Summary
Limitation of marine primary production by the availability of nitrogen or phosphorus is common. E miliania huxleyi, a ubiquitous phytoplankter that plays key roles in primary production, calcium carbonate precipitation and production of dimethyl sulfide, often blooms in mid‐latitude at the beginning of summer when inorganic nutrient concentrations are low. To understand physiological mechanisms that allow such blooms, we examined how the proteome of E . huxleyi (strain 1516) responds to N and P limitation. We observed modest changes in much of the proteome despite large physiological changes (e.g. cellular biomass, C, N and P) associated with nutrient limitation of growth rate. Acclimation to nutrient limitation did however involve significant increases in the abundance of transporters for ammonium and nitrate under N limitation and for phosphate under P limitation. More notable were large increases in proteins involved in the acquisition of organic forms of N and P, including urea and amino acid/polyamine transporters and numerous C‐N hydrolases under N limitation and a large upregulation of alkaline phosphatase under P limitation. This highly targeted reorganization of the proteome towards scavenging organic forms of macronutrients gives unique insight into the molecular mechanisms that underpin how E . huxleyi has found its niche to bloom in surface waters depleted of inorganic nutrients.
Introduction
Marine phytoplankton require the essential macronutrients C, N and P for biosynthesis, which on average are required in the molar ratio described by Redfield of C106 : N16 : P1 (Moore et al., 2013). Nitrogen is required for the biosynthesis of proteins, nucleic acids and other macromolecules including chlorophyll, as well as low molecular weight compounds such as glycine‐betaine. Phosphorus is required for the synthesis of metabolites, including those involved in energy transfer, phospholipids for membrane synthesis and nucleotides for nucleic acid synthesis.
Living in an environment with an abundance of dissolved inorganic carbon, it is typically the macronutrients N or P that limit or co‐limit growth in regions where resupply by vertical mixing is relatively slow (Moore et al., 2008; 2013), whereas the micronutrient Fe is often limiting where macronutrient supplies are high (Moore et al., 2006; 2013). Nitrogen availability tends to limit productivity throughout much of the surface low‐ and mid‐latitude ocean at least seasonally (Moore et al., 2013), although some regions such as North Atlantic are very close to N and P co‐limitation (Graziano et al., 1996; Moore et al., 2006), whereas P limitation may be restricted, for example to the eastern Mediterranean Sea (Thingstad et al., 2005) or to those organisms that are capable of fixing N2 (Moore et al., 2013).
In the oceans, dissolved inorganic nitrogen exists in various oxidation states (NH4 +, NO2 −, NO3 −) and is converted and removed by biological activity, while inorganic P exists in one oxidation state (orthophosphate, PO4 3−) that does not undergo biological reduction or oxidation. As well as inorganic nutrients, some phytoplankton utilize dissolved organic N (DON), which consists of compounds such as urea, amino acids, amides, methylamines and purines, and dissolved organic P (DOP), which exists in a many forms of P‐esters or phosphonates. Seasonal differences in nutrient availability occur, with lower concentrations of dissolved inorganic nutrients and higher concentrations of DON and DOP in summer.
One phytoplankter that has evolved to thrive under nutrient‐limited conditions is the ubiquitous coccolithophore Emiliania huxleyi found throughout the ocean except the polar regions. Emiliania huxleyi is the most abundant coccolithophore (Paasche, 2001), and is important in marine food webs and plays very significant roles in ocean biogeochemistry through its contributions to both primary production via the incorporation of CO2 into organic matter and production of CaCO3 coccolith plates that surround the cell, which after death and sedimentation contribute to the production of chalk and limestone sediments. The importance of this species led to it being the first haptophyte to have its genome sequenced (Read et al., 2013).
Emiliania huxleyi can bloom to high concentrations covering vast areas in excess of 200 000 km2 (Holligan et al., 1993; Sukhanova and Flint, 1998) that are easily detected by satellite remote sensing due to high reflectance of solar radiation by shed coccoliths (Brown and Yoder, 1994). Such blooms are typically found when the mixed surface layers are exposed to both high light and declining macronutrients (Iglesias‐Rodriguez et al., 2002). Blooms of E. huxleyi often occur after the demise of diatom blooms when NO3 −, PO4 3− and silicate have been depleted (Holligan et al., 1993). It appears that factors that contribute to its success [other than the ability to tolerate high light and a wide range of photon flux densities (PFDs); Suggett et al., 2007; McKew et al. 2013a] are its ability to thrive on numerous forms of DON (Antia et al., 1975; Ietswaart et al., 1994; Palenik and Henson, 1997), its very high affinity for inorganic P (Reigman et al., 2000) and its ability to utilize DOP (Dyhrman and Palenik, 2003).
The ability of E. huxleyi to thrive in high light experienced during blooms involves large changes in the components of the proteome that allowed cells to alter their capacities to absorb light and resist photooxidative stress, but relatively small changes in the remainder of the proteome (McKew et al., 2013a, 2013b). In this present study, we used liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) shotgun proteomics to investigate readjustment of the proteome in response to both N and P limitation. The most notable responses were in the abundance of proteins associated with nutrient assimilation, giving a unique, mechanistic insight into how E. huxleyi acclimates to low‐nutrient environments.
Results and discussion
Differences in the steady‐state growth rates between nutrient‐replete, P‐limited and N‐limited conditions were accompanied by differences in cell size and elemental composition (Figs 1 and 2). Relative to nutrient‐replete cells, there was a significant decrease in cellular biovolume of N‐limited cells, and in contrast a significant increase in the biovolume of the P‐limited cells (Fig. 1B). Significant differences in cellular Chl a content were observed between the three treatments, being lowest in N‐limited and highest in P‐limited cells (Fig. 1C). Both N‐limited and nutrient‐replete cells contained about 50% less particulate organic carbon (POC) than the larger P‐limited cells (Fig. 2A), consistent with previous reports (Reigman et al., 2000). Cellular biovolume was highly correlated with cellular POC content (r = 0.92; P < 0.0001) and particulate inorganic carbon (PIC) content (r = 0.89; P < 0.0001), less so with cellular N content (r = 0. 61; P = 0.045), but not with cellular phosphorus (r = −0.36; P = ns). Particulate inorganic carbon followed the same pattern as POC, and consequently the PIC : POC ratios did not vary significantly among the three conditions (PIC : POC nutrient‐replete 0.56 ± 0.10; N‐limited 0.70 ± 0.02; P‐limited 0.73 ± 0.10).
Figure 1.
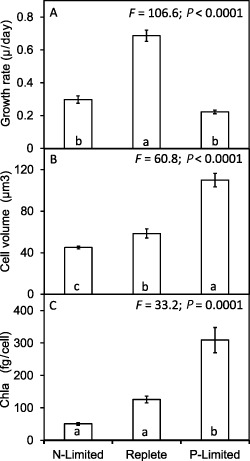
Growth rates of E miliania huxleyi 1516 and changes in cellular biovolume and Chl a content in response to N and P limitation. Significant differences are shown by analysis of variance and Tukey honestly significant difference tests (different letters a, b, c indicate significant differences at 0.05 confidence level).
Figure 2.
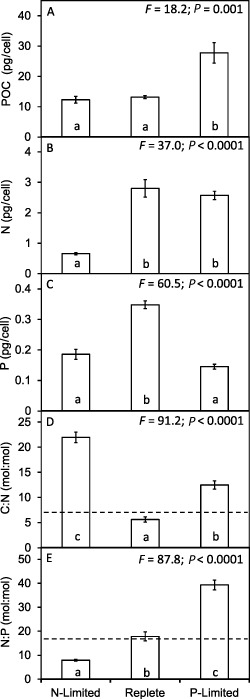
Changes in cellular POC, N and P content of E miliania huxleyi 1516 and shifts in molar stoichiometry of POC : N : P in response to N and P limitation. Significant differences are shown by analysis of variance and Tukey honestly significant difference tests (different letters a, b, c indicate significant differences at 0.05 confidence level). Dotted lines on D and E represent Redfield ratio stoichiometry.
While the P‐limited cells contained high levels of POC, they contained less than half the particulate P content of the replete cells (Fig. 2C), and while the N‐limited cells contained similar levels of POC to the replete cells the particulate N was some fourfold lower (Fig. 2B). The resulting molar ratios of cellular POC : N (Fig. 2D) and N : P (Fig. 2E) were also significantly different. The POC : N : P observed in the nutrient‐replete cells (C98 : N18 : P1) was similar to Redfield stoichiometry (C106 : N16 : P1). Significantly higher POC : N ratios were observed in the P‐limited (POC : N = 12.5 mole/mole) and N‐limited cultures (POC : N = 22) than in nutrient‐replete (POC : N = 5.6). The increase of POC : N in response to P limitation was primarily due to increase in cell size and POC, whereas the increase in response to N limitation was brought about by lower N. Relative to the nutrient‐replete cells, which had an N : P ratio of 17.8, the N‐limited cells had a significantly lower N : P ratio of 7.9, whereas P‐limited cells had a significantly higher ratio of 39. These deviations from Redfield C : N : P stoichiometry that occurred in response to nutrient limitation are consistent with previous observations for E. huxleyi (Leonardos and Geider, 2005; Borchard et al., 2011), and are consistent with N limitation placing a constraint on protein synthesis, while P limitation constrains RNA production (Loladze and Elser, 2011).
NH4 + was the preferred nitrogen source (confirmed by daily measurements of residual NO3 − and NH4 +) and NO3 − was consumed only after NH4 + had been depleted. This was also confirmed by nutrient uptake assays that involved incubating N‐limited cells in NO3 −‐replete, NH4 +‐replete and NH4 +NO3 −‐replete media and measuring N uptake at hourly intervals. Vmax was 2.8‐fold greater when supplied NH4 + (6.86 ± 0.18 fmol N cell–1 hr–1) compared with NO3 − (2.43 ± 0.18 fmol N cell–1 hr–1), and when supplied NH4 +NO3 − only NH4 + uptake was observed.
Cellular protein followed the same pattern as, and correlated with, cellular biovolume (r = 0.84; P = 0.001) and cellular N (r = 0.78; P = 0.005), where relative to the nutrient‐replete cells protein content per cell was significantly lower in the smaller N‐limited cells and significantly higher in the larger P‐limited cells (Fig. 3A). While the relative differences in protein content per cell between the treatments are realistic, the absolute cellular protein contents (e.g. approximately 3 pg cell–1 under nutrient‐replete conditions) appear to be low given the observed cellular N contents. We may have underestimated cellular protein either due to incomplete protein extraction and/or differences in sensitivity of the protein assay between E. huxleyi proteins and the BSA standards. However, we also note that the reported values are in line with other studies (e.g. Strom et al., 2003; Kaffes et al., 2010), suggesting underestimation of protein content of this organism may be common. RNA content per cell was two to three times greater in the nutrient‐replete cells than the P‐limited and N‐limited cells (data not shown), giving similar protein : RNA ratios under replete and N‐limited conditions but a significantly higher ratio under P limitation.
Figure 3.
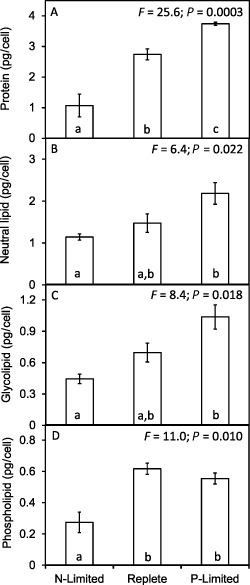
Changes in cellular protein and lipid contents of E miliania huxleyi 1516 in response to N and P limitation. Significant differences are shown by analysis of variance and Tukey honestly significant difference tests (different letters a, b, c indicate significant differences at 0.05 confidence level).
The lowest concentrations of neutral lipid (Fig. 3B), glycolipid (Fig. 3C) and phospholipid (Fig. 3D) were observed in the N‐limited cells, with significantly higher levels present in the P‐limited cells (approximately twofold higher in all cases). Intermediate concentrations of neutral lipids and glycolipids were present in the nutrient‐replete cells. Phospholipids did not follow this pattern as slightly higher levels were measured in the nutrient‐replete than in P‐limited cells, although the difference was not significant. Neutral lipids represented 53% of total lipid in the nutrient‐replete cells, and 61% and 58% of total lipid in the N‐limited and P‐limited cells respectively. Glycolipid as a percentage of total lipid was similar in all treatments (24% N‐limited, 25% nutrient‐replete and 27% in P‐limited cells). Phospholipid represented 22% of total lipid in the nutrient‐replete cells and 15% under both nutrient‐limited conditions. While the abundance of the lipid fractions differed between treatments, the ratios of the constituent fatty acids were highly similar. For example, docosahexaenoic acid (DHA C22:6n3) was the most abundant lipid, accounting for 34–35% of total lipid in all three treatments. Other abundant lipids were myristic acid (C14:00), palmitic acid (C16:00), oleic acid (C18:1n9c), linolenic acid (C18:3n3), docosatetraenoic acid (C22:4n6) and eicosapentaenoic acid (C20:5n3), indicating that while cellular contents of lipids changed significantly the fatty acid composition of the lipids remained highly similar. The observed lipid composition was similar to previously reported (Pond and Harris, 1996; Sayanova et al., 2011).
Neutral lipids are present in all growth phases of E. huxleyi, but these energy storage lipids have been shown to increase particularly during N‐starved stationary phase (Pond and Harris, 1996) as carbon incorporation into lipids is 40–60%, whereas carbon incorporation into protein is lower at 20% due to the high N demand (Fernández et al., 1994). It has been shown that increases in neutral lipid are observed 4–10 days after nutrient stress under both N and P limitation (e.g. Eltgroth et al., 2005). We also observe this response in N‐limited cultures in the early stage of N limitation (data not shown), but here we show that during steady‐state growth (cultures were in balanced growth and had been under nutrient limitation for 1 month before sampling) the levels of cellular lipid return to lower levels are in fact the lowest in the smaller N‐limited cells. However, the fact that protein was also much lower under N limitation is evident in the lipid : protein ratio (1 under replete and P‐limited conditions increasing to 1.8 under N limitation) as lipid represents a non‐nitrogenous pool. Lipid responses are also seen under P limitation, as many phytoplankton, including E. huxleyi, can reduce their cellular phosphorus requirements by substituting non‐phosphorus membrane sulfolipids for phospholipids (Van Mooy et al., 2009), and we observed that the phospholipid pool did not follow the same pattern as glyco‐ and neutral lipids of being higher in the larger P‐limited cells. Although lipid increased under P limitation, lipid represented 20% of cell C under replete, decreasing to 15% and 14% under N and P limitation respectively.
Proteomic analysis
Three technical replicates were run for each of the four biological replicates of each of the N‐limited, P‐limited and nutrient‐replete conditions, resulting in 613 505 total scans, of which 265 368 spectra were confidently assigned to 3012 different proteins. Spectral counts ranged from 1 to 6418 per protein, with over 500 proteins having 100 or more spectral counts assigned. Of the 500 proteins with the highest number of spectral counts, 36 were at least twofold higher under N limitation (relative to replete) and 53 were at least twofold higher under P limitation. Similar numbers of proteins were also twofold less abundant (52 under N limitation and 61 under P limitation, both relative to nutrient‐replete). Many proteins were detected with too few spectral counts to allow comparison among treatments. Approximately 2000 of the detected proteins had very low spectral counts, averaging 1 or less per run. Thus, it was necessary to sum the spectral counts for related proteins at the level of macromolecular complexes (e.g. chloroplast ATP synthase) or metabolic pathways (e.g. glycolysis) or by function (e.g. amino acid metabolism) to obtain meaningful information on changes of abundance among treatments (Table 1; for details of all individual proteins assigned to particular functions or pathways, see Table S1). Between 13% and 15% of the spectral counts were assigned to proteins with unknown functions.
Table 1.
Differences in abundance of normalized spectral counts assigned to proteins associated with specific functions or pathways
| Spectral counts | Tukey HSD | |||||||
|---|---|---|---|---|---|---|---|---|
| N‐Limited | Replete | P‐Limited |
F
(ANOVA) |
P
(ANOVA) |
N‐Limited | Replete | P‐Limited | |
| Light harvesting | 329 | 338 | 455 | 28 | < 0.001 | b | b | a |
| Photosynthetic electron transfer chain | 152 | 158 | 156 | 0.3 | 0.782 | a | a | a |
| Chloroplast ATP synthase | 321 | 343 | 330 | 2.7 | 0.131 | a | a | a |
| Calvin cycle | 338 | 392 | 375 | 4.4 | 0.052 | b | a | ab |
| Other photosynthesis | 26 | 52 | 31 | 42 | < 0.0001 | b | a | b |
| Carotenoids/porphyrin/pigment synthesis | 39 | 79 | 54 | 36 | 0.0001 | c | a | b |
| Thioredoxin | 2.6 | 1.4 | 3.5 | 32 | < 0.001 | b | c | a |
| FtSH | 8.8 | 14 | 9.0 | 27 | < 0.001 | b | a | b |
| Bicarbonate transport | 2.7 | 3.1 | 0.8 | 9.7 | 0.007 | a | a | b |
| Carbonic anhydrase | 3.9 | 6.2 | 5.0 | 1.9 | 0.213 | a | a | a |
| Glycolysis | 173 | 148 | 208 | 6.5 | 0.021 | ab | b | a |
| Krebs cycle | 127 | 101 | 129 | 7.2 | 0.016 | a | b | a |
| Other energy production and conversion | 123 | 97 | 113 | 6.9 | 0.018 | a | b | ab |
| Amino acid metabolism | 137 | 174 | 146 | 28 | < 0.001 | b | a | b |
| Nucleotide metabolism | 59 | 63 | 56 | 1.9 | 0.210 | a | a | a |
| Fatty acid synthesis | 119 | 130 | 123 | 4.5 | 0.049 | b | a | ab |
| Fatty acid degradation beta oxidation | 29 | 32 | 30 | 0.9 | 0.453 | a | a | a |
| Carbohydrate metabolism | 81 | 71 | 62 | 7.9 | 0.013 | a | ab | b |
| Isoprenoid biosynthesis | 7.2 | 14 | 13 | 20 | 0.001 | b | a | a |
| Other secondary metabolism | 110 | 122 | 94 | 11 | 0.005 | a | a | a |
| Heat shock proteins HSP90 | 46 | 63 | 51 | 13 | 0.003 | b | a | b |
| Heat shock proteins HSP70 | 100 | 108 | 100 | 1.4 | 0.298 | a | a | a |
| Heat shock proteins HSP60 | 22 | 36 | 33 | 20 | 0.001 | b | a | a |
| Heat shock proteins HSP40 (DnaJ) | 11 | 13 | 8 | 8.6 | 0.010 | ab | a | b |
| Heat shock proteins HSP TCP‐1 | 3.6 | 5.3 | 3.3 | 5.0 | 0.040 | ab | a | b |
| Clp protease | 22 | 27 | 19 | 8.2 | 0.012 | ab | a | b |
| Peroxidase | 14 | 7.1 | 9.8 | 9.6 | 0.007 | a | b | ab |
| Vitamin B6 synthesis | 0.7 | 2.5 | 2.0 | 5.2 | 0.036 | b | a | ab |
| Superoxide dismutase | 3.4 | 2.6 | 3.2 | 0.6 | 0.561 | a | a | a |
| Myosin and actin | 34 | 17 | 29 | 13 | 0.003 | a | b | a |
| Microtubules | 10 | 16 | 7.1 | 17 | 0.001 | b | a | b |
| Cell cycle | 44 | 43 | 36 | 8.1 | 0.012 | a | a | b |
| Ribosomal proteins | 204 | 241 | 144 | 19 | 0.001 | a | a | b |
| Chloroplast ribosomal proteins | 35 | 60 | 15 | 41 | < 0.0001 | b | a | c |
| Histones | 164 | 108 | 60 | 97 | < 0.0001 | a | b | c |
| Translation/transcription | 309 | 356 | 260 | 17 | 0.001 | ab | a | b |
| Protein modification and transport | 350 | 302 | 315 | 3.5 | 0.083 | a | a | a |
| Protein degradation (ubiquitin and proteasome) | 127 | 94 | 99 | 16 | 0.001 | a | b | b |
| Signal transduction | 31 | 17 | 21 | 12 | 0.004 | a | b | b |
| Urea cycle | 12 | 30 | 16 | 26 | < 0.001 | b | a | b |
| Nitrogen assimilation/metabolism | 100 | 41 | 40 | 335 | < 0.0001 | a | b | b |
| Phosphorous assimilation/metabolism | 25 | 6 | 267 | 37 | < 0.0001 | b | b | a |
| Sulfate assimilation/metabolism | 47 | 57 | 59 | 11 | 0.006 | b | a | a |
| Unknown/unassigned function | 690 | 602 | 603 | 4.9 | 0.040 | a | a | a |
Significant differences are shown by analysis of variance and Tukey honestly significant difference tests (different letters a, b, c indicate significant differences at 0.05 confidence level).
Nitrogen acquisition
Some of the most significantly upregulated proteins in response to nutrient limitation were those involved in nitrogen and phosphorus transport (Fig. 4). Numerous nitrogen transporters were detected in the N‐limited cells, but not in the replete and P‐limited conditions. Despite the fact that NH4 + was the preferred N source, and that NO3 − was not drawn down to any appreciable extent in nutrient‐replete or P‐limited treatments, NH4 + transporters were only detected under N limitation. This suggests that the abundance of NH4 + transporters under N‐replete conditions remains below the limit of detection of the shotgun proteomic approach, but that significant upregulation during N limitation allowed these transporters to be detected. The abundance of NO3 − transporters (Fig. 4) and nitrate reductase (Fig. 5) was significantly greater under N limitation, consistent with the preference of E. huxleyi for NH4 + relative to NO3 −. This suggests nitrate reductase was induced in the N‐limited cells as all available NH4 + was continuously assimilated, and thus 50% of their N uptake was from NO3‐, but under both P‐limited and nutrient‐replete conditions nitrate reductase was repressed due to abundant available NH4 +. Reduced expression of nitrate reductase when E. huxleyi was grown on NH4 + has been reported previously (Bruhn et al., 2010), while in diatoms both NO3 − reductase transcription (Song and Ward, 2004) and enzyme activity (Lomas and Gilbert, 1999) have been shown to be induced by NO3 − and inhibited by NH4 +.
Figure 4.
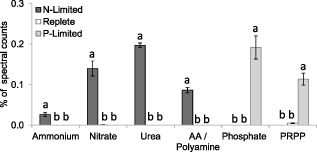
Changes in relative abundance of nutrient transporter proteins (for nutrients as labelled: AA, amino acid; PRPP, phosphate repressible phosphate permease) in E miliania huxleyi 1516 in response to N and P limitation. Significant differences are shown by analysis of variance (P < 0.0001 in all cases) and Tukey honestly significant difference tests (different letters a, b, c indicate significant differences at 0.05 confidence level).
Figure 5.
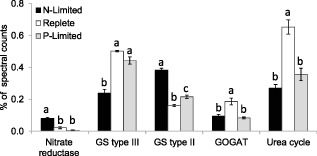
Changes in relative abundance of nitrate reductase, the GS/GOGAT (glutamine synthetase‐glutamate synthase) and urea cycle pathway proteins in E miliania huxleyi 1516 in response to N and P limitation. Significant differences are shown by analysis of variance (P < 0.002 in all cases) and Tukey honestly significant difference tests (different letters a, b, c indicate significant differences at 0.05 confidence level).
As well as the observed upregulation of inorganic nitrogen transporters under N limitation, the proteome was further modified for the uptake of DON. Urea and amino acid/polyamine transporters were observed in N‐limited cells, but were undetected in nutrient‐replete and P‐limited cells (Fig. 4). Urease, the enzyme that catalyses the hydrolysis of urea into carbon dioxide and NH4 +, was upregulated alongside the urea transporters (Fig. 6). The upregulation of all these inorganic and organic N transporters strongly suggests a strategy for scavenging all types of N sources from the surrounding environment, which is induced by N limitation. Numerous C‐N hydrolases were upregulated to an even greater extent (Fig. 6); the most abundantly detected of these was AMI1 formamidase (hydrolyses formamide into formate and NH4 +). This was despite the fact that the only form of N provided in our experiments was NH4NO3, indicating that expression was induced by N limitation not by the presence of urea or formamide. While it is possible that further regulation of expression of key genes may be brought about by the presence of organic compounds such as urea or formamide, it has previously been shown that large fold changes in formamidase expression are approximately equal when E. huxleyi is growing on formamide or under N limitation, when compared with growth under NO3‐replete conditions (Bruhn et al., 2010). Cytosolic hydrolases such as formamidase are upregulated when growing on amides transported from the environment, but a number of different hydrolases were upregulated, the exact functions of which are unknown, so it is possible that some may also play a role in internal remobilization of N from amino acids, proteins or other N containing molecules to provide glutamine synthetase (GS) with NH4 + under N limitation. Although internal remobilization may contribute to changes in cell composition when growth is unbalanced, such as what occurs when cells enter stationary phase in nutrient‐starved cultures, our cultures were in balanced growth, for which we do not expect internal remobilization to be significant. This is because internal remobilization would require that the molecule of interest be synthesized and then subsequently degraded without making a contribution to net growth; as such, it would represent a metabolic cost without an obvious benefit.
Figure 6.
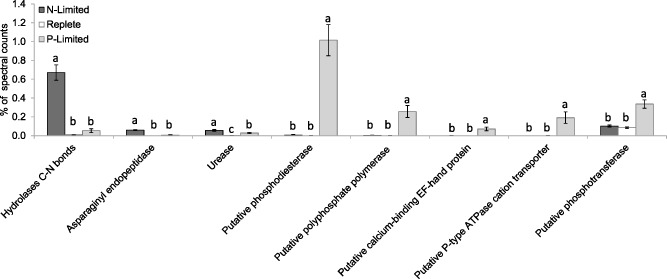
Changes in relative abundance of other proteins associated with organic N and P turnover/assimilation in E miliania huxleyi 1516 in response to N and P limitation. Significant differences are shown by analysis of variance (P < 0.01 in all cases) and Tukey honestly significant difference tests (different letters a, b, c indicate significant differences at 0.05 confidence level).
Our observations are consistent with previous reports that E. huxleyi can thrive on various forms of DON, including urea, purines, and amides such as acetamide and formamide (Antia et al., 1975; Ietswaart et al., 1994; Palenik and Henson, 1997; Bruhn et al., 2010). Formamidase activity has been shown to increase up to 31‐fold during N limitation, but formamidase expression is reduced when growing on NO3 − (Bruhn et al., 2010). While some other phytoplankton can also utilize DON (Wawrik et al., 2009), some organic sources of N such as formamide have been shown to be a poor N source for numerous phytoplankton, with many species unable to grow on it at all (Palenik and Henson, 1997). In contrast, for E. huxleyi, formamide was in fact a significantly better N source than NO3 −. Asparaginyl endopeptidase was also significantly more abundant under N limitation (Fig. 6) and may play a role in proteolysis and mobilization of N within proteins, as can occur in vacuolar bulk protein degradation in plants (Chen et al., 2004).
Phosphorus acquisition
Proteome modification during P limitation was even more pronounced than that observed in response to N limitation. Inorganic PO4 3− transporters were only detected during P limitation (Fig. 4) as were numerous other proteins with putative roles in P assimilation (Fig. 6). Proteins that were significantly higher under P limitation also included a phosphate repressible phosphate permease (Fig. 4), and putative phosphodiesterase, polyphosphate polymerase, EF‐hand protein, P‐type ATPase cation transporter, and phosphotransferase proteins (Fig. 6). Two hypothetical proteins (4739624 and 463656) detected under P limitation and absent under N limitation and nutrient‐replete conditions may also have a role in phosphorus metabolism. The most striking change in response to P limitation was a large increase in the abundance of alkaline phosphatase (AP), which in terms of spectral counts was the most abundantly detected protein under P limitation (Fig. 7). Alkaline phosphatase cleaves 5′ phosphate groups from DNA and RNA, nucleotides and proteins by hydrolysis of phosphate‐ester bonds. Thus, as in Aureococcus anophagefferens (Wurch et al., 2011) and Thalassiosira pseudonana (Dyhrman et al., 2012), acclimation to P limitation in E. huxleyi resulted in the upregulation of the ability to obtain inorganic P by cleaving phosphate from dissolved organic matter and enhancing transport of inorganic P into the cell by increasing the abundance of PO4 3− transporters.
Figure 7.
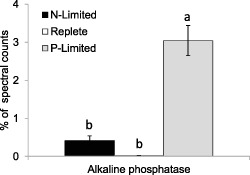
Large change in relative abundance of alkaline phosphatase enzymes (for dephosphorylation of organic P) in E miliania huxleyi 1516 in response to N and P limitation. Significant differences are shown by analysis of variance (P < 0.0001) and Tukey honestly significant difference tests (different letters a and b indicate significant differences at 0.05 confidence level).
Alkaline phosphatases are cell surface proteins in P‐limited E. huxleyi for scavenging external sources of organic P from the environment (Dyhrman and Palenik, 2003; Landry et al., 2006). Alkaline phosphatase is expressed by many phytoplankton in response to P limitation (Kuenzler and Perras, 1965; Sakshaug et al., 1984; Dyhrman and Palenik, 1997; Dyhrman and Palenik, 2003; Wurch et al., 2011), and AP activity has been suggested as an index of P limitation (Beardall et al, 2001). However, Kuenzler and Perras (1965) reported E. huxleyi had the highest AP production rates of 10 different algae, and Reigman and colleagues (2000) reported E. huxleyi's affinity for P to be the highest recorded for a phytoplankton species and can dominate in competition experiments with low P (Reigman et al., 1992). Diatoms in contrast may be poor competitors in low P environments (Egge, 1998). This evidence all suggests how blooms of E. huxleyi can occur after demise of diatom blooms when inorganic N and P are depleted, and particularly why it is hypothesized that P limitation is a key fact that allows E. huxleyi to outcompete other phytoplankton and bloom to large cell densities (Lessard et al., 2005). The AP activity in E. huxleyi has been shown to be induced approximately 6 days after P limitation, and its activity is rapidly lost when PO4 3− is added to P‐limited cultures (Dyhrman and Palenik, 2003). Expression of the gene ehap1 has been shown to increase in E. huxleyi over 1000‐fold within 24 h after cells were transferred from P‐replete to P‐deplete medium with its expression being regulated by extracellular and intracellular P concentrations (Xu et al., 2006).
The thickness of the diffusion boundary layer decreases as cells become smaller, meaning there is less likelihood of limitation of nutrient uptake by molecular diffusion. Thus, the decrease of cell size in E. huxleyi in response to N limitation could be interpreted as an acclimation that increases the potential for nutrient uptake. In fact, the reduction of transport limitation with decreasing cell size is commonly invoked as one of the factors that favours small cell size in the oligotrophic ocean. The increase of cell size in response to P limitation in E. huxleyi is inconsistent with this tendency and requires another explanation. Cell surface biotinylation experiments have shown that APs are dominant cell surface proteins in P‐limited E. huxleyi 1516 (Landry et al., 2006). The very large increase in abundance of the EHAP1 protein (433041) as well as another AP (414308), which together accounted for 3% of the spectral counts in P‐limited E. huxleyi, will require a large area of plasma membrane for attachment and may explain the increase in cell surface area relative to cell P content under P limitation. The larger P‐limited cells had about 40% more surface area than nutrient‐replete cells, which would be able to accommodate the additional cell surface AP. Some of the differences in cell sizes under N or P limitation could also possibly be attributed to the different forms of nutrient limitation affecting the progression of cells though the cell cycle. For example, if cells are arresting during the G1 phase (before much of the cells growth has occurred) under N limitation, but during the G2 + M phase under P limitation, differences in the average cells sizes of the populations would be seen. In both the diatom Thalassiosira weissflogii and the coccolithophorid Hymenomonas carterae, N‐starved cells arrested in the early part of the cell cycle at the G1 phase (Vaulot et al., 1987). The length of both G1 and G2 phases decrease with growth rate increase in the diatom Thalassiosira pseudonana (Claquin et al., 2002) under both N and P limitation, but with a lower percentage of cells in the S phase (when DNA is replicated) and a larger percentage of cells in the G2 + M phase under P limitation. In contrast, P starvation of Prochlorococcus spp. has been shown to induce accumulation of cells in the S phase as well as in the G2 phase in (Parpais et al., 1996).
N assimilation and amino acid synthesis
Most of the proteins involved in nitrogen assimilation and amino acid synthesis were less abundant in the slower growing nutrient‐limited cells (Fig. 5 and Table 1). Glutamate synthase (GOGAT) and the urea cycle enzymes (carbamoyl‐phosphate synthase (CPS), argininosuccinate synthase and argininosuccinate lyase) were approximately twofold less abundant in the nutrient‐limited conditions (Fig. 5). Glutamate synthase is required for the assimilation of NH4 + into amino acids, whereas the urea cycle enzyme CPS plays a key role in the synthesis of amido‐N from NH4 + (Allen et al., 2011). The urea cycle also plays a role in the recycling of NH4 + generated by protein turnover (Armbrust et al., 2004). The cycle appears to be essential for diatom growth, with knockdown of CPS found to impair the response of nitrogen‐limited diatoms to nitrogen addition (Allen et al., 2011); our data suggest that it also plays a role in the coccolithophore E. huxleyi. In contrast, GS, also required for NH4 + assimilation into amino acids via the GS‐GOGAT pathway, showed a more complicated pattern of response. Significantly, the decrease was evident under both N limitation and P limitation, suggesting that the reduced demand for N due to the lower rates of biosynthesis allowed reallocation of N to other proteins. In addition, under N limitation, the larger type III GS (436026; 72 kDa), which requires considerably more N to synthesize, was significantly less abundant (approximately twofold), whereas smaller type II GS (437187, 470023, 69253, 124007; 14–47 kDa) were upregulated and were approximately twofold more abundant (Fig. 5), although the sum of GS type II and GS type III remained a constant proportion of the spectral counts. Upregulation of the type II GS and nitrate reductase in the N‐limited cultures (where 50% of N uptake was from NO3 −) is consistent with the observation in diatoms of the role of II GS in assimilating NH4 + derived from NO3 − reduction (Takabayashi et al., 2005).
Glycolysis and Krebs cycle
The sum of spectral counts assigned to proteins involved in glycolysis were 17% greater under N limitation and 40% more abundant under P limitation. Proteins involved in the Krebs cycle were also some 50% greater in both P‐limited and N‐limited cells than in nutrient‐replete cells. This pattern is consistent with the high ratio of respiration to photosynthesis that is typically observed in nutrient‐limited cells. Isocitrate dehydrogenase was the exception, which was most abundant in nutrient‐replete cells, although the difference between P‐limited and nutrient‐replete conditions was not significant. The higher rate of amino acid synthesis required to support higher growth rates in nutrient‐replete conditions may account for this pattern. Isocitrate dehydrogenase catalyses the synthesis of 2‐oxoglutarate, which is one of the substrates used in amino acid synthesis via the GS‐GOGAT pathway. Also, among the mitochondrial proteins that were twofold less abundant under nutrient limitation was pyruvate carboxylase (B9X0T8), which catalyses the carboxylation of pyruvate to form oxaloacetate (OAA), and thus plays a role in the anapleurotic assimilation of bicarbonate, leading to synthesis of aspartate, other amino acids and pyrimidines.
Protein synthesis
Relative to nutrient‐replete cells, the sum of proteins involved in transcription and translation was approximately 13% lower under N limitation and 25% lower under P limitation (Table 1). Approximately 300 proteins were assigned to this group, although for most we detected too few spectral counts to determine any differences at the level of individual proteins.
Large differences were evident in abundance of ribosomal proteins, which were 20% less abundant under N limitation and almost 50% lower under P limitation than in nutrient‐replete cultures (Table 1), with greater reductions in the abundance of chloroplastic ribosomal proteins than of cytoplasmic ribosomal proteins. The large reduction under P limitation is consistent with the growth rate hypothesis, which links synthesis of RNA to P assimilation (Sterner and Elser, 2002). Assuming limited variability in the ribosomal efficiency, the growth rate hypothesis predicts proportionality between specific growth rate and rRNA : protein. This hypothesis has also been invoked to explain differences in organismal C : N : P ratios, which are assumed to be caused by differences in the amounts of RNA needed to meet the protein synthesis demands of rapid rates of biomass growth and development (Sterner and Elser, 2002).
Proteolysis
Total counts for proteins involved in protein degradation (the sum of counts of over 130 individual proteasome and ubiquitin proteins) were higher under N limitation (Table 1). The eukaryotic ubiquitin‐proteasome system plays an essential role in adaptation to N limitation (Sato et al., 2011), and the observed upregulation under N limitation suggests an increase in proteins being ubiquitylated for targeted degradation by the proteasome to remobilize nitrogen.
Histones
Histones are small, basic proteins that interact with the DNA double helix to form nucleosomes (Mcghee and Felsenfeld, 1980). Histones 2A and 2B differed significantly among treatments (Table 1). Relative to the nutrient‐replete cells, these proteins were 50% more abundant under N limitation and 44% less abundant under P limitation. The proportion of the proteome assigned to histones followed a similar pattern to DNA‐to‐protein ratio (not shown), being highest in N‐limited cells, intermediate in nutrient‐replete cells and lowest in P‐limited cells.
Conclusions
In this study we observed modest changes in much of the proteome of E. huxleyi despite the two to three‐fold reduction in growth rate and large physiological changes (e.g. cellular biomass and C, N and P), due to nutrient limitation. Similarly, relatively small changes in the abundance of proteins involved in many key pathways were observed in our previous study of acclimation of E. huxleyi to irradiance (McKew et al., 2013a, 2013b) and that of Jones and colleagues (2013) to elevated CO2 (1390 p.p.m. versus 390 p.p.m.). In contrast to the stability of most of the proteome to a range of abiotic factors, abiotic limitation or stress can induce large changes in subsets of proteins that are involved in counteracting the limitation or stress. For example, E. huxleyi's response to high irradiance involved increases in proteins involved in photoprotection at the expense of those involved in light harvesting (McKew et al., 2013b). In contrast, in the current study, we found that acclimation of E. huxleyi to nutrient limitation was most evident in marked increases in the abundance of proteins involved in inorganic nutrient transport and both the scavenging and internal remobilization of organic forms of N and P (e.g. C‐N hydrolases, urea and amino acid/polyamine transporters and AP). Nonetheless, these proteins that were upregulated under N or P limitation accounted for only 1.7% and 5.7% of the total spectral counts respectively. This highly targeted reorganization of the proteome towards scavenging of DON and DOP gives unique insight into how E. huxleyi has found its niche to bloom in highly oligotrophic surface waters.
Experimental procedures
Culture conditions
Cultures of the calcifying strain E. huxleyi 1516 were grown in filter sterilized (0.2 μm) artificial seawater enriched seawater, artificial water (ESAW) (Berges et al., 2001) with bicarbonate amended to 3 mM and enriched with f/8 vitamins and trace metals (Guillard and Ryther, 1962) and the addition of 1 nM selenium. Inorganic N and P additions were altered in the different treatments. Nutrient‐replete media contained 150 μM NH4NO3 and 20 μM PO4 3− (N : P = 15 : 1), N‐limited media contained 25 μM NH4NO3 and 20 μM PO4 3− (N : P = 2.5 : 1), whereas the P‐limited media contained 150 μM NH4NO3 and 7 μM PO4 3− (N : P = 43 : 1). Stock cultures used as inocula were pretreated with antibiotics to remove any contaminating bacteria. The cultures were grown in 3 l cylindrical borosilicate glass vessels and maintained at a constant temperature of 18°C. The cultures were gently and continuously stirred with a magnetic stirrer bar and aerated (100 cm3 min–1) via a borosilicate glass gas distribution tube (8 mm diameter, porosity 1; Fisher Scientific, Loughborough, UK). The cultures were illuminated with cool‐white fluorescent tubes (Lumilux CoolWhite, OSRAM, Munich, Germany) on a light : dark cycle of 16 : 8 h at a PFD of 300 μmol photons m–2 s–1 (measured in the centre of the culture vessels while containing distilled water with a 4π Biospherical Instruments QSL100 light sensor, San Diego, CA, USA).
Nutrient‐replete cultures were maintained in exponential steady‐state growth phase at cell concentrations of approximately 6 × 105 ml–1 by continuous dilution. In the nutrient‐limited cultures, the dilution rate was reduced to achieve specific growth rates that were two to three times lower than under nutrient‐replete conditions. The cells were grown into steady‐state nutrient limitation (confirmed by reduced growth rates and no measurable inorganic N or P in respective vessels) and maintained through 10 generations under these conditions before sampling. Four replicate cultures were grown for each treatment, and all were sampled 4 h into the light cycle. Cultures were monitored daily for cell abundance and to confirm the absence of bacterial contaminants by direct counts with an improved Neubauer hemocytometer (Fisher Scientific) and cell size with a Coulter Counter Z2 particle counter (Beckman Coulter, High Wycombe, UK). Growth rates were calculated from the cell counts and dilution rates as described previously (Brading et al., 2011). Residual dissolved NO3 − and PO4 3− in the culture medium were measured daily after filtering (0.2 μm), using the methods of Collos and colleagues (1999) and Solorzano and Sharp (1980) respectively.
Particulate composition
Samples for determination of POC, PIC, and particulate nitrogen, particulate phosphorus, chlorophyll a, DNA, RNA and protein quantification were collected and processed as described in McKew and colleagues (2013a).
Lipid analysis
Cells were filtered onto MF300 glass microfibre filters (Fisher Scientific) and flash‐frozen in liquid N2 until extraction. Total lipids were extracted from cells by a modified Bligh and Dyer (1959) method in 9.5 ml of high‐performance liquid chromatography (HPLC) grade methanol : chloroform : H2O (10 : 5 : 4 v\v\v) and sonicated for 30 min. A further 2.5 ml of chloroform and 2.5 ml of H2O was added (final ratio 10 : 10 : 9) and the extract centrifuged (700 g). The lower organic layer was to be separated and dried under N2 at 37°C. Lipid fractions were separated using 3 ml Supelco Discovery DSC‐Si Solid Phase Extraction cartridges (Sigma‐Aldrich, St Louis, MO, USA) with 0.5 g sodium sulfate added to the top of the cartridges as a drying agent. Neutral lipids were eluted with 10 ml chloroform, glycol lipids with 10 ml of acetone and phospholipids with 10 ml of methanol. All lipid fractions were evaporated to dryness under N2 at 37°C. Derivatization was performed by mild alkaline methanolysis (Dowling et al., 1986) by reconstituting dried lipids in 1 ml HPLC grade toluene : methanol (1 : 1 v/v dried on sodium sulfate), adding 1 ml of 0.2 M methanolic potassium hydroxide, gently mixing and incubated at 37°C for 30 min. The reaction was stopped with 0.25 ml of 1 M acetic acid. Of HPLC grade hexane : chloroform (4 : 1 v/v), 5 ml was added, sonicated for 30 min, centrifuged to separate the two phases, and the aqueous lower layer was removed and discarded. Of 0.3 M sodium hydroxide, 3 ml was added and the extract filtered via sodium sulfate‐packed glass syringes, and then evaporated to dryness under N2 at 25°C. The dried fatty acid methyl esters (FAMEs) were reconstituted with 200 μl of hexane (HPLC grade) containing an alpha‐cholestane injection standard. Fatty acid methyl esters were separated and analysed by a splitless injection of 1 μl onto a Thermo Trace gas chromatograph (injector temp 270°C) coupled to a Thermo DSQ mass spectrometer (transfer line 270°C, ion source 225°C) using a Supelco SP‐2380 fused silica capillary column (30 m length, 0.25 mm ID, 0.2 μm film; Sigma‐Aldrich). Helium was used as the carrier gas (1 ml min–1), and the FAMEs were separated by using a temperature programme, starting at 60°C for 1 min increasing at 5°C min–1 to 160°C, followed by 10°C min–1 to 240°C and 25°C min–1 until reaching 270°C. All peak areas were normalized to the injection standard, and the FAMEs were identified and quantified against Supelco FAME analytical standards (Sigma‐Aldrich) and by comparison of ion spectra against the NIST Atomic Spectra Database.
Proteomic analysis
Cells were collected from 300 ml of culture by vacuum filtration onto Whatman 3 μm pore polycarbonate filters (GE Healthcare, Little Chalfont, UK), which were flash‐frozen and stored in liquid N2 until proteomics analysis. Proteins were extracted, digested with trypsin, the constituent tryptic peptides concentrated and analysed by electrospray‐ionization tandem mass spectrometry on a hybrid high‐resolution LTQ/Orbitrap Velos instrument (Thermo Scientific, Hemel Hempstead, UK) using 2 μg peptides per injection onto 15 cm long pulled‐tip nanocolumn, as detailed in McKew and colleagues (2013b). The resulting MS/MS raw data files were converted to mzXML format using the ReAdW programme and uploaded onto the LabKey server. The open‐source search engine x! tandem was used for protein identification and acquisition of spectral count data. The primary statistical evaluation and filtering of the protein and spectral count data was performed using the PeptideProphet and ProteinProphet programmes (Nesvizhskii et al., 2003) integrated into the LabKey CPAS (Computational Proteomics Analysis System version 2.2). Peptides and proteins were filtered at 0.3% false discovery rate to obtain the final dataset. Proteins were quantified by counting the number of MS/MS spectra matched to corresponding proteins, and all counts were normalized to the total number of spectral counts. Each of the four biological replicates for each treatment was re‐injected and analysed three times (technical replicates) for higher confidence of the spectral counts for each biological replicate (i.e. the spectral counts assigned to each biological replicate for any individual protein is the mean of three technical replicates). Uniprot and the E. huxleyi 1516 genome (Read et al., 2013) databases were used for the identification of proteins.
Supporting information
Table S1. Normalized spectral counts for all detected proteins (each biological replicate presented is the mean of three technical replicates).
Acknowledgement
This research was supported by NERC Grant NE/G003688/1.
Subject category: microbial ecology and functional diversity of natural habitats: aquatic and sediment microbial ecology.
References
- Allen, A.E. , Dupont, C.L. , Oborník, M. , Horák, A. , Nunes‐Nesi, A. , McCrow, J.P. , et al (2011) Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473: 203–207. [DOI] [PubMed] [Google Scholar]
- Antia, N.J. , Berland, B.R. , Bonin, D.J. , and Maestrini, S.Y. (1975) Comparative evaluation of certain organic and inorganic sources of nitrogen for phototropic growth of marine microalgae. J Mar Biol Assoc UK 55: 519–539. [Google Scholar]
- Armbrust, E.V. , Berges, J.A. , Bowler, C. , Green, B.R. , Martinez, D. , Putnam, N.H. et al (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86 [DOI] [PubMed] [Google Scholar]
- Beardall, J. , Young, E. , and Roberts, S. (2001) Approaches for determining phytoplankton nutrient limitation. Aquat Sci 63: 44–69. [Google Scholar]
- Berges, J.A. , Franklin, D.J. , and Harrison, P.J. (2001) Evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the last two decades. J Phycol 37: 1138–1145. [Google Scholar]
- Bligh, E.G. , and Dyer, W.J. (1959) A rapid method for total lipid extraction and purification. Can J Biochem Physiol 37: 911–917. [DOI] [PubMed] [Google Scholar]
- Borchard, C. , Borges, A.V. , Händel, N. , and Engel, A. (2011) Biogeochemical response of Emiliania huxleyi (PML B92/11) to elevated CO2 and temperature under phosphorous limitation: a chemostat study. J Exp Mar Biol Ecol 410: 61–71. [Google Scholar]
- Brading, P. , Warner, M.E. , Davey, P. , Smith, D.J. , Achterberg, E.P. , and Suggett, D.J. (2011) Differential effects of ocean acidification on growth and photosynthesis among phylotypes of Symbiodinium (Dinophyceae). Limnol Oceanogr 56: 927–938. [Google Scholar]
- Brown, C.W. , and Yoder, J.A. (1994) Coccolithophorid blooms in the global ocean. J Geophys Res 99: 7467–7482. [Google Scholar]
- Bruhn, A. , LaRoche, J. , and Richardson, K. (2010) Emiliania huxleyi (Prymnesiophyceae): nitrogen‐metabolism genes and their expression in response to external nitrogen sources. J Phycol 46: 266–277. [Google Scholar]
- Chen, H.‐J. , Hou, W.‐C. , Liu, J.‐S. , Yang, C.‐Y. , Huang, D.‐J. , and Lin, Y.‐H. (2004) Molecular cloning and characterization of a cDNA encoding asparaginyl endopeptidase from sweet potato (Ipomoea batatas (L.) Lam) senescent leaves. J Exp Bot 55: 825–835. [DOI] [PubMed] [Google Scholar]
- Claquin, P. , Martin‐Jezequel, V. , Kromkamp, J.C. , Veldhuis, M.J.W. , and Kraay, G.W. (2002) Uncoupling of silicon compared with carbon and nitrogen metabolisms and the role of the cell cycle in continuous cultures of Thalassiosira pseudonana (Bacillariophyceae) under light, nitrogen, and phosphorus control. J Phycol 38: 922–930. [Google Scholar]
- Collos, Y. , Mornet, F. , Sciandra, A. , Waser, N. , Larson, A. , and Harrison, P.J. (1999) An optical method for the rapid measurement of micromolar concentrations of nitrate in marine phytoplankton cultures. J Appl Phycol 11: 179–184. [Google Scholar]
- Dowling, N.J.E. , Widdel, F. , and White, D.C. (1986) Phospholipid ester‐linked fatty acid biomarkers of acetate‐oxidizing sulphate‐reducers and other sulphide‐forming bacteria. J Gen Microbiol 132: 1815–1825. [Google Scholar]
- Dyhrman, S.T. , and Palenik, B. (2003) Characterization of ectoenzyme activity and phosphate‐regulated proteins in the coccolithophorid Emiliania huxleyi . J Plankton Res 25: 1215–1225. [Google Scholar]
- Dyhrman, S.T , and Palenik, B.P. (1997) The identification and purification of a cell‐surface alkaline phosphatase from the dinoflagellate Prorocentrum minimum (Dinophyceae). J Phycol 33: 602–612. [Google Scholar]
- Dyhrman, S.T. , Jenkins, B.D. , Rynearson, T.A. , Saito, M.A. , Mercier, M.L. , Alexander, H. , et al (2012) The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE 7: e33768. doi:10.1371/journal.pone.0033768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egge, J.K. (1998) Are diatoms poor competitors at low phosphate concentrations? J Marine Syst 16: 191–198. [Google Scholar]
- Eltgroth, M.L. , Watwood, R.L. , and Wolfe, G.V. (2005) Production and cellular localization of neutral long‐chain lipidsin the haptophyte algae Isochrysis galbana and Emiliania huxleyi . J Phycol 41: 1000–1009. [Google Scholar]
- Fernández, E. , Balch, W.M. , Marañon, E. , and Holligan, P.M. (1994) High rates of lipid biosynthesis in cultured, mesocosm and coastal populations of the coccolithophore Emiliania huxleyi . Mar Ecol Prog Ser 114: 13–22. [Google Scholar]
- Graziano, L.M. , Geider, R.J. , Li, W.K.W. , and Olaizola, M. (1996) Nitrogen limitation of North Atlantic phytoplankton: analysis of physiological condition in nutrient enrichment experiments. Aquat Microb Ecol 11: 53–64. [Google Scholar]
- Guillard, R.R.L. , and Ryther, J.H. (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea (Cleve) Gran. Can J Microbiol 8: 229–239. [DOI] [PubMed] [Google Scholar]
- Holligan, P.M. , Fernandez, E. , Aiken, J. , Balch, W.M. , Boyd, P. , Burkill, P.H. , et al (1993) A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North‐Atlantic. Global Biogeochem Cycles 7: 879–900. [Google Scholar]
- Ietswaart, T. , Schneider, P.J. , and Prins, R.A. (1994) Utilization of organic nitrogen‐sources by two phytoplankton species and a bacterial isolate in pure and mixed cultures. Appl Environ Microbiol 60: 1554–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias‐Rodriguez, M.D. , Brown, C.W. , Doney, S.C. , Kleypas, J. , Kolber, D. , Kolber, Z. , et al (2002) Representing key phytoplankton functional groups in ocean carbon cycle models: coccolithophorids. Global Biogeochem Cycles 16: 47‐1–47‐20. [Google Scholar]
- Jones, B.M. , Iglesias‐Rodriguez, M.D. , Skipp, P.J. , Edwards, R.J. , Greaves, M.J. , Young, J.R. , et al (2013) Responses of the Emiliania huxleyi proteome to ocean acidification. PLoS ONE 8: e61868. doi:10.1371/journal.pone.0061868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffes, A. , Thoms, S. , Trimborn, S. , Rost, B. , Langer, G. , Richter, K.‐U. et al (2010) Carbon and nitrogen fluxes in the marine coccolithophore Emiliania huxleyi grown under different nitrate concentrations. J Exp Mar Biol Ecol 393: 1–8. [Google Scholar]
- Kuenzler, E.J. , and Perras, J.P. (1965) Phosphatases of marine algae. Biol Bull 128: 271–286. [Google Scholar]
- Landry, D.M. , Gaasterland, T. , and Palenik, B.P. (2006) Molecular characterization of a phosphate‐regulated cell‐surface protein from the coccolithophorid, Emiliania huxleyi (Prymnesiophyceae). J Phycol 42: 814–821. [DOI] [PubMed] [Google Scholar]
- Leonardos, N. , and Geider, R.J. (2005) Elevated atmospheric carbon dioxide increases organic carbon fixation by Emiliania huxleyi (haptophyta), under nutrient‐limited high‐light conditions. J Phycol 41: 1196–1203. [Google Scholar]
- Lessard, E.J. , Merico, A. , and Tyrrell, T. (2005) Nitrate : phosphate ratios and Emiliania huxleyi blooms. Limnol Oceanogr 50: 1020–1024. [Google Scholar]
- Loladze, I. , and Elser, J.J. (2011) The origins of the Redfield nitrogen‐to‐phosphorus ratio are in a homoeostatic protein‐to‐rRNA ratio. Ecol Lett 14: 244–250. [DOI] [PubMed] [Google Scholar]
- Lomas, M.W. , and Gilbert, P.M. (1999) Interactions between NH4 + and NO3 − uptake and assimilation: comparison of diatoms and dinoflagellates at several growth temperatures. Mar Biol 133: 541–551. [Google Scholar]
- McGhee, J.D. , and Felsenfeld, G. (1980) Nucleosome Structure. Ann Rev Biochem 49: 1115–1156. [DOI] [PubMed] [Google Scholar]
- McKew, B.A. , Lefebvre, S.C. , Achterberg, E.P. , Metodieva, G. , Raines, C.A. , Metodiev, M.V. , and Geider, R.J. (2013a) Plasticity in the proteome of Emiliania huxleyi to extremes of light is highly targeted. New Phytol 200: 61–73. doi:10.1111/nph.12352. [DOI] [PubMed] [Google Scholar]
- McKew, B.A. , Davey, P. , Finch, S.J. , Hopkins, J. , Lefebvre, S.C. , Metodiev, M.V. , et al (2013b) The trade‐off between light‐harvesting and photoprotective functions of fucoxanthin‐chlorophyll proteins dominates light acclimation in Emiliania huxleyi (clone CCMP 1516). New Phytol 200: 74–85. doi:10.111/nph.12373. [DOI] [PubMed] [Google Scholar]
- Moore, C.M. , Mills, M.M. , Milne, A. , Langlois, R. , Achterberg, E.P. , Lochte, K. , et al (2006) Iron supply limits primary productivity during spring bloom development in the central North Atlantic. Glob Change Biol 12: 626–634. [Google Scholar]
- Moore, C.M. , Mills, M.M. , Languois, R. , Milne, A. , Achterberg, E.P. , La Roche, J. , and Geider, R.J. (2008) Relative influence of nitrogen and phosphorus availability on phytoplankton physiology and productivity in the oligotrophic sub‐tropical North Atlantic Ocean. Limnol Oceanogr 53: 291–305. [Google Scholar]
- Moore, C.M. , Mills, M.M. , Arrigo, K.R. , Berman‐Frank, I. , Bopp, L. , Boyd, P.W. , et al (2013) Processes and patterns of oceanic nutrient limitation. Nat Geosci 6: 701–710. doi:10.1038/ngeo1765. [Google Scholar]
- Nesvizhskii, A.I. , Keller, A. , Kolker, E. , and Aebersold, R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658. [DOI] [PubMed] [Google Scholar]
- Paasche, E. (2001) A review of the coccolithophorid Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification‐photosynthesis interactions. Phycologia 40: 503–529. [Google Scholar]
- Palenik, B. , and Henson, S.E. (1997) The use of amides and other organic nitrogen sources by the phytoplankton Emiliania huxleyi . Limnol Oceanogr 42: 1544–1551. [Google Scholar]
- Parpais, J. , Marie, D. , Partensky, F. , Morin, P. , and Vaulot, D. (1996) Effect of phosphorus starvation on the cell cycle of the photosynthetic prokaryote Prochlorococcus . Mar Ecol Prog Ser 132: 265–274. [Google Scholar]
- Pond, D.W. , and Harris, R.P. (1996) The lipid composition of the coccolithophore Emiliania huxleyi and its possible ecophysiological significance. J Mar Biol Assoc UK 76: 579–594. [Google Scholar]
- Read, B.A. , Kegel, J. , Klute, M.J. , Kuo, A. , Lefebvre, S.C. , Maumus, F. , et al (2013) Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499: 209–213. doi:10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- Reigman, R. , Noordeloos, A.A.M. , and Cadee, G.C. (1992) Phaeocystis blooms and eutrophication of the continental coastal zones of the North Sea. Mar Biol 112: 479–484. [Google Scholar]
- Reigman, R. , Stolte, W. , Noordeloos, A.A.M. , and Slezak, D. (2000) Nutrient uptake and alkaline phosphatase (EC 3:1:3:1) activity of Emiliania huxleyi (Prymnesiophyceae) during growth under N and P limitation in continuous cultures. J Phycol 36: 87–96. [Google Scholar]
- Sakshaug, E. , Graneli, E. , Elbrächter, M. , and Kayser, H. (1984) Chemical composition and alkaline phosphatase activity of nutrient saturated and P‐deficient cells of four marine dinoflagellates. J Exp Mar Biol Ecol 77: 241–254. [Google Scholar]
- Sato, T. , Maekawa, S. , Yasuda, S. , and Yamaguchi, J. (2011) Carbon and nitrogen metabolism regulated by the ubiquitin‐proteasome system. Plant Signal Behav 6: 1465–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayanova, O. , Haslam, R.P. , Calerón, M.V. , López, N.R. , Worthy, C. , Rooks, P. , et al (2011) Identification and functional characterisation of genes encoding the omega‐3 polyunsaturated fatty acid biosynthetic pathway from the coccolithophore Emiliania huxleyi . Phytochemistry 72: 594–600. [DOI] [PubMed] [Google Scholar]
- Solorzano, L. , and Sharp, J.H. (1980) Determination of total dissolved phosphorus and particulate phosphorus in natural waters. Limnol Oceanogr 25: 754–758. [Google Scholar]
- Song, B. , and Ward, B.B. (2004) Molecular characterization of the assimilatory nitrate reductase gene and its expression in the marine green alga Dunaliella tertiolecta (Chlorophyceae). J Phycol 40: 721–731. [Google Scholar]
- Sterner, R.W. , and Elser, J.J. (2002) Ecological stoichiometry: the biology of elements from molecules to the biosphere. Princeton, NJ, USA: Princeton University Press. [Google Scholar]
- Strom, S.L. , Wolfe, G.V. , Holmes, J.L. , Stecher, H.A. , Shimeneck, C. , Lambert, S. , and Moreno, E. (2003) Chemical defense in the microplankton I: Feeding and growth rates of heterotrophic protists on the DMS‐producing phytoplankter Emiliania huxleyi . Limnol Oceanogr 48: 217–229. [Google Scholar]
- Suggett, D.J. , Le Floc, H.E. , Harris, G. , Leonardos, N. , and Geider, R.J. (2007) Different strategies of photoacclimation by two strains of Emiliania huxleyi (Haptophyta). J Phycol 43: 1209–1222. [Google Scholar]
- Sukhanova, I.N. , and Flint, M.V. (1998) Anomalous blooming of coccolithophorids over the eastern Bering Sea shelf. Oceanology 38: 502–505. [Google Scholar]
- Takabayashi, M. , Wilkerson, F.P. , and Robertson, D. (2005) Response of glutamine synthetase gene transcription and enzyme activity to external nitrogen sources in the diatom Skeletonema costatum (Bacillariophyceae). J Phycol 41: 84–94. [Google Scholar]
- Thingstad, T.F. , Krom, M.D. , Mantoura, R.F.C. , Flaten, G.A.F. , Groom, S. , Herut, B. , et al (2005) Nature of phosphorus limitation in the ultraoligotrophic eastern Mediterranean. Science 309: 1068–1071. [DOI] [PubMed] [Google Scholar]
- Van Mooy, B.A. , Fredricks, H.F. , Pedler, B.E. , Dyhrman, S.T. , Karl, D.M. , Koblízek, M. , et al (2009) Phytoplankton in the ocean use non‐phosphorus lipids in response to phosphorus scarcity. Nature 458: 69–72. [DOI] [PubMed] [Google Scholar]
- Vaulot, D. , Olson, R.J. , Merkel, S. , and Chisholm, S.W. (1987) Cell‐cycle response to nutrient starvation in two phytoplankton species, Thalassiosira weissflogii and Hymenomonas carterae . Mar Biol 95: 625–630. [Google Scholar]
- Wawrik, B. , Callaghan, A.V. , and Bronk, D.A. (2009) Use of inorganic and organic nitrogen by Synechococcus spp. and diatoms on the West Florida Shelf as measured using stable isotope probing. Appl Environ Microbiol 75: 6662–6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurch, L.L. , Bertrand, E.M. , Saito, M.A. , Van Mooy, B.A.S. , and Dyhrman, S.T. (2011) Proteome changes driven by phosphorus deficiency and recovery in the brown tide‐forming alga Aureococcus anophagefferens . PLOS One 6: e28949. doi:10.1371/journal.pone.0028949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Wahlund, T.M. , Feng, L. , Shaked, Y. , and Morel, F.M.M. (2006) A novel alkaline phosphatase in the coccolithophore Emiliania huxleyi (Prymnesiophyceae) and its regulation by phosphorus. J Phycol 42: 835–844. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Normalized spectral counts for all detected proteins (each biological replicate presented is the mean of three technical replicates).


