Abstract
Background & objectives:
The role of cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations in congenital bilateral absence of vas deferens and unilateral renal agenesis (CBAVD-URA) has been controversial. Here, we report the cases of five Indian males with CBAVD-URA. The objective was to evaluate the presence or absence of CFTR gene mutations and variants in CBAVD-URA. The female partners of these males were also screened for cystic fibrosis (CF) carrier status.
Methods:
Direct DNA sequencing of CFTR gene was carried out in five Indian infertile males having CBAVD-URA. Female partners (n=5) and healthy controls (n=32) were also screened.
Results:
Three potential regulatory CFTR gene variants (c.1540A>G, c.2694T>G and c.4521G>A) were detected along with IVS8-5T mutation in three infertile males with CBAVD-URA. Five novel CFTR gene variants (c.621+91A>G, c.2752+106A>T, c.2751+85_88delTA, c.3120+529InsC and c.4375-69C>T), four potential regulatory CFTR gene variants (M470V, T854T, P1290P, Q1463Q) and seven previously reported CFTR gene variants (c.196+12T>C, c.875+40A>G, c.3041-71G>C, c.3271+42A>T, c.3272-93T>C, c.3500-140A>C and c.3601-65C>A) were detected in infertile men having CBAVD and renal anomalies
Interpretation & conclusions:
Based on our findings, we speculate that CBAVD-URA may also be attributed to CFTR gene mutations and can be considered as CFTR-related disorder (CFTR-RD). The CFTR gene mutation screening may be offered to CBAVD-URA men and their female partners undergoing ICSI. Further studies need to be done in a large sample to confirm the findings.
Keywords: Congenital bilateral absence of vas deferens, congenital absence of seminal vesicles, cystic fibrosis transmembrane conductance regulator gene, ectopic kidney, unilateral renal agenesis
The incidence of cystic fibrosis (CF) is considered to be low in Asian population as compared to Caucasians; however, the incidence of congenital bilateral absence of vas deferens (CBAVD) does not differ between these populations1. The CBAVD occurs in 2-6 per cent of infertile but otherwise healthy men and is responsible for as much as 25 per cent of obstructive azoospermia2. Isolated CBAVD is transmitted as autosomal recessive trait and is frequently associated with mutations in cystic fibrosis transmembrane conductance regulator (CFTR) gene3.
Geographical or ethnic differences have been demonstrated in CFTR gene mutations in males with CBAVD with a high frequency of F508del mutation in Caucasians but rarely observed in Japanese, Taiwanese and Chinese populations1,4,5. However, the IVS8-5T allele is reported to be higher or of same frequency in non-Caucasian populations1,6. CBAVD is known to be associated with renal anomalies in 12-21 per cent of cases3. The exact mechanism of pathogenesis of CBAVD contributed by CFTR gene mutations and variants is still poorly understood. The role of CFTR gene mutations in patients with CBAVD having unilateral renal agenesis (CBAVD-URA) is controversial as in majority of cases with CBAVD-URA reported in literature; CFTR gene mutations were not detected3,7,8. In view of these observations, such cases of CBAVD-URA are proposed as a distinct entity arising of genetic causes other than CFTR gene mutations probably due to paucity of such data3. Information about the frequency of CFTR gene mutations in Indian patients with CBAVD is limited. Further, there are no guidelines for CFTR gene mutations screening in India for males with CBAVD.
We describe here the detection of CFTR gene mutations and variants in five infertile males with CBAVD-URA. The female partners were also screened to assess the CF carrier status.
Material & Methods
The study was conducted at the National Institute for Research in Reproductive Health, (NIRRH) Mumbai, India, during 2011-2014. Written informed consent was obtained from cases, their female partners and healthy controls included in our study. The study was approved by the institutional ethics committee. None of the patients with CBAVD-URA reported in our study had associated clinical manifestations (pulmonary or gastrointestinal) or a family history suggestive of CF. The diagnosis of CBAVD was based on azoospermia, low seminal volume (<1.5 ml), absent/low fructose in seminal fluid and absence of bilateral vas deferens on scrotal examination by the andrologist. Testicular volume was evaluated using a Prader orchidometer (Holtain, UK). Ultrasonography (USG) of the abdomen and pelvis was done to detect renal anomalies. Transrectal USG (TRUS) was carried out to detect anomalies of seminal vesicles and ejaculatory ducts. Proven fertile men from general population attending Family Welfare Clinics of NIRRH, Mumbai, willing to participate in this study were included as controls (n=32). The inclusion criteria of controls were as follows: (i) Proven fertile males having normal semen analysis9 (ii) The vas deferens was bilaterally palpable; and (iii) No signs and symptoms of CF and any CF related disorders.
Peripheral venous blood (5 ml) was collected into EDTA tubes and genomic DNA was isolated with commercial blood genomic DNA purification kit (GenElute™ Blood Genomic DNA Kit, Sigma, USA). The essential promoter, entire coding regions and splice sites of 27 exons of the CFTR gene were amplified by polymerase chain reaction (PCR) using specific primers (designed manually, specificity checked using in silico PCR; https://genome.ucsc.edu/cgi-bin/hgPcr). PCR cycling conditions were as follows: initial denaturation at 94° C for four min, 35 cycles of denaturation at 94° C for one min, annealing at 56-58° C for one min (depending on the primer sequence), extension at 72° C for one min and final extension step at 72° C for five min. PCR products were sequenced on ABI 3730xl DNA Analyzer automated sequencer (Applied Biosystems, Foster City, USA). Female partners were also screened for CFTR gene mutations to assess their CF carrier status. The sequencing results were compared with the wild type CFTR gene sequence published in the Gen Bank (NM_000492.3). The original CFTR nucleotide numbering was used, i.e. from the 1st nucleotide of exon 1.
Statistical analysis: The data were evaluated with Fisher's extract test using SPSS software version 19 (SPSS, South Asia Pvt. Ltd, Bengaluru, India).
Results
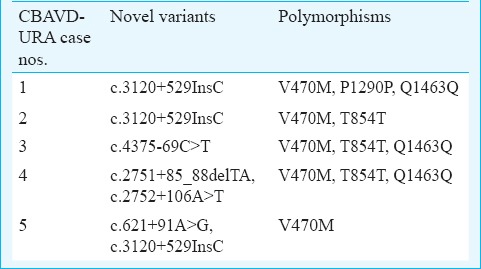
Case 1: A 37 yr old male from Maharashtra, married since 10 years presented with a history of infertility for evaluation in andrology clinic at NIRRH, Mumbai, in January 2011. There was no history of consanguineous marriage. The detailed semen parameters, reproductive hormones, scrotal examination, renal assessment are presented in Tables I and II. The testicular volume was within normal range (>15 ml). On scrotal examination, only head of the epididymis was palpable and bilateral vas deferens was found to be absent on both sides. Scrotal USG showed bilateral dilated tubules in head of epididymis suggestive of obstruction at the level of epididymis. Abdominal USG showed absence of left kidney. Testicular biopsy showed normal spermatogenesis. The sequence analysis of CFTR gene resulted in identification of one novel intronic variant c.3120+529InsC (KJ159564; homozygous) (Figure D), three previously reported potential regulatory coding CFTR gene variants (AGA haplotype) c.1540G>A M470V (homozygous), c.4002A>G P1290P (homozygous), c.4521G>A Q1463Q (homozygous)10 and two previously reported intronic variants c.3041-71G>C (heterozygous), c.3272-93T>C (homozygous)10. Intron 8 splice site variant [TG(n)-5T] analysis identified TG12-5T/TG10-9T (heterozygous) in intron 8. The female partner was not found to be CF carrier.
Table I.
Demographic characteristics, semen and hormonal parameters in infertile males with congenital bilateral absence of vas deferens and unilateral renal agenesis (CBAVD-URA)
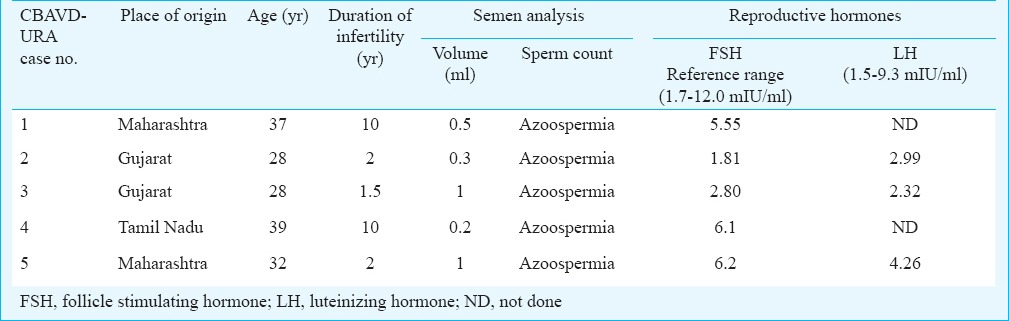
Table II.
Clinical characteristics of infertile males having CBAVD-URA
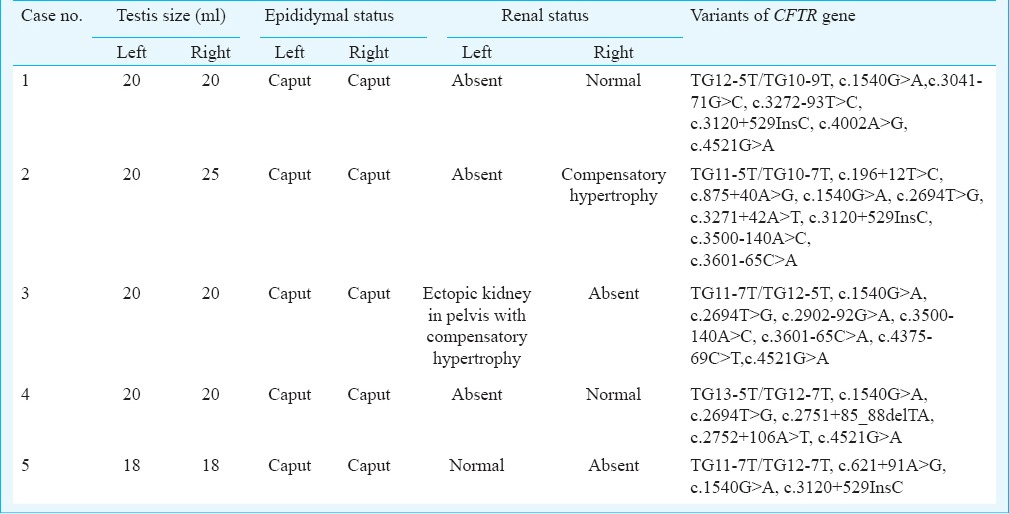
Figure.
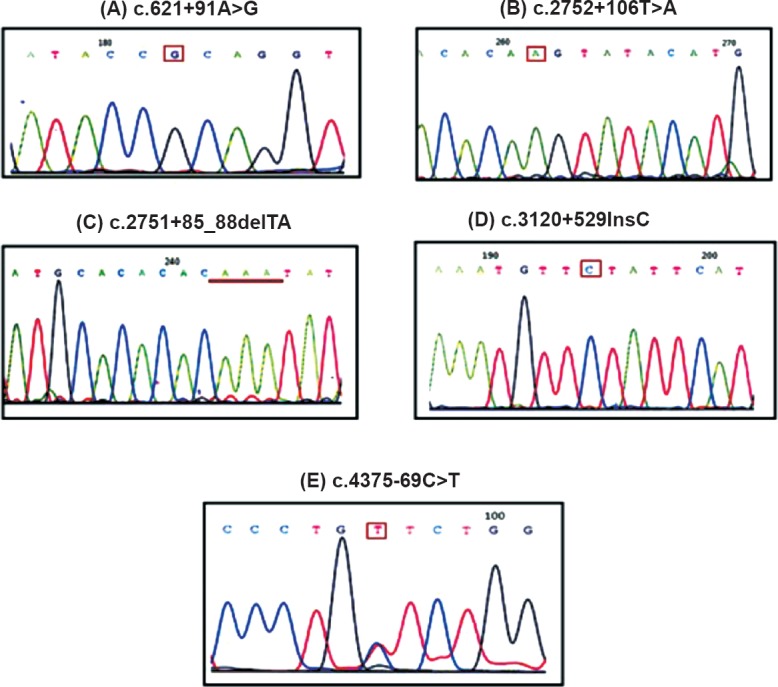
Identification of novel variants in infertile males with CBAVD and renal anomalies. (A) c.621+91A>G (intron 4); (B) c.2752+106T>A (intron 14A); (C) c.2751+85_88 delTA (intron 14A); (D) c.3120+529 InsC (intron 19); (E) c.4375-69C>T (intron 23).
Case 2: A 28 yr old male from Gujarat, married since last two years with non-consanguineous marriage presented with a history of infertility in May 2011. The semen parameters, reproductive hormones status, scrotal examination, renal assessments are represented in Tables I and II. The testicular volume was within normal range (>15 ml). On scrotal examination, only head of the epididymis was palpable and vas deferens was absent on both sides. Scrotal USG showed bilateral varicocele and an anechoic tubular lesion in the head of right epididymis due to dilated efferent tubules. Abdominal USG showed absent left kidney and there was compensatory hypertrophy of right kidney. Bilateral testicular biopsy showed normal spermatogenesis in 70 per cent of seminiferous tubules.
TG11-5T/TG10-7T (heterozygous) mutation, two previously reported potential regulatory coding CFTR gene variants c.1540G>A V470M (heterozygous), c.2694T>G T854T (homozygous) and five previously reported intronic variants c.196+12T>C (heterozygous)11, c.875+40A>G (heterozygous)12, c.3271+42A>T (heterozygous)12, c.3500-140A>C (heterozygous)13, c.3601-65C>A (heterozygous)14, one novel variant c.3120+529InsC (homozygous) (Figure D) were observed in this patient (Table III). His female partner was 26 years old whose CFTR gene sequencing analysis revealed that she was not a CF carrier.
Table III.
Identified CFTR gene variants in five cases having CBAVD-URA
Case 3: A 28 yr old male from Gujarat, married since one and half years in a non-consanguineous marriage was referred for andrology evaluation in June 2011. His 26 years old female partner was also evaluated for infertility. The semen parameters, reproductive hormones, scrotal examination, renal assessments are presented in Tables I and II. Semen analysis on two separate occasions showed azoospermia with absent fructose, low volume of ≤1 ml and low pH of 6.0. The testicular volume was within normal range (>15 ml). On palpation, only head of the epididymis was palpable and bilateral vas deferens was found to be absent. Scrotal USG showed a normal testicular structure and the epididymal heads of the right and left epididymis with the defect in epididymal bodies and tails. Scrotal USG also showed bilateral gross dilatation of epididymal tubules suggestive of distal obstruction. Doppler study of scrotum showed bilateral dilated and tortuous veins of pampiniform plexus suggestive of bilateral varicocele. TRUS revealed absence of bilateral seminal vesicles, bilateral absences of vas deferens and normal prostate. Transabdominal and pelvic USG showed absent right kidney and left was ectopic kidney in pelvis on left side of urinary bladder with mild compensatory hypertrophy. Serum creatinine level was normal.
The CFTR gene mutation analysis identified one novel intronic variant c.4375-69C>T (KJ159563; heterozygous) (Figure E), three previously reported potential regulatory coding CFTR gene variants (AGA haplotype) c.1540G>A V470M (heterozygous), c.2694T>G T854T (heterozygous), c.4521G>A Q1463Q (homozygous)10 and three previously reported intronic variants c.2909-92G>A (heterozygous)11, c.3500-140A>C (heterozygous)13, c.3601-65C>A (heterozygous)13 and TG12-5T/TG11-7T (heterozygous) (Table III). The female partner was not found to be a potential CF carrier. Percutaneous epididymal sperm aspiration (PESA) revealed good quality motile sperm. The couple had undergone one cycle of intra-cytoplasmic sperm injection (ICSI), however, there was no pregnancy.
Case 4: A 39 yr old male from Tamil Nadu, married since 10 years in non-consanguineous marriage presented with a history of infertility in May 2013. The semen parameters, reproductive hormones, scrotal examination, renal assessment are presented in Tables I and II. Abdominal USG showed normal right kidney and absent left kidney. The sequencing analysis resulted in identification of TG13-5T/TG12-7T (heterozygous) mutation, three previously reported potential regulatory coding CFTR gene variants (AGA haplotype) c.1540G>A M470V (homozygous), c.2694T>G T854T (homozygous), c.4521G>A Q1463Q (heterozygous)10 and two novel intronic variants c.2751+85_88delTA (KJ159561; homozygous) and c.2752+106A>T (KJ159562; homozygous). (Figure B, C) (Table III). No previously known intronic variants were identified in this patient. The female partner was not found to be a potential CF carrier.
Case 5: A 32 yr old Indian male from Maharashtra, married since two years with no consanguinity presented with history of infertility in February 2014. The semen parameters, reproductive hormones, scrotal examination, renal assessment are presented in Tables I and II. Abdominal USG showed absent right kidney and normal left kidney. The sequencing analysis resulted in identification of two novel intronic variants c.621+91A>G (heterozygous) and c.3120+529InsC (homozygous) (Figure A, D), and two of the previously reported potential regulatory coding CFTR gene variants (AGA haplotype) c.1540G>A M470V (homozygous)10 and TG11-7T/TG11-7T (homozygous) (Table III). No previously known intronic variants were identified in this patient. The female partner was not found to be a potential CF carrier.
CFTR gene screening in controls: A total of 64 chromosomes from 32 control subjects were screened for detection of CFTR gene mutation. None of the 32 controls were carriers of F508del or other CFTR gene mutations. However, the most common CFTR gene variant IVS8-5T was detected in 18.75 per cent (6 of 32). V470M variant was found in 46.87 per cent (15 of 32) controls. c.4521G>A Q1643Q SNP was found in 28.12 per cent (9 of 32) controls.
The sequencing results were compared with the sequence of the wild type CFTR gene published in the CFTR mutation databases such as Cystic Fibrosis Mutation Database (www.genet.sickkids.on.ca/), Universal Mutation Database (www.umd.be/CFTR/) and recently developed CFTR-2 database (http://www.cftr2.org/).
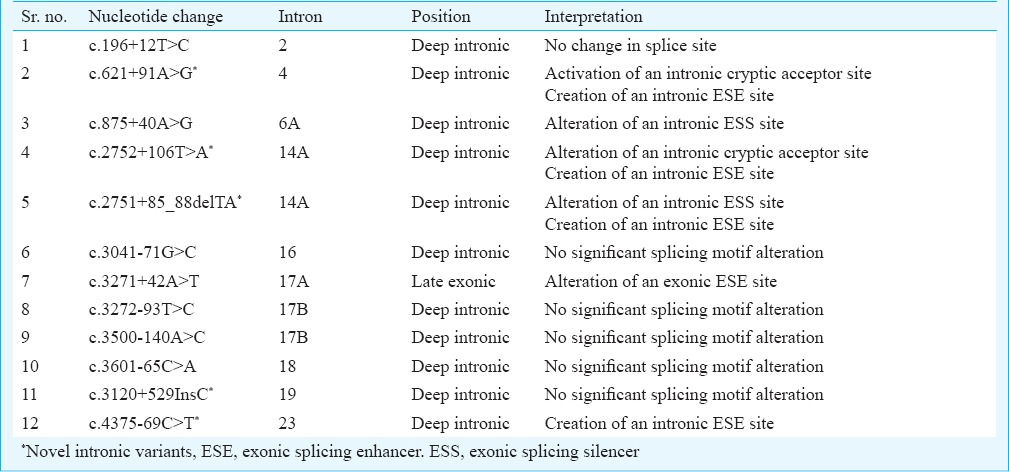
In-silico analysis: Five novel and seven previously reported intronic variants in the CFTR gene were detected in five males with CBAVD-URA and their female partner. To better understand the splicing defects due to the presence of intronic variants in these males, we analysed the novel and previously known intronic variants using Human Splicing Finder 3.0 (http://www.umd.be/HSF3/). In-silico splice site analysis using this software predicted four novel intronic variants (c.621+91A>G, c.2751+85_88delTA, c.4095+61T>A and c.4375-69C>T) to have potential to alter splicing in the CFTR gene (Table IV) and one novel intronic variants (c.3120+529InsC) was found to have no significant splicing motif alteration in the CFTR gene. However, two previously known intronic variants (c.875+40A>G, and c.3271+42A>T) were predicted to have potential to alter splicing in the CFTR gene and five previously known intronic variants (c.196+12T>C, c.3041-71G>C, c.3272-93T>C, c.3500-140A>C and c.3601-65C>A) were found to have no significant splicing motif alteration in the CFTR gene (Table IV).
Table IV.
Splice site analysis of CFTR gene intronic variants in infertile males with CBAVD-URA
Discussion
Role of CFTR gene mutations in males with CBAVD-URA has been controversial, as the previously reported studies did not detect any CFTR gene mutations14,15,16. In the present study, we found CFTR gene variants in five cases of CBAVD having renal malformations. Congenital bilateral absence of seminal vesicles (CASV) and CBAVD are uncommon anomalies and such patients usually have normal kidneys17. However, in the present study, case 3 was found to have bilateral absence of seminal vesicles and left ectopic kidney; which is a rare observation. Case 5 showed presence of hypoplastic seminal vesicles and absence of right kidney. Normally, in addition to prostatic secretions, seminal vesicular secretions also contribute to the alkalinity of the ejaculate and make up approximately 90 per cent of fluid in ejaculate. Thus, patients having CASV and CBAVD present with history of infertility and usually patients with URA remain undiagnosed until there is some pathology in the contralateral kidney. We detected IVS8-5T mutation in three out five CBAVD-URA patients, all the CBAVD-URA males had V470M all CBAVD-URA males harboured intronic variants and other SNPs T854T, P1290P and Q1463Q.
In this study, five novel CFTR gene variants (c.621+91A>G, c.2752+106A>T, c.2751+85_88delTA, c.3120+529InsC and c.4375-69C>T), other coding SNPs such as T854T, P1290P, Q1463Q and seven previously reported CFTR gene variants were identified in patients with CBAVD-URA. Two CBAVD-URA patients in our study were found to have longer IVS8-TG repeats (TG12 or TG13) in cis with 5T and M470V polymorphism. Previous studies reported that M470V along with short poly-T (5T) and long TG-repeat tracks (TG12, TG13) may contribute to CBAVD risk2,18. This genotype was not detected in normal male participants suggesting that longer TG-short T repeats in association with M470V and other variants might be responsible for CBAVD-URA phenotype. It is reported that IVS8-5T allele is found in higher frequencies in non-caucasian populations such as Indians and is involved in many cases of CBAVD even in populations where CF is rare3.
It has been reported that three common CFTR coding SNPs (c.1540A>G, c.2694T>G and c.4521G>A) might contribute to reduced levels of CFTR transcripts10. They also reported AGA haplotype (c.1540A>G, c.2694T>G and c.4521G>A) in association with CF in individual having chronic pancreatitis and this haplotype was found to have independent contribution to the phenotype irrespective of CF mutations10. In our study, three potential regulatory CFTR gene variants (c.1540A>G, c.2694T>G and c.4521G>A) were detected in cases 1, 3 and 4. Based on the information available in the literature, we can speculate that these common coding single nucleotide polymorphism (cSNPs) in the CFTR gene along with IVS-5T might be leading to decreased levels of normal transcript contributing to the variable phenotype as observed in these patients. It has been reported that patients with c.1540G>A M470V, c.2694T>G T854T and c.4521G>A Q1463Q haplotypes could have lower CFTR transcript levels than that expected from otherwise normal haplotypes. This might lead to a major and/or earlier clinical expressionof cystic fibrosis or cystic fibrosis-related disorders (CFTR-RD)10. Also, a combination of IVS8-5T mutation and M470V has been known to influence the efficiency of exon 919.
It is important to note that due to the large size of Indian population, the total burden of CF and CFTR gene carrier could be much larger than in many European countries. Thus, CFTR gene analysis should be offered to CBAVD-URA patients and their female partners if the couple is opting for ICSI. Further studies are in progress to investigate the complete spectrum of CFTR gene mutations in Indian CBAVD males.
Acknowledgment
The authors thank Dr Smita Mahale, Director, NIRRH and Dr S.D. Kholkute, former Director, NIRRH, Mumbai for encouragement and support and Dr Shahina Begum for statistical analysis. The authors acknowledge Drs Anurupa Maitra, Srabani Mukharjee and Parag Tamhankar for assistance in DNA sequencing, and Drs Vikas Dighe and Suparna Chatterjee for their help in initial primer designing. This study was partially funded by extramural grant of Board of Research in Nuclear Sciences (BRNS) Department of Atomic Energy (DAE), Government of India. Dr Nobhojit Roy (BARC) is acknowledged for co-ordinating BRNS funding.
Footnotes
Conflicts of Interest: None.
References
- 1.Ni WH, Jiang L, Fei QJ, Jin JY, Yang X, Huang XF. The CFTR polymorphisms poly-T, TG-repeats and M470V in Chinese males with congenital bilateral absence of the vas deferens. Asian J Androl. 2012;14:687–90. doi: 10.1038/aja.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radpour R, Gourabi H, Dizaj AV, Holzgreve W, Zhong XY. Genetic investigations of CFTR mutations in congenital absence of vas deferens, uterus, and vagina as a cause of infertility. J Androl. 2008;29:506–13. doi: 10.2164/jandrol.108.005074. [DOI] [PubMed] [Google Scholar]
- 3.Bombieri C, Claustres M, De Boeck K, Derichs N, Dodge J, Girodon E, et al. Recommendations for the classification of diseases as CFTR-related disorders. J Cyst Fibros. 2011;10:86–102. doi: 10.1016/S1569-1993(11)60014-3. [DOI] [PubMed] [Google Scholar]
- 4.Wu CC, Alper OM, Lu JF, Wang SP, Guo L, Chiang HS, et al. Mutation spectrum of the CFTR gene in Taiwanese patients with congenital bilateral absence of the vas deferens. Hum Reprod. 2005;20:2470–5. doi: 10.1093/humrep/dei077. [DOI] [PubMed] [Google Scholar]
- 5.Anzai C, Morokawa H, Okada H, Kamidono S, Eto Y, Yoshimura K. CFTR gene mutations in Japanese individuals with congenital bilateral absence of the vas deferens. J Cyst Fibros. 2003;2:14–8. doi: 10.1016/S1569-1993(02)00144-3. [DOI] [PubMed] [Google Scholar]
- 6.Steiner B, Truninger K, Sanz J, Schaller A, Gallati S. The role of common single-nucleotide polymorphisms on exon 9 and exon 12 skipping in nonmutated CFTR alleles. Hum Mutat. 2004;24:120–9. doi: 10.1002/humu.20064. [DOI] [PubMed] [Google Scholar]
- 7.Sharma N, Acharya N, Singh SK, Singh M, Sharma U, Prasad R. Heterogenous spectrum of CFTR gene mutations in Indian patients with congenital absence of vas deferens. Hum Reprod. 2009;24:1229–36. doi: 10.1093/humrep/den500. [DOI] [PubMed] [Google Scholar]
- 8.Sharma H, Mavuduru RS, Singh SK, Prasad R. Heterogeneous spectrum of mutations in CFTR gene from Indian patients with congenital absence of the vas deferens and their association with cystic fibrosis genetic modifiers. Mol Hum Reprod. 2014;20:827–35. doi: 10.1093/molehr/gau047. [DOI] [PubMed] [Google Scholar]
- 9.WHO laboratory manual for the examination and processing of human semen. Geneva: WHO; 2010. World Health Organization (WHO), Departmant of Reproductive Health and Research. [Google Scholar]
- 10.Cid R, Ramos M, Aparisi L, Garcia C, Mora J, Estivill X, et al. Independent contribution of common CFTR variants to chronic pancreatitis. Pancreas. 2010;39:209–15. doi: 10.1097/MPA.0b013e3181bab679. [DOI] [PubMed] [Google Scholar]
- 11.Maurya N, Awasthi S, Dixit P. Association of CFTR gene mutation with bronchial asthma. Indian J Med Res. 2012;135:469–78. [PMC free article] [PubMed] [Google Scholar]
- 12.Castellani C, Cuppens H, Macek M, Cassiman J, Kerem E, Durie P, et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J Cyst Fibros. 2008;7:179–96. doi: 10.1016/j.jcf.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parad RB, Gerard C. A sequence variation in intron 17B of the cystic fibrosis transmembrane conductance regulator gene. Hum Mutat. 1992;1:258–9. doi: 10.1002/humu.1380010313. [DOI] [PubMed] [Google Scholar]
- 14.Dörk T, Neumann T, Wulbrand U, Wulf B, Kälin N, Maass G, et al. Intra- and extragenic marker haplotypes of CFTR mutations in cystic fibrosis families. Hum Genet. 1992;88:417–25. doi: 10.1007/BF00215676. [DOI] [PubMed] [Google Scholar]
- 15.Schwarzer JU, Schwarz M. Significance of CFTR gene mutations in patients with congenital aplasia of vas deferens with special regard to renal aplasia. Andrologia. 2012;44:305–7. doi: 10.1111/j.1439-0272.2012.01281.x. [DOI] [PubMed] [Google Scholar]
- 16.de la Taille A, Rigot JM, Mahe P, Gervais R, Dumur V, Lemaitre L, et al. Correlation of genitourinary abnormalities, spermiogram and CFTR genotype in patients with bilateral agenesis of the vas deferens. Prog Urol. 1998;8:370–6. [PubMed] [Google Scholar]
- 17.Kim B, Kawashima A, Ryu JA, Takahashi N, Hartman RP, King BF. Imaging of the seminal vesicle and vas deferens. Radiographics. 2009;29:1105–21. doi: 10.1148/rg.294085235. [DOI] [PubMed] [Google Scholar]
- 18.Cuppens H, Lin W, Jaspers M, Costes B, Teng H, Vankeerberghen A, et al. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes: The polymorphic (TG)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J Clin Invest. 1998;101:487–96. doi: 10.1172/JCI639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claustres M. Molecular pathology of the CFTR locus in male infertility. Reprod Biomed Online. 2005;10:14–41. doi: 10.1016/s1472-6483(10)60801-2. [DOI] [PubMed] [Google Scholar]


