Abstract
Background:
Acute diarrhea remains the serious problem in developing countries, especially among children under 5 years of age. Currently, only two or three common diarrhea pathogens were screened at most hospitals in China. The aim of this study was to provide a wide variety of diarrhea pathogens and their antimicrobial resistance patterns in children under 5 years of age.
Methods:
Totally 381 stool samples collected from Tongji Hospital between July 1, 2014 and June 30, 2015 were tested by culture and/or polymerase chain reaction for eight kinds of bacteria and five kinds of viruses. An antimicrobial sensitivity test was performed using dilution method recommended by the Clinical and Laboratory Standards Institute.
Results:
Viral infections were mainly identified in infants (0–11 months), whereas bacterial infections were more prevalent in the age of 24–59 months. About 69.8% of samples were positive for at least one pathogen, 51.7% of samples were virus positive, followed by bacteria positive cases (19.4%), and 12.6% of cases displayed co-infections with two viruses or a virus and a bacterium. Rotavirus was the most prevalent pathogen, followed closely by norovirus, while Salmonella was the most commonly isolated bacteria, followed by diarrheagenic Escherichia coli (DEC) and Campylobacter. More than 40% of Salmonella spp. and DEC isolates were resistant to first-line antibiotics (ampicillin, trimethoprim-sulfamethoxazole, and tetracycline). Around 10% of Salmonella spp. isolates were resistant to ceftriaxone and ciprofloxacin simultaneously. Campylobacter spp. displayed high resistance to ciprofloxacin but kept low resistance to azithromycin and doxycycline.
Conclusions:
The etiology of acute diarrhea varies in children of different age groups. The high frequency of infection with viruses suggests the urgent demand for new viral vaccine development. Proper use of antibiotics in the treatment of acute diarrhea is crucial due to the high level of antibiotic resistance.
Keywords: Acute Diarrhea, Bacteria, Children, Etiology, Virus
INTRODUCTION
Diarrhea remains the serious problem globally due to its leading cause of death for children, especially among children under 5 years of age. Walker et al.[1] reported that there were 1.731 billion episodes of diarrhea leading to more than 700,000 deaths worldwide in 2011. In the past two decades, to reduce the burden of diarrhea, the Chinese government has made a great progress to improve public hygiene, which resulted in a remarkable decrease of mortality rate in children.[2] Despite these efforts, China is still 1 of 15 high-incidence countries (0.7 episodes per person-year).[1] Diarrhea ranked top 3 among the notifiable infectious disease by far.[3]
In China, diarrhea is the most common reason for children to visit health-care clinics, and the antibiotic resistance has become a growing problem due to misuse of antibiotics. However, not all the bacterial or viral diarrhea pathogens causing diarrhea can be detected due to the technical limitation. In most clinical laboratories, only culture or immunological technique was used to screen for common diarrheal pathogens, such as Salmonella, Shigella, and/or rotavirus. Data from these assays alone cannot show the full spectrum of diarrheal pathogens. In this study, following a literature review and based on experts’ opinions, we detected a panel of proven and/or plausible diarrheal pathogens to fully understand the main causes of acute diarrhea in children under 5 years of age in Wuhan, China.
METHODS
Sample collection
Totally, 381 children under 5 years of age, who visited the outpatient clinic of Tongji Hospital, Tongji Medical College of Huazhong University of Sciences and Technology (Wuhan, China) with acute diarrhea between July 1, 2014 and June 30, 2015, were enrolled in this general surveillance. Acute diarrhea was defined as the passing of liquid, loose, mucoid, or bloody stool for three or more times daily, with an episode lasting no longer than 14 days. The children's parents verbally agreed to participate in this study. All their fecal samples were collected in sterile containers and subjected to culture and/or polymerase chain reaction (PCR) analysis at the clinical laboratory of Tongji Hospital. For bacteriological testing, samples were inoculated within 2 h on arrival at the laboratory. For virological testing, the samples must be kept at −20°C without preservatives prior to processing within a week. All residues were kept at −70°C for further analysis.
Isolation and identification of bacteria
On arrival at the laboratory, samples were plated onto selective and differential culture media. For isolation of Salmonella, Shigella, diarrheagenic Escherichia coli (DEC), Vibrio, Yersinia, Aeromonas and Plesiomonas, MacConkey agar (Mac), xylose lysine desoxycholate medium, sorbitol Mac agar, thiosulfate citrate bile salt, and Columbia blood agar (Oxoid, UK) were used. Sulfa enrichment broth (SBG) (Qingdao HopeBio-Technology, China) and phosphate-buffered saline were used as enrichment media to improve the isolation of Salmonella and Yersinia, respectively. Cefoperazone deoxycholate agar and Skirrow's medium were used to culture Campylobacter spp. in a microaerophilic environment at 42°C.
After incubation, suspected colonies were submitted to biochemical test using Vitek 2-compact automated system (bioMe´rieux, Marcy-l’E’toile, France). For DEC and Campylobacter, bacteria colonies were pooled for DNA extraction. The DNA products were subjected to PCR analysis using the primers as reported previously.[4,5,6,7]
Serotyping of Salmonella spp. and Shigella spp. was performed by slide agglutination tests using commercially available antisera (Salmonella Diagnostic Antisera Kit, Statens Serum Institute SSI Diagnostica, Copenhagen, Denmark; and anti-Shigella antiserum from Lanzhou Institute of Biological Products Co., Ltd., Lanzhou, China).
Virus detection
The presence of viral pathogens including rotavirus (Groups A, B, and C), norovirus (GI and GII), sapovirus, astrovirus, and adenovirus were detected by molecular biological techniques. Nucleic acids were extracted from fecal suspensions in 0.9% physiological saline using High Pure Viral Nucleic Acid Kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. The products were then subjected to one-step reverse transcription-PCR with type-specific primers as reported previously.[8,9,10,11] All PCR-positive products were re-confirmed with DNA-sequencing.
Antimicrobial susceptibility tests
As recommended by the Clinical and Laboratory Standards Institute (CLSI), the minimum inhibitory concentration (MIC) of antibiotics was performed using standard agar dilution technique, except for Campylobacter, for which broth microdilution method was used. The antimicrobials were bought from China Food and Drug Research Institute, China. MIC results were interpreted according to CLSI M100, 2015 and CLSI M45, 2006 (for Campylobacter) recommendations. The reference strains, ATCC25922, ATCC 35218, and ATCC 29213 were used as controls.
Since CLSI did not recommend azithromycin breakpoints for Campylobacter or Salmonella spp. (except for Salmonella typhi), we used the breakpoints of CDC epidemiological cutoff values (ECOFFs)[12] for Campylobacter and interpreted the results for Salmonella spp. according to breakpoints for S. typhi, CLSI M100, 2015.
RESULTS
Characteristics of patients
Besides diarrhea, vomiting was the most common clinical manifestation, almost 32.3% of cases (123/381) presented with concomitant vomiting. Fever was the second most common clinical manifestation, with 27.5% (105/381) of children complaining of low- or mid-grade fever. Diarrhea lasted 1–4 days in two hundred and seventy children, and 5–10 days in another one hundred children, prior to the hospital visit. Only 11 children had 10–14 days of diarrhea [Figure 1].
Figure 1.
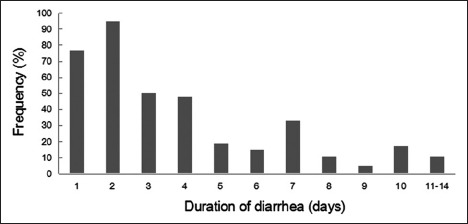
Duration of diarrhea in children under 5 years of age.
Prevalence of enteropathogens among different age groups and different seasons
The age distribution of 381 case patients in this study was as following: 64.6% were aged 0–11 months, 22.3% were 12–23 months, and 13.1% were 24–59 months [Figure 2a]. Viral agents (especially rotavirus and norovirus) were more likely to be identified in 0–11-month infants, while bacterial agents were more prevalent in the age group of 24–59 months [Figure 2b]. The seasonal distribution of the patients infected with virus or bacterium was also investigated. Viruses were prevalent in autumn months, while bacteria were prevalent in summer months [Figure 2c].
Figure 2.
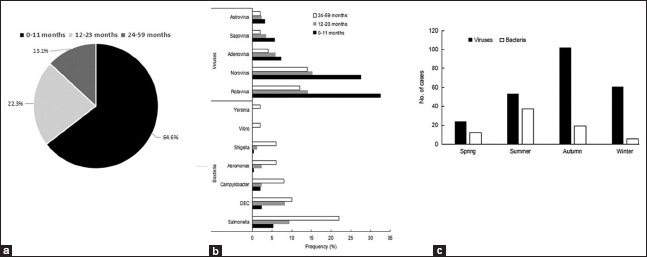
Positive percentages of enteropathogens among children aged 0–59 months in acute diarrheal illnesses. (a) Patients with age distribution; (b) Prevalence of enteropathogens among different age groups; (c) Prevalence of enteropathogens among different seasons.
Distribution of varied enteropathogens
As shown in Figure 3, at least one pathogen was tested positive in 266 (69.8%) samples from 381 children. Viruses took up the largest part of detectable enteropathogens with a positive rate of 51.7% (197/381), followed by bacteria with 19.4% of samples (74/381). There were 48 cases co-infected with two pathogens simultaneously. No specific pathogens were detected in 30.2% (115/381) of the samples [Figure 3a].
Figure 3.
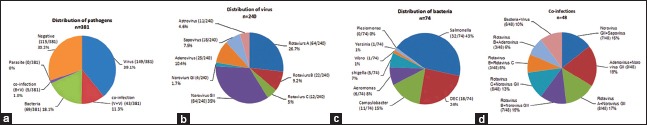
Distribution of diarrheal pathogens. (a) Distribution of pathogens; (b) Distribution of viruses; (c) Distribution of bacteria; (d) Co-infections.
Viral etiology
Rotavirus was the most commonly detected organism (40.8%, 98/240), followed by norovirus (36.7%, 88/240). These two viruses accounted for almost 80% of detectable viral diarrheal pathogens in this study. Among 98 rotavirus positive samples, Group A was the leading group identified, accounting for 65% (64/98), followed by Group B (22%, 22/98) and Group C (12%, 12/98). Among norovirus positive samples, GII was undoubtedly predominant, accounting for 95% (84/88) of these samples. Other viruses such as astrovirus, sapovirus, and adenovirus were detected less frequently (4% to 11%) [Figure 3b].
Bacterial etiology
Salmonella was the most frequently identified organism, accounting for 43% of 74 bacterium-positive samples, followed by DEC (24%, 18/74). Campylobacter took the third place in bacteria inducing diarrhea, accounting for 15% (11/74), followed by Aeromonas (8%, 6/74), Shigella (7%, 5/74), Vibrio (1%, 1/74), and Yersinia (1%, 1/74). All Campylobacter identified in the current study were Campylobacter jejuni [Figure 3c].
Co-infection
Forty-eight (12.6%, 48/381) cases displayed co-infection with two different enteropathogens. Co-infections with rotavirus and norovirus (44%, 21/48) were the most commonly identified, followed by co-infections of norovirus and adenovirus (19%, 9/48) and norovirus and sapovirus (15%, 7/48), successively [Figure 3d].
Serotyping
Of the 32 Salmonella isolates, the predominant serotype was Salmonella typhimurium which accounted for more than 40% (13/32) of isolates, followed by paratyphoid B and Salmonella enteritidis. Both of them accounted for 13%. Of the 18 DEC isolates, enteropathogenic Escherichia coli (EPEC) and Shiga-toxin-producing E. coli (STEC) were the leading genotypes identified, account for 50% and 33%, respectively. Enteroaggregative E. coli accounted for only 17%, while both enterotoxigenic E. coli and enteroinvasive E. coli were not identified. The predominant serotypes of Shigella isolates were Shigella flexneri (3/5) and Shigella sonnei (2/5). Neither Vibrio cholera nor O157:H7 was identified.
Antimicrobial resistance
All bacteria were subjected to antimicrobial susceptibility test; however, only the data of Salmonella, DEC, and Campylobacter were shown because too few strains of other bacterial species were isolated.
As shown in Table 1, Salmonella exhibited a high frequency of resistance to the first-line antibiotics: ampicillin (68%) and trimethoprim-sulfamethoxazole (53%). Resistance rates to conventional and/or first-line antibiotics such as ciprofloxacin, levofloxacin, tetracycline, chloramphenicol, nalidixic acid, ceftriaxone, and azithromycin were 12%, 9%, 56%, 41%, 26%, 22%, and 16% respectively; 9% (3/32) of isolates were resistant to ceftriaxone and ciprofloxacin simultaneously.
Table 1.
Antimicrobial resistance rate of Salmonella, DEC, and C. jejuni (%)
| Antibiotics | Salmonella (n = 32) | DEC (n = 18) | C. jejuni (n = 11) |
|---|---|---|---|
| Ampicillin | 68 | 67 | – |
| Ampicillin/sulbactam | – | 33 | – |
| Nalidixic acid | 26 | – | – |
| Cefazolin | – | 33 | – |
| Cefuroxime | – | 33 | – |
| Cefotaxime | – | 28 | – |
| Ceftriaxone | 22 | – | – |
| Imipenem | – | 0 | – |
| Amikacin | – | 17 | – |
| Gentamicin | – | 33 | – |
| Ciprofloxacin | 12 | 33 | 54 |
| Levofloxacin | 9 | 17 | – |
| Chloramphenicol | 41 | 33 | – |
| Tetracycline | 56 | 67 | 23 |
| Trimethoprim-sulfamethoxazole | 53 | 55 | – |
| Azithromycin | 16* | – | 9† |
| Erythromycin | – | – | 18 |
| Doxycycline | – | – | 0 |
–: Not tested. *According to breakpoints for Salmonella typhi, CLSI M100, 2015. †According to breakpoints of Centers for Disease Control ECOFFs. ECOFFs: Epidemiological cutoff value; DEC: Diarrheagenic Escherichia coli; CLSI: Clinical and Laboratory Standards Institute; C. jejuni: Campylobacter jejuni.
Of the 18 DEC isolates, more than 50% were resistant to ampicillin, tetracycline, and trimethoprim-sulfamethoxazole. Approximately, 30% of isolates were resistant to ampicillin/sulbactam, cefazolin, cefuroxime, cefotaxime, gentamicin, ciprofloxacin, and chloramphenicol, and about 20% of isolates were resistant to amikacin and levofloxacin. All 18 isolates were sensitive to imipenem.
Campylobacter isolates (n = 11) showed low resistant to azithromycin, erythromycin, and doxycycline, with the rates of 9%, 18%, and 0, respectively; 54% of isolates were resistant to ciprofloxacin and 23% to tetracycline [Table 1].
DISCUSSION
This study was conducted in central China to investigate a wide variety of diarrhea pathogens and their antimicrobial resistance patterns in children under 5 years of age. To get reliable data, both molecular and conventional techniques were used to detect each sample. Furthermore, this study covered a whole year including the peak infection seasons, May to September.
Consistent with other studies,[13] our data showed that the most commonly detected pathogen causing diarrhea in children under 5 years of age were viruses. Among 381 samples, 197 (51.7%) were tested positive for at least one virus. Notably, infants (0–11 months) were more likely to be infected with viruses, especially rotavirus and norovirus, while bacterial agents were more prevalent in the age group of 24–59 months. This diversity of age distribution might reflect a natural change in host immunity and dietary. The seasonal distribution of viruses and bacteria was also different. Our data suggest that viruses were prevalent in autumn months, while bacteria were prevalent in summer months, which was consistent with previous studies.[14]
In this study, rotavirus was the most common diarrheal pathogen as reported by the previous studies.[13,14,15,16] However, a study from Africa showed bacteria were the prominent pathogens associated with diarrhea.[17] Norovirus took the second place in the list of most frequently detected diarrheal virus, which was similar to reports from other provinces of China.[15,18] Of 88 norovirus positive samples, 84 were identified as subtype GII, only 4 were subtype GI, which was similar to Anders et al.'s report.[19] Furthermore, of the 84 norovirus GII positive cases, 37 (44%) were co-infected with other diarrheal pathogens, while none of the norovirus GI cases was found to be co-infected with any other pathogens. The most frequently detected co-infections were rotavirus and norovirus, accounting for around 50% (44%, 21/48). Notably, the proportion of norovirus identified in this study was around 23.1% (88/381), which was much higher than the results of other studies (10%).[15,18] However, it was consistent with the report by Ramani et al.,[20] which showed that norovirus had replaced rotaviruses in recent years as the predominant viral pathogen in diarrheal children. The economic burden induced by norovirus infection inspired the urgent demand for norovirus vaccine in China. National Vaccine and Serum Institute has started research and development on norovirus vaccine.
In the present study, almost one-fifth of participants were infected with or carried one bacterial pathogen. It was noteworthy that the distribution of bacterial pathogens in different age groups was not even. Children aged 24–59 months were more likely to be infected with bacterial enteropathogens than other age groups. Among all identified bacterial enteropathogens in this study, Salmonella was the most frequently isolated bacterial pathogen which is usually prevalent in the developing countries such as India and Pakistan, while it is rare in industrialized countries.[21,22] A 5-year surveillance conducted in Shanghai, a highly developed area in China, demonstrated that the isolation rate of Salmonella decreased year by year,[23] which was attributed to the improvement of hygiene condition. Among the Salmonella positive samples in this study, the most frequently detected serotype was S. typhimurium which accounted for more than 40% of all Salmonella isolates. This was in agreement with previous reports from Niger and from Guangdong province in China. S. typhimurium, a multidrug-resistant strain, has been of concern to both veterinary and public health officials as an important swine and human pathogen. What is more, S. typhimurium infection has even become an increased risk to public health in some developed countries, such as UK and USA, accounting for nearly one-half of human Salmonella infections.[24] In our study, Salmonella exhibited a high frequency of resistance to the conventional first-line antibiotics: ampicillin, trimethoprim-sulfamethoxazole, and tetracycline (resistance rate ≥50%). Salmonella also displayed increased resistance to third-generation cephalosporins, such as ceftriaxone (22%) and to fluoroquinolones, such as ciprofloxacin (13%) and levofloxacin (9%). Even worse is the growth of strains which were both resistant to third-generation cephalosporin and fluoroquinolones, accounting for around 10% (3/32). The rates of antibiotic resistance observed in our study were greater than that reported in a previous surveillance conducted in Shanghai, China.[25] Empirical antimicrobial therapy of severe Salmonella infection faces the challenge from the increasing resistance to extended-spectrum cephalosporins and ciprofloxacin.[26] However, 84% of Salmonella isolates were sensitive to azithromycin in this study, that is consistent with the previous studies[27,28] and suggests that azithromycin may be a good choice to treat bacterial gastroenteritis.
As shown in this study, DEC was the second most commonly isolated bacteria associated with children's diarrhea. EPEC was the most frequent isolated pathogen, which was consistent with other reports.[29,30] EPEC is usually prevalent in low-income countries where the poor hygiene condition contributes to its circulation. The prevalence of EPEC in central China suggests that the government needs to put further effort into improving the public health. STEC was the second frequently isolated pathogen in this study. Notably, STEC can lead to not only mild, self-limiting diarrhea but also hemorrhagic colitis and hemolytic uremic syndrome as a complication.[31] Its high isolation rate should be of concern to public health office. DEC also displayed high resistant to conventional and/or first-line-antibiotics (28–67%), low resistant to levofloxacin and amikacin (<20%), while all DEC strains were sensitive to imipenem. Campylobacter (2.8%, 11/381) was the third most frequently isolated bacteria causing diarrhea, even more than Shigella (1.0%, 4/381). However, this number might be still underestimated in China as a result of the limited bacterial culture in the primary health-care facilities or use of antibiotics prior to visiting the hospital. In some developed countries, Campylobacter has become the leading bacteria causing acute diarrhea in children.[30,31] Our antimicrobial susceptibility tests showed that Campylobacter isolates displayed low-level resistance to azithromycin, erythromycin, and doxycycline (<10%). However, the ciprofloxacin resistance rate in Campylobacter isolates reached 54%, which is higher than that reported by Riley et al.[32] Moreover, the resistance rate of these isolates to tetracycline was 20%. Therefore, we should avoid using these two antibiotics to treat acute diarrhea.
As shown in the current study, viruses were the most predominant pathogenic agents causing acute diarrhea in children under 5 years of age in this region. Virus was more frequently isolated from infants than from the age group of 24–59 months, which was consistent with previous surveys from other cities of China.[15,18] However, the proportions of the two leading viral agents had changed. There was a considerable increase in the percentage of samples those were norovirus positive, compared to data from Chen et al.'s or Sai et al.'s studies.[15,18] This suggests that norovirus could replace rotavirus as the main cause of acute diarrhea in children under 5 years of age. Our study also indicated that a wide range of bacteria is responsible for acute diarrhea in children under 5 years of age. Salmonella and DEC continue to be the leading bacterial agents. Antimicrobial susceptibility screening indicated that ampicillin, trimethoprim-sulfamethoxazole, and tetracycline should not be used as the first-line therapeutic drugs for Salmonella spp. and DEC strains.
In summary, etiologic diagnosis of diarrhea is valuable for public health interventions. Our data can inform the development and implementation of diarrhea control and prevention programs in China. Antibiotics to treat bacterial diarrhea are widely available, but there are few vaccines for diarrhea-associated pathogens. This might explain the high incidence of acute diarrhea in children under 5 years of age, especially in rural areas, where sanitation is still lacking.[33] Until 2001, there was no rotavirus vaccine available in China.[2] The control of diarrhea cases heavily relied on the use of antibiotics. However, the high level of antibiotics resistance has become a serious problem in central China, challenging the effectiveness of antibiotic use.
This study had several limitations. It was a single-center study, and the number of cases was not as many as those reported in multicenter studies due to restrictions on time and resources. Nevertheless, this study will enrich the epidemiology database of China and will be of great importance for guiding the proper use of antibiotics.
Financial support and sponsorship
This study was supported by a grant of the Infectious Diseases Control Project from Ministry of Health of China (No. 2012ZX10004207-004).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Yuan-Yuan Ji
REFERENCES
- 1.Walker CL, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, et al. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381:1405–16. doi: 10.1016/S0140-6736(13)60222-6. doi: 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin CL, Chen SC, Liu SY, Chen KT. Disease caused by rotavirus infection. Open Virol J. 2014;8:14–9. doi: 10.2174/1874357901408010014. doi: 10.2174/1874357901408010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu HX, Zhang J. Analysis of reported infectious diarrhea (other than cholera dysentery, typhoid and paratyphoid) in China in 2011 (in Chinese) Chin J Prevent Med. 2013;47:328–32. [PubMed] [Google Scholar]
- 4.Tobias J, Vutukuru SR. Simple and rapid multiplex PCR for identification of the main human diarrheagenic Escherichia coli. Microbiol Res. 2012;167:564–70. doi: 10.1016/j.micres.2011.11.006. doi: 10.1016/j.micres.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Kagambèga A, Martikainen O, Lienemann T, Siitonen A, Traoré AS, Barro N, et al. Diarrheagenic Escherichia coli detected by 16-plex PCR in raw meat and beef intestines sold at local markets in Ouagadougou, Burkina Faso. Int J Food Microbiol. 2012;153:154–8. doi: 10.1016/j.ijfoodmicro.2011.10.032. doi: 10.1016/j.ijfoodmicro.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 6.Denis M, Soumet C, Rivoal K, Ermel G, Blivet D, Salvat G, et al. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett Appl Microbiol. 1999;29:406–10. doi: 10.1046/j.1472-765x.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 7.Linton D, Lawson AJ, Owen RJ, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568–72. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asmah RH, Green J, Armah GE, Gallimore CI, Gray JJ, Iturriza-Gómara M, et al. Rotavirus G and P genotypes in rural Ghana. J Clin Microbiol. 2001;39:1981–4. doi: 10.1128/JCM.39.5.1981-1984.2001. doi: 10.1128/JCM.39.5.1981-1984.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan H, Nguyen TA, Phan TG, Okitsu S, Li Y, Ushijima H. Development of RT-multiplex PCR assay for detection of adenovirus and group A and C rotaviruses in diarrheal fecal specimens from children in China. Kansenshogaku Zasshi. 2004;78:699–709. doi: 10.11150/kansenshogakuzasshi1970.78.699. [DOI] [PubMed] [Google Scholar]
- 10.Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J Clin Microbiol. 1995;33:797–801. doi: 10.1128/jcm.33.4.797-801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan H, Yagyu F, Okitsu S, Nishio O, Ushijima H. Detection of norovirus (GI, GII), sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J Virol Methods. 2003;114:37–44. doi: 10.1016/j.jviromet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Humphries RM, Schuetz AN. Antimicrobial susceptibility testing of bacteria that cause gastroenteritis. Clin Lab Med. 2015;35:313–31. doi: 10.1016/j.cll.2015.02.005. doi: 10.1016/j.cll.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Nitiema LW, Nordgren J, Ouermi D, Dianou D, Traore AS, Svensson L, et al. Burden of rotavirus and other enteropathogens among children with diarrhea in Burkina Faso. Int J Infect Dis. 2011;15:e646–52. doi: 10.1016/j.ijid.2011.05.009. doi: 10.1016/j.ijid.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Wang J, Sun H, Xia S, Duan R, Liang J, et al. Etiology of childhood infectious diarrhea in a developed region of China: Compared to childhood diarrhea in a developing region and adult diarrhea in a developed region. PLoS One. 2015;10:e0142136. doi: 10.1371/journal.pone.0142136. doi: 10.1371/journal.pone.0142136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Li Z, Han D, Cui D, Chen X, Zheng S, et al. Viral agents associated with acute diarrhea among outpatient children in Southeastern China. Pediatr Infect Dis J. 2013;32:e285–90. doi: 10.1097/INF.0b013e31828c3de4. doi: 10.1097/INF.0b013e31828c3dne4. [DOI] [PubMed] [Google Scholar]
- 16.Lou JT, Xu XJ, Wu YD, Tao R, Tong MQ. Epidemiology and burden of rotavirus infection among children in Hangzhou, China. J Clin Virol. 2011;50:84–7. doi: 10.1016/j.jcv.2010.10.003. doi: 10.1016/j.jcv.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Saeed A, Abd H, Sandstrom G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum, Sudan. J Med Microbiol. 2015;64:432–7. doi: 10.1099/jmm.0.000043. doi: 10.1099/jmm.0.000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sai L, Sun J, Shao L, Chen S, Liu H, Ma L. Epidemiology and clinical features of rotavirus and norovirus infection among children in Ji’nan, China. Virol J. 2013;10:302. doi: 10.1186/1743-422X-10-302. doi: 10.1186/1743-422X-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders KL, Thompson CN, Thuy NT, Nguyet NM, Tu le TP, Dung TT, et al. The epidemiology and aetiology of diarrhoeal disease in infancy in Southern Vietnam: A birth cohort study. Int J Infect Dis. 2015;35:3–10. doi: 10.1016/j.ijid.2015.03.013. doi: 10.1016/j.ijid.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramani S, Atmar RL, Estes MK. Epidemiology of human Noroviruses and updates on vaccine development. Curr Opin Gastroenterol. 2014;30:25–33. doi: 10.1097/MOG.0000000000000022. doi: 10.1097/MOG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kothari A, Pruthi A, Chugh TD. The burden of enteric fever. J Infect Dev Ctries. 2008;2:253–9. doi: 10.3855/jidc.218. [DOI] [PubMed] [Google Scholar]
- 22.Hassing RJ, Goessens WH, Mevius DJ, van Pelt W, Mouton JW, Verbon A, et al. Decreased ciprofloxacin susceptibility in Salmonella typhi and paratyphi infections in ill-returned travellers: The impact on clinical outcome and future treatment options. Eur J Clin Microbiol Infect Dis. 2013;32:1295–301. doi: 10.1007/s10096-013-1878-9. doi: 10.1007/s10096-013-1878-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhao Y, Ding K, Wang X, Chen X, Liu Y, et al. Analysis of bacterial pathogens causing acute diarrhea on the basis of sentinel surveillance in Shanghai, China 2006-2011. Jpn J Infect Dis. 2014;67:264–8. doi: 10.7883/yoken.67.264. [DOI] [PubMed] [Google Scholar]
- 24.Parsons BN, Crayford G, Humphrey TJ, Wigley P. Infection of chickens with antimicrobial-resistant Salmonella enterica typhimurium DT193 and monophasic Salmonella typhimurium-like variants: An emerging risk to the poultry industry? Avian Pathol. 2013;42:443–6. doi: 10.1080/03079457.2013.822469. doi: 10.1080/03079457.2013.822469. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Xie X, Xu X, Wang X, Chang H, Wang C, et al. Nontyphoidal Salmonella infection in children with acute gastroenteritis: Prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis. 2014;11:200–6. doi: 10.1089/fpd.2013.1629. doi: 10.1089/fpd2013.1629. [DOI] [PubMed] [Google Scholar]
- 26.Yu F, Chen Q, Yu X, Li Q, Ding B, Yang L, et al. High prevalence of extended-spectrum beta lactamases among Salmonella enterica typhimurium isolates from pediatric patients with diarrhea in China. PLoS One. 2011;6:e16801. doi: 10.1371/journal.pone.0016801. doi: 10.1371/journal.pone.0016801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girgis NI, Butler T, Frenck RW, Sultan Y, Brown FM, Tribble D, et al. Azithromycin versus ciprofloxacin for treatment of uncomplicated typhoid fever in a randomized trial in Egypt that included patients with multidrug resistance. Antimicrob Agents Chemother. 1999;43:1441–4. doi: 10.1128/aac.43.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frenck RW, Jr, Mansour A, Nakhla I, Sultan Y, Putnam S, Wierzba T, et al. Short-course azithromycin for the treatment of uncomplicated typhoid fever in children and adolescents. Clin Infect Dis. 2004;38:951–7. doi: 10.1086/382359. doi: 10.1086/382359. [DOI] [PubMed] [Google Scholar]
- 29.Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–13. doi: 10.3201/eid0805.010385. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langendorf C, Le Hello S, Moumouni A, Gouali M, Mamaty AA, Grais RF, et al. Enteric bacterial pathogens in children with diarrhea in Niger: Diversity and antimicrobial resistance. PLoS One. 2015;10:e0120275. doi: 10.1371/journal.pone.0120275. doi: 10.137/journal.pone.0120175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster K, Schnitzler E. Hemolytic uremic syndrome. Handb Clin Neurol. 2014;120:1113–23. doi: 10.1016/B978-0-7020-4087-0.00075-9. doi: 10.1016/B978-0-7020-4087-0.00075-9. [DOI] [PubMed] [Google Scholar]
- 32.Riley A, Eshaghi A, Olsha R, Allen VG, Patel SN. Antibiotic susceptibility of clinical isolates of Campylobacter jejuni and Campylobacter coli in Ontario, Canada during 2011-2013. Diagn Microbiol Infect Dis. 2015;83:292–4. doi: 10.1016/j.diagmicrobio.2015.07.020. doi: 10.1016/j.diagmicrobio.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 33.Feng XL, Guo S, Yang Q, Xu L, Zhu J, Guo Y. Regional disparities in child mortality within China 1996-2004: Epidemiological profile and health care coverage. Environ Health Prev Med. 2011;16:209–16. doi: 10.1007/s12199-010-0187-5. doi: 10.1007/s12199-010-0187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


