Abstract
Background
Spinal muscular atrophy (SMA) is the most common autosomal recessive disease in children, and the diagnosis is complicated and difficult, especially at early stage. Early diagnosis of SMA is able to improve the outcome of SMA patients. In our study, Real-time PCR was developed to measure the gene mutation or deletion of key genes for SMA and to further analyse genotype-phenotype correlation.
Methods
The multiple real-time PCR for detecting the mutations of survival of motor neuron (SMN), apoptosis inhibitory protein (NAIP) and general transcription factor IIH, polypeptide 2 gene (GTF2H2) was established and confirmed by DNA sequencing and multiplex ligation-dependent probe amplification (MLPA). The diagnosis and prognosis of 141 hospitalized children, 100 normal children and further 2000 cases of dry blood spot (DBS) samples were analysed by this multiple real-time PCR.
Results
The multiple real-time PCR was established and the accuracy of it to detect the mutations of SMN, NAIP and GTF2H2 was at least 98.8 % comparing with DNA sequencing and MLPA. Among 141 limb movement disorders children, 75 cases were SMA. 71 cases of SMA (94.67 %) were with SMN c.840 mutation, 9 cases (12 %) with NAIP deletion and 3 cases (4 %) with GTF2H2 deletion. The multiple real-time PCR was able to diagnose and predict the prognosis of SMA patients. Simultaneously, the real-time PCR was applied to detect trace DNA from DBS and able to make an early diagnosis of SMA.
Conclusion
The clinical and molecular characteristics of SMA in Southwest of China were presented. Our work provides a novel way for detecting SMA in children by using real-time PCR and the potential usage in newborn screening for early diagnosis of SMA.
Electronic supplementary material
The online version of this article (doi:10.1186/s12883-016-0651-y) contains supplementary material, which is available to authorized users.
Keywords: Spinal muscular atrophy, Multiplex real-time PCR, Newborn screening
Background
Spinal muscular atrophy (SMA) is one of the most common autosomal recessive disorders, with an incidence of 1 in 10,000 births [1]. The disease is characterized by the degeneration of the anterior horn cells of the spinal cord, resulting in symmetrical limb muscle atrophy and weakness. SMA is classified into three clinical subtypes: Type I SMA (Werdnig-Hoffmann disease, MIM253300), Type II SMA (MIM253550) and Type III (Kugelberg-Welander disease, MIM253400) [2]. The phenotypes of these three subtypes of SMA are based on previous reports [3].
Previously, a clinical diagnosis of SMA is confirmed by muscle biopsy and electromyography (EMG). These procedures are time-consuming, affected by cross-talk and post-processing artefacts and yield non-conclusive results in young infants [4, 5]. Thus, the diagnosis of SMA is usually dependent on the doctor’s experience. When a diagnosis is made, these infants often suffer from irreversible loss of neurological function. Early diagnosis of SMA can improve outcomes, and the efficacy of early diagnosis of SMA is also motivated by progress in therapeutic development [6].
Due to the shortcomings of the traditional diagnosis of SMA, molecular diagnosis for SMA has gradually been developed in recent years. For the molecular genetics of SMA, all three clinical subtypes of SMA are associated with mutations in the survival of motor neuron (SMN) gene, which is mapped to chromosome 5q13. Normally, this region contains one telomeric SMN1 gene (Genbank: NG_008691.1) and one centromeric paralogue SMN2 gene (Genbank: NG_008728.1), which differs in exon 7 at cDNA residue 840 (C for SMN1 and T for SMN2) [7, 8]. Patients with a homozygous deletion of SMN1 and a high SMN2 copy number have a phenotype [8–10] due to the small fraction of normal transcripts, which indicates that infants with a homozygous mutation on c.840 of C > T will have symptoms of SMA. Moreover, the phenotype is correlated with the exon loss of several genes in the 5q13 region, such as apoptosis inhibitory protein (NAIP) (Genbank: NG_008724.1) and general transcription factor IIH, polypeptide 2 gene (GTF2H2) (Genbank: NT_187651.1) [11]. The absence of exon 4 and 5 of NAIP may result from an unequal crossing over, leading to severe SMA [12]. The GTF2H2 gene and/or exon deletion might be related to the severity of the disease, but its clinical significance is still unclear [13].
The “gold standard” to detect a single nucleotide difference is DNA sequencing, and the best method to test for the exon loss of several genes is multiplex ligation-dependent probe amplification (MLPA) [11]. Only trace DNA can be extracted from a dry blood spot (DBS), which limits the application of DNA sequencing and MLPA during newborn screening, when is the best time to make an early diagnosis of SMA and treat in time, based on DBS being the only specimen could be obtained during newborn screening. Moreover, both of these methods are time-consuming. For restriction fragment length polymorphism (RFLP), which was established by Van’s group [14], it is also time-consuming, taking nearly 20 h to get the result, because the procedure contains PCR and enzyme digestion; on the other hand, it is hard to judge the results in daily work only by 22 base pairs differences through electrophoresis. Multiple real-time PCR has been used to detect single nucleotide differences and exon loss in adults and newborn screening [15, 16], which means multiple real-time PCR diagnosing SMA is very likely to be successful. Moreover, the technique is high sensitivity, specificity and rapid, so we chose it to establish a new method for SMA diagnosis. We screened and chose the best multiple pairs of primers and probes to establish the new real-time PCR method, which was fast (<3 h), cheap (<US$2), accurate, to analyse exons deletion of SMN, NAIP and GTF2H2 genes.
In our study, the clinical and molecular characteristics of a pediatric population with SMA from Southwest China were presented. Real-time PCR was developed to measure the gene mutation or deletion of key genes for SMA and to further analyse genotype-phenotype correlation; subsequently, 2000 cases of DBS were randomly selected for trace DNA, and real-time PCR was performed.
Methods
Patients
This retrospective and prospective study was performed at the Children’s Hospital of Chongqing Medical University. In this study, 1613 children with limb movement disorders were screened, of which 141 children were confirmed to exhibit the inclusion criteria listed in the “Interpretation” section. These 141 children and an additional 100 normal children were enrolled in a retrospective study, and their blood specimens and medical records were collected. Simultaneously, 2000 DBS samples were randomly selected from the Newborn Screening Center at the Children’s Hospital of Chongqing Medical University to carry out the prospective study. This study was approved by the Ethics Committee of Children’s Hospital of Chongqing Medical University, and the reference number was CHCMU-20110005. Written informed consent was obtained from the parents or legal guardians of the patients. The datasets supporting the conclusions of this article are included within the article and its additional files.
Interpretation
The inclusion criteria for the retrospective study were patients confirmed with a normal nutritional status; disorder on sitting, standing or walking; physical examination showing myasthenia or muscle atrophy; sequencing, multiplex PCR and MLPA detected disorder; and hospitalized. There was no inclusion criteria for the prospective study because the samples were selected randomly. The guidelines for SMA diagnosis and typing were based on a previous report [3] and were described in Additional file 1: Table S1.
Real-time PCR for SMN, NAIP and GTF2H2 genes in clinical specimens
All 141 patients and 100 normal children were enrolled. DNA was purified according to the manufacturer’s instructions (DP318-03, Tiangen Biotech, Beijing, China). Next, 1 μl of DNA template (20 ng/μl) and 19 μl of a real-time PCR master mix (FP203-02, Tiangen Biotech, Beijing, China), as shown in Additional file 2: Table S2, were used to perform real-time PCR. One sample should be detected by two real-time PCR reactions: tube I and tube II. Tube I detected the mutation of SMN gene and the deletion of NAIP exon5 and tube II detected internal control and the deletion of NAIP exon4 and GTF2H2 exon 10.
The primers and probes used in this study are shown in Additional file 3: Table S3 and were synthesized by Invitrogen (Life Technologies Corporation, Shanghai, China). Real-time PCR was performed on the Applied Biosystem 7500 real-time PCR system (Applied Biosystems, CA, USA) using the following conditions: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 55 °C for 40 s. All standards and samples were performed in triplicate.
Sequencing for SMN gene in clinical specimens
All 141 patients and 100 normal children were analysed using Sanger DNA sequencing for the point mutation at c.840 C > T of the SMN gene. The DNA template was the same template used in real-time PCR. Each PCR reaction was performed in a 50-μl volume, as shown in Additional file 4: Table S4. PCR was performed on an ABI-Verity Thermal Cycler (Applied Biosystems, Foster City, California, USA) using the following conditions: 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 53 °C for 30 s and 72 °C for 30 s, and a final extension step at 72 °C for 5 min. All samples were referred to Sunny (Sunny Biotechnology Co. Ltd., Shanghai, China) for sequencing.
Multiplex PCR and MLPA for NAIP and GTF2H2 genes in clinical specimens
All 141 patients and 100 normal children were analysed using multiplex PCR and MLPA for the NAIP and GTF2H2 genes as previously reported [11, 17]. The DNA template was the same template that was used in real-time PCR. All of the conditions of each PCR reaction for multiplex PCR were the same as those used for sequencing. All of the samples were amplified in duplicate.
The PCR products of the NAIP and GTF2H2 genes were used for DNA sequencing at Sunny (Sunny Biotechnology Co., Ltd., Shanghai, China) to confirm the expected products. If there were exons missing, as determined by electrophoresis, MLPA was performed on the DNA samples to confirm the results at Genesky (Genesky Biotechnologies Inc., Shanghai, China).
Real-time PCR for SMN, NAIP and GTF2H2 genes in DBS
Real-time PCR was performed on 2000 randomly selected DBS samples. DNA was purified by Chelex-100 (1422842, Bio-Rad Laboratories Co., Ltd., Shanghai, China). Briefly, one dried blood spot (diameter of 3 mm) was clipped and mixed with 500 μl of nuclease-free water to wash and was then centrifuged, and the supernatants were discarded. Next, 5 % Chelex-100 was mixed before the addition of 100 μl to each pellet, and the mixture was incubated at 56 °C for 10 min. The mixture was then mixed and centrifuged to harvest the supernatants.
Real-time PCR was performed on the DNA templates of the 6 μl and 14 μl PCR master mix, as shown in Additional file 5: Table S5. All of the real-time PCR conditions were the same as those described in the “Real-time PCR for SMN, NAIP and GTF2H2 genes on clinical specimens” section.
Statistics
The categorical data were expressed as percentages. Differences in the clinical characteristics of the various subtypes of SMA were determined using Fisher’s exact test. The survival rates of the different groups were analysed using log-rank. McNemar’s test was performed to analyse the paired results of diverse methods, and p < 0.05 was considered statistically significant. Data were analysed using software SAS 9.13 (SAS Institute, Cary, NC, USA).
Results
Patients and general characteristics
A total of 1613 children with limb movement disorders were initially investigated, and 141 child patients suspected to have SMA were enrolled in this study according to the inclusion criteria. The age of the children with limb movement disorders ranged from 1 month (m) to 15 years. The ratio of boys to girls was 1.660:1. The subtype of SMA was based on the onset age and clinical phenotypes [3], and the children in the study were divided into groups of ≤6 m, 6–18 m and ≥18 m. The general and clinical characteristics of the SMA-suspected children enrolled in the study are listed in Table 1.
Table 1.
The general and clinical characteristics of children in the retrospective study
| General and clinical characteristics | N (%) |
|---|---|
| All children | 141 (100 %) |
| Gender | |
| Boy | 88 (62.41) |
| Girl | 53 (37.59) |
| Age | |
| < 6 m | 60 (42.55) |
| 7-18 m | 45 (31.91) |
| > 18 m | 36 (25.53) |
| Clinical characteristics | |
| Congenital heart disease | 11 (7.80) |
| Respiratory failure | 14 (9.93) |
| Muscular atrophy | 38 (26.95) |
| EMG abnormalities | 117 (82.98) |
| Disorder on sitting/standing/walking | 64 (45.39)/39 (27.66)/29 (20.57) |
| Prenatal | |
| Ultrasonography abnormalities | 3 (2.13) |
| Decreased fetal movement | 6 (4.26) |
| Creatine kinase value | |
| Normal | 50 (35.46) |
| Elevation | 91 (64.54) |
| Diseases | |
| SMA | 75 |
| DMD/BMD | 59 |
| ME | 6 |
| CMT | 1 |
There were 75 SMA children based on the SMA diagnosis guideline [3], and most of these children were grouped as Type I and Type II (Table 2). From this table, children with Type I SMA were more susceptible to congenital heart disease and respiratory failure and more prone to decreased fetal movement compared to children with Type II SMA. However, there were no significant differences between the two groups (Table 2). The remaining 66 children had Duchenne or Becker muscular dystrophy (DMD/BMD), mitochondrial encephalomyopathy (ME) and Charcot-Marie-Tooth (CMT) (Table 1).
Table 2.
The general and clinical characteristics of children in different subtypes of SMA
| Type I (%) (n = 41) |
Type II (%) (n = 29) |
Type III (%) (n = 5) | Total (%) (n = 75) | P Value* | |
|---|---|---|---|---|---|
| Gender* | |||||
| Boy | 24 (58.54) | 19 (65.52) | 3 (60) | 46 (61.33) | |
| Girl | 17 (41.46) | 10 (34.48) | 2 (40) | 29 (38.67) | |
| Clinical symptoms* | |||||
| Congenital heart disease | 3 (7.32) | 0 | 0 | 3 (4) | 0.5137 |
| Respiratory failure | 6 (14.63) | 2 (6.90) | 0 | 8 (10.67) | 0.7308 |
| Muscular atrophy | 20 (48.78) | 13 (44.83) | 2 (40) | 35 (46.67) | 0.9835 |
| EMG abnormalities | 39 (95.12) | 28 (96.55) | 4 (80) | 71 (94.67) | 0.4405 |
| Prenatal* | |||||
| Ultrasonography abnormalities | 0 | 0 | 0 | 0 | - |
| Decreased fetal movement | 3 (7.32) | 2 (6.90) | 0 | 5 (6.67) | 1.0000 |
| CK value* | |||||
| Normal | 6 (14.63) | 8 (27.59) | 2 (40) | 16 (21.33) | 0.3391 |
| Elevation | 35 (85.37) | 21 (72.41) | 3 (60) | 59 (78.67) | |
| Molecular detection | |||||
| Homozygous mutation at c.840 C > T in SMN gene | 40 (97.56) | 26 (89.66) | 5 (100) | 71 (94.67) | |
| Deletion of NAIP exon4 and 5 | 6 (14.63) | 0 | 0 | 6 (8) | |
| Deletion of NAIP exon4 | 3 (7.32) | 0 | 0 | 3 (4) | |
| Deletion of GTF2H2 exon10 | 2 (4.88) | 1 (3.45) | 0 | 3 (4) | |
*Fisher’s exact test
Molecular diagnosis and prognosis of SMA using novel real-time PCR methods
There were no significant differences of abnormal physical examination syndromes and laboratory detection between SMA and other neurological disorders, for instance, DMD/BMD [18]. The basic molecular diagnosis for SMA was performed by detecting the presence of a homozygous exon 7 deletion in the SMN gene, as previously reported [8, 9]. Deletions in the NAIP and GTF2H2 genes were correlated with the severity and phenotype of SMA [12, 13], which contributed to SMA prognosis and typing. These real-time PCR results demonstrated a typical “S-shape” curve in different colors, and the threshold of relative fluorescent units (RFU) was set as 102. The meaning of the colorful curves were shown in Fig. 1, and the real-time PCR results are shown in Fig. 1, indicating that this new real-time PCR was able to detect the exons with a homozygous loss of SMN, NAIP and GTF2H2.
Fig. 1.
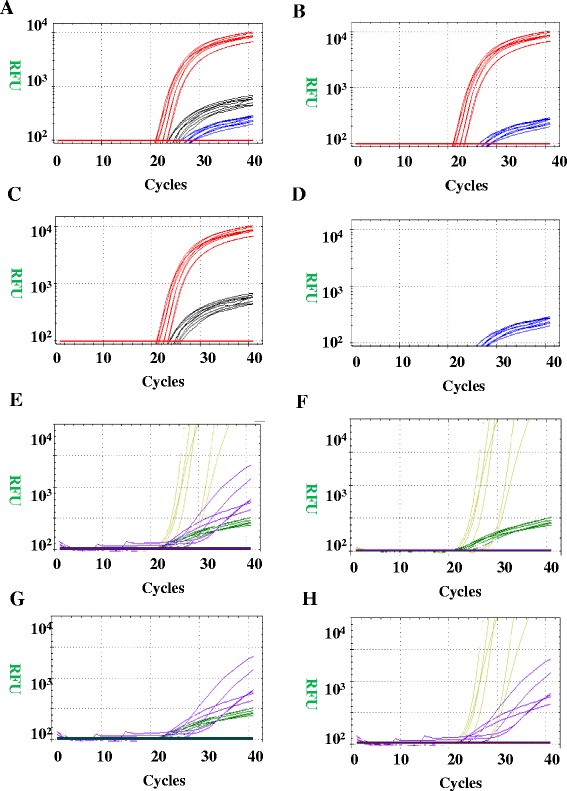
The amplification curves of real-time PCR. The threshold of RFU was set to be 102. The red curve represented NAIP exon5; the black curve represented wild type of SMN gene; the blue curve represented mutation type of SMN; the yellow curve represented GTF2H2 exon10, the purple curve represented NAIP exon4 and the green curve represented internal control. a The image represented that the patient was only with heterozygous mutation of SMN and no mutation on NAIP exon5; (b) The image represented that the patient was with homozygous mutation of SMN and no mutation on NAIP exon5; (c) The image represented that the patient was normal for SMN and NAIP exon5; (d) The image represented that the patient was with homozygous mutation of SMN and the deletion of NAIP exon5; (e) The image represented that there was no mutation on GTF2H2 exon10 and NAIP exon4, and internal control was normal; (f) The image represented that the patient was with the deletion of NAIP exon4; (g) The image represented that the patient was with the deletion of GTF2H2 exon10; (h) The image represented that there was no amplification for internal control, and sample should be detected again
There were 71 (94.67 %) SMA patients with homozygous deletion on SMN1 among 75 SMA patients; 6 SMA patients with a homozygous deletion of NAIP (exon 4 and exon 5), 3 SMA patients with a homozygous deletion of NAIP (exon 4) and 3 SMA patients with a homozygous deletion of GTF2H2 (exon 10). Eleven of the patients were diagnosed as having Type I SMA, and the remaining patients were diagnosed as having Type II SMA (Table 2). The percentage of the NAIP and GTF2H2 homozygous deletions were only 12 % (9/75) and 4 % (3/75), respectively, among all of the SMA children (Table 2).
Consistency of SMN, NAIP and GTF2H2 gene detection among Sanger DNA sequencing, MLPA and real-time PCR
The “gold standard” for detecting a single nucleotide difference and exon loss of several genes [11, 17] is DNA sequencing and MLPA, which were simultaneously used to confirm the results of our newly designed real-time PCR. Wild-type SMN and the homozygous mutation at c.840 C > T in the SMN gene were confirmed using Sanger DNA sequencing (Fig. 2a, b). The electrophoresis results of this new multiplex real-time PCR, including normal samples, samples with homozygous mutations in NAIP in exon 4, homozygous deletions of NAIP in exon4 and exon5 and homozygous deletions of GTF2H2 in exon 10, were shown in Fig. 2c. The wild-type and patients with the deletion of NAIP (exon4 or 5) or GTF2H2 (exon10) detected by single PCR were shown in Fig. 2d.
Fig. 2.
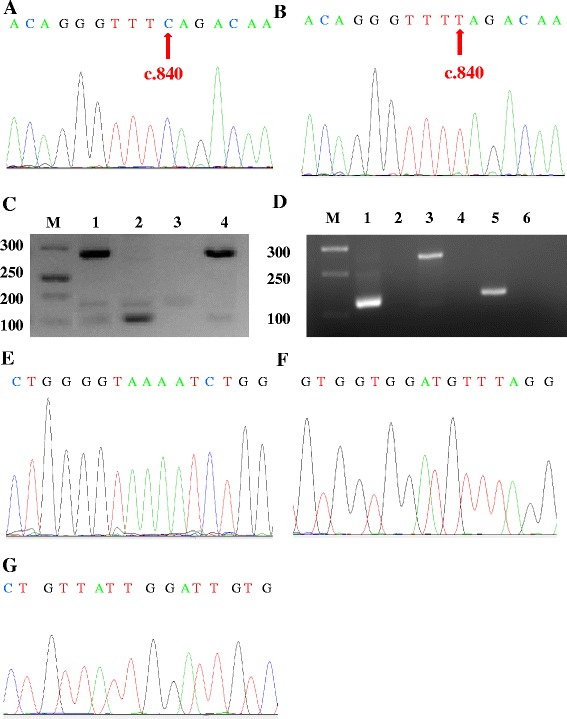
Detection SMN, NAIP and GTF2H2 genes by multiplex PCR and DNA Sanger sequencing. a The DNA sequence result of wild-type at c.840 of SMN (exon7); (b) The DNA sequence result of homozygous mutation of C > T at c.840 of SMN (exon7); (c) The multiplex PCR image for NAIP and GTF2H2 gene deletions. M, Marker; a, NAIP exon4; b, GTF2H2 exon10; c, NAIP exon5; (d): The single PCR image for wild type and NAIP or GTF2H2 gene deletions. M, Marker; 1, 3, 5, samples of wild type of NAIP and GTF2H2; 2, patients with NAIP exon5 deletion; 4, patients with NAIP exon4 deletion; 6, patients with GTF2H2 exon10 deletion; (e) The DNA sequence map of NAIP exon4; (f) The DNA sequence map of NAIP exon5; (g) The DNA sequence map of GTF2H2 exon10
If there was no exon deletion in SMN, NAIP and GTF2H2, then all of the PCR products were confirmed using DNA Sanger sequencing (Fig. 2e, f and g). If any exon homozygous deletions were detected, then the abnormal and normal samples were confirmed using MLPA. There were four main peaks for normal samples, which indicated no deletion of the three exons of NAIP (exon 4 and exon 5) and GTF2H2 (exon 10) (Fig. 3a). For the three different types of homozygous deletions of the NAIP or GTF2H2 gene, one or more peaks were observed to be missing (Fig. 3b, c and d).
Fig. 3.
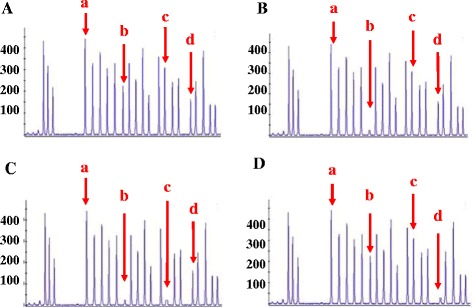
MLPA detection for NAIP and GTF2H2 genes. a The representative MLPA figures of normal sample; (b) The representative MLPA figures of NAIP exon4 deletion; (c) The representative MLPA figures of deletion of NAIP exon4 and 5; (d) The representative MLPA figures of GTF2H2 exon10 deletion. a: Internal control; b: Exon4 of NAIP; c: Exon5 of NAIP; d: Exon10 of GTF2H2
The results of this novel real-time PCR were compared with those obtained from DNA Sanger sequencing and multiplex PCR plus MLPA (Table 3). The accuracy of real-time PCR for the homozygous exon 7 deletion in the SMN gene was 99.6 % (240/241), and the false positive and negative rates of real-time PCR were 0 and 0.6 % (1/171), respectively, based on the results of DNA Sanger sequencing, indicating that this new real-time PCR was an ideal sequencing alternative for the detection of the SMN point mutation (Table 3). Simultaneously, the heterozygous SMN exon 7 deletion was found in no SMA patients, but was found in 62 children among the DMD/BMD, ME and normal patients, as confirmed using DNA Sanger sequencing (Table 4).
Table 3.
The correlation of SMN, NAIP and GTF2H2 in different methods
| MLPA + multiplex PCR/Real-time PCR (%) | Sequencing/Real-time PCR (%) | χ 2 | P value* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| +/+a | −/+ b | +/− c | −/− d | +/+ e | −/+ f | +/− g | −/− h | |||
| NAIP (exon4) | 9 (3.7) | 3 (1.2) | 0 | 229 (95.1) | 3.0 | 0.0833 | ||||
| NAIP (exon5) | 6 (2.5) | 2 (0.8) | 0 | 233 (96.7) | 2.0 | 0.1573 | ||||
| GTF2H2 (exon10) | 3 (1.2) | 2 (0.8) | 0 | 236 (98.8) | 2.0 | 0.1573 | ||||
| SMN | 70 (29.1) | 0 | 1 (0.4) | 170 (70.5) | 1.0 | 0.3173 | ||||
aThere were exons deletion of NAIP or GTF2H2 detected for both methods, meaning true positive for real-time PCR compared with MLPA + multiplex PCR
bThere was no exon deletion of NAIP or GTF2H2 detected by MLPA + multiplex PCR but an exon deletion detected by real-time PCR, meaning false positive for real-time PCR compared with MLPA + multiplex PCR
cThere was exon deletion of NAIP or GTF2H2 detected by MLPA + multiplex PCR but not detected by real-time PCR, meaning false negative for real-time PCR compared with MLPA + multiplex PCR
dThere was no exon deletion for both MLPA + multiplex PCR and real-time PCR, meaning true negative for real-time PCR compared with MLPA + multiplex PCR
eThere was homozygous mutation on SMN c.840 C > T for both methods, meaning true positive for real-time PCR compared with DNA sequencing
fThere was no homozygous mutation on SMN c.840 C > T detected by Sanger DNA sequencing but detected by real-time PCR, meaning false positive for real-time PCR compared with DNA sequencing
gThere was homozygous mutation on SMN c.840 C > T for Sanger DNA sequencing but not detected by real-time PCR, meaning false negative for real-time PCR compared with DNA sequencing
hThere were normal and heterozygous mutation on SMN c.840 C > T for both Sanger DNA sequencing and real-time PCR, meaning true negative for real-time PCR compared with DNA sequencing
*McNemar test
Table 4.
Heterozygous mutations of SMN, NAIP and GTF2H2 genes among children
| SMN | NAIP | GTF2H2 | ||
|---|---|---|---|---|
| Exon7 | Exon4 | Exon5 | Exon10 | |
| SMA | 0 | 0 | 0 | 0 |
| DMD/BMD | 38 | 1 | 1 | 0 |
| ME | 1 | 0 | 0 | 0 |
| CMT | 0 | 0 | 0 | 0 |
| Normal | 23 | 7 | 3 | 4 |
Interestingly, the accuracy of real-time PCR for NAIP exon 4 and exon 5 was 98.8 % (238/241) and 99.2 % (239/241), respectively. The false positive rates of this new real-time PCR for NAIP exon 4 and 5 were 1.3 % (3/232) and 0.9 % (2/235), respectively, and the false negative rates of this new PCR for NAIP exon 4 and 5 were both 0. The accuracy of this novel real-time PCR for GTF2H2 exon 10 was 99.2 % (239/241), and the false positive and negative rates were 0.8 % (2/238) and 0, respectively, based on the results of MLPA plus multiplex PCR in groups of SMA children and normal children. These results revealed that this new real-time PCR was an ideal substitution of MLPA for NAIP and GTF2H2 mutation detection (Table 3). In addition, heterozygous deletions of NAIP or GTF2H2 were found in 16 children, but no deletions were found in SMA patients using MLPA (Table 4).
Prognosis of Type I SMA patients via the detection of SMN, NAIP and GTF2H2 genes
The prognosis of patients is helpful to guide clinical treatment. Survival rate analysis was performed to evaluate the prognosis of patients with or without a homozygous gene deletion. There was only 1 patient with a homozygous deletion in GTF2H2 diagnosed with Type II SMA, and thus, only the survival rate of Type I SMA patients was analysed. Among the 11 Type I SMA children, there was no patient with a complex gene deletion of NAIP or GTF2H2. The survival rates between Type I SMA patients with a homozygous deletion of NAIP or GTF2H2 and patients without a deletion in NAIP or GTF2H2 were analysed (Fig. 4a). There was a much lower survival rate in patients with a homozygous deletion in NAIP or GTF2H2 (p < 0.01), particularly in the first 2 months after suffering from pneumonia (Fig. 4a).
Fig. 4.
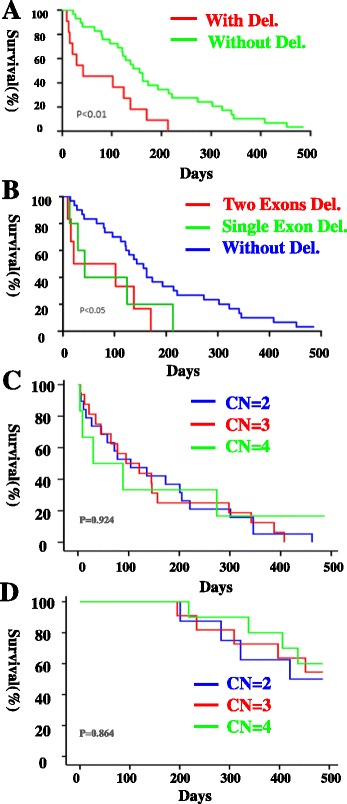
The survival analysis of type I SMA patients. a There were two groups of SMA type I patients: (1) patients with NAIP or GTF2H2 deletion, shown as red curve, n = 11; (2) patients without NAIP or GTF2H2 deletion, shown as green curve, n = 30. (b) SMA type I patients were classified into three groups: (1) patients with exon4 and 5 of NAIP deletion, shown as red curve, n = 6; (2) patients with one exon deletion in NAIP or GTF2H2, shown as green curve, n = 5; (3) patients without NAIP or GTF2H2 deletion, shown as blue curve, n = 30. (c) SMA type I patients were classified into three groups: (1) patients’ copy number (CN) of SMN2 = 2, shown as blue curve, n = 19; (2) patients’ copy number (CN) of SMN2 = 3, shown as red curve, n = 16; (3) patients’ copy number (CN) of SMN2 = 4, shown as green curve, n = 6. (d) SMA type II patients were classified into three groups: (1) patients’ copy number (CN) of SMN2 = 2, shown as blue curve, n = 8; (2) patients’ copy number (CN) of SMN2 = 3, shown as red curve, n = 11; (3) patients’ copy number (CN) of SMN2 = 4, shown as green curve, n = 10
Due to the different types of gene deletions, we determined if there was a difference in the prognosis among patients with a homozygous deletion in one exon, two exons, or no exons in SMA children. Type I SMA patients were divided into three groups: patients without a deletion in NAIP or GTF2H2, patients with a homozygous deletion in one exon of NAIP or GTF2H2, and patients with a homozygous deletion in exon 4 and 5 of NAIP (Fig. 4b). There was a much lower survival rate of patients with a homozygous deletion in one or two exons compared with patients without a deletion (p < 0.05), but there was no significant difference between patients with a homozygous deletion in one exon and those with a homozygous deletion in exon 4 and 5 of NAIP in Type I SMA patients.
According to He’s report [11], the copy number of SMN2 was related to prognosis. In our study the copy number of SMN2 varied from 2–4 of our SMA patients, as shown in Table 5. The relationship between the copy number of SMN2 and patients’ prognosis was analysed, as shown in Fig. 4c, d. There were no significant differences among patients with different copy number of SMN2 in Type I and Type II SMA patients (p > 0.05) in our study. All the patients with Type III SMA were still alive.
Table 5.
SMN2 copy number in different sub-types of SMA
| SMA sub-types | SMN2 copy number | Total | ||
|---|---|---|---|---|
| 2 | 3 | 4 | ||
| Type I | 19 (46.34) | 16 (39.02) | 6 (14.63) | 41 |
| Type II | 8 (27.59) | 11 (37.93) | 10 (34.48) | 29 |
| Type III | 1 (20) | 2 (40) | 2 (40) | 5 |
| Total | 28 (37.33) | 29 (38.67) | 18 (24) | 75 |
Novel real-time PCR for trace DNA
Although real-time PCR can replace DNA Sanger sequencing and MLPA for detecting mutations of SMN, NAIP and GTF2H2, it is necessary to determine whether it can function in newborn screening with trace DNA samples. We randomly selected 2000 DBS samples from 2011 to 2014 using the new real-time PCR method for the detection gene mutations (Table 6).
Table 6.
Real-time PCR for Newborn screening
| 2011 | 2012 | 2013 | 2014 (up to July) | |
|---|---|---|---|---|
| DBS screened/Total | 213/33499 | 637/99841 | 706/110592 | 444/69224 |
| Positive number | 0 | 0 | 1 | 0 |
All trace DNA samples were successfully collected and purified from the 2000 DBS samples. There were 23 samples detected as positive in the first run and all of them were reconfirmed by our Real-time PCR and then DNA sequenced. Twenty-two of them were proven to be false-positive, so there was only one sample, whose ID number was NS-13050012. This patient was first hospitalized due to pneumonia and myasthenia when he was 1 month old in May 2013, who was homozygous for the SMN exon 7 deletion, and the remaining 1999 samples were negative for SMN, NAIP and GTF2H2 deletion. For the negative samples, 1997 of the patients had no symptoms related to SMA and 2 of the patients had limb movement disorders after a follow-up investigation. The 2 children that had limb movement disorders were subsequently diagnosed with progressive muscular dystrophy.
On the other hand, we collected peripheral blood from new SMA patients to make DBS from January 1st to May 16th. There were 36 SMA patients. The results were shown in Table 7. It was obvious that the ability to detect DNA from DBS of our novel real-time PCR was the same as detecting DNA from peripheral blood. The low positive rate of the method in newborn screening DBS was related to the samples, which only contained 1 SMA patient, not because of the limit of our real-time PCR.
Table 7.
Detection of SMA through DNA from peripheral blood or from DBS
| SMN deletion | NAIP deletion | GTF2H2 deletion | ||
|---|---|---|---|---|
| Exon7 | Exon4 | Exon5 | Exon10 | |
| Peripheral blood (n = 36) | 35 | 4 | 1 | 0 |
| DBS (n = 36) | 35 | 4 | 1 | 0 |
Discussion
Here, we established a novel real-time PCR method for detecting mutations in the SMN, NAIP and GTF2H2 genes. The accuracy of this real-time PCR method for detecting the presence of a SMN point mutation or a homozygous deletion in NAIP, GTF2H2 exons was at least 98.8 % compared with DNA Sanger sequencing and MLPA. Real-time PCR is a fast and an ideal method to replace DNA Sanger sequencing, multiplex PCR, and MLPA for the diagnosis of SMA and requires small amounts of DNA [19]. Real-time PCR is also suitable for application in newborn screening for SMA, similar to Somech’s report of T cell receptor excision circles in combined T and B cell immunodeficiency [20]. However, real-time PCR was not able to distinguish between different sub-types of SMA patients. This may be due to the correlation between the genotypes and phenotypes of SMA with hundreds of genes that are not located on chromosome 5q [21, 22], and thus, whole genome sequencing should be further explored.
The incidence of SMA was 4.65 % (75/1613) in children with limb movement disorders in the southwest part of China. Interestingly, most of our SMA patients were classified as Type I and II (Table 1). Type I SMA children were more susceptible to congenital heart diseases and respiratory failure and were more prone to decreased fetal movement compared with Type II SMA children, consistent with previous reports [23–25]. Neither the proportion of muscular atrophy abnormalities nor EMG were found in different subtypes of SMA patients.
In our SMA children, 94.67 % (71/75) displayed a homozygous SMN1 exon 7 deletion, which was similar to the findings obtained in a previous report [26]. However, the odds ratios of NAIP and GTF2H2 homozygous deletions were only 12 % and 4 %, respectively, among SMA patients, which was much lower than the report in east China [11], indicating the different inheritance characteristics for SMA in southwest China. In our study, there was no heterozygous mutations or deletions of SMN, NAIP and GTF2H2 found in SMA patients. Indeed, the multiplex real-time PCR detected a SMN heterozygous exon 7 deletion, as shown in Fig. 1a. In addition, real-time PCR detected heterozygous deletions of the NAIP or GTF2H2 gene when a normal sample with no deletion of the genes was used as an internal control and an equal amount of DNA from patient was amplified (data not shown). Not only patients with DMD/BMD or ME and normal children, as shown in Table 4, but also 20 pairs of the parents of SMA patients (data not shown), who had no symptom, were found to have heterozygous mutations of SMN1, NAIP and GTF2H2. Some of them had no clinical symptom and some of them had clinical symptoms not related to mutations of SMN1, NAIP and GTF2H2. There was no clinical significance for heterozygous mutations of SMN1, NAIP and GTF2H2.
Among the Type I SMA patients, the development of SMA was much faster in patients with a homozygous deletion in NAIP or GTF2H2 compared to patients without this type of deletion. For the 11 Type I SMA patients, 3 patients demonstrated decreased fetal movement during pregnancy. Among these 3 patients, the patient with a homozygous deletion in exon 4 of NAIP had a poor prognosis (the child died within the first 2 months). The amount of samples was too small to obtain a conclusion that decreased fetal movement may signal a bad prognosis for SMA patients. Thus, more cases of SMA children should be further investigated.
The relationship between the prognosis of patients and SMN2 copy number in our study was different from others [11, 27]. There were no significant differences between the possibility of developing to different sub-types of SMA and SMN2 copy numbers in our study according to Table 5, whereas an inverse relationship between SMN2 copy number and possibility of developing to Type I SMA in He J’s research. Maybe different genetic background result in this different prognosis comparing with He J’s research. The onset age of most of our type I SMA patients (38/41, 92.7 %) was <2 months and 72.4 % (21/29) of type II SMA patients carried more than 3 copies of SMN2, whereas the onset age of Qu’s type I patients was 31.1 % (33/106) and copies of SMN2 of Qu’s type II patients was 96 %. As Qu’s research, the clinical and genetic characters of our patients were more severe, leading to worse survival rate.
Only one DBS-positive sample, whose ID number was NS-13050012, was analysed in our study. The doctor diagnosed that the baby suffered from Type I SMA using DNA sequencing and MLPA. The child was nursed as a Type I SMA patient and was still alive until September 2014, with no obvious symptoms except non-sitting. If we performed the newborn screening for SMA for newborn babies, children with SMA will benefit from being diagnosed as Type I SMA at an early age and can begin an early specific nursing program. Thus, their prognosis will improved, similar to the child in our study.
There are four main methods to detect SMA: Sanger DNA sequencing, MLPA, RFLP and multiple routine PCR. No one was used in newborn screening. Sanger DNA sequencing and MLPA are both time consuming (>2 weeks), expensive (>US$21) and high amount of DNA required (amount of DNA is trace in newborn screening); RFLP is also time consuming (20 h) and not able to detect exons deletion of NAIP and GTF2H2; multiplex routine PCR is not able to detect exon 7 deletion of SMN1 and it is more subjective because of being judged by electrophoresis gel image; even the method of RFLP with multiplex routine PCR is also time consuming and subjective to judge results. Our new approach is fast (<3 h), cheap (<US$2), objective and able to analyse exons deletion of SMN, NAIP and GTF2H2 genes simultaneously with trace DNA.
According to Prior’s and Stabley’s studies [28, 29], there are several limitations of real-time PCR. At the same time, digital PCR (dPCR) is able to detect SMN mutation. Moreover, for the coefficients of variation, dPCR is even better than real-time PCR.
For limitations of real-time PCR, (1) we recommended eathylene diamine tetraacetic acid (EDTA) as anticoagulant for periphery blood sample, avoiding heparin for its inhibiting Taq polymerase activity. Our method detected more than 200 EDTA peripheral blood and 2000 DBS samples, and the amplification efficiency was excellent, so the compounds in specimen did not shown inhibition; (2) it is possible that DNA sequence variants located under the primer binding sites may be a problem to influence the results. To avoid the question, we need to add another pair of primers to cover outside of binding sites of the primers we used to amplify samples in future; (3) for data analysis, repeating reactions twice and evaluating the capability of equipment could ensure the quality of results to a certain extent.
The recent new method of dPCR is suitable and promising for detecting SMN mutation. However, the ability of dPCR to simultaneously detect NAIP and GTF2H2 mutation and for DBS is not available and need further evaluation. Moreover, dPCR is more expensive (>US$15) in China and specific equipment is required. In a word, the Real-time PCR for SMN, NAIP and GTF2H2 is cheaper and feasibility for clinical usage at present in China.
Conclusions
In summary, we have provided a better understanding of the incidence, clinical and laboratory characteristics of SMA in southwest China and established a new fast, cheap and accurate real-time PCR method for detecting SMA during newborn screening, which will be helpful in maintaining the motor function of Type I and II SMA patients in the near future (ClinicalTrial.gov. Safety and efficacy of olesoxime (TRO19622) in 3–25 years SMA patients. From http://clinicaltrials.gov/show/NCT01302600. Accessed October 8, 2013). Combined with SMA therapy development, SMA patients may also be specifically treated in a more timely fashion in the future [6]. Furthermore, whole genome sequencing should be explored to determine the genotype-phenotype correlation and typing of SMA.
Abbreviations
DBS, dry blood spot; dPCR, digital PCR; EDTA, eathylene diamine tetraacetic acidEMG, electromyography; GTF2H2, general transcription factor IIH, polypeptide 2 gene; MLPA, multiplex ligation-dependent probe amplification; NAIP, NLR family, apoptosis inhibitory protein; RFLP, restriction fragment length polymorphism; RFU, relative fluorescent units; SMA, spinal muscular atrophy; SMN, survival of motor neuron
Acknowledgements
We thank all patients and guardians involved in the study.
Funding
This work was supported in part by grants from the National Natural Science Foundation of China (81373444, 90919013).
Availability of data and materials
All the data on which the conclusions of our manuscript rely were presented in the main paper and additional supporting files.
Authors’ contributions
LZ, LJ and SH participated in the conception and design of the study. ZL, PZ and BP performed the statistical analysis and interpretation of data. ZL, PZ, ST, XH and RZ carried out PCR. XW, SL and JT collected medical records. ZL, LZ, XH, LJ and SH drafted the manuscript. All authors read and approved the final version of the manuscript.
Authors’ information
Corresponding Author: LZ
Position:
Professor & Director, Center for Clinical Molecular Medicine, Children’s Hospital, Chongqing Medical University, China
Education Background:
Sep 2001 - Jul 2004
M. D. & Ph. D., Faculty of Laboratory Medicine, Chongqing Medical University
Sep 1999 - Jul 2001
Master, Faculty of Laboratory Medicine, Chongqing Medical University
Sep 1996 - Jul 1998
Junior College of Chongqing Medical University
Competing interests
The authors declare that they have no competing interest.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Children’s Hospital of Chongqing Medical University, and the reference number was CHCMU-20110005. We have received consents, which were written by the parents and guardians.
Additional files
The guideline for SMA diagnosis and typing. (DOC 34 kb)
The components of each real-time PCR reaction. (DOC 36 kb)
The sequence of primers and probes. (DOC 37 kb)
The reaction systems for DNA sequencing and multiplex PCR. (DOC 38 kb)
The master mix of real-time PCR for DBS. (DOC 36 kb)
Contributor Information
Zhidai Liu, Email: 1052955698@qq.com.
Penghui Zhang, Email: zhangph7203@sina.com.
Xiaoyan He, Email: hexiaoyanaa@163.com.
Shan Liu, Email: 447380047@qq.com.
Shi Tang, Email: 544756431@qq.com.
Rong Zhang, Email: 316892372@qq.com.
Xinbin Wang, Email: w4wxb@163.com.
Junjie Tan, Email: 11602368@qq.com.
Bin Peng, Email: cellepy@gmail.com.
Li Jiang, Email: dr.jiangl@hotmail.com.
Siqi Hong, Email: 455261055@qq.com.
Lin Zou, Phone: 86-23-6362-2128, Email: 2660622115@qq.com.
References
- 1.Hendrickson BC, Donohoe C, Akmaev VR, Sugarman EA, Labrousse P, Boguslavskiy L, et al. Differences in SMN1 allele frequencies among ethnic groups within North America. J Med Genet. 2009;46(9):641–644. doi: 10.1136/jmg.2009.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zerres K, Davies KE. (1999) 59th ENMC international workshop: spinal muscular atrophies: recent progress and revised diagnostic criteria 17–19 April 1998, Soestduinen, The Netherlands. Neuromuscul Disord. 1998;9(4):272–278. doi: 10.1016/S0960-8966(99)00016-4. [DOI] [PubMed] [Google Scholar]
- 3.Wang CH, Finkel RS, Bertini ES, Schroth M, Simonds A, Wong B, et al. Consensus statement for standard of care in spinal muscular atrophy. J Child Neurol. 2007;22:1027–1049. doi: 10.1177/0883073807305788. [DOI] [PubMed] [Google Scholar]
- 4.DerakhshandehPeykar P, Esmaili M, OusatiAshtiani Z, Rahmani M, Babrzadeh F, Farshidi S, et al. Molecular analysis of the SMN1 and NAIP genes in Iranian patients with spinal muscular atrophy. Ann Acad Med Singapore. 2007;36:937–941. [PubMed] [Google Scholar]
- 5.Sartori M, Farina D, Lloyd DG. Hybrid neuromusculoskeletal modeling to best track joint moments using a balance between muscle excitations derived from electromyograms and optimization. J Biomech. 2014;47(15):3613–3621. doi: 10.1016/j.jbiomech.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Rothwell E, Anderson RA, Swoboda KJ, Stark L, Botkin JR. Public attitudes regarding a pilot study of newborn screening for spinal muscular atrophy. Am J Med Genet A. 2013;161A(4):679–686. doi: 10.1002/ajmg.a.35756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 8.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 10.Mailman MD, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Wirth B, et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med. 2002;4(1):20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 11.He J, Zhang QJ, Lin QF, Chen YF, Lin XZ, Lin MT, et al. Molecular analysis of SMN1, SMN2, NAIP, GTF2H2, and H4F5 genes in 157 Chinese patients with spinal muscular atrophy. Gene. 2013;518(2):325–329. doi: 10.1016/j.gene.2012.12.109. [DOI] [PubMed] [Google Scholar]
- 12.Roy N, Mahadevan MS, McLean M, Shutler G, Yaraghi Z, Farahani R, et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80(1):167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 13.Arkblad E, Tulinius M, Kroksmark AK, Henricsson M, Darin N. A population-based study of genotypic and phenotypic variability in children with spinal muscular atrophy. Acta Paediart. 2009;98(5):865–872. doi: 10.1111/j.1651-2227.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 14.van der Steege G, Grootscholten PM, van der Vlies P, Draaijers TG, Osinga J, Cobben JM, et al. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995;345(8955):985–986. doi: 10.1016/S0140-6736(95)90732-7. [DOI] [PubMed] [Google Scholar]
- 15.Martins FT, Ramos PZ, Svidnicki MC, Castilho AM, Sartorato EL. Optimization of simultaneous screening of the main mutations involved in non-syndromic deafness using the TaqMan® OpenArray™ Genotyping platform. BMC Med Genet. 2013;14:112. doi: 10.1186/1471-2350-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saavedra-Matiz CA, Isabelle JT, Biski CK, Duva SJ, Sweeney ML, Parker AL, et al. Cost-effective and scalable DNA extraction method from dried blood spots. Clin Chem. 2013;59(7):1045–1051. doi: 10.1373/clinchem.2012.198945. [DOI] [PubMed] [Google Scholar]
- 17.Liang YH, Chen XL, Yu ZS, Chen CY, Bi S, Mao LG, et al. Deletion analysis of SMN1 and NAIP genes in Southern Chinese children with spinal muscular atrophy. J Zhejiang Univ Sci B. 2009;10(1):29–34. doi: 10.1631/jzus.B0820125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglas AG, Wood MJ. Splicing therapy for neuromuscular disease. Mol Cell Neurosci. 2013;56:169–185. doi: 10.1016/j.mcn.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozbek SM, Ozbek A, Yavuz MS. Detection of human cytomegalovirus and Epstein-Barr Virus in symptomatic and asymptomatic apical periodontitis lesions by real-time PCR. Med Oral Patol Oral Cir Bucal. 2013;18(5):e811–e816. doi: 10.4317/medoral.18905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker MW, Grossman WJ, Laessig RH, Hoffman GL, Brokopp CD, Kurtycz DF, et al. Development of a routine newborn screening protocol for severe combined immunodeficiecy. J Allergy Clin Immunol. 2009;124(3):522–527. doi: 10.1016/j.jaci.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Peeters K, Chamova T, Jordanova A. Clinical and genetic diversity of SMN1-negative proximal spinal muscular atrophies. Brain. 2014;137(Pt11):2879–2896. doi: 10.1093/brain/awu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen A, Dimlich DN, Guruharsha KG, Kankel MW, Hori K, Yokokura T, et al. Genetic circuitry of Survival motor neuron, the gene underlying spinal muscular atrophy. Proc Natl Acad Sci U S A. 2013;110(26):E2371–E2380. doi: 10.1073/pnas.1301738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudnik-Schöneborn S, Heller R, Berg C, Betzler C, Grimm T, Eggermann T, et al. Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J Med Genet. 2008;45(10):635–638. doi: 10.1136/jmg.2008.057950. [DOI] [PubMed] [Google Scholar]
- 24.Finkel RS, Weiner DJ, Mayer OH, McDonough JM, Panitch HB. Respiratory muscle function in infants with spinal muscular atrophy type I. Pediatr Pulmonol. 2014;49(12):1234–1242. doi: 10.1002/ppul.22997. [DOI] [PubMed] [Google Scholar]
- 25.MacLeod MJ, Taylor JE, Lunt PW, Mathew CG, Robb SA. Prenatal onset spinal muscular atrophy. Eur J Paediatr Neurol. 1999;3(2):65–72. doi: 10.1016/S1090-3798(99)80015-4. [DOI] [PubMed] [Google Scholar]
- 26.Alías L, Bernal S, Fuentes-Prior P, Barceló MJ, Also E, Martínez-Hernández R, et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum Genet. 2009;125(1):29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- 27.Qu YJ, Ge XS, Bai JL, Wang LW, Cao YY, Lu YY, et al. Association of copy numbers of survival motor neuron gene 2 and neuronal apoptosis inhibitory protein gene with the natural history in a Chinese spinal muscular atrophy cohort. J Child Neurol. 2015;30(4):429–436. doi: 10.1177/0883073814553271. [DOI] [PubMed] [Google Scholar]
- 28.Prior TW, Nagan N, Sugarman EA, Batish SD, Braastad C. Technical standards and guidelines for spinal muscular atrophy testing. Genet Med. 2011;13(7):686–694. doi: 10.1097/GIM.0b013e318220d523. [DOI] [PubMed] [Google Scholar]
- 29.Stabley DL, Harris AW, Holbrook J, Chubbs NJ, Lozo KW, Crawford TO, et al. SMN1 and SMN2 copy numbers in cell lines derived from patients with spinal muscular atrophy as measured by array digital PCR. Mol Genet Genomic Med. 2015;3(4):248–257. doi: 10.1002/mgg3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data on which the conclusions of our manuscript rely were presented in the main paper and additional supporting files.


