Abstract
Virus populations may be challenged to evolve in spatially heterogeneous environments, such as mixtures of host cells that pose differing selection pressures. Spatial heterogeneity may select for evolved polymorphisms, where multiple virus subpopulations coexist by specializing on a narrow subset of the available hosts. Alternatively, spatial heterogeneity may select for evolved generalism, where a single genotype dominates the virus population by occupying a relatively broader host niche. In addition, the extent of spatial heterogeneity should influence the degree of divergence among virus populations encountering identical environmental challenges. Spatial heterogeneity creates environmental complexity that should increase the probability of differing adaptive phenotypic solutions, thus producing greater divergence among replicate virus populations, relative to counterparts evolving in strictly homogeneous host environments. Here, we tested these ideas using experimental evolution of RNA virus populations grown in laboratory tissue culture. We allowed vesicular stomatitis virus (VSV) lineages to evolve in replicated environments containing BHK-21 (baby hamster kidney) cells, HeLa (human epithelial) cells, or spatially heterogeneous host cell mixtures. Results showed that generalist phenotypes dominated in evolved virus populations across all treatments. Also, we observed greater variance in host-use performance (fitness) among VSV lineages evolved under spatial heterogeneity, relative to lineages evolved in homogeneous environments. Despite measurable differences in fitness, consensus Sanger sequencing revealed no fixed genetic differences separating the evolved lineages from their common ancestor. In contrast, deep sequencing of evolved VSV populations confirmed that the degree of divergence among replicate lineages was correlated with a larger number of minority variants. This correlation between divergence and the number of minority variants was significant only when we considered variants with a frequency of at least 10 per cent in the population. The number of lower-frequency minority variants per population did not significantly correlate with divergence.
Keywords: experimental evolution, environmental heterogeneity, quasispecies, niche breadth, Vesicular stomatitis virus, adaptation
1. Introduction
RNA viruses frequently navigate heterogeneous environments, and environmental heterogeneity is predicted to have consequences for viral evolution. One aspect of environmental heterogeneity that presents a particular challenge for viruses is heterogeneity in the host community. Viruses encounter heterogeneity in their host communities at a variety of levels (Wasik and Turner 2013). Within an individual multicellular host, viruses navigate heterogeneous cell and tissue types; at a larger scale, viruses navigate host communities and landscapes that are heterogeneous in terms of host species, genotypes, and resistance phenotypes. Understanding the evolutionary consequences of such environmental heterogeneity is relevant to understanding the evolution of niche breadth and the maintenance of genetic diversity in viral populations.
Evolution in heterogeneous environments may result in more genetically diverse populations than evolution in homogeneous environments. Theoretical models show that it is possible for heterogeneous environments to support balanced polymorphism, in which multiple alleles are actively maintained in the gene pool at frequencies above that of mutation (Levene 1953; Roughgarden 1972; Gliddon and Strobeck 1975; Strobeck 1979; Czochor and Leonard 1982). This multiple-niche balanced polymorphism can occur when the environment is heterogeneous and no one genotype has the highest fitness in all environments. Genetic diversity is more likely to be maintained by spatial heterogeneity than by temporal heterogeneity in the environment, and is more likely when there is habitat selection or limited gene flow between niches (Hedrick 2006).
Experimental work in microbial systems supports the prediction that spatial environmental heterogeneity can produce polymorphic populations (Rainey and Travisano 1998; Brisson and Dykhuizen 2004; Dykhuizen and Dean 2004; Zhong et al. 2004). Environmental heterogeneity has also been proposed as an explanation for the maintenance of polymorphism in several eukaryotic systems (Hedrick 1976, 1986, 2006). Similarly, heterogeneous host communities might support polymorphic virus populations if no virus genotype has the highest fitness on both hosts. The likelihood of a stable virus polymorphism in a mixed host environment is expected to be influenced by the relative frequencies of the host types as well as the relative fitness of each viral genotype on each host type (Dykhuizen and Dean 2004).
Environmental heterogeneity may also result in more divergent evolutionary outcomes among populations that have evolved in replicate environments as opposed to populations that have evolved in homogeneous environments (Cooper and Lenski 2010). When populations evolve in homogeneous environments, they are expected to converge on the most successful strategy within that environment. This hypothesis that populations evolved in replicate homogeneous environments tend to converge on parallel phenotypes has been supported by experimental evolution (Bennet et al. 1992; Bull et al. 1997; Reboud and Bell 1997; Kassen and Bell 1998; Weaver et al. 1999; Wichman et al. 1999; Cuevas et al. 2002; Agudelo-Romero et al. 2008; Remold et al. 2008; Rokyta et al. 2009; Dessau et al. 2012; Alto et al. 2013).
In contrast, when populations evolve in spatially heterogeneous environments, several outcomes are theoretically conceivable. Spatial heterogeneity may select for a monomorphic generalist population capable of using all available environmental patches, a polymorphic population consisting of subpopulations specialized on different environmental patches, or a monomorphic population specialized on the more productive or more frequently encountered patch. The evolutionary outcome is expected to depend on the relative abundance and productivity of patches, the strength of interphenotypic competition, the amount of gene flow between patches, the heritability of niche width, and whether there is a trade-off between patches (Roughgarden 1972). If one patch is much more frequently encountered or is more productive, the population may evolve to specialize on that patch (Jasmin and Kassen 2007). Polymorphic populations are most readily maintained when dispersal among patches is low, and the number of individuals contributed by each patch to the total population is relatively balanced (Kassen 2002).
In the current study, we experimentally tested whether fine-grained spatial heterogeneity in host communities would influence the evolution of host-use strategies, genetic diversity, and the repeatability of evolution in RNA virus populations. We experimentally evolved vesicular stomatitis virus (VSV) in homogeneous single-host environments or in heterogeneous mixed-host environments. VSV is an arthropod-borne Rhabdovirus (-ssRNA) that naturally infects a wide range of vertebrate and invertebrate hosts, and it is an important disease agent in domesticated animals such as horses and cattle. VSV has an ∼11.2 kb genome encoding five proteins: the nucleocapsid (N) protein that encapsidates the genomic RNA, phosphoprotein (P) and large (L) protein that make up the polymerase, glycoprotein (G) involved in cell-surface binding, and matrix (M) protein important both for virion formation and inhibition of host antiviral gene expression (Lyles and Rupprecht 2007). VSV has been used as an experimental model for studying evolution of specialism and generalism in RNA viruses (Novella et al. 1999; Turner and Elena 2000; Cuevas et al. 2002; Remold et al. 2008).
We allowed VSV populations to evolve in replicated tissue-culture environments that varied in host community (cell type) composition for twenty-five passages (∼100 generations of VSV evolution). We chose two host cell types for this study: BHK-21, a hamster kidney derived cell line, and HeLa, a human cancer derived cell line. In previous experiments where VSV was passaged on BHK or HeLa cells, some studies reported a trade-off between the two host types (Turner and Elena 2000; Remold et al. 2008), while in other studies VSV evolved a generalist phenotype with no evidence of a trade-off (Smith-Tsurkan et al. 2010). In the current study, VSV lineages were evolved in either homogeneous single-host environments, consisting of monolayers of BHK or HeLa cells, or heterogeneous environments consisting of mixed monolayers of the two host cell types. We tested three hypotheses: (1) that populations evolved in heterogeneous environments would have more within-population genetic diversity than populations evolved in homogeneous environments, (2) when virus populations were assayed for fitness on a single host type, greater phenotypic variance for fitness would be observed among populations that evolved in heterogeneous environments compared with populations evolved in homogeneous environments. This is because a multi-host environment allows for the success of multiple strategies ranging from monomorphic generalist populations to polymorphic populations made up of specialists, potentially leading to phenotypic divergence among populations evolved in replicate environments, and (3) that populations evolved in heterogeneous environments would tend to evolve generalist phenotypes and populations evolved in homogeneous environments would tend to evolve specialist phenotypes.
2. Methods
2.1 Viruses and cell culture
Host cells for infection were BHK-21 cells and HeLa cells, kindly provided by John J. Holland and Isabel Novella. All viruses in the current study were derived from a plaque-purified clone isolated from a MARMU (Monoclonal Antibody Resistant Mutant) VSV population. MARMU, originally derived from the Mudd-Summer strain of VSV Indiana serotype, is a mutant resistant to neutralization by the mouse I1-monoclonal antibody (Vandepol et al. 1986; Holland et al. 1991). The original laboratory host of this VSV strain was BHK-21 cells. Viruses and host cells were cultured in Dulbecco’s modified Eagle’s minimum essential medium with 10 per cent fetal bovine serum and 1 per cent penicillin and streptomycin. Cells for infection were grown in 25-cm2 tissue culture flasks at 37 °C, 95 per cent relative humidity, and 5 per cent CO2 atmosphere.
2.2 Experimental evolution of virus populations
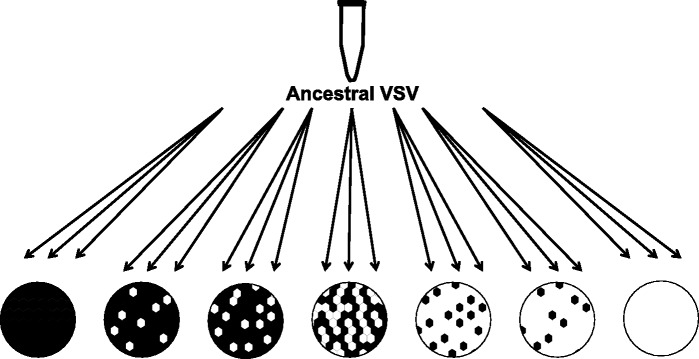
The MARMU virus stock (hereafter ancestor) was used to found three replicate lineages in each of seven treatments (twenty-one lineages in total). The treatments varied by the composition of the host cell community (Fig. 1). Two treatments represented homogenous single-host environments, comprised either exclusively of BHK cells or HeLa cells. The remaining five treatments represented heterogeneous host communities varying in the relative frequency of the two host types. Mixed host treatments had the following ratios of BHK to HeLa: 1:10, 1:5, 1:1, 5:1, 10:1. Twenty-four hours before each passage, BHK and HeLa cells were trypsinized and combined at a specified ratio; ∼1.5 × 106 total cells were seeded in 25-cm2 flasks and allowed to reach confluence in 24 h. Cell lines were maintained separately and only mixed the day before every infection. At each passage, cells were infected at multiplicity-of-infection (MOI; ratio of viruses to host cells) of roughly 0.001. The MOI was estimated at the beginning of the evolution experiment and adjusted for each replicate every five passages through the experiment. Supernatant containing viral progeny was collected at 24 h post-infection and diluted to infect the subsequent passage. Virus populations were passaged for 25 days, corresponding to ∼100 generations of VSV replication. Live cells were still visible at the 24-h time point when virus was harvested, with proportion of cell death increasing over the course of the experiment as the virus populations adapted.
Figure 1.
Experimental design. The ancestral VSV strain was used to found three replicate virus lineages in each experimental treatment, which were then serially passaged for roughly 100 VSV generations. Experimental treatments varied in the composition of the host cell community. Here the relative frequency of each cell type is represented, with black representing HeLa cells and white representing BHK cells.
2.3 Cell imaging
Each cell type was stained separately with a different fluorescent dye, either CellTracker Green CMFDA Dye (Life Technologies C2925) or Orange CMFDA Dye (C2927). The stained cells were then combined to form mixed cell monolayers on glass slides placed in tissue culture dishes. Monolayers were fixed using paraformaldehyde and imaged using confocal microscopy.
2.4 Plaque assays
Viral titers (plaque-forming units [pfu] per milliliter) were estimated using plaque assays, in which serially diluted samples were plated on BHK cells under DMEM medium with 10 per cent fetal bovine serum and solidified with 1 per cent agarose, and incubated for 36 h using the above-described conditions. After incubation, cells were fixed with 10 per cent formaldehyde, media and agarose were removed, and plates were stained with crystal violet to visualize plaques. Each plaque was assumed to have originated from a single infecting virus.
2.5 Fitness assays
Each evolved (passage 25) population was competed against antibody-sensitive wildtype VSV (Holland et al. 1991). Fitness was assayed on BHK cell monolayers and on HeLa cell monolayers. An antibody-resistant test virus was mixed 1:1 with the antibody-sensitive competitor, and the mixture was immediately sampled and diluted to perform parallel plaque assays in the presence and absence of monoclonal antibody to gauge the true initial ratio (R0) of test virus to MARM virus at time t0 (Turner and Elena 2000). After 24 h, the supernatant containing virus progeny was collected and diluted, and similar plaque assays gauged the R1 ratio of test virus to competitor virus at t1. The fitness (W) of a test virus relative to the common competitor was estimated as R1/R0 (Turner and Elena 2000). Fitness values were expressed as the natural logarithm (ln W). Before fitness assays were performed, a plaque assay was performed with each test virus to ensure that antibody resistance had been maintained over the course of the evolution experiment (data not shown).
2.6 Whole genome consensus sequencing and analysis
Genomic viral RNA was isolated from twenty-two lineages (twenty-one evolved populations at the final passage [passage 25] plus the ancestor) using the QiaAmp Viral RNA mini kit (Qiagen). cDNA was generated by reverse-transcription with Superscript II (Invitrogen) using random hexamer primers. The majority of the genome sequence was amplified via polymerase chain reaction (PCR) using eight primer pairs (listed in supplement), generating overlapping PCR fragments 1–2.5 kb in length. Following purification with ExoI and Antarctic Phosphatase (New England BioLabs Inc.), the fragments were sequenced via dye termination (Sanger) at the DNA Analysis Facility at Yale University. Coverage was generated at two- to four-fold using overlapping fragments and multiple directional sequencing primers. This approach generates a consensus genomic sequence for each population, rather than a genome sequence derived from representative clones. All sequences were reviewed following genome assembly using DNA Workbench 6 (CLC Bio). Consensus sequences from the evolved populations were aligned to the ancestral sequence generated in this study.
2.7 Library preparation and deep sequencing
Genomic viral RNA was isolated from twenty-two lineages (twenty-one evolved populations at the final passage [passage 25] plus the ancestor) using the QIAamp Viral RNA mini kit (Qiagen). All subsequent steps, from reverse transcription through sequencing, were completed in duplicate, resulting in two technical replicates of each sample. cDNA was generated by reverse-transcription with Superscript II (Life Technologies) using random hexamer primers. The majority of the genome sequence was amplified via PCR using eight primer pairs, generating overlapping PCR fragments 1–2.5 kb in length. Amplified genome fragments were purified using the QIAquick PCR Purification Kit (Qiagen), and the eight fragments from each virus population were combined to create an equimolar mixture. Libraries were prepared from the pooled viral amplicons using the Nextera XT Kit (Illumina Inc.) and the Nextera Index Kit (Illumina Inc.). Samples were sequenced via paired-end, 75-bp read Illumina HiSeq 2000 (Illumina Inc.) at the Yale Center for Genome Analysis. Quality control of reads was conducted using FastQC (Andrews 2010) with a minimum quality score of 20 and a minimum length of 30 bp.
2.8 Genome assembly and alignment
Initially, sequences were mapped to the publicly available VSV genome (GI:9627229) using BWA v0.7.12 (Li and Durbin 2009) using default parameters. Single nucleotide polymorphism (SNP) calling was conducted using the Genome Analysis Toolkit (GATK v3.3) (McKenna et al. 2010) with the minimum PHRED-scaled variant confidence set to 30. Consensus sequences for each sample are shown in Supplementary Appendix S1. To further investigate minority variation within treatments, we undertook minority variant detection using the QUASR v6.08 pipeline (Watson et al. 2013) using the default settings. Detection was undertaken at various levels—10 per cent, 5 per cent, and 1 per cent of the population. Only polymorphisms detected in both technical sequencing replicates of each sample were retained to reduce false-positive results due to sequencing error.
2.9 Statistical analysis
Mixed linear models were used to assess the effects of fixed effect of evolutionary history and random effects of population and block on viral fitness (PROC MIXED, SAS Institute 2004). To test for a correlation between the variation in fitness among replicates within each treatment and the number of minority variants within each population, the variance in fitness on BHK cells for each treatment was regressed against the average number of polymorphisms detected in 10 per cent, 5 per cent, and 1 per cent of each treatment, respectively. Regression calculations were carried out using the base package in R v3.1.2 (R Core Development Team, 2013). We also conducted χ2 tests to determine if any specific minority variants (i.e. SNPs) were significantly associated with fitness variation observed in treatments at the 10 per cent, 5 per cent, and 1 per cent minority detection level using the base package in R v3.1.2 (R Core Development Team 2013). Variances in fitness between heterogeneous treatments and homogeneous treatments were contrasted by two-tailed t-test in R v3.1.2 (R Core Development Team 2013).
3. Results
3.1 Imaging reveals evenly mixed host environments
We assumed that mixing of BHK and HeLa host cells in combined monolayers (heterogeneous environments) would not produce clumping among host cells of either type. We tested this by conducting a preliminary experiment to examine the spatial composition of mixed host environments. HeLa and BHK cells were fluorescently labeled, and mixed cultures were imaged across a range of relative cell proportions (Supplementary Fig. S1). Visual inspection of the images showed the mixed cells formed an evenly mixed monolayer, with no evidence of higher order structure (i.e. cells did not seem to preferentially bind to like cells). These images demonstrated that the infecting virus populations experienced expected probabilities of host encounter in passages containing mixed host environments (e.g. in a 1:1 monolayer, there should be roughly equal probability that progeny viruses exiting a cell encounter HeLa versus BHK host cells).
3.2 Treatment-specific patterns of fitness change
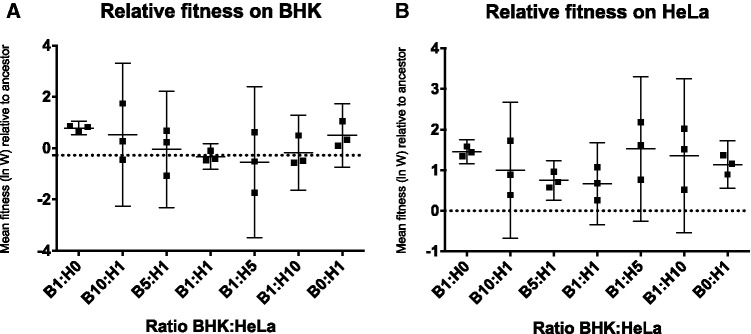
Evolved virus populations were assayed for fitness relative to a common competitor on BHK cells and on HeLa cells. Results showed that all evolved virus populations improved on HeLa cells relative to the ancestral virus (Fig. 2). There was no significant difference in performance on HeLa cells among experimental treatments (mixed linear model, P = 0.31); however variance was significantly greater in heterogeneous treatments than in homogeneous treatments (two-way F-test: F = 0.157, num df = 5, denom df = 14, P-value = 0.05). When fitness was assayed on BHK cells, all virus populations either showed improved fitness on BHK relative to the ancestor or had fitness on BHK that was not statistically different than that of the ancestor. Evolved fitness on BHK cells was not significantly different among treatments (mixed linear model, P = 0.06); however this P-value comes close to our threshold for significance (P < 0.05), and it is possible that significance would have been reached with a larger sample size. Variance was greater on average in heterogeneous treatments than in homogeneous treatments, but not significantly (two-way F-test: F = 0.185, num df = 5, denom df = 14, P-value = 0.07286).
Figure 2.
Relative fitness (ln(W)) of evolved virus populations. Each point represents the fitness of an evolved population relative to the common competitor virus (mean of three trials). The grand mean (solid line) and 95 per cent CI are shown for each treatment. The dotted line represents the fitness of the ancestral virus relative to the common competitor. (A) Relative fitness on BHK cells. (B) Relative fitness on HeLa cells.
3.3 Consensus sequences
None of the evolved populations showed changes in their consensus sequences relative to the ancestor (data not shown). A consensus sequence gives the most frequent base for each nucleotide position in the virus genome for an entire population, and a lack of changes to the consensus genome demonstrated that no substitutions went to fixation over the course of the experiment. To rule out the possibility of contamination during sequencing, we conducted a second round of sequencing with a subset of seven virus populations from passage 23, which had not been handled since their initial storage. We observed no changes in the consensus sequences of the passage-23 isolates (data not shown), which was consistent with the initial sequencing of passage-25 populations. We note that in this sequencing strategy, the leader and trailer (portions of the 3′ and 5′ UTR) of the virus genome are not sequenced.
3.4 Minority variant detection via high-throughput sequencing
To examine the hypothesis that phenotypic differences among populations were driven by minority variants within the evolved populations, we conducted whole-genome high-throughput sequencing of all evolved virus populations and the ancestral virus stock. We recovered an average of 1,974,256 reads per sample of which an average of 98.8 per cent aligned to the reference genome (average coverage of 6,348× per sample) (Supplementary Appendix S1 and S3). While SNP calling indicated that there were 114 polymorphisms in the samples when compared with the reference sequence, these polymorphisms were fixed across all samples and represent differences between our ancestral VSV strain and the reference. This lack of changes to the consensus genome confirmed our results from Sanger sequencing, and the data demonstrated that no substitutions went to fixation in any sample over the course of the experiment.
To further examine the structure of the viral mutant spectra, we identified minority variants in the virus populations existing above various frequency thresholds. An average of 5, 13, and 186 minority variant polymorphisms were detected per population at the 10 per cent, 5 per cent, and 1 per cent levels, respectively (Fig. 3, Supplementary Appendix S2).
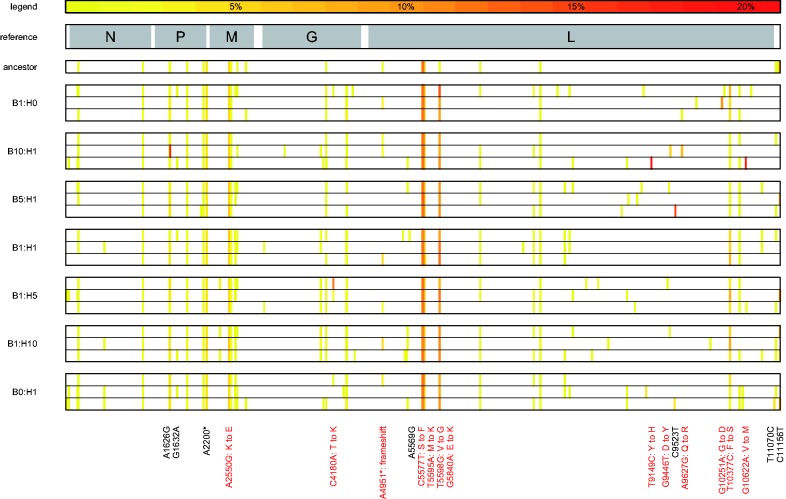
Figure 3.
Map of polymorphic loci. Polymorphic sites for which a minority allele was detected at a frequency of 1 per cent or greater are shown mapped onto the VSV genome. Allele frequency is determined as the mean of two technical replicates, and only sites with depth of coverage >30 were considered for this analysis. Each row represents one virus population, with the experimental treatment (ratio of BHK to HeLa cells) shown on the left. A reference map of the VSV genome is displayed above, with gene coding regions shaded, with overlaid gene labels. The frequency of minority alleles in each population is coded by color (see legend). The location of minority alleles that appear in any population >5 per cent are labeled below. Nonsynonymous changes are labeled in red, and the amino acid substitution is listed.
3.5 Analysis of intra-population genetic diversity
We found a significant (P = 0.03) correlation between variance in growth rate on BHK among replicate populations within a treatment and the average number of minority variants present above the 10 per cent threshold in those populations; however, the relationship was not significant when polymorphisms present at below 10 per cent were included (Table 1). This indicates that differences at polymorphic sites among populations may explain the observed phenotypic divergence of replicate populations, but only when those variants are present in at least 10 per cent of the population. We did not find a significant relationship between treatment and the specific polymorphisms found in a sample (Table 2). Many of the minority alleles were highly parallel among the majority of populations, whereas some were singletons, and a small number of alleles were parallel but not widespread (Fig. 3). Highly parallel minority alleles were frequently also observed in the ancestral virus stock used to found the experimental lines; this result was unsurprising, as the mutant swarm recovers rapidly after bottlenecks including plaque purification (Schommer and Kleiboeker 2006). A highly parallel nonsynonymous substitution in the L protein (T10377C) was observed in all evolved lineages at 2–10 per cent frequency, but was below our level of detection (1%) in the ancestor. We identified at least one mutation (C4180A) that was previously described in VSV experimental evolution (Remold et al. 2008). In the Remold et al. study, this mutation was identified both in populations evolved on HeLa cells and in populations that alternated between two cell types (HeLa and MDCK). In the current study, we observed this mutation at ∼14 per cent frequency in one lineage evolved in a mixed environment, at ∼1.5 per cent in one lineage evolved on HeLa cells, and ∼1 per cent in one lineage evolved on BHK. Additionally, one population in the B1:H5 treatment had another minority substitution at the same site (C4180G). Both the C4180A and C4180G substitutions are nonsynonymous, changing the ancestral threonine to a lysine and an arginine, respectively. There was no significant difference between the number of polymorphisms present in populations from heterogeneous versus homogeneous treatments at >10 per cent (two-tailed t-test: t = −1.09, df = 11.63, P-value = 0.30), >5 per cent (two-tailed t-test: t = −1.48, df = 16.09, P-value = 0.16), or 1 per cent (two-tailed t-test: t = −1.15, df = 8.43, P-value = 0.28).
Table 1.
Results of χ2 analysis testing whether specific minority variants detected in at least 10 per cent, 5 per cent, and 1 per cent of each population, respectively, were associated with particular treatments. No significant relationships between the specific minority variants detected and treatment were detected. df—degrees of freedom, P—significance value.
| SNP % | χ2 | df | P |
|---|---|---|---|
| 10% | 54.24 | 84.00 | 0.99 |
| 5% | 125.45 | 186.00 | 0.99 |
| 1% | 1,738.89 | 2,406.00 | 0.99 |
Table 2.
Results of regression analysis testing for a significant correlation between the variance of growth on both BHK and HeLa cells within a treatment, and the average number of minority variants detected in at least 10 per cent, 5 per cent, and 1 per cent within each treatment. A significant correlation was detected between minority variants detected in at least 10 per cent of the population and variance in growth on BHK cells. r2—regression coefficient, P—significance value.
| SNP % | BHK |
HeLa |
||
|---|---|---|---|---|
| R2 | P | r2 | P | |
| 10 | 0.57 | 0.03* | 0.25 | 0.14 |
| 5 | −0.17 | 0.74 | 0.06 | 0.29 |
| 1 | −0.14 | 0.64 | −0.17 | 0.72 |
*indicates significance at P < 0.05.
4. Discussion
We experimentally tested whether the relative availability of host types in a spatially heterogeneous environment versus homogenous single-host environments influenced the evolution of virus host-use strategies, within-population genetic diversity, and evolutionary divergence among virus populations. We found that generalist phenotypes dominated for virus populations evolved in all treatments with widespread correlated improvements on the two host cell types. We found support for the hypothesis that replicate populations evolved in heterogeneous environments have higher variance in their fitness on each host cell type than populations evolved in homogeneous environments. We observed no changes to the consensus sequences of evolved virus populations relative to the ancestor; however, we detected polymorphic loci in all evolved virus populations, and these minority allele variants appeared to influence the measured population phenotypes. We did not find support for the hypothesis that populations evolved in heterogeneous host environments would select for higher levels of polymorphism than homogeneous host environments.
In the current experiment, we observed that VSV populations evolved in either single-host treatment (BHK or HeLa) showed improvement on the selected host and correlated fitness gains on the unselected host, resulting in generalist phenotypes. In heterogeneous environments, populations either showed correlated improvement on both host cells, or improved on one host while remaining statistically similar to the ancestral fitness on the other. These data indicated that populations did not experience an evolutionary trade-off across the two host cell lines, and generalist phenotypes dominated across all experimental treatments. Our observations here were similar to a previous experiment in which VSV populations evolved on BHK or HeLa cells showed correlated improvement on the alternate host cell type (Smith-Tsurkan et al. 2010). However, other experiments have shown that VSV populations can experience trade-offs when specializing on these host cell types (Turner and Elena 2000; Remold et al. 2008).
In the current experiment no mutations went to fixation, despite the fact that VSV populations improved on the selected host cell types. VSV is a generalist virus, and although there is room for increasing fitness, the ancestral strain we used to found our experimental virus populations already grew relatively robustly on the experimental cell types. It is possible that the selective pressures in this experiment were not strong enough to drive new variants to fixation in the time allowed, but minority variants may have fixed had the experiment been continued for further generations.
Prior studies using RNA viruses have empirically demonstrated that the phenotype of a viral population can change measurably without changes to the consensus sequence, presumably owing to shifts in the genetic composition of the population (Bordería et al. 2010; Coffey and Vignuzzi 2011). Bordería et al. (2010) reported that populations of the retrovirus HIV-1 passaged in vitro showed increased fitness in tissue culture without changes in the consensus sequence. However, these populations did show increased diversity in their mutant spectra as determined by sequencing individual virus clones from the populations. Additionally, Coffey and Vignuzzi (2011) observed that serially passaged populations of the positive-sense ssRNA virus chikungunya virus showed increased growth on novel cell types without changes in the consensus sequences. Sequencing of viral clones showed increased diversity in evolved virus populations, which the authors hypothesized was responsible for the changes in viral fitness.
Similarly, in the current study, we observed measurable phenotypic changes in evolved populations of the negative-sense ssRNA virus VSV, but did not observe changes in the consensus sequences. We hypothesized that polymorphic alleles that were below the level of detection through Sanger sequencing produced the observed changes in phenotype. To examine this hypothesis, we used high-throughput sequencing to analyze intra-population genetic variation in ancestral and evolved virus populations. As expected, we found that all viral populations harbored a set of low-frequency minority variants occurring below the threshold of detection by consensus sequencing. In all sequenced populations, the distribution of minority variant frequencies was roughly exponential, with low-frequency variants being more common than higher-frequency minority variants (Supplementary Fig. S2). We detected more minority variants above the threshold of 10 per cent frequency in treatments for which replicate virus populations showed the greatest variation in their growth on BHK cells. Observed differences in niche breadth may in fact be governed by variability at polymorphic sites. Interestingly, this analysis suggests that while variants are most numerous at low frequencies, variants may need to reach a higher frequency threshold before they are relevant for population fitness and host-use strategies. Future studies with larger sample sizes will be required to analyze the importance of low-frequency variants for virus populations.
Next-generation sequencing data also allowed us to test our hypothesis that heterogeneous environments support more genetically diverse virus populations than do homogeneous environments. While higher levels of polymorphism were correlated with greater divergence among replicates, we did not generally find a significant difference in levels of polymorphism between heterogeneous and homogeneous environments. All treatments had qualitatively similar distributions of minority variant frequencies.
Our observation that homogeneous and heterogeneous environments support similar levels of genetic diversity is not surprising. While heterogeneous environments have the potential to support balanced polymorphisms, they can only do so when specific conditions are met. Balanced polymorphism can emerge only when no one genotype has the highest fitness in all environments. In the current experiment we did not observe evidence of a trade-off between the two host types, which confirmed that generalist genotypes could dominate in heterogeneous environments composed of BHK and HeLa host cells. The heterogeneity in our mixed-host environments was also fine-grained, which in the absence of habitat selection resulted in high gene flow between environmental patches. High gene flow between patches reduces the likelihood of balanced polymorphism. Our results are consistent with the idea that while balanced polymorphisms are possible in heterogeneous environments, they are unlikely to be stable unless specific criteria are met (Levene 1953; Roughgarden 1972; Gliddon and Strobeck 1975; Strobeck 1979; Czochor and Leonard 1982; Hedrick 1986).
We observed variance in fitness is higher in heterogeneous host cell treatments than in homogeneous host cell treatments, and that there was a further correlation between variance in fitness and the number of polymorphisms (>10%). Transitivity should lead to the conclusion that heterogeneous populations had more polymorphisms; however, this was not the case. We believe that the most likely cause of this transitivity issue was small sample size, which limited our statistical power. We also acknowledge that the number of the polymorphic sites alone gives limited information; a similar distribution but of different alleles could result in different fitness effects. Unraveling the complex interactions between the genetic diversity within RNA virus populations and population-level fitness and phenotype remains a major challenge for modern evolutionary virology.
Experimental evolution has played a critical role in defining our expectations for the dynamics of adaptation in homogeneous environments. Much exciting recent and ongoing work in experimental evolution is expanding the ecological scenarios explored to include environments that are heterogeneous in space and time. These experiments bring us a step closer to understanding how natural populations contend with complexity in their environments, while maintaining the experimental advantages of controlled and replicated environments. The current study adds to this body of work by exploring the evolution of RNA viruses in single-host versus mixed-host environments. This study also highlights the importance of further work to understand how environmental complexity shapes adaptive dynamics and the role of minority variants in determining phenotypes of virus populations.
Data Availability
Raw sequencing data is available through the Sequencing Read Archive (SRA) project accession #SRP066436. Data from fitness assays is available in the online supplement to this manuscript.
Supplementary data
Supplementary data are available at Virus Evolution online.
Acknowledgements
We thank two anonymous reviewers, associate editor Rafael Sanjuan, and members of the Turner research group for valuable feedback on this study, Nadya Morales for technical assistance, and Aaron Mertz for assistance with cell imaging. This work was supported by grant #DEB-1021243 from the US National Science Foundation. V.J.M. was supported by the US National Science Foundation (NSF) Graduate Research Fellowship grant #DGE-1122492, and P.E.T. by the NSF BEACON Center for Study of Evolution in Action.
References
- Agudelo-Romero P., de la Iglesia F., Elena S. F. (2008) ‘The Pleiotropic Cost of Host-Specialization in Tobacco etch potyvirus’, Infection, Genetics, and Evolution, 8: 806–14. [DOI] [PubMed] [Google Scholar]
- Alto B. W., et al. (2013) ‘Stochastic Temperatures Impede RNA Virus Adaptation’, Evolution, 67: 969–79. [DOI] [PubMed] [Google Scholar]
- Andrews S. (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data <http://www.bioinformatics.babraham.ac.uk/projects/fastqc/2010>. [Google Scholar]
- Bennet A. F., Lenski R. E., Mittler J. E. (1992) ‘Evolutionary Adaptation to Temperature. 1. Fitness Responses of Escherichia coli to Changes in its Thermal Environment’, Evolution, 46: 16–30. [DOI] [PubMed] [Google Scholar]
- Bordería A. V., et al. (2010) ‘Initial Fitness Recovery of HIV-1 is Associated with Quasispecies Heterogeneity and Can Occur Without Modifications in the Consensus Sequence’, PLoS One, 5: e10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D., Dykhuizen D. E. (2004) ‘ospC Diversity in Borrelia Burgdorferi’, Genetics, 168: 713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J. J., et al. (1997) ‘Exceptional Convergent Evolution in a Virus’, Genetics, 147: 1497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey L. L., Vignuzzi M. (2011) ‘Host Alternation of Chikungunya Virus Increases Fitness While Restricting Population Diversity and Adaptability to Novel Selective Pressures’, Journal of Virology, 85: 1025–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T., Lenski R. E. (2010) ‘Experimental Evolution with E. coli in Diverse Resource Environments. I. Fluctuating Environments Promote Divergence of Replicate Populations’, BMC Evolutionary Biology, 10: 1471–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas J. M., Elena S. F., Moya A. (2002) ‘Molecular Basis of Adaptive Convergence in Experimental Populations of RNA Viruses’, Genetics, 612: 533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czochor R. J., Leonard K. J. (1982) ‘Multiple-Niche Polymorphism in Haploid Microorganisms’, The American Naturalist, 119: 293–6. [Google Scholar]
- Dessau M., et al. (2012) ‘Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme’, PLoS Genetics, 8: e1003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen D. E., Dean A. M. (2004) ‘Evolution of Specialists in an Experimental Microcosm’, Genetics, 167: 2015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliddon C., Strobeck C. (1975) ‘Necessary and Sufficient Conditions for Multiple-Niche Polymorphism in Haploids’, The American Naturalist, 109: 233–5. [Google Scholar]
- Hedrick P. W. (1976) ‘Genetic Polymorphism in Heterogeneous Environments’, Annual Review of Ecology and Systematics, 7: 1–32. [Google Scholar]
- Hedrick P. W. (1986) ‘Genetic Polymorphism in Heterogeneous Environments: A Decade Later’, Annual Review of Ecology and Systematics, 17: 535–66. [Google Scholar]
- Hedrick P. W. (2006) ‘Genetic Polymorphism in Heterogeneous Environments: The age of Genomics’, Annual Review of Ecology, Evolution, and Systematics, 37: 67–93. [Google Scholar]
- Holland J. J., et al. (1991) ‘Quantitation of Relative Fitness and Great Adaptability of Clonal Populations of RNA Viruses’, Journal of Virology, 65: 2960–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmin J., Kassen R. (2007). On the Experimental Evolution of Specialization and Diversity in Heterogeneous Environments’, Ecology Letters, 10: 272–81. [DOI] [PubMed] [Google Scholar]
- Kassen R. (2002) ‘The Experimental Evolution of Specialists, Generalists, and the Maintenance of Diversity’, Journal of Evolutionary Biology, 15: 173–90. [Google Scholar]
- Kassen R., Bell G. (1998). Experimental Evolution in Chlamydomonas. IV. Selection in Environments that Vary Through Time at Different Scales’, Heredity, 80: 732–41. [Google Scholar]
- Levene H. (1953) ‘Genetic Equilibrium when more than One Ecological Niche is Available’, The American Naturalist, 87: 331–3. [Google Scholar]
- Li H., Durbin R. (2009) ‘Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform’, Bioinformatics, 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles D. S., Rupprecht C. E. (2007) ‘Rhabdoviridae', in Knipe D. M., Howley P. M. (eds). Fields Virology, pp. 1363–408. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins. [Google Scholar]
- McKenna A., et al. (2010) ‘The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-generation DNA Sequencing Data’, Genome Research, 20: 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella I. S., et al. (1999) ‘Lack of Evolutionary Stasis During Alternating Replication of an Arbovirus in Insect and Mammalian Cells’, Journal of Molecular Biology, 287: 459–65. [DOI] [PubMed] [Google Scholar]
- Rainey P. B., Travisano M. (1998) ‘Adaptive Radiation in a Heterogeneous Environment’, Nature, 394: 69–72. [DOI] [PubMed] [Google Scholar]
- Reboud X., Bell G. (1997) ‘Experimental Evolution in Chlamydomonas. 3. Evolution of Specialist and Generalist Types in Environments that Vary in Space and Time’, Heredity, 78: 507–14. [Google Scholar]
- Remold S. K., Rambaut A., Turner P. E. (2008) ‘Evolutionary Genomics of Host Adaptation in Vesicular Stomatitis Virus’, Molecular Biology and Evolution, 25: 1138–47. [DOI] [PubMed] [Google Scholar]
- Rokyta D. R., Abdo Z., Wichman H. A. (2009) ‘The Genetics of Adaptation for Eight Microvirid Bacteriophages’, Journal of Molecular Evolution, 69: 229–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughgarden J. (1972) ‘Evolution of Niche Width’, The American Naturalist, 106: 683–718. [Google Scholar]
- Schommer S. K., Kleiboeker S. B. (2006) ‘Use of a PRRSV Infectious Clone to Evaluate in vitro Quasispecies Evolution’, Advances in Experimental Medicine and Biology, 581: 435–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Tsurkan S. D., Wilke C. O., Novella I. S. (2010) ‘Incongruent Fitness Landscapes, Not Tradeoffs, Dominate the Adaptation of Vesicular Stomatitis Virus to Novel Host Types’, Journal of General Virology, 91: 1484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck C. (1979) ‘Haploid Selection with n Alleles in m Niches’, The American Naturalist, 113: 439–44. [Google Scholar]
- R Core Development Team. (2013) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Turner P. E., Elena S. F. (2000) ‘Cost of Host Radiation in an RNA Virus’, Genetics, 156: 1465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepol S. B., Lefrancois L., Holland J. J. (1986) ‘Sequences of the Major Antibody Binding Epitopes of the Indiana Serotype of Vesicular Stomatitis Virus’, Virology, 148: 312–25. [DOI] [PubMed] [Google Scholar]
- Wasik B. R., Turner P. E. (2013) ‘On the Biological Success of Viruses’, Annual Review of Microbiology, 67: 519–41. [DOI] [PubMed] [Google Scholar]
- Watson S. J., et al. (2013) ‘Viral Population Analysis and Minority-variant Detection Using Short Read Next-generation Sequencing’, Philosophical Transactions of the Royal Society B, 368: 20120205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S. C., et al. (1999) ‘Genetic and Fitness Changes Accompanying Adaptation of an Arbovirus to Vertebrate and Invertebrate Cells’, Journal of Virology, 73: 4316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman H. A., et al. (1999) ‘Different Trajectories of Parallel Evolution During Viral Adaptation’, Science, 285: 422–4. [DOI] [PubMed] [Google Scholar]
- Zhong S., et al. (2004) ‘Evolutionary Genomics of Ecological Specialization’, Proceedings of the National Academy of Sciences, 101: 11719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data is available through the Sequencing Read Archive (SRA) project accession #SRP066436. Data from fitness assays is available in the online supplement to this manuscript.