an undescribed lipopeptide, ralsolamycin, produced by an R. solanacearum non-ribosomal peptide synthetase-polyketide synthase hybrid. Inactivation of the hybrid non-ribosomal peptide synthetase-polyketide synthase gene, rmyA, abolished ralsolamycin synthesis. R. solanacearum mutants lacking ralsolamycin no longer induced chlamydospore development in fungal coculture and invaded fungal hyphae less well than wild-type. We propose that ralsolamycin contributes to the invasion of fungal hyphae and that the formation of chlamydospores may provide not only a specific niche for bacterial colonization but also enhanced survival for the partnering fungus.
Introduction
Ralstonia solanacearum is the causative agent of lethal bacterial wilt and is a globally distributed soil-borne plant pathogen known to infect well over 200 different plant hosts (Hayward, 1991). The broad distribution of this bacterium in a wide variety of soil environments suggests that R. solanacearum likely encounters a diversity of other soil microbes, including fungi. Although the molecular interactions between R. solanacearum and host plants have been well studied (Genin and Denny, 2012), there has been little exploration of R. solanacearum's interactions with soil cohabitants.
Soil is a heterogeneous, complex microcosm replete with inter-organismal interactions, which restructure the physical and chemical composition of the shared environment. Small molecule signals are increasingly recognized as important communication signals in microbial interactions (Tarkka et al., 2009). Historically, many of these specialized metabolites (Davies, 2013) have been harnessed for pharmaceutical purposes, and there has been a resurgence of interest in natural products based on microbial crosstalk (Cragg and Newman, 2013). Genome mining of R. solanacearum strain GMI1000 shows that it has the potential to produce a variety of secondary metabolites. A few of these metabolites have been characterized including ralfuranone (Wackler et al., 2011) and the yersinabactin-like siderophore, micacocidin (Kreutzer et al., 2011). Micacocidin has moderate antimycoplasma and antifungal effects in vitro because of its metal chelating properties (Kobayashi et al., 2000). These and other compounds produced by R. solanacearum likely impact how it interacts with microbes in the soil.
Small molecule-mediated antibiosis has been documented in both bacterial and fungal systems (Haas and Défago, 2005; Burlinson et al., 2011), but antagonistic interactions are only one potential interaction outcome. Recent advances in the understanding of bacterial–fungal interactions indicate that bacterial natural products can stimulate morphological transitions in many fungi. In particular, bacterial metabolites commonly induce fungal sporulation. For example, Paenibacillus validus produces trisaccharides that induce hyphal elongation and sporulation in the arbuscular mycorrhizal fungus Glomus intraradices (Hildebrandt et al., 2006). Similarly, the fungal plant pathogen Rhizopus microsporus sporulates only when it is infected with its endosymbiotic bacterial partner, Burkholderia rhizoxinica (Lackner et al., 2011a).
Other research has shown that bacterial volatile organic compounds and diffusible small molecules can induce developmental shifts in fungi aside from sporulation. For example, reciprocal volatile signaling between R. solanacearum and the soil fungus Aspergillus flavus represses A. flavus sporulation and inhibits production of R. solanacearum virulence factors (Spraker et al., 2014). Fengycin A, a lipopeptide produced by Bacillus subtilis, causes diverse fungal taxa to form chlamydospores, thick-walled spores associated with fungal survival in harsh environmental conditions (Li et al., 2012). In addition, phenazine redox metabolites produced by Pseudomonas aeruginosa trigger Aspergillus spp. to sporulate along a diffusion gradient (Zheng et al., 2015). These developmental shifts may contribute to microbial niche securement in the soil by triggering hard-wired defense or survival mechanisms, however, these ecological mechanisms are largely unexplored.
Here we characterize a small molecule-mediated interaction between the common soil-borne bacterial pathogen R. solanacearum and a taxonomic variety of plant- and soil-associated fungi that results in fungal chlamydospore development. Using matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry (MALDI-IMS) and tandem mass spectrometry (MS/MS) technologies, we identified a novel diffusible R. solanacearum lipopeptide, here named ralsolamycin. We observed a developmental shift in all fungi ranging from chlamydospore formation to growth inhibition along an increasing gradient of ralsolamycin. Inactivating rmyA, a gene that encodes a hybrid non-ribosomal peptide synthetase-polyketide synthase abolished ralsolamycin production. Further, the rmyA mutant did not induce chlamydospore formation. Remarkably, we also demonstrate that R. solanacearum invades the chlamydospores, a previously undescribed niche that may contribute to the noted environmental persistence of this devastating plant pathogen.
Materials and methods
Media and growth conditions and coculture experiments
All strains used in this study are listed in Supplementary Table S1. R. solanacearum strains were routinely grown at 30 °C on casamino acids-peptone-glucose (CPG) agar (Hendrick and Sequeira, 1984), and mucoid colonies were selected for subsequent experiments. Liquid bacterial cultures were grown overnight in CPG at 30 °C and 180 r.p.m. When appropriate, the antibiotic gentamycin (15 mg l–1) was added to the media. Overnight liquid cultures of R. solanacearum were pelleted by centrifugation, washed twice in equal volumes of sterilized, double-distilled water, quantified using OD600 values and adjusted to ~2 × 108 cells ml–1. Then 5 μl (1 × 106 cells) of bacterial suspension was spotted at each point.
Aspergillus spp. were routinely grown at 30 °C on glucose minimal medium (Shimizu and Keller, 2001), and conidia were harvested by applying a 0.01% Tween 80 solution to the plate followed by agitation with a plastic cell spreader. Conidial suspensions were quantified using a hemocytometer and diluted in sterile water to 2 × 105 spores ml–1. Then 5 μl containing 1 × 103 spores was applied to a spot 2 cm away from the R. solanacearum spot on CPG plates. The droplets were allowed to dry in a NUAIRE biological safety cabinet (NU425-400), then plates were wrapped with parafilm once and incubated at room temperature. Owing to variable sporulation habits, all other fungi were routinely grown on potato dextrose agar and allowed to grow at room temperature. For assays, mycelial plugs were removed from actively growing cultures using the wide end of a 1 ml pipette tip and transferred to the assay plate.
For coculture experiments, R. solanacearum and fungi were plated as described above and incubated for 14 days at room temperature. Chlamydospore formation for all fungi was assessed as the fungal colonies made contact with the R. solanacearum cultures. As the colonies expanded and grew into one another, sections of the interaction zone were excised for microscopic evaluation. For IMS, experimentation colonies were grown for 3 days at room temperature before being excised and processed. For assessment of endofungal bacteria, green fluorescent protein (GFP) strains of R. solanacearum were similarly cocultured with A. flavus, but the colonies were inoculated in direct contact to allow for physical interactions.
Microscopy of chlamydospores
Fungal mycelia were harvested from coculture plates by cutting 5 mm × 5 mm sections from the interaction zone between the two cultures and examined for chlamydospore formation. Cell walls were stained with calcofluor white at a final concentration of 1 mg ml–1 for 1 min before microscopy. DAPI (4,6-diamidino-2-phenylindole) staining of nuclei was carried out by submerging these sections in 1 mg ml–1 DAPI solution for 30 min followed by a wash in 0.1 m phosphate-buffered saline pH 7.0. Lipid bodies were stained by submerging sections in 10 μg ml–1 Nile Red, followed by a wash with 0.1 m phosphate-buffered saline. Stained samples were wet mounted and imaged on a Zeiss Axio Imager A10 microscope (Carl Zeiss, Oberkochen, Germany) equipped with a Zeiss EC Plan- NEOFLUAR 40 × /1.3 Oil DIC/∞/0.17 and Zeiss EC Plan- NEOFLUAR 100 × /1.3 Oil DIC/∞/0.17 objective and a series 120 X-Cite light source (EXFO). When different filters could be used, samples were stained with multiple stains to provide comparable images for the different features. Images were collected with AxioVision Release 4.7 software (Carl Zeiss).
For counts of chlamydospores of all fungi, interaction zones from coculture with GMI1000 and the rmyA mutant were collected and stained with calcofluor as described above. Three separate images of hyphae collected from these interaction zone were captured and chlamydospores per high-powered field were enumerated manually using the cell-counter plug-in of the Fiji software package (Schindelin et al., 2012).
Time-lapse microscopy experiments were carried out on a Nikon Eclipse Ti inverted fluorescent microscope (Nikon Instruments, Melville, NY, USA) with a PlanFluor 10 × objective attached to a Nikon DS-Qi1Mc CCD camera (Nikon Instruments). Four filters for four different excitation/emission wavelengths were available: ex-350 nm/em-400 nm; ex-492 nm/em-517 nm; ex-572 nm/em-600 nm; ex-647 nm/em-700 nm. Images were collected with NIS-Elements AR version 4.30.01 software (Nikon Instruments).
Confocal laser-scanning microscopy experiments were prepared as above but performed on a Zeiss 510 Meta microscope (Carl Zeiss). To verify endohyphal localization of bacteria, 0.5 μm Z-sections were taken across multiple planes of view. An argon ion laser was used to excite GFP-labeled cells and linear unmixing calculations were carried out in the Zeiss LSM software to reduce fungal autofluorescence. A helium laser was used to excite the FM4-64-labeled membranes. All confocal images were processed in the open source Fiji software package and all overlays of multiple fluorescence channels are of single confocal Z-planes.
Plasmid construction and genetic manipulation
The primers and plasmids used in this work are listed in Supplementary Table S2. PCR amplification was carried out on a C1000 Thermal Cycler from Bio-Rad (Hercules, CA, USA). For creation of the rmyA-targeted disruption cassette, flanking regions of the targeted promoter and fatty acyl-AMP ligase (FAAL) domain were amplified from GMI1000 genomic DNA using primers RSp0641_5′F and RSp0641_5′R for the upstream sequence and RSp0641_3′F and RSp0641_3′R for the downstream sequence, each amplifying ~1-kb fragments. Both RSp0641_5′R and RSp0641_3′F were designed with overlapping sequences to the 0.8-kb gentamycin resistance cassette, Gmr, which was amplified from the plasmid pUCgm using the primer set, GentR_F and GentR_R. These PCR products were purified with a QIAquick gel extraction kit (Qiagen, Valencia, CA, USA) and quantified before being fused using standard double joint PCR protocols (Yu et al., 2004). The fusion product was blunt-end ligated into the Zero Blunt PCR cloning kit (Invitrogen, Carlsbad, CA, USA) to create pJES12.3. R. solanacearum strain GMI1000 was transformed with pJES12.3 by electroporation and plated on gentamycin media to select for appropriate transformants. Colony PCR using RSp0641_5′F and RSp0641_3′R were used to screen for loss of the targeted rmyA promoter and FAAL domain. All PCR steps were carried out with Pfu Ultra II DNA Polymerases (Agilent, Santa Clara, CA, USA).
The GMI1000 strain constitutively expressing GFP was made by using natural transformation to move the Tn5::gfp (Liu et al., 2001) insertion from genomic DNA of strain K60GFP (Yao and Allen, 2006) into the GMI1000 chromosome and transformants were selected on tetracycline plates at a concentration of 15 mg ml–1 and confirmed with fluorescence microscopy. Subsequently, the ΔrmyA strain constitutively expressing GFP was made by using natural transformation to move the Tn5::gfp insertion from the GMI1000-GFP strain. These strains were selected for tetracycline and gentamycin resistance, confirmed with fluorescence microscopy, and verified phenotypically to not induce chlamydospores.
Imaging mass spectrometry
Liquid bacterial cultures were grown overnight in CPG at 30 °C and 180 r.p.m. Overnight liquid cultures of R. solanacearum were pelleted by centrifugation, washed 2 × in equal volumes of sterilized, double-distilled water, quantified using OD600 values and adjusted to 2 × 108 cells ml–1. Then 5 μl (~1 × 106 cells) of this cell suspension was spotted at each point. GMI1000 or K60 (5 μl) were spotted next to A. flavus (1 μl) on yeast extract/malt extract ISP2 agar (10 ml) in 100 × 25 mm Petri dishes (Yang et al., 2012). All organisms were inoculated on separate Petri dishes as controls. Samples were incubated at room temperature for 72 h. The region of interest and controls were excised from the agar and transferred to the same MALDI MSP 96 anchor plate (Bruker Daltonics, Billerica, MA, USA). A photograph was taken and the aerial hyphae of A. flavus were removed gently with a cotton swab dampened with acetonitrile (Moree et al., 2012). Following the removal of the aerial hyphae, another photograph as taken and Universal MALDI matrix (Sigma-Aldrich, St Louis, MO, USA) was applied using a 53 μm stainless steel sieve (Hogentogler & Co., INC., Columbia, MD, USA). The plate was then transferred to an oven and desiccated at 37 °C overnight.
Following adherence to the MALDI plate, another photographic image was taken and the sample was subjected to MALDI-TOF mass spectrometry (Microflex from Bruker Daltonics) for IMS acquisition. Data were acquired in positive reflectron mode, with a 500 μm laser interval in the XY and a mass range of 250–2500 Da. The resulting data were analyzed using FlexImaging software v. 3.0 (Bruker Daltonik GmbH, Bremen, Germany).
Extraction and semi-purification
R. solanacearum 24-h liquid cultures were filter sterilized using a 0.2 μm pore-size filter and applied to actively growing hyphae of A. flavus to determine if direct bacterial contact was necessary for the formation of the chlamydospore-like structures. Within <12 h, a large number of mature hyphal cells differentiated into chlamydospores, indicating that direct physical contact between the microbes was unnecessary for chlamydospore development suggesting diffusible metabolite was responsible for chlamydospore development.
R. solanacearum GMI1000, K60 and A. flavus and their interaction zones were excised from thin agar (1 × 1 cm) and transferred to separate glass scintillation vials. Acetonitrile (1 ml) was added to the samples and sonicated for 10 min. These samples were partitioned against both 50:50 ACN:H2O and butanol and the resulting mixture was centrifuged at 10 000 r.p.m. for 1 min. The soluble organic fraction was removed and dried in vacuo. Samples were then re-suspended in 100 μl of MeOH for further analysis.
Bulk extraction of the metabolite was done from liquid culture by growing GMI1000 for 4 days in 1 liter of CPG with 100 g Amberlite XAD-16 resin (Sigma-Aldrich). The resin was separated from the culture broth via vacuum filtration and rinsed with 500 ml double-distilled H2O to remove excess salts and cellular debris. The adsorbed compounds were eluted in 1:1 MeOH/DCM and organic fractions were dried in vacuo. Subsequently, these fractions were subjected to solid phase extraction using a Waters Sep-Pak C18 cartridge (5 g) eluting with a 20% step gradient of MeOH/H2O, ranging from 20 to 100% MeOH.
MS/MS data acquisition
Samples were directly infused into the mass spectrometer using a Triversa nanomate-electrospray ionization source (Advion Biosystems, Ithaca, NY, USA) coupled to a 6.42 T Thermo LTQ-FT-ICR mass spectrometer (Thermo-Electron Corporation, San Jose, CA, USA). For nanomate, samples were diluted in 50:50 MeOH:H2O with 0.1% formic acid and then directly infused using a back pressure of 0.35–0.5 psi and a spray voltage of 1.3–1.45 kV. FT-MS and ion trap MS/MS spectra were acquired using Tune Plus software version 1.0 and Xcalibur software version 1.4 SR1 (Thermo-Electron Corporation). The instrument was tuned on m/z 816, the 15+ charge state of cytochrome C.
The instrument scan cycle consisted of one 10-min segment, during which a profile FT scan with a resolution of 25 000 was cycled with four data-dependent scans in the ion trap. MS/MS data were collected in a data-dependent manner, during which profile mode FT-MS scans (150–1600 m/z) cycled with four MS/MS scans in the ion trap. The four most abundant peaks were fragmented (2 m/z isolation width, a normalized collision energy of 35%, and an activation time of 30 ms) and then added to an exclusion list for 600 s.
Network analyses
The resulting spectra were converted to mzXML format using msconvert in the ProteoWizard software package (http://proteowizard.sourceforge.net/tools.shtml). The data were then subjected to molecular networking (Watrous et al., 2012). The data were then clustered with MS-Cluster with a parent mass tolerance of 1.0 Da and a MS/MS fragment ion tolerance of 0.3 Da to create consensus spectra. Further, consensus spectra that contained <1 spectrum were discarded. A network was then created where edges were filtered to have a cosine score above 0.65 and >6 matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other's respective top 10 most similar nodes.
Phylogenetic analysis of bacteria
Bacterial 16S DNA sequences were obtained from GenBank and aligned using Muscle in the MEGA6 software package (Tamura et al., 2013) using default parameters. The evolutionary history was inferred using the maximum parsimony method. Tree 1 out of 2 most parsimonious trees (length=1648) is shown. The consistency index is (0.480386), the retention index is (0.730577) and the composite index is 0.372381 (0.350959) for all sites and parsimony-informative sites (in parentheses). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985). The maximum parsimony tree was obtained using the Subtree–Pruning–Regrafting algorithm (Nei and Kumar, 2000) with search level 1 in which the initial trees were obtained by the random addition of sequences (10 replicates). The analysis involved 29 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1211 positions in the final data set.
Results
A diffusible compound produced by R. solanacearum induces chlamydospore development across diverse fungal taxa
R. solanacearum inhabits a variety of soils and infects over 200 plant hosts. Soils and plants are both associated with many different fungal species, so we hypothesized that R. solanacearum utilizes chemical signals to mediate interactions with a diversity of fungi. Using the characterized and sequenced R. solanacearum strain GMI1000, we examined the interaction of cocultures of the bacterium and 34 species of fungi: 30 Ascomycota species, 1 Basidiomycota species (Rhizoctonia solani) and 3 Zygomycota species (Phycomyces blakesleeanus, Mucor bacilliformis and M. hiemalis). We found that a distinct zone of fungal inhibition formed between 2 and 14 days of coculture. In addition, all fungi formed distinct hyphal swellings near (~1.5 cm) the R. solanacearum colony, referred to hereafter as the ‘interaction zone' (Figure 1a).
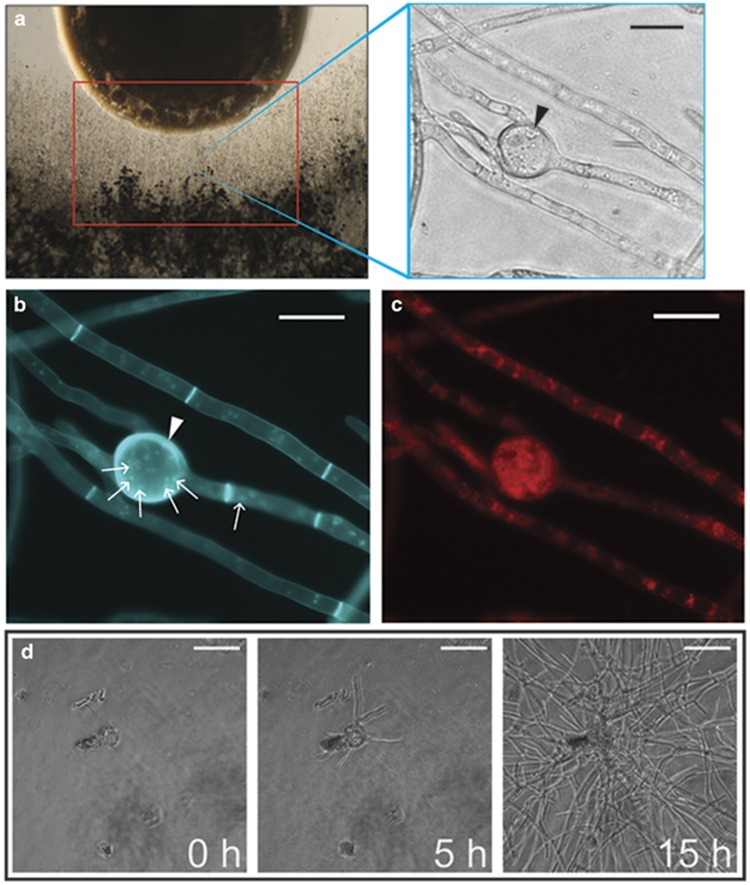
Figure 1.
Examination of the interaction zone of the R. solanacearum/fungal coculture revealed fungal hypertrophy resembling chlamydospores produced by other fungi (a) left—coculture set-up of bacteria (top) and fungi (bottom). Red box indicates the ‘interaction zone' where many intercalated chlamydospore-like structures (right—black arrows) developed in response to coculture with R. solanacearum strain GMI1000. Control images of non-chlamydospore-inducing bacterial cocultures can bee seen in Supplementary Figure S1. Histological analysis of chlamydospores shows that they are (b) polynucleate (small white arrows), have thickened cell walls (white arrowheads) and (c) accumulate lipid bodies. (d) Time-lapse microscopy of isolated chlamydospores placed in fresh media shows these structures are independently viable, and they can germinate to form a new fungal colony. Complete time-lapse video can be viewed in Supplementary Video S1.
The hyphal swellings resembled chlamydospores, thick-walled spores associated with certain fungal genera and thought to aid in fungal survival under harsh environments (Couteaudier and Alabouvette, 1990b; Li et al., 2012). The rates of chlamydospore formation varied considerably between species, but all of the fungi tested formed chlamydospores within 14 days of coculture with R. solanacearum. Several fungi only formed chlamydospores after being in direct contact with the R. solanacearum colony for multiple days: the necrotrophic plant pathogens Botrytis cinerea and Sclerotinia sclerotiorum, the coprophilous fungus Sordaria fimicola, one Fusarium species (verticillioides) and the Basidiomycete Rhizoctonia solani.
To study the hypertrophic structures in detail, the interaction zones of A. flavus were excised and characterized. We chose A. flavus because volatile-mediated interactions between A. flavus and R. solanacearum do not result in chlamydospore formation (Spraker et al., 2014) and also because the genus is generally not known to produce chlamydospores. Micromanipulation and fluorescent histological microscopy confirmed that the hyphal swellings were thick walled, chlamydospores. Hoescht staining showed that most chlamydopsores harbored multiple nuclei (Figure 1b) although some lacked nuclei. A fungal cell wall-specific stain, calcofluor white, demonstrated that chlamydospore cell walls were thicker than normal hyphae cell walls (Figure 1b), and Nile Red staining showed chlamydospores accumulated lipid bodies (Figure 1c). Together, the histological evidence suggested that A. flavus chlamydospores could be viable structures. To test this hypothesis, chlamydospores were isolated via micromanipulation, washed and placed in fresh media and monitored for germination. After 5 h, the chlamydospores germinated with multiple germ tubes (Figure 1d,Supplementary Video S1).
Endofungal lifestyle of R. solanacearum
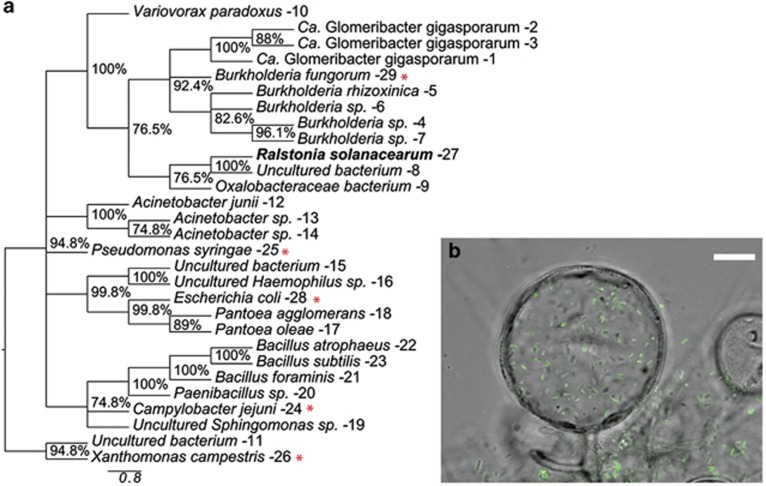
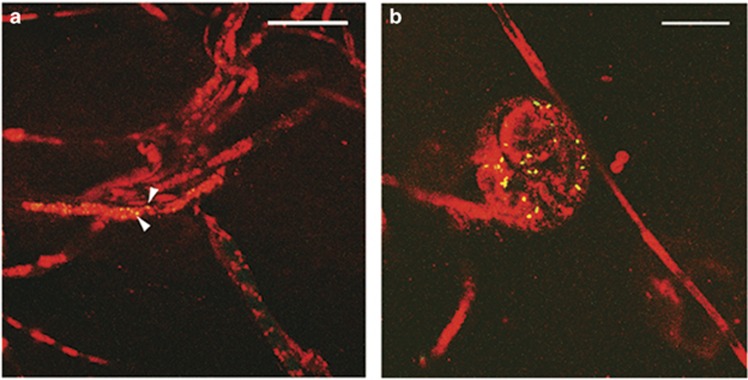
Many bacteria live as endofungal symbionts in a variety of fungal hosts. Ralstonia spp. are nested within a larger clade of identified endofungal bacteria (Hoffman and Arnold, 2010; Figure 2a,Supplementary Table S3). In order to test the hypothesis that R. solanacearum was capable of invading fungal hyphae, a constitutive GFP-producing R. solanacearum strain was cocultured with A. flavus for 72 h. After harvesting the resulting mycelial mat and thoroughly washing the hyphae to remove extracellular bacteria, samples were imaged with a fluorescent microscope. Many bacterial cells remained associated with the fungal hyphae, and bacterial aggregates were often associated with chlamydospores, many appearing to be within the chlamydospores. Confocal laser-scanning microscopy confirmed that a substantial number of bacteria were localized to the intracellular space of some chlamydospores (Figure 2b, Supplementary Video S2). Bacteria were rarely found within the non-differentiated hyphae. These data supported our hypothesis that R. solanacearum can invade fungal hyphae with specificity to the chlamydospores.
Figure 2.
R. solanacearum is nested among other known endofungal bacteria and can be found within fungal cells. (a) Phylogenetic analysis of endofungal bacteria based on maximum parsimony analysis of 16S ribosomal RNA gene sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Branches with <70% support values were collapsed. GenBank accession numbers are listed in Supplementary Table S3. Well-known bacterial species not characterized as endofungal symbionts were inserted for reference and are marked with a red asterisk (*). (b) Confocal laser-scanning microscopy image of GFP-labeled GMI1000 colonizing A. flavus chlamydospore. Scale bar (white) represents 10 um. Z-stack scan can be viewed in Supplementary Video S2.
Identification of the chlamydospore-inducing compound
To identify Ralstonia-specific metabolites that induced chlamydospore formation, we cocultured A. flavus with a phylogenetically diverse set of R. solanacearum strains to identify a non-chlamydospore-producing isolate. Under these conditions, strain GMI1000 was the only isolate to induce substantial chlamydospore formation in A. flavus (Supplementary Figure S1). We selected the non-chlamydospore-inducing R. solanacearum strain, K60 for comparative metabolite studies because like GMI1000, the K60 genome was sequenced and publicly available. GMI1000 and K60 interaction zones were excised at 72 h. At this time point, chlamydospores were plentiful but the cultures were not in physical contact. The zones were coated with universal MALDI matrix and subjected to MALDI-TOF IMS (Yang et al., 2012) to explore the spatial distribution of secreted metabolites and to identify differential signals between these isolates. In total, 22 ions were detected in the bacterial fungal interactions. We attributed production of most metabolites to the bacterium or the fungus based on their spatial distributions with respect to the optical image and their presence in individual cultures.
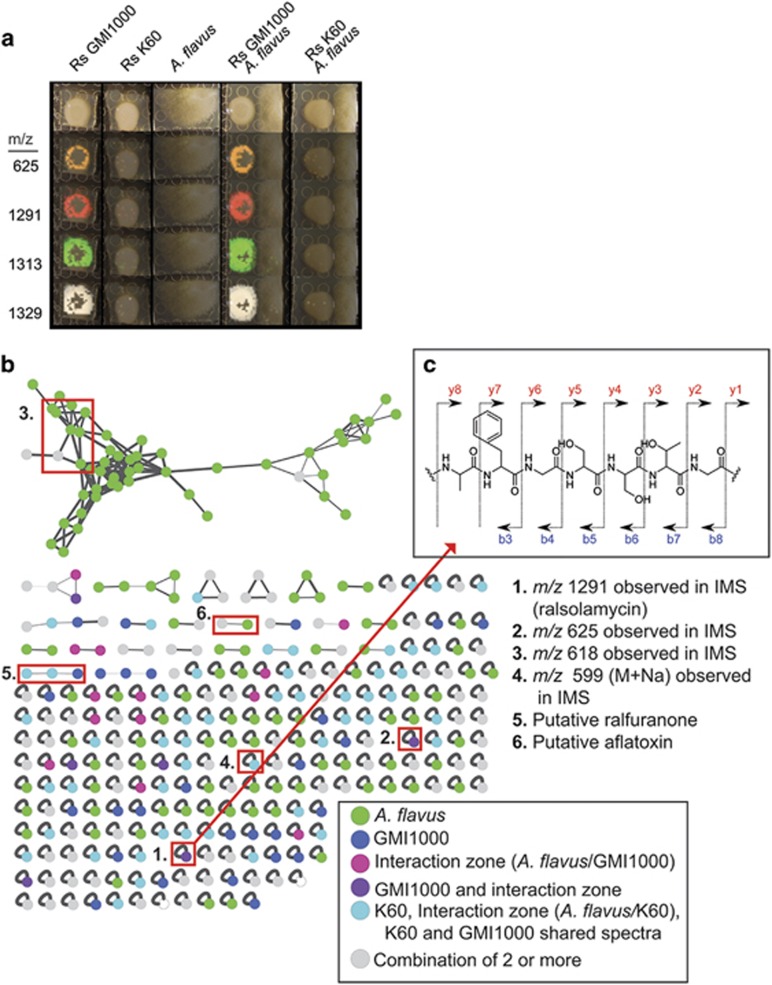
Isolate GMI1000 produced high amounts of four ions that K60 did not produce. Three of these ions clearly diffused beyond the margins of the bacterial colony and reached the margins of the fungal colony. These ions were different ionized forms (protonated, sodiated and potassiated species) of one compound with the exact mass of 1291.7142 (Figure 3a,Supplementary Figure S2). The diffusible nature of this compound suggested it was the chlamydospore-inducing metabolite. This compound was further characterized by tandem mass spectrometry (MS/MS) coupled with molecular networking analysis. The molecular network analyses showed that the fragmentation pattern of this compound was unique to GMI1000 (Figure 3b, Supplementary Figure S3).
Figure 3.
Analysis of differential IMS data sets, MS-MS networks indicate a single lipopeptide produced by R. solanacearum strain GMI1000 is responsible for initiating chlamydospore formation. (a) IMS analysis of strains GMI1000 and K60 showing only differential signals from this data set. Complete data set is in Supplementary Figure S4. (b) Network analyses of microbial metabolites detected in the MS-MS studies. The network is composed of nodes representing ions associated with the microbial colonies. Nodes are connected by edges, which represent the relatedness of the fragmentation patterns of each spectrum. Nodes only associated with A. flavus colonies are green. Nodes only associated with only GMI1000 are dark blue. Nodes associated with the ‘interaction zone' are labeled in pink, Nodes found in the GMI1000 cultures and ‘interaction zone' (A. flavus/GMI1000) are labeled purple. Nodes associated with any of the K60 culture conditions or that are shared between k60 and GMI1000 are colored in light blue. Nodes common to two or more of the previous node classes are labeled in gray. Nodes of interest from the IMS study (1–4) and those that are known products of the microbes (5–6) are highlighted with red boxes and listed on the right of the figure. (c) MS/MS spectra of extracted metabolites shows a unique fragmentation pattern of G-T-S-S-G-F-A for the compound with a m/z of 1291(top right).
Mapping compound to gene cluster and assessing bacterial colonization
Using tandem mass spectrometry, an automated peptidogenomics analysis (Kersten et al., 2011) and the program Pep2Path (Medema et al., 2014), we identified characteristic partial peptide fragmentation patterns associated with the signal of interest and identified one fragment exhibiting the following residue sequence tag: Gly-Thr-Ser-Ser-Gly-Phe-Ala (Figure 3b). Although this sequence tag does not explain the entire mass of the metabolite, it does strongly indicate that a non-ribosomal peptide synthase (NRPS) was involved in the synthesis of this compound. Using the in silico analysis capabilities of Pep2Path, in combination with AntiSMASH (Medema et al., 2011), we examined the R. solanacearum GMI1000 and K60 genomes to identify putative secondary metabolic gene clusters where a major criterion was to find NRPS clusters unique to the GMI1000 genome that matched the partial sequence tag described above.
One such gene cluster contained two large genes encoding a non-ribosomal peptide synthetase-polyketide synthase hybrid (RSp0641) and a NRPS (RSp0642). Several lines of evidence suggested this gene cluster produced the chlamydospore-inducing metabolite: these genes were absent from the K60 genome, and the domain architecture of both RSp0641 and RSp0642 predicted a product best matched to the 1291.7 m/z of the signal identified in the IMS studies. In addition, the NRPS adenylation domains were predictive of the amino-acid residues detected in our MS/MS studies (Supplementary Table S4). We termed this new compound ralsolamycin and named RSp0641 and RSp0642 ralsolamycin synthase genes rmyA and rmyB, respectively (Figure 4a). The protein products of rmyA-rmyB appear to a large secondary metabolite megasynthase containing both PKS and NRPS domains (Figure 4b).
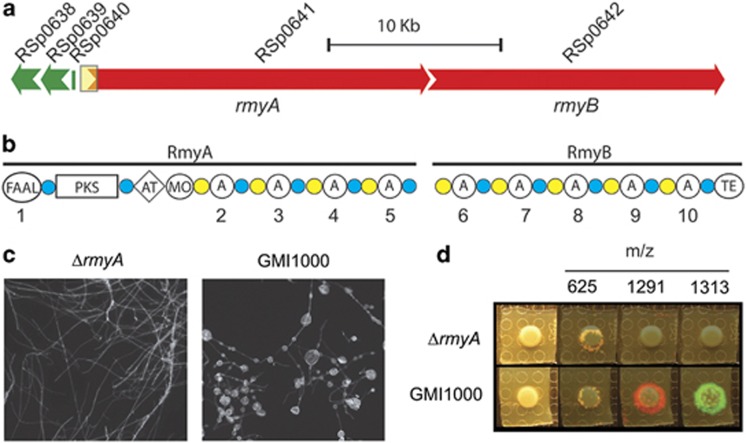
Figure 4.
(a) Graphical representation of the predicted biosynthetic gene cluster as predicted by comparative genomics. Yellow box indicates the region (putative promoter sequence and FAAL encoding sequence) targeted for creation of the rmyA mutant using a gentamycin resistance cassette. Genes and their closest protein homologs are listed in Supplementary Table S6. (b) Predicted domain architecture of the hybrid PKS megasynthases RmyA and RmyB. FAAL, fatty acid acyl ligase domain; PKS, polyketides synthase domain; AT, aminotransferase domain; MO, monooxygenase domain; A, adenylation domain; TE, thioesterase domain; blue circles, phosphopantetheine attachment site; yellow circles, condensation domains. Adenylation domains are numbered for reference to their predicted substrates listed in Supplementary Table S4. (c) rmyA mutant does not induce fungal chlamydospores. (d) rmyA mutant does not produce the ralsolamycin signals (m/z of 1291 and 1313) but does produce the other GMI1000-specific compound (m/z 625).
In silico analysis using a PKS-NRPS predictor (Bachmann and Ravel, 2009) and domain-specific BLAST searches, suggested that RmyA contains an initiating FAAL domain at its N-terminus followed by a keto-synthase domain, and two acetyl ornithine aminotransferase domains coupled to luciferase-like monooxygenases. The remainder of the rmyA gene product contains four canonical NRPS modules containing adenylation (A), peptide carrier protein (PCP) and condensation (C) domains. The predicted peptide product of rmyA terminates with a PCP domain. Conventional knowledge of PCP domains suggests that the product would remain tethered to RmyA. Similar analysis of RmyB showed that it initiates with a C domain, suggesting that RmyB functionally associates with the C-terminal PCP domain of RmyA. Further, rmyB encodes five NRPS modules, ending with a predicted thioesterase (TE) domain. We predict the TE domain cyclizes and releases the final product. The FAAL and PKS domains of RmyAB suggest the product is a lipopeptide (a fatty acid-like moiety attached to an amino-acid chain). Supplementary Table S4 lists all predicted domain substrates and their closest identified homologs.
We disrupted the putative promoter and FAAL domain of rmyA (Liu et al., 2013). The rmyA mutant had significantly reduced chlamydospore-inducing activity (P<0.05) in 28 of the 34 assayed fungal cocultures (Table 1), including A. flavus (Figure 4c), and lost all ralsolamycin IMS signals (Figure 4d). Furthermore, addition of semi-purified ralsolamycin to A. flavus triggers chlamydospore formation (Supplementary Figure S4). These data show that rmyA contributes to ralsolamycin biosynthesis and that ralsolamycin induces fungal chlamydospore formation.
Table 1. Chlamydospore counts per high-power field (magnification: x200) from interactions with R. solanacearum strains GMI1000 and ΔrmyA.
| Fungal species |
GMI1000 |
ΔrmyA |
P-value | ||
|---|---|---|---|---|---|
| Mean | s.d. | Mean | s.d. | ||
| Alternaria alternataa | 28.00 | 8.49 | 0.33 | 0.47 | 1.00E–02 |
| Alternaria solani | 88.00 | 47.77 | 12.67 | 5.25 | 9.09E–02 |
| Aspergillus aculeatusa | 80.00 | 17.57 | 6.33 | 2.87 | 4.25E–03 |
| Aspergillus brasiliensisa | 136.00 | 24.39 | 85.33 | 8.26 | 4.97E–02 |
| Aspergillus carbonariusa | 36.00 | 11.52 | 4.33 | 1.70 | 1.84E–02 |
| Aspergillus clavatusa | 97.33 | 22.60 | 0.00 | 0.00 | 3.68E–03 |
| Aspergillus flavusa | 105.00 | 17.91 | 0.00 | 0.00 | 1.15E–03 |
| Aspergillus fumigatusa | 11.33 | 0.47 | 0.00 | 0.00 | 4.46E–06 |
| Aspergillus nidulansa | 42.33 | 2.05 | 0.00 | 0.00 | 8.26E–06 |
| Aspergillus nigera | 9.67 | 2.87 | 0.33 | 0.47 | 1.05E–02 |
| Aspergillus oryzaea | 48.33 | 4.64 | 1.00 | 0.82 | 1.43E–04 |
| Aspergillus terreusa | 207.00 | 62.98 | 5.33 | 1.70 | 1.06E–02 |
| Aspergillus tubingensisa | 32.33 | 11.47 | 7.67 | 0.94 | 3.87E–02 |
| Aspergillus zonatusa | 126.00 | 58.76 | 0.00 | 0.00 | 3.87E–02 |
| Botrytis cinerea | 2.33 | 1.25 | 0.00 | 0.00 | 5.72E–02 |
| Colletotrichum graminicolaa | 58.67 | 18.26 | 16.67 | 5.31 | 3.54E–02 |
| Fusarium fujikuroia | 31.33 | 3.09 | 1.33 | 0.47 | 1.71E–04 |
| Fusarium graminearuma | 180.67 | 39.85 | 0.67 | 0.94 | 3.09E–03 |
| Fusarium oxysporuma | 47.33 | 4.78 | 1.00 | 1.41 | 1.94E–04 |
| Fusarium solania | 21.67 | 8.73 | 0.00 | 0.00 | 2.47E–02 |
| Fusarium sporotrichioidesa | 40.33 | 13.89 | 12.00 | 3.74 | 4.95E–02 |
| Fusarium verticillioidesa | 16.00 | 7.07 | 0.00 | 0.00 | 3.29E–02 |
| Morchella esculentaa | 21.00 | 4.55 | 0.00 | 0.00 | 2.84E–03 |
| Mucor bacilliformis | 58.00 | 30.01 | 17.67 | 2.87 | 1.31E–01 |
| Mucor hiemalisa | 26.00 | 3.56 | 0.67 | 0.47 | 5.67E–04 |
| Neosartorya fischeria | 51.00 | 15.77 | 0.67 | 0.94 | 1.08E–02 |
| Penicillium italicuma | 102.00 | 32.26 | 1.67 | 1.25 | 1.17E–02 |
| Phycomyces blakesleeanusa | 6.67 | 1.70 | 0.00 | 0.00 | 5.17E–03 |
| Rhizoctonia solani | 27.00 | 11.86 | 7.33 | 8.99 | 1.35E–01 |
| Sclerotinia sclerotioruma | 84.00 | 11.86 | 5.67 | 3.09 | 8.30E–04 |
| Sordaria fimicolaa | 102.33 | 6.65 | 22.00 | 4.55 | 1.47E–04 |
| Trichoderma hazarianum | 64.00 | 38.74 | 0.00 | 0.00 | 7.97E–02 |
| Trichothecium roseum | 24.33 | 9.88 | 7.33 | 3.30 | 8.22E–02 |
| Verticillium albo-atruma | 173.00 | 37.98 | 10.00 | 0.82 | 3.73E–03 |
Indicates strains with significantly greater (P<0.05) chlamydospore counts in coculture with GMI1000 relative to strain ΔrmyA. Mean, s.d. and P-values are listed to show variations between strains.
Ralsolamycin contributes to bacterial invasion of fungal cells
As we saw an abundance of R. solanacearum cells internalized in the chlamydospores but not in undifferentiated hyphae, we hypothesized that ralsolamycin is necessary for invasion of fungal tissues. To test this hypothesis, GFP-producing GMI1000 wild-type and rmyA mutant were cocultured with A. flavus. Samples were examined with confocal microscopy using amphiphilic styryl dye FM4-64 as a membrane counterstain. The ΔrmyA bacteria rarely invaded the hyphae. In contrast, the wild-type cells were abundant in the chlamydospores (Figure 5a). These findings suggest that ralsolamycin enhances R. solanacearum's invasion of fungal tissues with specificity to chlamydospores.
Figure 5.
Confocal laser-scanning microscopy shows that (a) the rmyA disruption does not completely inhibit bacterial invasion of fungal hyphae (arrowheads indicate GFP-expressing bacteria) but (b) much larger populations of wild-type bacteria can be found in chlamydospores. White bar indicates 10 μm.
Discussion
Fungal chlamydospore development in response to ralsolamycin
Many fungi produce chlamydospores under adverse conditions. However, because not all fungi are known to produce chlamydospores, traditional taxonomic studies use these structures as a trait to delineate evolutionary relationships. Our histological data suggest that chlamydospores formed in response to ralsolamycin are similar to those described in the literature (van Eck, 1978). We provide the first evidence of chlamydospores formation in many of these fungi including all of the Aspergillus species tested. Our results also indicate that these structures may be more common across the kingdom Fungi than is currently recognized.
The finding that all tested Aspergillus species can produce chlamydospores was unexpected as only A. parasiticus chlamydospores have been putatively described (Abou-Gabal and Fagerland, 1981). These data suggest a novel mode of persistence for this well-studied genus. Although most of the assayed fungi formed abundant chlamydospores in response to ralsolamycin, a subset formed relatively few chlamydospores (Table 1). We have not ruled out that these taxa may form chlamydospores more abundantly under other conditions. Perhaps differences in plasma membrane composition or signaling cascades among these fungi temper the conserved chlamydospore formation response to ralsolamycin. Alternatively, these fungi may respond to specific cues shown to affect chlamydospore formation in other fungi including nutrients (Barran et al., 1977; Ohara and Tsuge, 2004), light (Kües et al., 1998), osmolarity (Eisman et al., 2006), pH (Regúlez et al., 1980), air (Barran et al., 1977), temperature (Ohara and Tsuge, 2004) and other interkingdom signals (Li et al., 2005; Staib and Morschhäuser, 2005; Li et al., 2012). Regardless of quantity, the finding that all of the fungi developed chlamydospores when exposed to ralsolamycin supports a conserved differentiation process in the kingdom Fungi in response to this lipopeptide.
Endofungal lifestyle of R. solanacearum
Our data support the hypothesis that R. solanacearum colonizes fungal cells, and that formation of the chlamydospores facilitates bacterial entry. Recently, diverse fungal lineages have been shown to harbor endofungal bacteria, most of which are Gram-negative taxa (Hoffman and Arnold, 2010). Of these related genera, Burkholderia rhizoxinica and other closely related species have been particularly well studied for their endofungal lifestyle and their role in the production of bioactive secondary metabolites (Partida-Martinez et al., 2007; Lackner et al., 2009; Lackner et al., 2011a, b). Our data analyzing ΔrmyA and GMI1000 suggest that although ralsolamycin production is not essential for bacterial invasion of fungal hyphae, it does contribute significantly to this process and/or that chlamydospores may provide a more suitable site for bacterial growth upon invasion.
Many bacterial lipopeptides such as iturin (Maget-Dana and Peypoux, 2003), fengycin (Patel et al., 2011), lichenysin (Madslien et al., 2013) and syringomycin (Hutchison et al., 1995) form pores and permeabilize membranes. We presume that ralsolamycin-mediated membrane restructuring contributes to the initiation of chlamydospore formation and may facilitate the passage of R. solanacearum into fungal cells. R. solanacearum possesses other factors that could contribute to fungal endosymbioses such as chitinolytic enzymes, known to be important in fungal cell wall reorganization (Adams, 2004), and genes for all of the major Gram-negative protein secretion systems (Genin and Boucher, 2004). The R. solanacearum type III secretion machinery shows significant conservation of gene order and primary sequence to that of B. rhizoxinica, a system that has been shown to be important for maintaining a stable endofungal symbiosis of B. rhizoxinica with the fungus R. microsporus (Lackner et al., 2011a) and possibly a similar mechanism contributes to R. solanacearum invasion of fungal cells.
R. solanacearum is known to persist in soils for remarkably long periods of time relative to other bacterial plant pathogens (Álvarez et al., 2010). The traits underlying environmental persistence are poorly studied, but external factors such as non-host reservoir plants, permissive soil temperatures and high soil moisture content may contribute to persistence. Our findings suggest that R. solanacearum colonization within chlamydospores is a novel persistence mechanism for bacterial survival in the absence of host plants. The large numbers of bacteria within colonized chlamydospores indicates that they multiply within chlamydospores, which may contribute to increasing inoculum load once plants are introduced. The chlamydospores contain substantial amounts of lipid droplets, which could contribute to bacterial replication in these specialized structures. Currently, little is known regarding nutritional interactions between bacterial/fungal symbioses (Burlinson et al., 2011) and efforts toward elucidating the nutritional interactions and survival outcomes of the R. solanacearum/chlamydospore complex are currently underway in our lab.
As homologs of the ralsolamycin biosynthetic cluster are present in a subset of the R. solanacearum strains sequenced to date, it is intriguing to consider that the invasion of chlamydospores may be important to these strains. We did not find any phylogenetic or geographic trends that suggest that ralsolamycin biosynthesis is restricted to a single phylotype, geographic locale or host plant (Supplementary Table S5); however, as more isolates are sequenced a pattern of occurrence may be uncovered. In addition, CMR15 homologs of RmyA and RmyB showed 94% and 93% identity although this strain did not induce chlamydospore formation. These data suggest that either this cluster makes a different metabolite; that the genes do not produce functional proteins; or that the cluster may be differentially regulated under the conditions tested here. Continued research into these strain differences as well as increasing genomic information will help elucidate the impact of ralsolamycin biosynthesis on R. solanacearum biology.
Ralsolamycin identification, characterization and biosynthetic genes
The combination of spatial data from our IMS experiments and the MS/MS network analysis indicates a single compound, ralsolamycin, is largely responsible for the bacterial induction of fungal chlamydospore formation. Multiple in silico analyses tools indicate that ralsolamycin is produced by the megasynthase enzyme encoded by the genes rmyA-rmyB. These two backbone genes, rmyA (a PKS-NRPS hybrid) and rmyB (a NRPS), are the two largest ORFs in the GMI1000 genome, approximately 20.7 and 17.9 kb, respectively. The bipartite genomes of strains GMI1000 and K60 were analyzed via AntiSMASH (Medema et al., 2011) to look for canonical backbone secondary metabolite genes. This analysis identified 19 putative secondary metabolic clusters in GMI1000. Of these, only three were predicted to contain putative NRPSs: the micacocidin-producing RSc1806-1811 (Kreutzer et al., 2011), the ralfuranone-producing RSp1419-1434 (Wackler et al., 2011) and the ralsolamycin-producing RSp0638-0642 cluster characterized here. Identification of the 1290 Da metabolite by IMS as the key metabolite was supported by the Pep2Path program, which can link NRPS-products to their biosynthetic genes based on predictions of adenylation substrate recognition domains (Medema et al., 2014). This program mapped the partial amino-acid sequence tag G-T-S-S-G back to the hybrid non-ribosomal peptide synthetase-polyketide synthase backbone gene RSp0641-0642, which encodes 10 predicted adenylation domains in total.
The domain architecture of RmyA is very similar to that responsible for the production of the antifungal lipopeptide mycosubtilin, MycA, produced by Bacillus subtilis (Duitman et al., 1999). The presence of the N-terminal FAAL domain followed by a keto-synthase and aminotransferase domain is a known motif in bacterial lipopeptide biosynthesis (Du et al., 2001). These domains cooperate to convert carboxylic acid moieties into activated β-amino acids, which are utilized by subsequent NRPS modules. The remaining modules in RmyA-B appear to have a canonical NRPS adenylation, peptide carrier, condensation domain structure suggesting normal non-ribosomal peptide biosynthesis beyond the initial FAAL domain. We are currently working toward complete characterization of this compound to verify its amino-acid composition.
Although it appears that RmyA and RmyB constitute a single operon, examination of this region suggests that there may be up to five genes involved in its biosynthesis and transport including rmyA, rmyB and three additional genes: RSp0638, RSp0639 and RSp0640 (Figure 4a) encoding a transmembrane cyclic peptide transporter, a metallo-beta-lactamase and an MbtH-like protein, respectively (Figure 5b,Supplementary Table S6). Comparative analysis of the available R. solanacearum strain genomes showed that these five genes are located within a copper resistance cluster common to most R. solanacearum strains. Although we have not verified the functions of RSp0638-40, we predict that they participate in the production and transport of ralsolamycin as they are all common components of secondary metabolic clusters.
Conclusions
We have demonstrated that the bacterial plant pathogen R. solanacearum strain GMI1000 induces chlamydospore formation in a wide range of fungal taxa, many of which were not known to produce chlamydospores. We used IMS to rapidly identify potential mediators of this diffusible interaction phenotype. Tandem mass spectrometry coupled with in silico peptidogenomics tools and molecular genetics identified a putative R. solanacearum lipopeptide, ralsolamycin and the PKS-NRPS responsible for its production.
We provide the first evidence that R. solanacearum can induce and invade specialized fungal cells, chlamydospores, which may have significant implications for environmental persistence of R. solanacearum. Chlamydospores enhance persistence of soil-borne fungi and related organisms (Couteaudier and Alabouvette, 1990a; Crone et al., 2013) and endosymbiotic mutualisms provide survival benefits (Nikoh et al., 2014; O'Connor et al., 2014). R. solanacearum has an unusually long survival time in soil, even when host plants are removed (Álvarez et al., 2010). Our data suggest ralsolamycin-induced chlamydospores contributes to bacterial invasion of fungal cells, but ralsolamycin is not the sole factor contributing to this process. We speculate that endofungal colonization benefits R. solanacearum survival in soil in the absence of plant–host tissue, and R. solanacearum co-opts the accumulated lipids in chlamydospores. Further research in our lab aims to determine if this symbiotic phenotype impacts the survival of R. solanacearum and if there is any impact on the fungi it interacts with.
Acknowledgments
This research was funded by an NSF Graduate Research Fellowship under grant no. DGE-1256259 to JES and in part by National Science Foundation Grant Emerging Frontiers in Research and Innovation 1136903 to NPK. This work was supported in part by GM097509 (PCD). PCD further acknowledges Bruker and NIH grant GMS10RR029121 for the support of the shared instrumentation infrastructure that enabled this work. LMS is supported by National Institutes of Health IRACDA K12 GM068524 grant award. TML is funded by NIH National Research Service Award T32 GM07215 and by Agriculture and Food Research Initiative Competitive grant # 2015-67011-22799 from the USDA National Institute of Food and Agriculture. We appreciate GFP-labeled GMI1000 from Tuan Tran and critical feedback from Dr Caitilyn Allen, both of the UW-Madison Department of Plant Pathology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation. Confocal laser-scanning microscopy work was done at the UW-Madison Plant Imaging Center with the help of Sara Swanson.
Author contributions
The majority of the experiments were designed and executed by JES under the guidance of NPK. The IMS and chemical characterization of ralsolamycin was done by LMS under the guidance of PCD. R. solanacearum transformations were done by JES and TML.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abou-Gabal M, Fagerland J. (1981). Ultrastructure of the chlamydospore growth phase of Aspergillus parasiticus associated with higher production of aflatoxins. Mykosen 24: 307–311. [DOI] [PubMed] [Google Scholar]
- Adams DJ. (2004). Fungal cell wall chitinases and glucanases. Microbiology 150: 2029–2035. [DOI] [PubMed] [Google Scholar]
- Álvarez B, Biosca EG, López MM. (2010). On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen. Technol Educ Top Appl Microbiol Microb Biotechnol 1: 267–279. [Google Scholar]
- Bachmann BO, Ravel J. (2009) Chapter 8: Methods for In Silico Prediction of Microbial Polyketide and Nonribosomal Peptide Biosynthetic Pathways from DNA Sequence Data, 1st edn. Academic Press: San Diego, CA, USA doi:10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- Barran L, Schneider E, Seaman WL. (1977). Requirements for the rapid conversion of macroconidia of Fusarium sulphureum to chlamydospores. Can J Microbiol 23: 148–151. [DOI] [PubMed] [Google Scholar]
- Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A. (2011). Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev 75: 583–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteaudier Y, Alabouvette C. (1990. a). Quantitative comparison of Fusarium oxysporum competitiveness in relation with carbon utilization. FEMS Microbiol Ecol 74: 261–268. [Google Scholar]
- Couteaudier Y, Alabouvette C. (1990. b). Survival and inoculum potential of conidia and chlamydospores of Fusarium oxysporum f.sp. lini in soil. Can J Microbiol 36: 551–556. [DOI] [PubMed] [Google Scholar]
- Cragg GM, Newman DJ. (2013). Natural products: a continuing source of novel drug leads. Biochim Biophys Acta - Gen Subj 1830: 3670–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone M, McComb JA, O'Brien PA, Hardy GESJ. (2013). Survival of Phytophthora cinnamomi as oospores, stromata, and thick-walled chlamydospores in roots of symptomatic and asymptomatic annual and herbaceous perennial plant species. Fungal Biol 117: 112–123. [DOI] [PubMed] [Google Scholar]
- Davies J. (2013). Specialized microbial metabolites: functions and origins. J Antibiot (Tokyo) 66: 361–364. [DOI] [PubMed] [Google Scholar]
- Du L, Sánchez C, Shen B. (2001). Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab Eng 3: 78–95. [DOI] [PubMed] [Google Scholar]
- Duitman EH, Hamoen LW, Rembold M, Venema G, Seitz H, Saenger W et al. (1999). The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc Natl Acad Sci USA 96: 13294–13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisman B, Román E, Arana D, Pla J, Roma E, Nombela C. (2006). The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell 5: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Genin S, Boucher C. (2004). Lessons learned from the genome analysis of Ralstonia solanacearum. Annu Rev Phytopathol 42: 107–134. [DOI] [PubMed] [Google Scholar]
- Genin S, Denny TP. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol 50: 67–89. [DOI] [PubMed] [Google Scholar]
- Haas D, Défago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3: 307–319. [DOI] [PubMed] [Google Scholar]
- Hayward A. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 29: 65–87. [DOI] [PubMed] [Google Scholar]
- Hendrick CA, Sequeira L. (1984). Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl Environ Microbiol 48: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt U, Ouziad F, Marner FJ, Bothe H. (2006). The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiol Lett 254: 258–267. [DOI] [PubMed] [Google Scholar]
- Hoffman MT, Arnold AE. (2010). Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microbiol 76: 4063–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison ML, Tester MA, Gross D. (1995). Role of biosurfactant and ion channel-forming activities of syringomycin in transmembrane ion flux: a model for the mechanism of action in the plant-pathogen interaction. Mol Plant Microbe Interact 8: 610–620. [DOI] [PubMed] [Google Scholar]
- Kersten RD, Yang Y-L, Xu Y, Cimermancic P, Nam S-J, Fenical W et al. (2011). A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol 7: 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Ikenishi Y, Takinami Y, Takema M, Sun WY, Ino A et al. (2000). Preparation and antimicrobial activity of micacocidin. J Antibiot (Tokyo) 53: 532–539. [DOI] [PubMed] [Google Scholar]
- Kreutzer MF, Kage H, Gebhardt P, Wackler B, Saluz HP, Hoffmeister D et al. (2011). Biosynthesis of a complex yersiniabactin-like natural product via the mic locus in phytopathogen Ralstonia solanacearum. Appl Environ Microbiol 77: 6117–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües U, Granado JD, Hermann R, Boulianne RP, Kertesz-Chaloupková K, Aebi M. (1998). The A mating type and blue light regulate all known differentiation processes in the basidiomycete Coprinus cinereus. Mol Gen Genet 260: 81–91. [DOI] [PubMed] [Google Scholar]
- Lackner G, Moebius N, Hertweck C. (2011. a). Endofungal bacterium controls its host by an hrp type III secretion system. ISME J 5: 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. (2011. b). Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics 12: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner G, Partida-Martinez LP, Hertweck C. (2009). Endofungal bacteria as producers of mycotoxins. Trends Microbiol 17: 570–576. [DOI] [PubMed] [Google Scholar]
- Li L, Ma M, Huang R, Qu Q, Li G, Zhou J et al. (2012). Induction of chlamydospore formation in Fusarium by cyclic lipopeptide antibiotics from Bacillus subtilis C2. J Chem Ecol 38: 966–974. [DOI] [PubMed] [Google Scholar]
- Li L, Qu Q, Tian B, Zhang KQ. (2005). Induction of chlamydospores in Trichoderma harzianum and Gliocladium roseum by antifungal compounds produced by Bacillus subtilis C2. J Phytopathol 153: 686–693. [Google Scholar]
- Liu H, Kang Y, Genin S, Schell M A, Denny TP. (2001). Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147: 3215–3229. [DOI] [PubMed] [Google Scholar]
- Liu Z, Ioerger TR, Wang F, Sacchettini JC. (2013). Structures of Mycobacterium tuberculosis FadD10 protein reveal a new type of adenylate-forming enzyme. J Biol Chem 288: 18473–18483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madslien EH, Rønning HT, Lindback T, Hassel B, Andersson MA, Granum PE. (2013). Lichenysin is produced by most Bacillus licheniformis strains. J Appl Microbiol 115: 1068–1080. [DOI] [PubMed] [Google Scholar]
- Maget-Dana R, Peypoux F. (2003). Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology 87: 151–174. [DOI] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, De Jager V, Zakrzewski P, Fischbach MA et al. (2011). AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Paalvast Y, Nguyen DD, Melnik A, Dorrestein PC, Takano E et al. (2014). Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput Biol 10: e1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree WJ, Phelan V V, Wu C, Bandeira N, Cornett DS, Duggan BM. (2012). Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc Natl Acad Sci USA. 109: 13811–13816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. (2000) Molecular Evolution and Phylogenetics. Oxford University Press: New York. [Google Scholar]
- Nikoh N, Hosokawa T, Moriyama M, Oshima K, Hattori M, Fukatsu T. (2014). Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci USA 111: 10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Tsuge T. (2004). FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, fungal plant pathogen Fusarium oxysporum. Eukaryot Cell 3: 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor RM, Fung JM, Sharp KH, Benner JS, McClung C, Cushing S et al. (2014). Gill bacteria enable a novel digestive strategy in a wood-feeding mollusk. Proc Natl Acad Sci USA 111: E5096–E5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Martinez LP, Groth I, Schmitt I, Richter W, Roth M, Hertweck C. (2007). Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporous. Int J Syst Evol Microbiol 57: 2583–2590. [DOI] [PubMed] [Google Scholar]
- Patel H, Tscheka C, Edwards K, Karlsson G, Heerklotz H. (2011). All-or-none membrane permeabilization by fengycin-type lipopeptides from Bacillus subtilis QST713. Biochim Biophys Acta - Biomembr 1808: 2000–2008. [DOI] [PubMed] [Google Scholar]
- Regúlez P, Pontón J, Domínguez JB, Goñi FM, Uruburu F. (1980). Lipid composition and the transition from yeast-like to chlamydospore cells of Pullularia pullulans. Can J Microbiol 26: 1428–1437. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Keller NP. (2001). Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraker JE, Jewell K, Roze L V, Scherf J, Ndagano D, Beaudry R et al. (2014). A volatile relationship: profiling an inter-kingdom dialogue between two plant pathogens, Ralstonia solanacearum and Aspergillus flavus. J Chem Ecol 40: 502–513. [DOI] [PubMed] [Google Scholar]
- Staib P, Morschhäuser J. (2005). Liquid growth conditions for abundant chlamydospore formation in Candida dubliniensis. Mycoses 48: 50–54. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkka MT, Sarniguet A, Frey-Klett P. (2009). Inter-kingdom encounters: recent advances in molecular bacterium-fungus interactions. Curr Genet 55: 233–243. [DOI] [PubMed] [Google Scholar]
- van Eck WH. (1978). Lipid body content and persistence of chlamydospores of Fusarium solani in soil. Can J Microbiol 24: 65–69. [DOI] [PubMed] [Google Scholar]
- Wackler B, Schneider P, Jacobs JM, Pauly J, Allen C, Nett M et al. (2011). Ralfuranone biosynthesis in Ralstonia solanacearum suggests functional divergence in the quinone synthetase family of enzymes. Chem Biol 18: 354–360. [DOI] [PubMed] [Google Scholar]
- Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY. (2012). Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA 109: 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Phelan V V, Simkovsky R, Watrous JD, Trial RM, Fleming TC et al. (2012). Primer on agar-based microbial imaging mass spectrometry. J Bacteriol 194: 6023–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Allen C. (2006). Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J Bacteriol 188: 3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41: 973–981. [DOI] [PubMed] [Google Scholar]
- Zheng H, Kim J, Liew M, Yan JK, Herrera O, Bok JW et al. (2015). Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus. Curr Biol 25: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.