Abstract
Microbes are well-recognized members of the coral holobiont. However, little is known about the short-term dynamics of mucus-associated microbial communities under natural conditions and after disturbances, and how these dynamics relate to the host's health. Here we examined the natural variability of prokaryotic communities (based on 16S ribosomal RNA gene amplicon sequencing) associating with the surface mucus layer (SML) of Porites astreoides, a species exhibiting cyclical mucus aging and shedding. Shifts in the prokaryotic community composition during mucus aging led to the prevalence of opportunistic and potentially pathogenic bacteria (Verrucomicrobiaceae and Vibrionaceae) in aged mucus and to a twofold increase in prokaryotic abundance. After the release of aged mucus sheets, the community reverted to its original state, dominated by Endozoicimonaceae and Oxalobacteraceae. Furthermore, we followed the fate of the coral holobiont upon depletion of its natural mucus microbiome through antibiotics treatment. After re-introduction to the reef, healthy-looking microbe-depleted corals started exhibiting clear signs of bleaching and necrosis. Recovery versus mortality of the P. astreoides holobiont was related to the degree of change in abundance distribution of the mucus microbiome. We conclude that the natural prokaryotic community inhabiting the coral SML contributes to coral health and that cyclical mucus shedding has a key role in coral microbiome dynamics.
Introduction
Corals live in a well-described mutualism with photoautotrophic endosymbiotic dinoflagellates of the genus Symbiodinium, frequently referred to as zooxanthellae (reviewed by Muscatine, 1990). More recently, the concept of the coral holobiont (Rohwer et al., 2002) has been proposed to describe the association of the coral host with a diverse microbial community including representatives of fungi, endolithic algae, bacteria and archaea, of which certain associations are species-specific (Ritchie and Smith, 1997; Rohwer et al., 2002; Koren and Rosenberg, 2006; Carlos et al., 2013). The sum of these microorganisms and their combined genetic material forms the coral microbiome, whose core composition is determined by the host but whose presence allows for metabolic adaptations to local environmental conditions by selection of beneficial genes (Kelly et al., 2014; Ainsworth et al., 2015).
The putative functions of the coral microbiome comprise, among others, the protection against pathogens (Rohwer et al., 2002; Shnit-Orland and Kushmaro, 2009) and the supply and cycling of nutrients (Lesser et al., 2004; Wegley et al., 2007). Gates and Ainsworth (2011) propose that all taxonomic components should be considered as important because of their potential to stimulate the holobiont's functioning. However, the extent of the microbiome's contribution to the health of the coral holobiont and to coral reef resilience remains largely unknown. Whereas the association between the coral and its eukaryotic symbionts of the genus Symbiodinium is fairly well characterized (Rowan, 2004; Frade et al., 2008), relatively little is known about the spatial and temporal variation of the coral's prokaryotic microbiome. Recent studies have shown shifts in the microbiome from healthy to diseased corals and under altered environmental conditions (Bourne et al., 2007; Mao-Jones et al., 2010), supporting the idea that both the disturbance of the native microbiota and the direct infection by specific pathogens threaten the well-being of corals.
The coral host provides several distinct habitats for its microbial inhabitants—the tissue (Lesser et al., 2004), the gastrovascular cavity (Herndl and Velimirov, 1985), the skeleton (Sharshar et al., 1997) and the surface mucus layer (SML) (Rohwer et al., 2002; Kooperman et al., 2007), each harboring a distinct microbial community (Sweet et al., 2011a). The SML is particularly important for the biology of corals not only as a habitat for a distinctive suite of coral-associated microbes but also due to its nutritional, protective and cleansing roles (Brown and Bythell, 2005). Consisting of polymeric glycoproteins and lipids (Bythell and Wild, 2011), coral mucus provides a nutritious medium on which a diverse assemblage of microbes thrives, many of which are highly host specific (Rohwer et al., 2002). Although coral mucus is in constant contact with the adjacent seawater, their prevalent microbial communities exhibit almost no overlap (Rohwer et al., 2001, 2002; Frias-Lopez et al., 2002). Furthermore, it has been hypothesized that the microbial community in the coral SML operates as a defense barrier and therefore protects the coral against invasive microbes either because of the production of antimicrobial substances or simply because of the occupation of this interface niche (Rohwer et al., 2002; Reshef et al., 2006; Rosenberg et al., 2007; Shnit-Orland and Kushmaro, 2009).
The SML is a very dynamic system whereby its molecular organization and composition vary between coral species and over time (Brown and Bythell, 2005). In addition, the SML experiences sloughing, either continuously or periodically, from the coral surface into the reef environment (Bythell and Wild, 2011). This cycle of structural changes creates habitat dynamics to which microbial communities are likely responding (Nelson et al., 2013).
Corals of the genus Porites sp. provide an ideal model system to study natural short-term dynamics of the mucus microbiota because of a clearly recognizable and well-described aging process that precedes periodical sloughing of the entire mucus layer and its reformation (Coffroth, 1991). The SML initially appears as a transparent surface film whose visual appearance slowly changes over the course of a few days into a conspicuous aged mucus sheet. After the release of this aged sheet of mucus into the water column, new fluidic mucus is produced at the surface of the coral leading to a new cycle, suggested to follow a lunar periodicity (Coffroth, 1991). Our rationale is that the temporal transformation and periodical release of mucus from the surface of poritid corals may trigger or relate to the establishment of a microbial succession analogous to the one shown for bacterioplankton after phytoplankton blooms (Teeling et al., 2012).
To better understand the role of the mucus microbiota on the health and resilience of coral holobionts, we apply an indicator species approach aiming (1) to unravel the natural short-term dynamics of mucus-associated prokaryotic communities coupled to the cyclical aging and release of SML of Porites astreoides and (2) to follow the successional steps taking place after disruption of the coral's microbiome.
Materials and methods
Species, study location and sampling approach
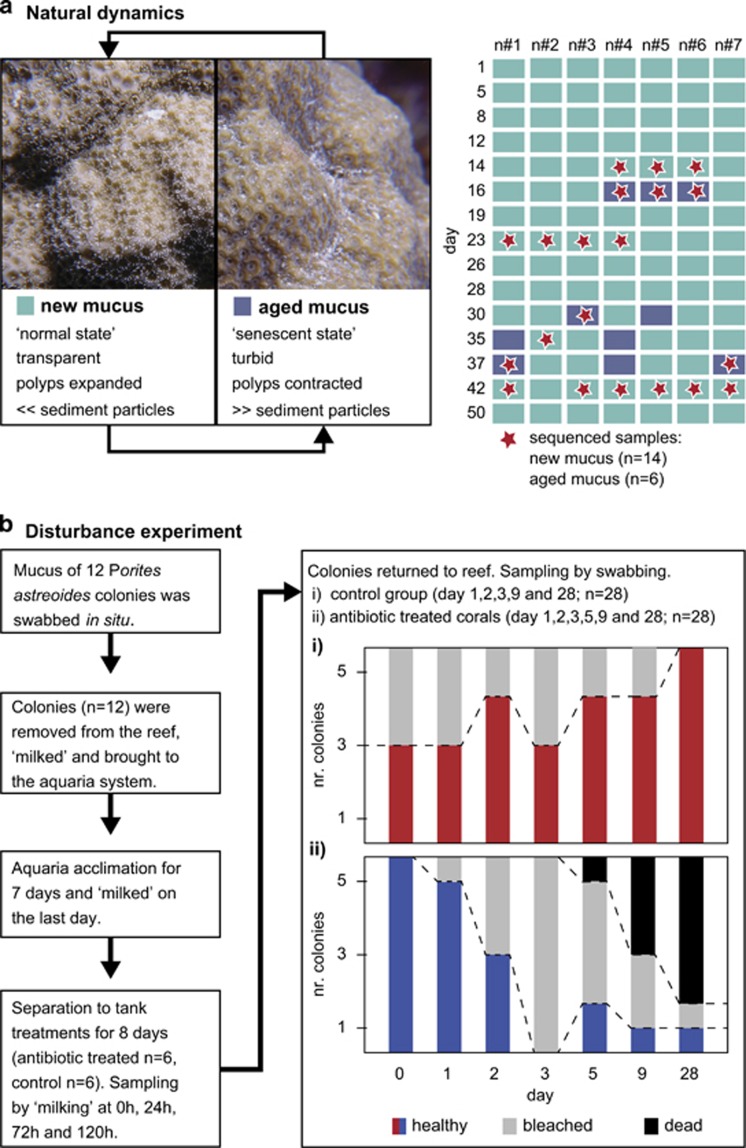
Porites astreoides Lamarck 1816, a common coral species on Caribbean reefs (Bak, 1975), exhibits the typical mucus aging behavior of poritid corals (Figure 1a and Supplementary Figure S1) and was chosen as a model species. Fieldwork was conducted between February and March 2014 on Curaçao (former Netherlands Antilles). Samples were collected by SCUBA diving at 5 m depth on the reef flat of the CARMABI Buoy One reef location (12°7.46'N, 68°58.31'W). In situ light irradiance (765±178 μmol photon m−2 s−1 at noon) and temperature (26.6 °C±0.4 °C) were measured at 5 m depth with a Hydrolab DS5 Sonde and with HOBO Pendant Temperature/Light Data Loggers, respectively, for the entire duration of fieldwork. Our approach consisted of two parts: an in situ description of the ‘natural dynamics of coral mucus-associated prokaryotes' related to the cyclical change between new and aged mucus over time, and a manipulative approach in which colonies were removed from the reef, their microbiome disturbed in the aquarium, and ‘prokaryotic mucus re-colonization after antibiotics disturbance' monitored after re-introduction of the colonies to the reef environment (Figure 1).
Figure 1.
Diagram depicting the two experimental approaches. (a) Natural dynamics of coral mucus-associated prokaryotes: surface mucus of seven Porites astreoides colonies was regularly sampled in situ over a 2-month period and mucus aging state (‘new mucus' versus ‘aged mucus') was visually assessed (following Coffroth, 1991). Insert depicts an in situ impression of the SML for the same P. astreoides colony at two different time points: with new mucus (left side) and with a conspicuous and aged mucus sheet (right side). A selection of new (n=14) and aged mucus (n=6) samples was later used for 16S ribosomal RNA (rRNA) gene amplicon sequencing. (b) Prokaryotic mucus re-colonization after antibiotics disturbance: mucus of 12 P. astreoides colonies exhibiting new mucus was sampled in situ and whole colonies later removed from the reef and brought to the aquaria system of the CARMABI station. Six colonies were incubated in a mix of antibiotics and the other six colonies were incubated as controls. After 8 days in the aquaria, corals were again sampled and thereafter brought back to the reef and installed on a rack (day 0). Over the next 28 days, the health of the colonies was visually assessed and mucus samples were regularly collected. No visual signs of mucus aging were observed throughout the experiment. The 16S rRNA gene was sequenced for all collected mucus samples (n=80) to follow microbial community composition during re-colonization of mucus after antibiotics disturbance.
Natural dynamics of coral mucus-associated prokaryotes
To assess in situ the microbial dynamics in the SML of P. astreoides, seven colonies were regularly monitored by collecting mucus samples with sterile cotton swabs (‘swabbing') every second to third day for a total period of 2 months (one sample per colony per day; see Figure 1a). On each sampling occasion, the mucus aging state was visually categorized as either ‘new' or ‘aged' following published approaches (Coffroth, 1991). At the same time, 50 ml of seawater were collected into Greiner tubes at approximately 20 cm distance from one of the sampled corals and later filtered onto 0.2 μm Millipore GTTP filters (Cork, Ireland). Sediment was collected from the upper sediment layer next to sampled corals on three distinct occasions during the fieldwork period. Immediately after each dive, all the samples were flash frozen in liquid nitrogen and kept at −80 °C.
Twelve other conspecific colonies exhibiting either conspicuously aged (n=6) or clearly new mucus (n=6) were removed from the reef on 2 consecutive days to collect large quantities of mucus through air exposure, thereafter referred to as ‘milking' (Garren and Azam, 2010). Removed corals were of similar size (diameter of approximately 10 cm) and all colonies were ‘milked' for 5 min before being brought back to the reef. Released mucus (range: 8–10 ml) was collected into sterile 50 ml Greiner tubes before fixing with formaldehyde (2% final concentration) at 4 °C for at least 4 h. In all, 1–2 ml of each fixed sample was then passed through an 0.2 μm GTTP filter (Millipore; supported by a 0.45 μm HAWP filter, Millipore). This filter was used to enumerate microbial abundance by epifluorescence microscopy (minimum of 30 fields of view and 300 cells) after 4′,6-diamidin-2-phenylindol (DAPI) staining. The microbial abundance associated with mucus is given below per mucus volume (Garren and Azam, 2010).
Prokaryotic mucus re-colonization after antibiotics disturbance
To assess the impact of antibiotic treatment and subsequent microbial re-colonization on coral health, 12 additional P. astreoides colonies (diameter of approximately 6 cm) with conspicuous new mucus were freshly collected from the reef and transferred to the reef-water flow-through aquaria system of the CARMABI station (Figure 1b and Supplementary Figure S2). Before removal of the colonies, their SML was sampled by ‘swabbing', and used as baseline for comparison with the microbial parameters determined in the subsequent experiments.
After an acclimation period of 7 days, coral colonies were transferred to transparent plastic beakers filled with aquarium seawater (600 ml) that was continuously agitated. Beakers were kept in the flow-through system at stable temperature conditions (26.5 °C±0.3 °C). Six of these colonies were incubated in an antibiotic mix (Supplementary Table S1) added in a dilution of 1:100 to seawater replaced every 12 h for a total incubation period of 8 days.
Corals were ‘milked' for a period of 2 min directly after removal from the reef, after the 7 days acclimation phase (t=0 h), and three times during the incubation (t=24, 72 and 120 h). Before ‘milking', each colony was rinsed with 0.2 μm-filtered aquarium seawater to remove loosely attached prokaryotes from the coral surface. Mucus samples were fixed with formaldehyde (2% final concentration) and processed to monitor prokaryotic abundance in the SML as described for the natural dynamics experiment.
After the aquaria incubation, all colonies, the antibiotic-treated and control colonies, were brought back to the CARMABI Buoy One reef and installed on a rack in an alternating order (Supplementary Figure S2). Colonies were ‘swabbed' directly after the re-introduction, at the following 3 consecutive days and then every second to third day for a total period of 28 days. In addition, the health status of each coral was visually monitored for occurrence of bleaching, lesions and mortality over the study period. Coral health was in general categorized in the three following groups: healthy—no visual signs of bleaching or necrosis, bleached—visual observation of bleaching and/or necrosis, dead—>80% of tissue loss. Throughout the disturbance experiment (incubation and re-introduction), none of the colonies exhibited visual signs of mucus aging.
DNA extraction, 16S ribosomal RNA gene sequencing and taxonomic annotation
DNA was extracted from 120 samples representing a selection of swabbed mucus (Figure 1), seawater and sediment samples using the FastDNA SPIN Kit for Soil (MP Biomedicals, Heidelberg, Germany) according to the manufacturer's instructions.
DNA extracts were sent on dry-ice to the IMGM laboratories GmbH (Martinsried, Germany) for sequencing a 728-bp fragment of the 16S ribosomal RNA (rRNA) gene (see Supplementary Information) using 454 GL FLX+ technology (Roche, Martinsried, Germany). Barcoded 16S rRNA gene PCR amplicons were denoised in Acacia (version 1.52.b0; Bragg et al., 2012) and analyzed using Qiime (version 1.8.0; Caporaso et al., 2010a). Obtained sequences were clustered into OTUs0.02 (operational taxonomic units) based on⩾98% sequence similarity excluding singletons. Representative OTUs0.02 were picked and aligned with PyNAST (version 1.2.2, Caporaso et al., 2010b) using the Greengenes database (version 13.5). Taxonomy was assigned using the Ribosomal Database Project Classifier (version 2.2, Wang et al., 2007) with a confidence minimum of 85%. OTU tables, based on the taxonomic hierarchical levels were generated in Qiime. Diversity estimates were calculated using R (R Development Core Team, 2008). To take the different sequencing efforts into account, 801 sequences were randomly picked from each sample as suggested by Qiime and chloroplast OTUs were removed. The rarefied OTU table based on family level and their relative abundances per sample was used for further analyses. Demultiplexed 16S rRNA gene raw reads and respective metadata are available in the NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP069317.
Statistics
The effect of sampling group on prokaryotic abundance as well as on diversity indices (Shannon Weaver Index, richness and evenness) was tested with repeated-measures analysis of variance (rANOVA) and further pairwise comparisons with the Tukey HSD test at 95% confidence level. Data were log transformed if required. Venn diagrams were used to depict numbers of unique, shared and ubiquitous OTUs.
The variation in prokaryotic community composition among different sampling groups (beta-diversity) was visualized by non-metric multidimensional scaling ordination (nMDS; after 10 000 permutations) based on Bray–Curtis dissimilarity matrices. Differences were further tested with analysis of similarity (ANOSIM; 10 000 permutations). Furthermore, the homogeneity of multivariate dispersions was tested using a resemblance-based permutation test (PERMDISP) before confirming differences in community structuring between sampling groups by applying a permutational multivariate analysis of variance (PERMANOVA) using Bray–Curtis dissimilarity matrices (Anderson et al., 2006).
The explanatory power of factors such as habitat, time (that is, sampling day) and treatment on the observed prokaryotic community assembly was determined by canonical correspondence analysis (CCA) and the significance of factors was further verified using an ANOVA-like permutation test based on 1000 permutations.
OTUs contributing up to a cumulative value of 70% of the total divergence in prokaryotic community assembly among different sampling groups were shown by similarity percentages (SIMPER).
In this study the indicator value analysis (IndVal; De Cáceres and Legendre, 2009) was applied to detect prokaryotic families significantly associated (P<0.05, when both specificity and fidelity have probabilities >0.5) to different sampling habitats: new mucus, aged mucus, seawater and sediment. Specificity is the probability that the specific taxon belongs to the habitat group given the fact that the species has been found, whereas fidelity is the probability of finding the taxon in assemblages belonging to the habitat group. Prokaryotic families identified by IndVal, here assumed to represent prokaryotic indicators of each particular habitat, have the highest probability of occurrence for that particular habitat. IndVal has been used in the past to identify strict habitat specialists of particular coral compartments (Li et al., 2014).
All statistical tests and graphs were compiled in R (R Development Core Team, 2008).
Results
Sequencing and sample overview
A total of 639 196 reads were retrieved from the 110 successfully sequenced and further analyzed samples (see Table 1 for details) and clustered into 353 OTUs (based on family level). The required minimum of 801 sequences per sample led to the removal of one mucus and two sediment samples. Observed richness within sampling groups was about two-thirds of the estimates obtained with Chao (Table 1).
Table 1. Overview of number of samples, number of 16S rRNA gene sequences retrieved and corresponding diversity indices (average±s.d.) for each sampling group within two experiments (natural dynamics and disturbance experiment).
| Sampling group | Total no. of samples | No. of sequencesa | Richness | Evenness | Shannon index | OTUs in totala | Chao estimatea |
|---|---|---|---|---|---|---|---|
| Natural dynamics | |||||||
| New mucus | 14 | 4882±2534 | 50±20 | 0.49±0.19 | 1.95±0.38 | 174 | 256 |
| Aged mucus | 6 | 5867±2770 | 82±29 | 0.81±0.05 | 3.48±0.90 | 178 | 248 |
| Seawater | 6 | 7709±3758 | 28±8 | 0.59±0.15 | 2.07±0.52 | 66 | 88 |
| Sediment | 4 | 923±116 | 70±12 | 0.80±0.05 | 3.40±0.30 | 137 | 210 |
| Disturbance experiment | |||||||
| Initial composition | 12 | 5941±2962 | 54±12 | 0.55±0.14 | 2.23±0.72 | 170 | 244 |
| Aquaria treated | 6 | 3837±2436 | 50±12 | 0.75±0.07 | 2.33±0.45 | 109 | 155 |
| Aquaria control | 6 | 4122±3249 | 84±19 | 0.81±0.05 | 3.32±0.47 | 176 | 230 |
| 1–3 Days treatedb | 18 | 7000±3728 | 67±17 | 0.68±0.10 | 2.84±0.54 | 201 | 316 |
| 1–3 Days controlb | 18 | 5494±1563 | 81±18 | 0.71±0.11 | 3.42±0.34 | 231 | 309 |
| 4–28 Days treatedb | 10 | 5528±3499 | 46±20 | 0.70±0.13 | 2.64±0.67 | 138 | 186 |
| 4–28 Days controlb | 10 | 8003±2583 | 71±15 | 0.78±0.06 | 3.17±0.43 | 183 | 315 |
All data are based on a rarefied OTU table at the family level from which chloroplast-affiliated OTUs were removed.
Abbreviations: OTU, operational taxonomic unit; rRNA, ribosomal RNA.
‘No. of sequences', ‘OTUs in total' and ‘Chao estimate' were calculated from the original non-rarefied OTU data set (including chloroplasts).
1–3 and 4–28 days represent the number of days after re-introduction on the reef. Data were pooled for each of those two periods.
Natural dynamics of coral mucus-associated prokaryotes
Each P. astreoides colony exhibited an aged mucus sheet on up to two occasions during the 2 months of fieldwork (Figure 1a). Mucus aging, however, was neither synchronized among different colonies nor related to any of the measured environmental parameters. Aged mucus remained for up to 3 days on the coral's surface before it was released into the surrounding environment. Prokaryotic abundance in the SMLs of P. astreoides colonies ranged from 3.2±0.5 × 105 cells ml−1 in newly produced mucus to 8.1±0.6 × 105 cells ml−1 in aged mucus sheets, representing a twofold increase in cell abundance (ANOVA, P<0.01). Mucus (aged and new), seawater and sediment prokaryotes comprised in total 255 OTUs, of which 32 OTUs were ubiquitously present in all habitats (Supplementary Figure S3). Approximately 18% and 21% of the OTUs associated with aged and new mucus layers, respectively, were unique to that particular SML aging state (Supplementary Table S2 and Supplementary Figure S3). Furthermore, mucus shared about two times more members with the prokaryotic community associated with sediments than with ambient seawater. Alpha diversity, richness and evenness were highest in aged mucus and sediment samples (Table 1), albeit statistically not significant (see Supplementary Table S3).
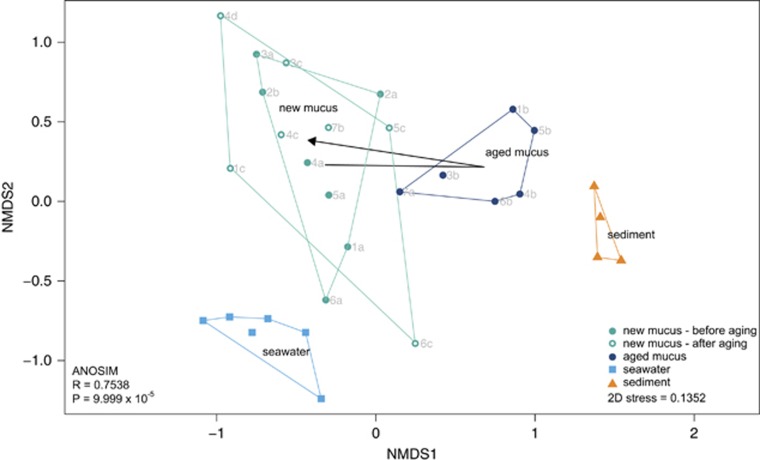
Prokaryotic community composition revealed by nMDS ordination (Figure 2) was structured based on habitat (ANOSIM, R=0.7538, P<0.001). Communities present in aged mucus were intermediate in their composition between those typically found in new mucus and in sediments. However, after the release of the aged mucus layers, the prokaryotic community reverted to its initial state in new mucus. A homogeneous multivariate dispersion among sampling groups (PERMDISP, P>0.05, Supplementary Table S4) allowed applying PERMANOVA to further confirm this habitat-based structuring of prokaryotic communities (P<0.001, Supplementary Table S5). CCA (Supplementary Figure S4) depicted that 81% of the variation in the prokaryotic community assembly was explained by the factor habitat (P<0.001), whereas the effect of time and colony was negligible (P>0.05).
Figure 2.
Natural dynamics of coral mucus-associated prokaryotes: non-metric multidimensional scaling (nMDS) plot of the Bray–Curtis-based dissimilarity matrix of prokaryotic communities colonizing the mucus of Porites astreoides and the adjacent reef environment (sediment and ambient water). Mucus samples are divided into ‘new mucus' (samples that were collected up to 2 weeks before and after an aging event) and into ‘aged mucus'. Arrow indicates shift in the centroid of the mucus community before aging, in aged mucus and after aged mucus was released. Each code depicted in the figure corresponds to an individual colony (1–6) throughout the experiment in a chronological order (a–d).
The relative abundances of those OTUs responsible for the observed changes in community structure between aged and new mucus (Supplementary Table S6) were compared (Supplementary Figure S5). Endozoicimonaceae was the most abundant bacterial family in new mucus of P. astreoides (41.6%±32.4% of the overall prokaryotic abundance), followed by Pelagibacteraceae (12.4%±8.8%) and Oxalobacteraceae (7.3%±9.9%). In contrast, aged mucus layers exhibited a low relative abundance of Endozoicimonaceae (4.1%±6.5%), however, an up to 10-fold increase was observed in Verrucomicrobiaceae (9.4%±4.1%), Vibrionaceae (7.6%±6.0%), Flammeovirgaceae (7.0%±3.8%) and Rhodobacteraceae (6.4%±2.4%) compared with new mucus. The archaeal domain generally exhibited a very low relative abundance in new and aged mucus (1.1%±1.7% and 1.4%±1.2%, respectively; Supplementary Figure S6) and did not significantly contribute to the variation in community structure between mucus aging stages.
Prokaryotic mucus re-colonization after antibiotics disturbance
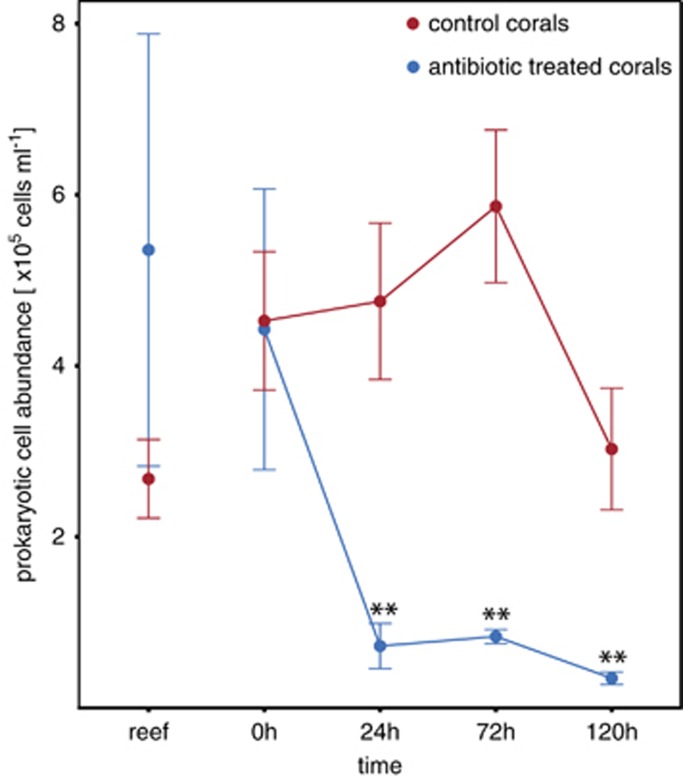
Antibiotic-treated colonies exhibited no visible signs of health deterioration during the 8 days of aquaria incubation. Control colonies, in contrast, were negatively affected, as half (n=3) of them showed bleaching and necrosis during the incubation period (Figure 1b). Prokaryotic abundance in mucus of the colonies was highly variable, with an average of 3.9±4.5 × 105 cells ml−1 before the experiment (n=12). After 24 h of incubation with antibiotics (n=6), prokaryotic abundance was only 5.4±4.6 × 104 cells ml−1 (Figure 3) representing a reduction to 14% of the original in situ abundance (rANOVA, P<0.001; Tukey HSD, P<0.01, see Supplementary Table S7). There was no apparent change in the overall alpha diversity of the coral mucus community (Table 1) concomitant with this reduction. This low abundance remained relatively constant until the end of the incubation period. In contrast, prokaryotic abundance in mucus of control colonies (n=6) remained relatively high (4.1±1.7 × 105 cells ml−1 at 24 h) throughout the entire incubation period (see Figure 3 and Supplementary Table S7), accompanied with a significant increase in alpha diversity (rANOVA, P<0.001; Tukey HSD, P<0.01; see Supplementary Table S8–S10) compared with the original in situ alpha diversity.
Figure 3.
Prokaryotic cell abundance in coral mucus of Porites astreoides throughout the aquaria incubation for antibiotic-treated and non-treated control colonies. Samples were collected directly after corals were removed from the reef (reef), after 8 days acclimatization to the aquaria environment (0 h) and 24, 72 and 120 h after the beginning of incubation with antibiotics. Antibiotic treatment resulted in a significant decrease (Tukey HSD; **P<0.01) in prokaryotic abundance relative to in situ values (reef). Error bars indicate ±s.d.
Once coral colonies were brought back to the reef they exhibited health response patterns opposite to those observed in the aquaria incubation (Figure 1b). Within the first 3 days, all antibiotic-treated colonies (n=6) showed rapid health deterioration and exhibited clear signs of bleaching, with only two surviving the following weeks. Non-treated corals, in contrast, recovered from aquarium incubations within 28 days of re-introduction to the natural reef environment (Figure 1b).
In total, 39% of the prokaryotic OTUs originally associated with mucus were present throughout the antibiotic treatment, as well as after re-introduction of treated corals into their natural habitat (Supplementary Figure S7a). Here, the families Endozoicimonaceae and Oxalobacteraceae were numerically dominant (Supplementary Figure S8). The control group, however, harbored 58% of the initial number of OTUs until the end of the incubation experiment (Supplementary Figure S7b). Furthermore, the prokaryotic community present in mucus of the control colonies (see Table 1) exhibited a significant increase in alpha diversity once transferred back to the reef (rANOVA, P<0.001; Tukey HSD, P<0.01 see Supplementary Table S8–S10). Re-introduced antibiotic-treated colonies, in contrast, were not significantly different in their alpha diversity from the original in situ alpha diversity (Tukey HSD, P>0.5, see Supplementary Table S8–S10).
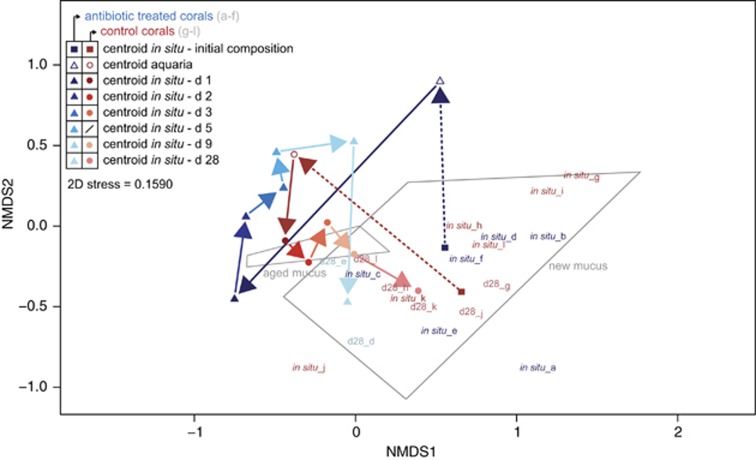
Prokaryotic succession in coral mucus, as visualized by nMDS ordination (Figure 4 and Supplementary Figure S9), revealed contrasting successional paths for antibiotic-treated and non-treated corals (ANOSIM, R=0.5082, P<0.001). Initially, during the aquarium incubation, both groups diverged from the natural community typical of new mucus layers on healthy corals. After being deployed back on the reef, antibiotic-treated corals exhibited a major change in their community assembly within the first 24 h becoming more similar to that found in sediments and in aged mucus layers. Within the next days, their community became similar to the one of aged mucus layers and of unhealthy-looking colonies such as the control group during the aquaria incubation. This shift in the prokaryotic community happened concomitantly with an increase in bleaching and mortality. After 28 days, the only two surviving colonies harbored a community similar to that in new mucus. The control group, however, after suffering of necrosis and bleaching during aquarium incubation, rapidly regained a prokaryotic community similar to the original one and concurrently exhibited again a healthy appearance (Figure 4).
Figure 4.
Prokaryotic mucus re-colonization after antibiotics disturbance: non-metric multidimensional scaling (nMDS) plot of the Bray–Curtis-based dissimilarity matrix of prokaryotic communities colonizing the mucus of Porites astreoides and their shifts after a disturbance event. The successional path, here represented by arrows, was reconstructed based on the position of group centroids and follows each group of corals (antibiotic-treated, a–f; control, g–l) at a particular time (indicated by color gradient). The starting point ‘in situ - initial community composition' represents the original in situ state, followed by the shift in the community composition in the aquaria incubation (dashed arrows) and finally describing the successional path over 28 days observed in the field after the two experimental groups of corals were brought back to the reef (normal arrows). Prokaryotic community structure of individual colonies is only given for mucus samples collected before the experiment started (in situ_a–l) and on the last day of the experiment (d28_a–l). Polygons indicate the relative position of prokaryotic communities associated with different mucus aging stages (‘new mucus' versus ‘aged mucus') characterized during the natural dynamics experiment and are shown as reference. Individual sample points throughout the whole experiment are given in detail in Supplementary Figure S9.
Differences in the successional path of antibiotic-treated and control corals throughout the monitoring period were revealed by multivariate community composition analysis (PERMDISP, P>0.05, Supplementary Table S11, PERMANOVA, P<0.01, Supplementary Table S12). Moreover, CCA (Supplementary Figure S10) attributed 67% of the variation in the prokaryotic community structure to differences in treatment (antibiotics versus control) and time (ANOVA-like permutation test, P<0.001 and P<0.01, respectively), whereas the colony effect was negligible (P>0.05).
Out of the prokaryotic families responsible for the divergence among treatment groups (SIMPER, Supplementary Table S13), Endozoicimonaceae was dominant in mucus (relative abundance up to 80%) of corals in their natural environment (Supplementary Figure S8). However, it decreased in relative abundance once corals suffered from bleaching and necrosis when kept in the aquaria but also in the reef. Within 24 h after re-introduction to the reef, the mucus of antibiotic-treated corals became dominated by Verrucomicrobiaceae (35.9%±20.2%) and Vibrionaceae (13.5%±14.7%). Within the next days, Rhodobacteraceae, Oceanospirillaceae, Vibrionaceae, Flammeovirgaceae, Verrucomicrobiaceae and Colwelliaceae dominated the mucus of antibiotic-treated colonies. Mucus of the control group became evenly colonized by various prokaryotic families such as Verrucomicrobiaceae, Vibrionaceae, Rhodobacteraceae, Alteromonadaceae, Colwelliaceae, Pelagibacteraceae and Synechococceae (Supplementary Figure S8). Archaea did not contribute to significant community variation and showed very low relative abundance in mucus throughout the disturbance experiment (overall 1.25%±4.28%) with the exception of two samples collected at the end of the antibiotic treatment (34.33% and 17.48%, respectively; Supplementary Figure S11).
Prokaryotic indicators associated with the coral mucus microbiome
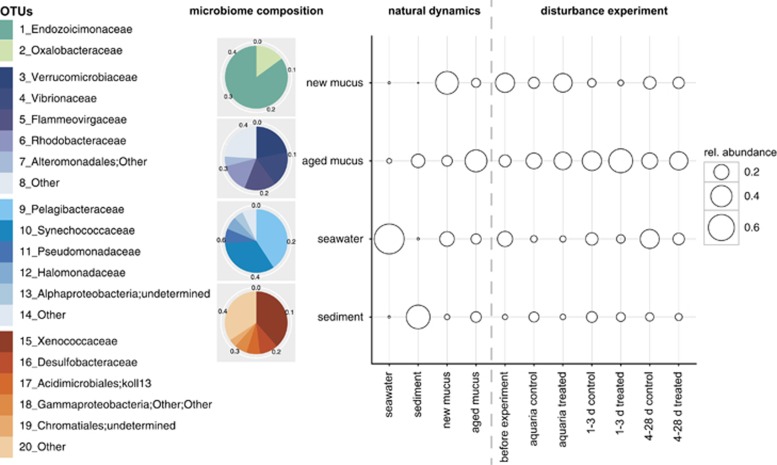
Identified prokaryotic indicators were distinct among the three habitats but individual members not strictly confined to the respective habitat (Figure 5 and Supplementary Table S14). Oxalobacteraceae and Endozoicimonaceae were identified as prokaryotic indicators associated with new mucus of P. astreoides. Together they accounted for 50% of all prokaryotes found in new coral mucus in situ. Similarly high contributions to the mucus microbiome were observed in the population of corals used for the disturbance experiment (before collection), as well as after treatment with antibiotics and, to a minor extent, within colonies surviving the disturbance experiment after 28 days. Indicators for new mucus, however, showed a reduction in their relative abundance in aged mucus, whose typical indicators consisted of Verrucomicrobiaceae, Vibrionaceae, Flammeovirgaceae, Rhodobacteraceae and Alteromonadales. Furthermore, mucus of disturbed corals showed increased relative abundances of aged mucus indicators, particularly dominant in antibiotic-treated colonies after re-introduction to the reef environment. Prokaryotic indicators identified for seawater and sediment did not exhibit major variations in dominance throughout the experiment, with the exception of a high contribution of typical seawater OTUs to the microbiome of control colonies at the end of the experiment.
Figure 5.
Prokaryotic indicator assemblages identified for the microbiome of SML of Porites astreoides (‘new mucus' versus ‘aged mucus'), the adjacent seawater and the sediment, and their abundance distribution throughout the natural dynamics and the disturbance experiment. Pie charts represent the relative abundance of indicator prokaryotic families (given on the left) that are significantly associated with a particular microbiome as revealed by the indicator value analysis (IndVal). Numbers around pie charts depict, for each habitat studied, the proportion of the total community represented by the indicator assemblages. The balloon plot on the right displays the relative abundance of the identified indicator assemblages in the various natural habitats and throughout the disturbance experiment. Note that prokaryotic indicator assemblages identified for a particular habitat are not restricted to that habitat and are also present (in lower abundance) in other habitats.
Discussion
Natural dynamics of coral mucus-associated prokaryotes
Although the cyclic aging and shedding of the SML in colonies of Porites sp. is fairly well documented (see Coffroth, 1991 and Supplementary Information), the associated dynamics of its mucus-dwelling prokaryotic community remained to be resolved. We demonstrate that the prokaryotic community undergoes significant changes throughout the mucus aging cycle, both in terms of cell abundance and community composition. Generally, the SML of P. astreoides was dominated by the bacterial families Oxalobacteraceae and Endozoicimonaceae, the latter of which has commonly been found in healthy corals (Apprill et al., 2013; Lema et al., 2014; Meyer et al., 2014) and suggested to have coevolved with specific host species (Bayer et al., 2013).
In contrast, aged mucus sheets exhibited a high relative abundance of Verrucomicrobiaceae, Flammeovirgaceae, Rhodobacteraceae and Vibrionaceae (see Figure 5 and Supplementary Figure S5). The latter two bacterial families include coral pathogens and are commonly associated with coral diseases (Ben-Haim et al., 2003; Sunagawa et al., 2010). After the detachment of the aged mucus sheet, however, the microbial community reverted to its original composition within 3–5 days (see Figure 2), supporting the idea that periodical mucus shedding in poritid corals generates a natural, rather deterministic fluctuation of the mucus-dwelling prokaryotic community, taking place within a temporal scale of weeks. As the prokaryotic indicator assemblages associated with aged mucus sheets showed surprisingly high similarity to the community associated with mucus of disturbed coral colonies (Figures 4 and 5), some of which died off, we propose that periodic mucus shedding (Coffroth, 1991) is an important mechanism supporting coral health by periodically removing undesirable prokaryotes from the surface of the colony leading to the maintenance of a beneficial mucus microbiome.
Once coral mucus is detached from the colony, it functions as particle trap before sinking to the seafloor, where it acts as energy and nutrient source for benthic organisms in coral reefs (Wild et al., 2004; Naumann et al., 2009). Alongside, aged mucus sheets frequently harbor sediment particles (Figure 1a), suggesting that prokaryotes colonizing upper sediment layers in coral reefs could ‘hitch a ride' to the nutrient-rich mucus layer via sediment resuspension. Thus, sediments may serve as a ‘seed-bank' for coral mucus-associated microbes as proposed by Carlos et al. (2013). Both, the resuspension of sediment particles (and its associated microbes) onto the coral's surface and the rapid sedimentation of detached coral mucus would lead to a high connectivity between the microbial communities of aged mucus and coral reef sediments, and consequently, may explain the similarity in their prokaryotic community composition (Figure 5). In contrast, microbial communities of the ambient seawater exhibited only minor overlap with those found in mucus. This confirms earlier findings (Rohwer et al., 2001; Frias-Lopez et al., 2002) and stresses the importance of the SML as a selective medium for the microbial pool in the adjacent seawater.
Prokaryotic mucus re-colonization after antibiotics disturbance
Antibiotic treatment led to a significant reduction of prokaryotic abundance in the SML and concurrent changes in community composition. Although the full extent of the influence of antibiotics on the holobiont's fitness remains elusive, no visual cues of a negative impact on coral health were noted. This confirms previous reports of minimal effect of antibiotic treatment on zooxanthellae photosynthetic efficiency and host tissue protein content (Gilbert et al., 2012). These findings suggest that coral hosts are not strictly dependent on their mucus-associated prokaryotic symbionts, at least for such short periods of up to 8 days and in the absence (or deactivation) of pathogens.
Control colonies kept in the aquaria without the addition of antibiotics exhibited obvious signs of bleaching and necrosis and underwent a significant shift in their mucus prokaryotic community (Figure 4). The bacterial family Endozoicimonaceae, which showed the highest relative abundances in newly produced SMLs and is associated with mucus of healthy P. astreoides colonies (Meyer et al., 2014), showed a significant decrease in its relative abundance within the control group. Concomitant with this decrease, many bacterial groups such as Rhodobacteraceae, Verrucomicrobiaceae, Colwelliaceae, Oceanospirillaceae and Flavobacteriaceae increased in their (relative and absolute) abundance (see Figure 3 and Supplementary Figure S8). These findings are consistent with a previous study attributing the expression of lesions in P. astreoides colonies to the loss of Endozoicimonaceae and the proliferation of an opportunistic bacterial community (Meyer et al., 2014). Compositional shifts in the microbial community associated with the SML have been observed under stressful environmental conditions (Thurber et al., 2008). Based on these findings, the observed health deterioration of the untreated control group may have been caused by aquaria conditions leading to a destabilization of the natural mucus community. This interpretation is in agreement with the hypothesis that disturbances in the dynamic equilibrium of the coral's native microbiota may result in health deterioration (Lesser et al., 2007).
Both, antibiotic-treated and control colonies, once brought back to their natural habitat, exhibited rapid changes in their mucus-associated prokaryotic community (Figure 4 and Supplementary Figure S8). For the microbe-depleted (treated) colonies, the increase in Vibrionaceae was dominated by Vibrio sp., a genus harboring well-known coral pathogens such as Vibrio shilonii (Kushmaro et al., 1997) and Vibrio corallilyticus (Ben-Haim et al., 2003; Garren et al., 2014). The latter is reported to use coral-produced sulfur compounds as a cue to target stressed corals (Garren et al., 2014). Curiously, a recent study has shown that P. astreoides (among other coral species) produces copious amounts of the organic sulfur compound dimethylsulfoniopropionate, particularly upon stress (Frade et al., 2015). Furthermore, Verrucomicrobiaceae, Flammeovirgaceae and Rhodobacteraceae, other families associated with necrotic and diseased P. astreoides colonies, have been found to be overrepresented in poritid corals suffering from White Band Disease (Séré et al., 2013; Roder et al., 2014). Although non-treated colonies slowly recovered from tissue lesions and regained a mucus community very similar to their original community, antibiotic-treated colonies suffered from mortality (from day 3 onward) and exhibited increased relative abundances of bacterial families described as early colonizers of marine biofilms, such as Rhodobacteraceae and Oceanospirillacae (O'Toole et al., 2000; Sweet et al., 2011b). Although we cannot exclude other synergistic effects on the host's health, these results suggest that microbe-depleted SML provides an open niche, which gets rapidly colonized by opportunistic bacteria. Thus, we hypothesize that the re-establishment of both coral health and the native prokaryotic community after disturbance of the P. astreoides holobiont depends on the initial degree of disruption of the microbiome. Total recovery of control colonies under in situ environmental conditions in contrast to mortality of microbe-depleted colonies suggests that, upon disturbance, the remnant prokaryotic community in the mucus/tissue may act as ‘seed-bank'.
The mucus microbiome and its influence on coral health and survival
Host-associated bacteria form unique microbiomes highly adapted to a particular host niche (Ainsworth et al., 2015). Although being generally considered to comprise commensals, microbiomes can fulfill important biological needs of their hosts, for example, immune development or nutrient acquisition (Round and Mazmanian, 2009; Shin et al., 2011). We identified Endozoicimonaceae and Oxalobacteraceae as significant indicators for the mucus microbiome of healthy P. astreoides colonies (Figure 5). The reduction in the relative abundance of these microbiome members observed before coral necrosis and bleaching suggests that the loss of beneficial bacteria can result in a serious health threat for the holobiont, often associated with an increase in opportunistic and potentially pathogenic bacteria (Meyer et al., 2014). Although our study does not aim at determining the metabolic function of particular microbiome members, it reveals that an intact mucus microbiome may function as a barrier against potentially harmful bacteria. This defense barrier could be based on commensal-like prokaryotes, which prevent harmful colonization by successfully outcompeting pathogens (Reid et al., 2001) and/or depend on stimulating host immune response via the recognition of commensal-derived signals such as microbial-associated molecular patterns (Mackey and McFall, 2006). Our results suggest that the mucus microbiome acts as defense barrier against pathogenic microbes, therefore facilitating homeostasis and contributing to the survival of the coral holobiont.
In summary, we demonstrate that the previously documented periodical mucus aging and shedding cycle in P. astreoides is provoking predictable shifts in the mucus microbiome, leading to changes between a beneficial community and a potentially opportunistic/pathogenic one. The periodical release of mucus seems to be part of a life strategy that supports the maintenance of a beneficial mucus microbiome and the resilience of coral health in shallow water habitats characterized by frequent sediment resuspension. However, severe disruption of the natural microbial community upon external stress could negatively and irreversibly affect the fate of the coral holobiont. Further investigations on the functional capacities of the mucus prokaryotic community are warranted to better understand the role of the mucus microbiome in the dynamic equilibrium of the coral holobiont. Finally, we have shown that specific bacterial members can be used as indicators of coral microbiome disruption, paving the way to the development of early diagnostic tools to monitor the health status of corals.
Acknowledgments
We thank Raphael Zimmermann for assistance with sample collection. Financial support for fieldwork, lab consumables and sequencing costs was provided by Marie Curie fellowship FP7-299320 and fellowship M1363-B20 from the Lise Meitner Program of the Austrian Science Fund (FWF) to PRF, and the FWF projects I486-B09 and P23234-B11 to GJH. We are grateful to the editor and three anonymous reviewers for comments, which greatly improved the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ainsworth TD, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9: 2261–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Ellingsen KE, McArdle BH. (2006). Multivariate dispersion as a measure of beta diversity. Ecol Lett 9: 683–693. [DOI] [PubMed] [Google Scholar]
- Apprill A, Hughen K, Mincer T. (2013). Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ Microbiol 15: 2063–2072. [DOI] [PubMed] [Google Scholar]
- Bak RPM. (1975). Ecological aspects of the distribution of reef corals in the Netherlands Antilles. Bijdragen tot de Dierkunde 45: 181–190. [Google Scholar]
- Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T et al. (2013). The Microbiome of the Red Sea coral stylophora pistillata is dominated by tissue-associated endozoicomonas bacteria. Appl Environ Microbiol 79: 4759–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim Y, Thompson F, Thompson C, Cnockaert M, Hoste B, Swings J et al. (2003). Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53: 309–315. [DOI] [PubMed] [Google Scholar]
- Bourne D, Iida Y, Uthicke S, Smith-Keune C. (2007). Changes in coral-associated microbial communities during a bleaching event. ISME J 2: 350–363. [DOI] [PubMed] [Google Scholar]
- Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. (2012). Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 9: 425–426. [DOI] [PubMed] [Google Scholar]
- Brown BE, Bythell JC. (2005). Perspectives on mucus secretion in reef corals. Mar Ecol Prog Ser 296: 291–309. [Google Scholar]
- Bythell JC, Wild C. (2011). Biology and ecology of coral mucus release. J Exp Mar Biol Ecol 408: 88–93. [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. (2010. b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010. a). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos C, Torres TT, Ottoboni LMM. (2013). Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Sci Rep 3: 1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffroth MA. (1991). Cyclical mucus sheet formation on poritid corals in the San Blas Islands, Panama. Mar Biol 109: 35–40. [Google Scholar]
- De Cáceres M, Legendre P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90: 3566–3574. [DOI] [PubMed] [Google Scholar]
- Frade PR, De Jongh F, Vermeulen F, Van Bleijswijk J, Bak RPM. (2008). Variation in symbiont distribution between closely related coral species over large depth ranges. Mol Ecol 17: 691–703. [DOI] [PubMed] [Google Scholar]
- Frade PR, Schwaninger V, Glasl B, Sintes E, Hill RW, Simó R et al. (2015). Dimethylsulfoniopropionate in corals and its interrelations with bacterial assemblages in coral surface mucus. Envir Chem e-pub ahead of print 27 August 2015 doi:10.1071/EN15023.
- Frias-Lopez J, Zerkle AL, Bonheyo GT, Fouke BW. (2002). Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl Environ Microbiol 68: 2214–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M, Azam F. (2010). New method for counting bacteria associated with coral mucus. Appl Environ Microbiol 76: 6128–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M, Son K, Raina J-B, Rusconi R, Menolascina F, Shapiro OH et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J 8: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates RD, Ainsworth TD. (2011). The nature and taxonomic composition of coral symbiomes as drivers of performance limits in scleractinian corals. J Exp Mar Biol Ecol 408: 94–101. [Google Scholar]
- Gilbert J, Hill R, Doblin M, Ralph P. (2012). Microbial consortia increase thermal tolerance of corals. Mar Biol 159: 1763–1771. [Google Scholar]
- Herndl GJ, Velimirov B. (1985). Bacteria in the coelenteron of Anthozoa: control of coelomic bacterial density by the coelenteric fluid. J Exp Mar Biol Ecol 93: 115–130. [Google Scholar]
- Kelly LW, Williams GJ, Barott KL, Carlson CA, Dinsdale EA, Edwards RA et al. (2014). Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc Natl Acad Sci USA 111: 10227–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooperman N, Ben-Dov E, Kramarsky-Winter E, Barak Z, Kushmaro A. (2007). Coral mucus-associated bacterial communities from natural and aquarium environments. FEMS Microbiol 276: 106–113. [DOI] [PubMed] [Google Scholar]
- Koren O, Rosenberg E. (2006). Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl Environ Microbiol 72: 5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushmaro A, Rosenberg E, Fine M, Loya Y. (1997). Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol Prog Ser 147: 159–165. [Google Scholar]
- Lema KA, Willis BL, Bourne DG. (2014). Amplicon pyrosequencing reveals spatial and temporal consistency in diazotroph assemblages of the Acropora millepora microbiome. Environ Microbiol 16: 3345–3359. [DOI] [PubMed] [Google Scholar]
- Lesser M, Mazel C, Gorbunov M, Falkowski P. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305: 997–1000. [DOI] [PubMed] [Google Scholar]
- Lesser MP, Bythell JC, Gates RD, Johnstone RW, Hoegh-Guldberg O. (2007). Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J Exp Mar Biol Ecol 346: 36–44. [Google Scholar]
- Li J, Chen Q, Long L-J, Dong J-D, Yang J, Zhang S. (2014). Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci Rep 4: 7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, McFall AJ. (2006). MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol Microbiol 61: 1365–1371. [DOI] [PubMed] [Google Scholar]
- Mao-Jones J, Ritchie KB, Jones LE, Ellner SP. (2010). How microbial community composition regulates coral disease development. PLoS Biol 8: e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JL, Paul VJ, Teplitski M. (2014). Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS One 9: e100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscatine L. (1990). The role of symbiotic algae in carbon and energy flux in reef corals. Coral Reefs 25: 1–29. [Google Scholar]
- Naumann MS, Richter C, el-Zibdah M, Wild C. (2009). Coral mucus as an efficient trap for picoplanktonic cyanobacteria: implications for pelagic–benthic coupling in the reef ecosystem. Mar Ecol Prog Ser 385: 65–76. [Google Scholar]
- Nelson CE, Goldberg SJ, Kelly LW, Haas AF, Smith JE, Rohwer F et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J 7: 962–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G, Kaplan HB, Kolter R. (2000). Biofilm formation as microbial development. Annu Rev Microbiol 54: 49–79. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, Available from http://www.R-project.org/. [Google Scholar]
- Reid G, Howard J, Gan BS. (2001). Can bacterial interference prevent infection? Trend Microbiol 9: 424–428. [DOI] [PubMed] [Google Scholar]
- Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. (2006). The coral probiotic hypothesis. Environ Microbiol 8: 2068–2073. [DOI] [PubMed] [Google Scholar]
- Ritchie KB, Smith GW. (1997). Physiological comparison of bacterial communities from various species of scleractinian corals. Proc 8th Int Coral Reef Sym 1: 521–526. [Google Scholar]
- Roder C, Arif C, Bayer T, Aranda M, Daniels C, Shibl A et al. (2014). Bacterial profiling of white plague disease in a comparative coral species framework. ISME J 8: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N. (2001). Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20: 85–91. [Google Scholar]
- Rohwer F, Seguritan V, Azam F, Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243: 1–10. [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5: 355–362. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan R. (2004). Coral bleaching: thermal adaptation in reef coral symbionts. Nature 430: 742–742. [DOI] [PubMed] [Google Scholar]
- Séré MG, Tortosa P, Chabanet P, Turquet J, Quod J-P, Schleyer MH. (2013). Bacterial communities associated with Porites White Patch Syndrome (PWPS) on three Western Indian Ocean (WIO) coral reefs. PLoS One 8: e83746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharshar N, Banaszak AT, Lesser MP, Amrami D. (1997). Coral endolithic algae: life in a protected environment. Pac Sci 51: 167–173. [Google Scholar]
- Shin SC, Kim S-H, You H, Kim B, Kim AC, Lee K-A et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334: 670–674. [DOI] [PubMed] [Google Scholar]
- Shnit-Orland M, Kushmaro A. (2009). Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol 67: 371–380. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Woodley CM, Medina M. (2010). Threatened corals provide underexplored microbial habitats. PloS One 5: e9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet MJ, Croquer A, Bythell JC. (2011. a). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30: 39–52. [Google Scholar]
- Sweet MJ, Croquer A, Bythell JC. (2011. b). Development of bacterial biofilms on artifical corals in comparison to surface-associated microbes of hard corals. PLoS One 6: e21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611. [DOI] [PubMed] [Google Scholar]
- Thurber RLV, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C et al. (2008). Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci USA 105: 18413–18418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9: 2807–2719. [DOI] [PubMed] [Google Scholar]
- Wild C, Rasheed M, Werner U, Franke U, Johnstone R, Huettel M. (2004). Degradation and mineralization of coral mucus in reef environments. Mar Ecol Prog Ser 267: 159–171. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.