Abstract
Members of the Picornaviridae are important and often zoonotic viruses responsible for a variety of human and animal diseases. However, the evolution and spatial dissemination of different picornaviruses circulating in domestic animals are not well studied. We examined the rate of evolution and time of origin of porcine enterovirus G (EV-G) and porcine kobuvirus species C lineages (PKV-C) circulating in pig farms in Vietnam and from other countries. We further explored the spatiotemporal spread of EV-G and PKV-C in Southwest Vietnam using phylogeographic models. Multiple types of EV-G are co-circulating in Vietnam. The two dominant EV-G types among isolates from Vietnam (G1 and G6) showed strong phylogenetic clustering. Three clades of PKV-C (PKV-C1-3) represent more recent introductions into Vietnam; PKV-C2 is closely related to PKV-C from Southwest China, indicating possible cross-border dissemination. In addition, high virus lineage migration rates were estimated within four districts in Dong Thap province in Vietnam for both EV-G types (G1, G6) and all PKV-C (C1-3) clades. We found that Chau Thanh district is a primary source of both EV-G and PKV-C clades, consistent with extensive pig trading in and out of the district. Understanding the evolution and spatial dissemination of endemic picornaviruses in pigs may inform future strategies for the surveillance and control of picornaviruses.
Keywords: porcine picornavirus, enterovirus, kobuvirus, evolution, phylogeography
1. Introduction
Picornaviruses are single-stranded, positive-sense RNA viruses that can cause a variety of diseases. The Enterovirus and Kobuvirus genera contain the most clinically and economically significant viruses in the picornavirus family (Jacques et al. 2008; Lewis-Rogers and Crandall 2010; Khamrin et al. 2014). A number of newly discovered types within each (e.g., identified on the basis of >25% VP1 nucleotide sequence divergence for enterovirus) in recent years (Tapparel et al. 2013; Van Dung et al. 2014) highlight the risk of outbreaks in both humans and animals. In addition, host jumps of enterovirus have been identified from humans into domestic pigs (Kadoi, Suzuki, and Nishio 2001; Van Dung et al. 2014). Cross-species transmission of kobuvirus has also been reported between domestic pigs, cattle, and sheep (Reuter et al. 2008; 2012).
Pigs have long been known to serve as reservoirs for many pathogens with zoonotic potential (Nowotny et al. 1997; Smith et al. 2011). Emerging and re-emerging swine-related zoonotic pathogens have important health and economic impacts, such as swine influenza viruses (SIV), hepatitis E virus (HEV), Nipah virus, and Japanese encephalitis (Meng et al. 1997; Mohd Nor, Gan, and Ong 2000; Cauchemez et al. 2013; Lindahl et al. 2013). To investigate viruses in pigs with zoonotic potential, a large-scale human cohort has been established within farming communities in Vietnam, with regular sampling in animal populations, thus providing a baseline survey of enteric viruses on pig farms in four different districts in Dong Thap province in Vietnam (Rabaa 2015). A diverse range of picornaviruses was identified in both asymptomatic pigs and pigs with diarrhoea on farms, included viruses from the genera enterovirus and kobuvirus (Van Dung et al. 2014).
Beyond Vietnam, porcine enterovirus and porcine kobuvirus are known to be endemic in pig farms across Asia, Europe, and North America (Reuter, Kecskemeti, and Pankovics 2010; Okitsu et al. 2012; Van Dung et al. 2014; Wang et al. 2014; Yang et al. 2014). However, no comprehensive phylogenetic study has investigated the evolution of porcine picornaviruses. In addition, relatively little is known about the spatial spread of picornaviruses in swine populations, which is important for understanding the interplay of co-circulating different picornaviruses and also for predicting future patterns of spread.
The aim of this study was to explore the evolution and spatial dissemination of endemic picornaviruses (EV-G and PKV-C) in pigs. Therefore, VP1 gene nucleotide sequence datasets were collected and analysed to infer the phylogeny, evolutionary rates, nucleotide substitution, and signs of selection. Here, we describe the spread of EV-G and PKV-C across their evolutionary histories, and outline the dissemination patterns of co-circulating PK and EV-G lineages in Vietnam over short time scales.
2. Methods
2.1 Sequence data used for picornavirus phylogenetic analysis
Faecal samples from pigs (both asymptomatic and with diarrhoea) were collected from 102 farms in four districts of Dong Thap province from February to May 2012. The partial capsid coding P1 genes (VP1) of 144 EV-G (848 nt) and 166 PKV-C (819 nt) from these Vietnamese viruses were sequenced as described in our study previously (Van Dung et al. 2014), and have been submitted to GenBank (accession number KT265800-KV265894) (Nguyen et al. 2015). No sign of recombination was detected in those VP1 sequences of EV-G and PKV-C using Recombination Detection Program v4.0 (Martin et al. 2015).
The sequence dataset additionally included the full-length VP1 sequences from other countries that were available in GenBank (www.ncbi.nlm.nih.gov/genbank/) up to September 2014. The final datasets were composed of 155 EV-G viruses and 240 PKV-C sequences. The GenBank accession numbers of sequences used in this study are listed in Supplementary Table S1. The VP1 genes of EV-G and PK virus were aligned independently using MUSCLE v3.5 (Edgar 2004). The mean percentage of nucleotide difference among sequences was calculated using DnaSP 5.10.01 (Librado and Rozas 2009).
2.2 Estimation of time-scaled phylogenies
The phylogenies of EV-G and PKV-C were generated using the Bayesian Monte Carlo Markov Chain (MCMC) method implemented in BEAST v.1.8.2 (Drummond and Rambaut 2007; Drummond et al. 2012). Different combinations of substitution models, clock models, and population size models were evaluated by the Bayes Factor (BF) test using the harmonic mean estimator (HME) to estimate marginal likelihoods. For both datasets, the best-fit model incorporated a SRD06 nucleotide substitution model (Shapiro, Rambaut, and Drummond 2006) with uncorrelated lognormal relaxed molecular clocks and a constant-population coalescent process prior over the phylogenies for analysing the total datasets (including sequences from different types) while exponential growth model for analysing the same types. Root-to-tip regression in Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/) was not able to detect temporal structure for either PKV-C or EVG. Therefore, strong priors of substitution rate were chosen according to published evolution rates of the VP1 gene from related genera in the picornavirus family, including 17 seventeen enteroviruses (species/types) and 10 ten non-enteroviruses (species/types) (Supplementary Table S2). While rates were in general, comparable between picornavirus genera, we applied the prior of EVG to enterovirus sequences analysed in our study, and the prior of PKV to other picornaviruses. For EVG, we considered priors ranging from 3 × 10−3 to 5 × 10−3 nucleotide substitutions per site per year (ns/s/y), with standard deviations (SD) from 5 × 10−4 to 3 × 10−3 ns/s/y. For PKV, we considered priors ranging from 1.5 × 10−3 to 3 × 10−3 ns/s/y, with SD from 5 × 10−4 to 2 × 10−3 ns/s/y. The estimated evolution rates and time to the most recent common ancestor (TMRCA) range were overlapped and also agreed with a previous study (Hicks and Duffy 2011). The reported analysis used a prior for the mean rate for EVG of 4 × 10−3 ns/s/y with SD = 1 × 10−3 and a prior for the mean rate for PKV-C of 2 × 10−3 ns/s/y with SD = 5 × 10−4.
MCMC chains were run for 100 million steps, sampled every 10,000 states with 10% burn-in. MCMC convergence and effective sample size of parameter estimates were evaluated using Tracer 1.5 (http://beast.bio.ed.ac.uk). Maximum clade credibility (MCC) trees were summarized using Tree Annotator and visualized using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). The overall rates of evolutionary change (ns/s/ynucleotide substitutions per site per year) and tree root ages were estimated simultaneously. Single likelihood ancestor counting (SLAC) in HyPhy (http://www.datamonkey.org/) was applied to estimate dN/dS for EV-G and PKV-C separately. The HKY85 nucleotide substitution model (Hasegawa, Kishino, and Yano 1985) was used in single-likelihood ancestor counting SLAC based analyses.
2.3 Discrete trait mapping of spatial spread
For both EV-G and PKV-C, major clades with adequate numbers of Vietnamese sequences identified from the global phylogenies were selected for further study to examine the spatial and temporal variation present in Vietnam. EV-G types G1 and G6 (Fig. 1), as well as the 3 clades of PKV-C (Fig. 2), were analysed independently to further compare different viral dissemination patterns. To determine the viral dissemination network between Vietnam and other countries, an asymmetric discrete phylogeography model was applied, using country as a trait for the discrete states. We also explored local viral lineage migration within Dong Thap province in Vietnam using the four districts (Cao Lanh, Chau Thanh, Hong Ngu, and Thanh Binh) as the discrete states. The analysis was performed and evaluated as described above with the addition of implementing Bayesian Stochastic Search Variable selection (BSSVS) to identify statistically significant transition rates between locations. For the rates calculated from Bayesian Stochastic Search Variable selection BSSVS, a BF test was adopted to identify non-zero transition rates with BF support >3. The dissemination network was inferred independently by using an asymmetric discrete traits model to reconstruct the ancestral location states of the internal nodes in the time-scaled tree phylogenies of EV-G and PKV-C.
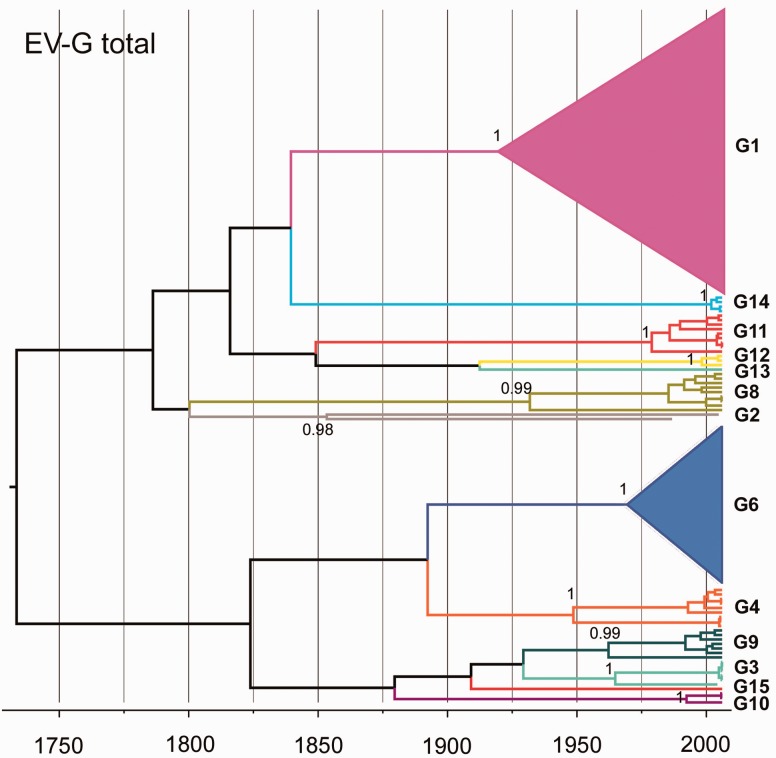
Figure 1.
Simplified Bayesian MCC tree of 155 VP1 sequences from EV-G isolated in pigs. Branches are coloured at the serotype level. The two predominant types in Vietnam: G1 (pink) and G6 (dark blue) are collapsed into triangles. The posterior support values for different EVG types were labelled on the nodes, except for type G13 and G15 (one sequence only). The tree labelled with sequences accession numbers is shown in Supplementary Fig. S1.
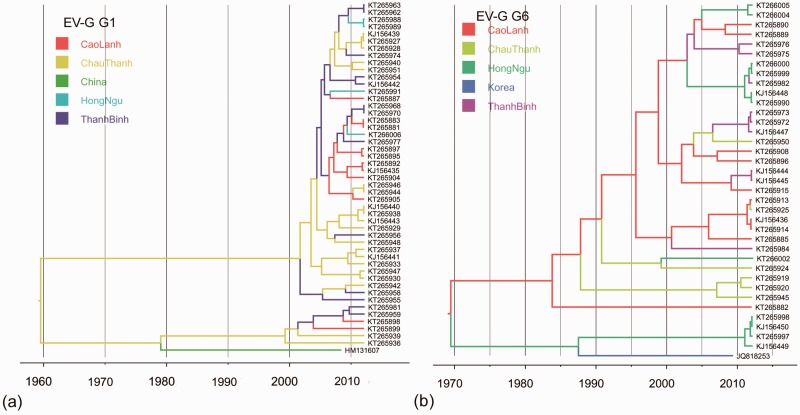
Figure 2.
MCC trees from the discrete phylogeography analysis of EV-G. (a) EV-G G1 tree phylogeny. (b) EV-G G6 tree phylogeny. Branches are coloured according to their descendent nodes annotated by the different sampled areas, with the key for colours shown on the left.
In addition, a joint analysis for discrete trait models of different clades of Vietnamese PKV-C was performed, where each clade was treated as an independent dataset with an individual relaxed clock model and tree prior, but shared the discrete trait model with all three clades giving a single transition matrix for each discrete trait.
3. Results
3.1 The phylogenetic structures of EV-G and PKV-C
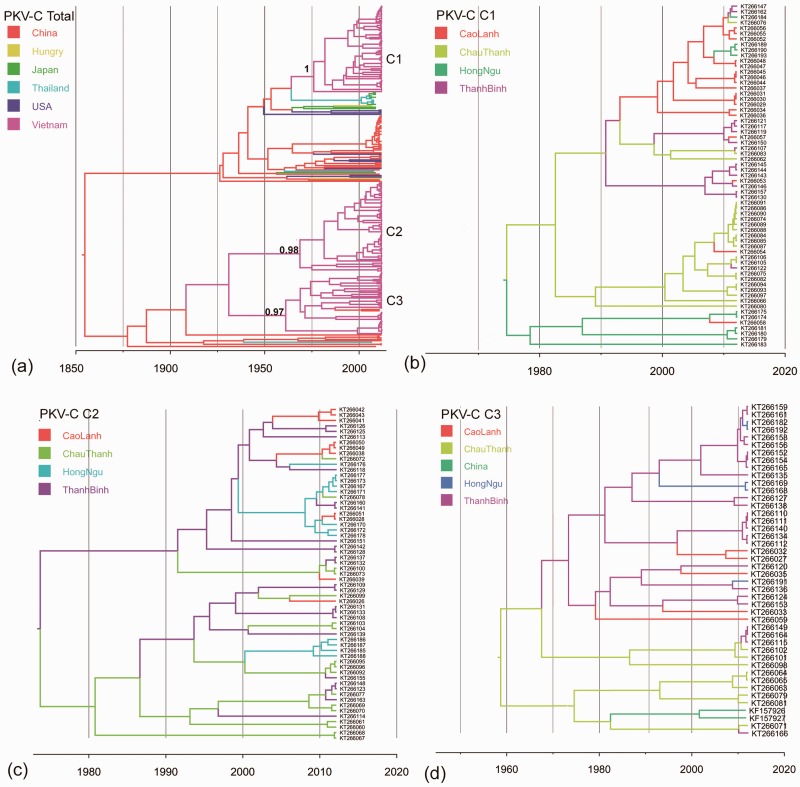
An initial phylogenetic analysis of EV-G and PKV-C was performed using VP1 sequences obtained from pigs in Vietnam, combined with a background dataset downloaded from NCBI. The overall phylogeny of EV-G VP1 identified 13 different clusters (>25% nucleotide sequence divergence) (Fig. 1). The two other recognised EV-G types (G5 and G7) have been isolated from sheep only and were not included in this analysis. The 13 types clustered into two distinct lineages: one composed of EV-G types G1, G2, G8, G11, G12, G13, G14, and G15; the other composed of G3, G4, G6, G9, and G10 and G15. Among the Vietnamese viruses, the most common EV-G type was EV-G1 (N = 54), followed by EV-G6 (N = 37). In contrast, the overall PKV-C phylogeny showed shorter branch lengths than the overall EV-G phylogeny. No diversity at type level was identified for PKV-C overall, but viruses belonging to three well-supported clades (C1: N = 63; C2: N = 61; and C3: N = 42) were found to be co-circulating in Vietnam (with nucleotide sequence divergence lower than 15% in VP1) (Fig. 3a).
Figure 3.
MCC trees from the discrete phylogeography analysis of PKV-C. (a) Simplified Bayesian MCC tree of 240 VP1 sequences of PKV-C. The posterior support for C1 to C3 is labelled on the nodes. The tree labelled with sequences accession numbers is shown in Supplementary Fig. S2. (b) PKV-C Clade 1 tree phylogeny. (c) PKV-C Clade 2 tree phylogeny. (d) PKV-C Clade 3 tree phylogeny. Branches are coloured according to their descendent nodes annotated by the different sampled areas, with the key for colours shown on the left.
We next estimated and compared the evolution of VP1 regions from EV-G and PKV-C using Bayesian coalescent analyses in order to determine the Time to the Most Recent Common Ancestor (TMRCA) for the phylogeny reconstructed from all available VP1 genes of EV-G strains as well as the TMRCA of the nodes adjoining the branch that represents the emergence of different EV-G types. These analyses were repeated for PKV-C.
The MRCA of the 13 EV-G types was estimated to have existed approximately 300 years ago with a 95% highest posterior density (HPD) interval of 201 to 411 years (Table 1). The total G1 type of EV-G sequences (with early G1 isolates in the 1,973) had a deeper root than the other clades (Fig. 1). The TMRCA of EVG-G1 in Vietnam was 1,959 with 95% HPD (1,935, 1,985) (Fig. 2a, Table 2). The estimated TMRCA for total EV-G G6 was 1,970 with 95% HPD (1,952, 1,984) (Fig. 2b, Table 2). In comparison to the deep divergence of EV-G types, the time-scaled phylogeny of the total PKV-C dataset predicted an MRCA for this species around 170 years ago with 95% HPD (83, 212) (Table 1). The three clades of Vietnamese PKV-C sequences showed similar origin times to EV-G; between 1960s and 1970s (Table 2). It is possible that the three clades had been independently introduced from elsewhere or the ancestral PKV-C lineage have circulated in Vietnam for some time before evolving into three different clades; the lack of earlier time point samples from Vietnam prevents investigation of these possibilities. The results indicate persistence of the two porcine picornaviruses in this region of Vietnam over time periods of decades.
Table 1.
Estimates of evolutionary parameters for EV-G and PKV-C.
| Virus | TMRCA (year) |
Evolutionary rate (×10-3 subs/s/y) |
dN/dS | ||
|---|---|---|---|---|---|
| Mean | (95% HPD lower, upper) | Mean | (95% HPD lower, upper) | ||
| EV-G | 1,715 | (1,602, 1,813) | 4.02 | (2.93, 4.82) | 0.093 |
| PKV-C | 1,843 | (1,738, 1,930) | 2.19 | (1.27, 3.08) | 0.096 |
subs/s/y, nucleotide substitutions per site per year.
Table 2.
Estimated TMRCAs of different clades of EV-G and PKV-C in Vietnam.
| Virus | TMRCA (year) |
|
|---|---|---|
| Mean | (95% HPD lower, upper) | |
| EV-G G1 | 1,959 | (1,935, 1,985) |
| EV-G G6 | 1,970 | (1,952, 1,984) |
| PKV-C C1 | 1,976 | (1,952, 1,998) |
| PKV-C C2 | 1,971 | (1,946, 1,994) |
| PKV-C C3 | 1,960 | (1,940, 1,990) |
The mean overall evolutionary rate was higher for EV-G [4.02 × 10−3 nucleotide substitutions per site per year (ns/s/y), 95% HPD (2.93 × 10−3, 4.82 × 10−3)] than for PKV-C [2.19 × 10−3 ns/s/y, 95% HPD (1.27 × 10−3, 3.08 × 10−3)] (Table 1). However, with overlapping confidence intervals, this difference was not statistically significant. The estimated evolutionary rate was [4.37 × 10−3 ns/s/y, 95% HPD (2.18 × 10−3, 7.08 × 10−3)] for EV-G G1 and was [4.05 × 10−3 ns/s/y, 95% HPD (2.04 × 10−3, 6.94 × 10−3)] for G6, respectively. No sites indicative of positive selection could be identified in the VP1 codon sites of either EV-G or PKV-C and the dN/dS ratios were similarly low for the VP1 gene of both viruses, suggesting most or all codons are under strong negative selection.
3.2 Phylogeography of EV-G and PKV-C
For EV-G type G1, Vietnamese sequences clustered with a sequence isolated from southeast China in 2008 (Zhang et al. 2012). In addition, the first EV-G G1 sequence identified in the United States (in 2013) was also closely related to G1 Vietnam sequences (Anbalagan, Hesse, and Hause 2014) (Fig. 2a and Supplementary Fig. S1). Vietnamese EV-G G6 clustered with a G6 strain isolated in South Korea in 2009 (Moon et al. 2012) (Fig. 2b). In comparison with the large number and diversity of EV-G sequences from Vietnam, few EV-G VP1 sequences were available from other countries (total N = 11). For this reason, we did not estimate the cross-border virus lineage migration rates of EV-G VP1.
The PKV-C sequences isolated in Vietnam segregated into three independent clades (C1-C3) (Fig. 3a). PKV-C C1 contained sequences originating in Japan and Thailand in 2007-2009. These sequences grouped into one major lineage with other sequences from China, USA, and Hungary in 2007-2012 (Fig. 3a, and 3b); another major lineage was comprised of PKV-C C2 and PKV-C C3, plus strains from China and Thailand (Fig. 3a, 3c, and 3d). Notably, two sequences originating from Southwest China in 2011 (Chen et al. 2013) fell into the Vietnam C3 clade, indicating possible cross-border movement events (Fig. 3d). We further explored the cross-border dissemination pattern using a discrete trait model. The overall between-country PKV-C viral virus lineage migration rates (expressed as exchanges year−1) were relatively low (mean rates = 0.002 to 0.007 exchanges year−1) (Table 3). Our phylogeographic analyses provided strong support for China as the recent source of PKV-C found circulating in Vietnam (mean rate = 0.002 exchanges year−1, BF = 40), Thailand (mean rate = 0.003 exchanges year−1), BF = 27), and USA (mean rate = 0.005 exchanges year−1, BF = 53) (Table 3). Strongly supported between-country movement was also found from Thailand to Japan (mean rate = 0.007, BF > 100); while moderate support was provided for spread from Vietnam back into China (mean rate = 0.002 exchanges year−1, BF = 6).
Table 3.
Estimated rates of PKV-C lineage migration among locations.
| PKV-C | ||||
|---|---|---|---|---|
| Donor | Recipient | BFa | Rateb | 95% HPDc |
| China | Thailand | 27 | 0.003 | (0.000, 0.007) |
| China | USA | 53 | 0.005 | (0.001, 0.01) |
| China | Vietnam | 40 | 0.002 | (0.000, 0.005) |
| Vietnam | China | 6 | 0.002 | (0.000, 0.005) |
| Japan | Hungry | 6 | 0.004 | (0.000, 0.01) |
| Thailand | Japan | 196 | 0.007 | (0.001, 0.03) |
aPairs of locations with Bayes Factor >3 are shown.
bMean rate of viral lineage migration between locations (exchanges year−1).
cNine-five per cent HPD intervals of viral lineage migration rate (exchanges year−1).
3.3 Spatial dissemination of EV-G and PKV-C in Dong Thap province in Vietnam
We further described the dissemination networks of different picornaviruses (EV-G G1/G6 and PKV-C C1-3) co-circulating across pig farms within Dong Thap province in Vietnam (Fig. 4). Generally, we found evidence of the movement of EV-G and PKV-C between the four districts over short time scales but with variable virus lineage migration rates and BF support. Evidence of virus imports and exports were found in Cao Lanh, Chau Thanh, and Than Binh, while Hong Ngu was more likely to import viruses from the other three districts than to export virus to them.
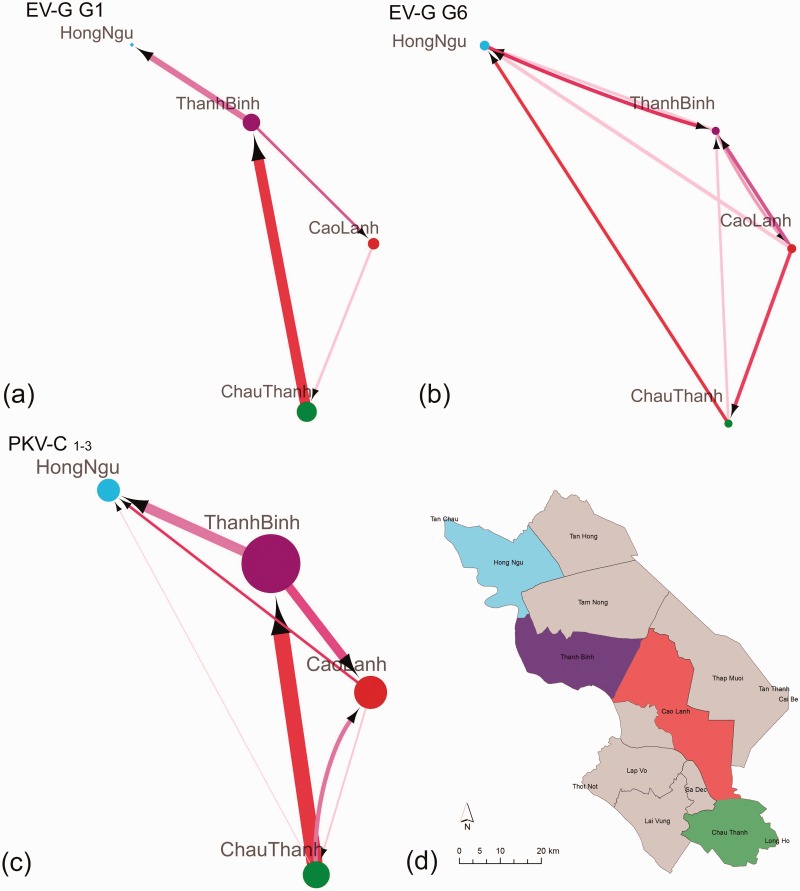
Figure 4.
Dissemination networks of co-circulating EV-G and PKV-C in Dong Thap province, Vietnam. Virus lineage migration rates (exchange year−1) between districts and BF support were estimated using an asymmetric discrete trait model on phylogenies of EV-G and PKV-C types separately and jointly. Node size reflects the sample number for each area. Arrows show the direction of transmission between the two states; the edge weight indicates the per capita transmission rate. The colour of edge from pink to red indicates BF support from low to high. Only rates with BF > 3 are shown: (a) EV-G G1; (b) EV-G G6; (c) joint PKV-C C1-3; (d) the location of the four districts in Dong Thap province. The rates and BF support are summarized in Supplementary Tables S3–S5.
Specifically, different dissemination patterns were found for EV-G G1 and EV-G G6 (Fig. 4a, and b, Supplementary Tables S3, and S4). For EV-G G1, the highest virus lineage migration rate and the strongest support was found for movements from Chau Thanh to Than Binh (mean rate 0.07 exchanges year−1, BF = 21) (Fig. 4a, Supplementary Table S3). In contrast, EV-G G6 showed more equal bi-directional dissemination (mean rate = 0.02 to 0.03 exchanges year−1) among all the four districts (Fig. 4b, Supplementary Table S4).
In contrast to different dissemination patterns found for EV-G G1 and G6, similar patterns were found among the three clades of PKV-C. This justified a joint analysis of the three clades to obtain the best estimate of PKV-C dissemination patterns in Dong Thap province. Of the four districts, Chau Thanh was found to contribute significantly to PKV-C spread within Dong Thap, which showed evidence of movement of PKV-C to the other three districts Cao Lanh, HongNgu, and Thanh Binh. (Fig. 4c and Supplementary Table S4). As for EV-G G1, the highest rate for PKV-C and the strongest support was found for the link from Chau Thanh to Than Binh (mean rate 0.05 exchanges year−1, BF > 300). The most likely recipient district was Hong Ngu, which showed evidence of spread of PKV-C from the other three districts: Chau Thanh (mean rate = 0.01 exchanges year−1, BF = 5), Cao Lanh (mean rate = 0.01 exchanges year−1, BF = 21), and Thanh Binh (mean rate = 0.02 exchanges year−1, BF = 21).
4. Discussion
In this study, we analysed VP1 sequences obtained from two genera of porcine picornaviruses that are endemic in pig populations in Southeast Asia.
Enterovirus is by far the largest and most diverse genus within the picornavirus family. We found EV-G has a much deeper inferred phylogenetic root than that of PKV-C, which suggests a longer evolutionary history and earlier diversification of multiple EV-G types. Similarly, a recent study of the evolution of the enterovirus C species (EVC) VP1 gene found distant ancestors of different EVC-types and suggested that the type-specific amino acids have remained conserved for hundreds of years, which might imply selection favouring these type-specific amino acids in the given genetic context, or indicate that there are no selection pressure driving sequence change away from the current sequences (Smura et al., 2014). The dN/dS values in our study are consistent with an earlier study, which also showed the dN/dS scores for both enterovirus and non-enterovirus were very low (<0.1) (Hicks and Duffy 2011).
We found that the origin time of total 240 PKV-C VP1 sequences from different countries went back to 169 (years) and the estimated origin of the Vietnam PKV-C clades was during 1960s to 1970s. In comparison, a much shorter TMRCA 17.2 (years) for all kobuvirus species and much higher evolutionary rate (2.73 × 10−2 ns/s/y) were described in another study (Cho et al. 2015). However, their estimation was based on a phylogeny which composed of only 4 PKV-C sequences (length = 132 amino acids) and other kobuvirus sequences from different host species (including feline, canine, and human), the results of PKV-C might have been influenced by the number and length of sequences and the type of samples included.
We have revealed the movement of porcine picornaviruses between geographic locations and compared movement patterns between and within different virus genera. In our study, although cross-border spread of PKV-C was detected, the between-country virus lineage migration rates (mean = 0.002 to 0.007 exchanges year−1) were lower than those within Dong Thap province in Vietnam (mean rates = 0.01 to 0.07 exchanges year−1), indicating a far higher level of gene flow over shorter distances. Spatial movement at smaller geographic scales rather than long distance was also been observed in other endemic viruses such as dengue (Raghwani et al. 2011) and influenza (Nelson et al. 2011).
We observed multiple between-district dissemination events within the same virus/type and distinct dissemination patterns between different viruses/types. Among the two genera of picornaviruses that were co-circulating in the same pig population in Dong Thap, PKV-C and EV-G G1 were transmitted directionally at unequal rates; EV-G G6 had more homogenous virus lineage migration rates, but the estimates were uncertain (wide HPD intervals) due to the limited number of G6 sequences (N = 37) to address the geographical movement with asymmetric model (Number of possible location exchange pairs = 12).
Our phylogeographic analysis suggested that the Chau Thanh district is a more important source of EV-G G1 and PKV-C dissemination than the other sampled districts in Dong Thap province. This is consistent with Chau Thanh being the region’s major pig trading district; pigs from Chau Thanh are industrially bought from and sold to other districts within the province, and exported to other provinces in Vietnam (such as Long An and An Giang). In addition, pigs in Dong Thap province are occasionally exported to and imported from neighbour countries (Cambodia and Thailand) due to price fluctuation. However, more detailed virus sampling and data on pig trading would be needed to confirm whether the spatial dissemination of picornaviruses is wholly or largely explained by pig trading routes.
The total EVG and PKV datasets available are small and may have been temporally and spatially biased. Therefore, we specified prior distributions of evolution rates to address this problem. We highlight here the importance of undertaking surveillance to fill gaps in viral sampling of endemic viruses. We note that our phylogeographical study was intended to compare the dissemination patterns of different animal picornaviruses in Vietnam, based on dense sampling from the same areas over the same time period, and this would not be greatly affected by the uncertainty of the complete evolution history.
In conclusion, the results of this study contribute to understanding the evolution and spatial dissemination of picornavirus in pig populations. Knowledge of the geographical sources, locations of entry, and mode of dispersal of these viruses may inform targeted surveillance and control in regions with high pig density. In the future, it would be helpful to routinely sequence picornavirus isolates in order to build-up a clearer and more complete picture of the evolution and spatial spread of pig-related picornaviruses and their relationship to human infections.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
Acknowledgements
This work was supported by VIZIONS, a strategic award from the Wellcome Trust (ref. WT/093724). Stephen Baker is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (Ref. 100087/Z/12/Z).
References
- Anbalagan S., Hesse R. A., Hause B. M. (2014) ‘First Identification and Characterization of Porcine Enterovirus G in the United States’, PLoS One, 9: e97517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchemez S., et al. (2013) ‘Using Routine Surveillance Data to Estimate the Epidemic Potential of Emerging Zoonoses: Application to the Emergence of US Swine Origin Influenza A H3N2v Virus’, PLoS Medicine, 10: e1001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., et al. (2013) ‘Molecular and Phylogenetic Analysis of the Porcine Kobuvirus VP1 Region Using Infected Pigs from Sichuan Province, China’, Virology Journal, 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. Y., et al. (2015) ‘Molecular Evolution of Kobuviruses in Cats’, Archives of Virology, 160: 537–41. [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007) ‘BEAST: Bayesian Evolutionary Analysis by Sampling Trees’, BMC Evolutionary Biology, 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., et al. (2012) ‘Bayesian Phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T. (1985) ‘Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA’, Journal of Molecular Evology, 22: 160–74. [DOI] [PubMed] [Google Scholar]
- Hicks A. L., Duffy S. (2011) ‘Genus-Specific Substitution Rate Variability Among Picornaviruses’, Journal of Virology, 85: 7942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques J., et al. (2008) ‘Epidemiological, Molecular, and Clinical Features of Enterovirus Respiratory Infections in French Children Between 1999 and 2005’, Journal of Clinical Microbiology, 46: 206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoi K., Suzuki H., Nishio O. (2001) ‘Isolation of Coxsackievirus B5 from Pigs’, New Microbiologica, 24: 217–22. [PubMed] [Google Scholar]
- Khamrin P., et al. (2014) ‘Epidemiology of Human and Animal Kobuviruses’, Virus Disease, 25: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Rogers N., Crandall K. A. (2010) ‘Evolution of Picornaviridae: An Examination of Phylogenetic Relationships and Cophylogeny’, Molecular Phylogenetics and Evolution, 54: 995–1005. [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009) ‘DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data’, Bioinformatics, 25: 1451–2. [DOI] [PubMed] [Google Scholar]
- Lindahl J. F., et al. (2013) ‘Circulation of Japanese Encephalitis Virus in Pigs and Mosquito Vectors Within Can Tho City, Vietnam’, PLoS Neglected Tropical Diseases, 7: e2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P., et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. J., et al. (1997) ‘A Novel Virus in Swine is Closely Related to the Human Hepatitis E Virus’, Proceedings of the National Academy of Sciences of the United States of America 94: 9860–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Nor M. N., Gan C. H., Ong B. L. (2000) ‘Nipah Virus Infection of Pigs in Peninsular Malaysia’, Revue Scietifique et Technique, 19: 160–5. [DOI] [PubMed] [Google Scholar]
- Moon H. J., et al. (2012) ‘Complete Genome Analysis of Porcine Enterovirus B Isolated in Korea’, Journal of Virology, 86: 10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. I., et al. (2011) ‘Spatial Dynamics of Human-Origin H1 Influenza A Virus in North American Swine’, PLoS Pathogens, 7: e1002077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. V., et al. (2015) ‘Large-Scale Screening and Characterization of Enteroviruses and Kobuviruses Infecting Pigs in Vietnam’, Journal of General Virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny N., et al. (1997) ‘Prevalence of Swine Influenza and Other Viral, Bacterial, and Parasitic Zoonoses in Veterinarians’,Journal of Infectious Diseases, 176: 1414–5. [DOI] [PubMed] [Google Scholar]
- Okitsu S., et al. (2012) ‘Sequence Analysis of Porcine Kobuvirus VP1 Region Detected in Pigs in Japan and Thailand’, Virus Genes, 44: 253–7. [DOI] [PubMed] [Google Scholar]
- Rabaa M. (2015) ‘The Vietnam Initiative on Zoonotic Infections (VIZIONS): A Strategic Approach to Studying Emerging Zoonotic Infectious Diseases’, EcoHealth, 12: 726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghwani J., et al. (2011).‘Endemic Dengue Associated with the Co-circulation of Multiple Viral Lineages and Localized Density-Dependent Transmission’, PLoS Pathogens, 7: e1002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Kecskemeti S., Pankovics P. (2010) ‘Evolution of Porcine Kobuvirus Infection, Hungary’, Emerging Infectious Diseases, 16: 696–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., et al. (2008) ‘Candidate New Species of Kobuvirus in Porcine Hosts’, Emerging Infectious Diseases, 14: 1968–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., et al. (2012) ‘Two Closely Related Novel Picornaviruses in Cattle and Sheep in Hungary from 2008 to 2009, Proposed as Members of a New Genus in the Family Picornaviridae’, Journal of Virology, 86, 13295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B., Rambaut A., Drummond A. J. (2006) ‘Choosing Appropriate Substitution Models for the Phylogenetic Analysis of Protein-Coding Sequences’, Molecular Biology and Evolution, 23: 7–9. [DOI] [PubMed] [Google Scholar]
- Smith T. C., et al. (2011) ‘Emerging Swine Zoonoses’, Vector Borne and Zoonotic Diseases, 11: 1225–34. [DOI] [PubMed] [Google Scholar]
- Smura T., et al. (2014) ‘The Evolution of Vp1 Gene in Enterovirus C Species Sub-group that Contains Types CVA-21, CVA-24, EV-C95, EV-C96 and EV-C99’, PLoS One, 9: e93737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C., et al. (2013) ‘Picornavirus and Enterovirus Diversity with Associated Human Diseases’, Infection, Genetics and Evolution, 14: 282–93. [DOI] [PubMed] [Google Scholar]
- Van Dung N., et al. (2014) ‘Prevalence, Genetic Diversity and Recombination of Species G Enteroviruses Infecting Pigs in Vietnam’, Journal of General Virology, 95: 549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., et al. (2014) ‘Complete Sequencing and Phylogenetic Analysis of Porcine Kobuvirus in Domestic Pigs in Northwest China’, Archives of Virology, 159: 2533–5. [DOI] [PubMed] [Google Scholar]
- Yang Z., et al. (2014) ‘Genetic Characterization of Porcine Kobuvirus and Detection of Coinfecting Pathogens in Diarrheic Pigs in Jiangsu Province, China’, Archives Virology, 159: 3407–12. [DOI] [PubMed] [Google Scholar]
- Zhang W., et al. (2012) ‘Complete Genome Sequence of a Novel Porcine Enterovirus Strain in China’, Journal of Virology, 86: 7008–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.