Abstract
Human influenza viruses are rapidly evolving RNA viruses that cause short-term respiratory infections with substantial morbidity and mortality in annual epidemics. Uncovering the general principles of viral coevolution with human hosts is important for pathogen surveillance and vaccine design. Protein regions are an appropriate model for the interactions between two macromolecules, but the currently used epitope definition for the major antigen of influenza viruses, namely hemagglutinin, is very broad. Here, we combined genetic, evolutionary, antigenic, and structural information to determine the most relevant regions of the hemagglutinin of human influenza A/H3N2 viruses for interaction with human immunoglobulins. We estimated the antigenic weights of amino acid changes at individual sites from hemagglutination inhibition data using antigenic tree inference followed by spatial clustering of antigenicity-altering protein sites on the protein structure. This approach determined six relevant areas (patches) for antigenic variation that had a key role in the past antigenic evolution of the viruses. Previous transitions between successive predominating antigenic types of H3N2 viruses always included amino acid changes in either the first or second antigenic patch. Interestingly, there was only partial overlap between the antigenic patches and the patches under strong positive selection. Therefore, besides alterations of antigenicity, other interactions with the host may shape the evolution of human influenza A/H3N2 viruses.
Keywords: Influenza A, viral evolution, antigenic evolution, positive selection, antibody binding
1. Introduction
Influenza viruses are pathogens with single-stranded RNA genomes that cause short-term infections of the respiratory tract (Molinari et al. 2007; World Health Organisation (WHO) 2009). Three genera of viruses (A–C) circulate in humans, with influenza A viruses causing the most infections. Influenza A viruses possess up to fourteen genes encoded on eight genome segments (Wise et al. 2009, 2012; Das et al. 2010; Jagger et al. 2012). They are further classified into subtypes based on the surface proteins hemagglutinin (HA) and neuraminidase (NA). Eighteen and eleven variants of HA and NA exist, respectively, which can infect a wide range of different hosts (Webster et al. 1992; Medina and García-Sastre 2011; Tong et al. 2012, 2013). Currently, the subtypes H1N1 and H3N2 are endemic in the human population. The H1N1 subtype descends from the swine-origin influenza A/H1N1 virus, which, since its introduction into the human population in 2009, has replaced the previously circulating H1N1 subtype in annual epidemics (Garten et al. 2009).
The surface proteins HA and NA are of particular importance for the evolution and adaptation of human influenza viruses (McHardy and Adams 2009). Amino acid changes on the surface of the globular head region of NA and in the epitope regions of HA result in alterations of antigenicity and reduced recognition by the host’s immune response, which is known as antigenic drift (Shih et al. 2007; Cattoli et al. 2011; Sandbulte et al. 2011). Reduced recognition by antibodies improves attachment of the viral receptor binding site (RBS) to sialic acid residues on the host cell surface, which initiates an infection (Wiley and Skehel 1987). The antigenic drift of type A and B influenza viruses necessitates almost annual updates of the human influenza virus vaccine (Dormitzer et al. 2011) so that it includes antigenically similar strains to the predominant circulating antigenic variants (Russell et al. 2008). Sites where alterations directly affect receptor avidity have been described for influenza A/H1N1 viruses, which change as part of viral adaptation to hosts with different levels of immune protection (Hensley et al. 2009).
Measures of positive selection can indicate sites that are relevant for the adaptation of human influenza viruses to variable environmental conditions, which include escape from immune recognition by antibodies generated in response to previous infections or vaccinations (Bush et al. 1999; Medina and García-Sastre 2011). Thus sites under positive selection have been previously used as a proxy to determine the antigenically relevant sites of HA. Protein sites with a significantly increased ratio of non-synonymous to synonymous mutations on HA (Fitch et al. 1997; Bush et al. 1999; Suzuki 2004; Suzuki and Gojobori 1999) and regions under positive selection including T- and B-cell epitope sites (Suzuki 2006) have been described. Sophisticated maximum likelihood approaches have been developed that estimate the degree of positive selection for individual sites (Nielsen and Yang 1998; Yang 2000; Pond, Frost, and Muse 2005); extensions of these approaches allow the dN/dS ratio to vary along both sites and lineages (Yang and Nielsen 2002; Kosakovsky Pond et al. 2008, 2011; Nozawa, Suzuki, and Nei 2009;; Yang and dos Reis 2011; Murrell et al. 2012). Application of these techniques to HA has determined overlapping sets of sites under positive selection (Yang 2000; Nei 2005; Kosakovsky Pond et al. 2008; Murrell et al. 2012). Alternative approaches to measure positive selection exist; for example, Meyer and Wilke (2012) devised a method that identifies sites in protein-coding sequences based on the exposure of an amino acid combined with its dN/dS (Meyer and Wilke 2012), and Bhatt, Holmes, and Pybus (2011) described a site frequency-based method to calculate the rates of adaptive evolution (Bhatt, Holmes, and Pybus 2011). Finally, pairs of sites where changes might be affected by epistatic interactions can be identified based on evolutionary distances in a phylogenetic tree (Kryazhimskiy et al. 2011).
Determination of the sites under selection can be improved by consideration of the protein structure, as the interaction of flexible macromolecules such as human antibodies and the viral surface proteins is likely to include multiple proximal residues of both interacting proteins. For instance, as well as sequentially linear epitopes, conformational epitopes have been described as having sequentially discontinuous residues which are in close three-dimensional proximity (Barlow, Edwards, and Thornton 1986; Huang and Honda 2006). We have recently described a graph-cut based clustering method to detect areas of sites under positive selection with arbitrary shapes on the structures of several human influenza A virus proteins (Tusche, Steinbrück, and McHardy 2012).
The antigenic differences between viral strains can be assessed with hemagglutination inhibition (HI) assays. These determine the strength of an interaction between a viral isolate and an antiserum elicited against another viral strain based on the concentration-dependent inhibition of red blood cell agglutination by a viral isolate with an antiserum, which is determined from a series of dilution steps (Hirst 1943). HI assays are routinely used in the global surveillance program of human influenza viruses by the WHO, as the antigenic novelty of circulating strains relative to previous predominating and vaccine strains is a relevant factor for their future epidemic potential. Using antigenic cartography, high-dimensional distance matrices between pairs of antigens and antisera derived from HI titers can be projected into a lower-dimensional space and the structure of the data can be visualized. This has revealed that clusters of antigenically similar strains predominate in epidemics for several years before being replaced by strains of a novel ‘antigenic cluster’ or ‘variant’ (Lapedes and Farber 2001; Smith et al. 2004). The original definition of the epitopes recognized by antibodies (Wiley, Wilson, and Skehel 1981) is broad and includes 131 out of 328 sites of the entire HA1 chain of HA. Recent computational studies therefore aimed to identify the key antigenicity-altering sites of the epitopes. Information gain (Huang, King, and Yang 2009), multivariate linear models (Lee et al. 2007) and similar scoring schemes (Liao et al. 2008; Liao, Lin, and Lin 2013) have been used to estimate the association between amino acid changes and changes in antigenic type. Lees, Moss, and Shepherd (2011) used the genetic variability in ‘cells’ on a three-dimensional grid on the protein structure, with similar regression models being used to weigh substitutions in preselected clusters as predictors for antigenic distances (Lees, Moss, and Shepherd 2011). Sun et al. (2013) used ridge regression to infer antigenic weights for amino acid changes (Sun et al. 2013). We recently used HI assay data to infer antigenic weights for individual branches of a phylogenetic tree (Steinbrück and McHardy 2012). The antigenic weight of amino acid changes and the average impact of changes at individual protein sites can be determined from this antigenic tree. Koel et al. (2013) experimentally quantified the antigenic impact of changes at individual and pairs of sites involved in antigenic cluster transitions for HA in H3N2 viruses (Koel et al. 2013). They found that seven positions that had altered in past antigenic cluster transitions have had a substantial antigenic impact. However, not all permutations of the observed changes could be tested due to the effort required, and the authors noted that changes at other sites may have had collective effects on antibody binding or they may have been compensatory mutations that were necessary to retain function. Computational methods that link genetic to phenotypic information are not limited to exploring subsets of sites but can also return predictions that might include ‘antigenic hitchhiker’ changes. Hitchhikers are (near) neutral changes that are introduced into a strain shortly before or after an antigenicity-altering change. As the strain then shows a significant change in antigenicity relative to other strains, the contributions of the individual amino acid changes to this antigenic difference cannot be distinguished from one another. Thus ‘antigenic hitchhikers’ may falsely be determined as being relevant for the antigenic evolution of the virus. Similarly, epistatic effects may lead to a suppression or enhancement of the antigenic impact of individual changes and therefore to problems in determining the most relevant sites.
We provide a description of the most relevant areas (patches) for alteration of antigenicity of the viral HA protein from human influenza A/H3N2 viruses, as measured by HI assay data. We derived these by following a computational approach combining genetic, phylogenetic, antigenic, and structural information. The antigenic weights for the individual HA sites were first inferred by antigenic tree inference. We then determined antigenicity-altering patches of sites on the three- dimensional structure of HA using a graph-cut clustering of sites, based on their spatial proximity and antigenic weights. Our method considers all HA1 residues and is not restricted to the study of a shorter list of candidate sites. Consideration of the spatial arrangement of sites on the protein structure allowed the removal of potential hitchhiking changes without real antigenic effect, which were identified in the inference of site- specific antigenic weights using the antigenic tree, as the removed sites with antigenic weight that were located far from other such sites on the HA structure. We show that the patches provide an alternative to the historically broad definition of epitope sites (Meyer and Wilke 2015) in characterizing the antigenic evolution of human influenza A/H3N2 viruses. The overall method was implemented in a software package named AntiPatch.
2. Materials and methods
Our method comprises two steps: First, antigenic weights were inferred for individual protein sites from the HA sequences and HI data using antigenic trees, with the data and methodology adapted from Steinbrück and McHardy (2012). Next, clusters of residues on the protein structure with large antigenic impact were identified with a graph-cut clustering approach (Tusche, Steinbrück, and McHardy 2012), using a novel parameter optimization procedure. In the following sections, we describe each step in more detail.
2.1 Inference of antigenic weights
We inferred antigenic weights for the HI assay dataset and associated sequences from Smith et al. (2004), following the methodology described in Steinbrück and McHardy (2012), with modifications as detailed below. Briefly, the HI assay data of Smith et al. (2004) were used and the associated HA1 sequences were downloaded from the GISAID EpiFlu database (Bogner et al. 2006). The collection comprised 258 seasonal human influenza A/H3N2 virus isolates from 1968 to 2003; the complete list of GISAID identifiers can also be found in Steinbrück and McHardy (2012). The sequence data were used to infer a phylogenetic tree with PhyML (Guindon and Gascuel 2003) and Garli (Zwickl 2006) under the GTR+I+ model selected with Modeltest (Posada 2008). To root the tree, a related avian sequence, A/duck/33/1980, was used and subsequently removed from the study. Ancestral sequences were reconstructed for all internal nodes of the tree using PAML v4.5 under the model (Jones, Taylor, and Thornton 1992; Yang 2007) inferred with ProtTest (Darriba et al. 2011). Amino acid changes between parent and child node sequences were then mapped to the branches of the tree. Note that this approach does not allow us to account for uncertainty in state reconstruction. However, we previously found that the level of variation in state reconstruction is very low for the data used here (Steinbrück and McHardy 2012), probably because of the temporal nature of the data, where the paths between internal nodes and leaf nodes tend to be short.
Assay data were normalized as described by Smith et al. (2004). The normalized antigenic distances between pairs of influenza antigens and antisera were used to infer an ‘antigenic tree’ (Steinbrück and McHardy 2012), with antigenic weights for the branches of the phylogeny being determined using the non-negative least squares optimization. To account for the asymmetric nature of HI assay data, both antigenic ‘up’ and ‘down’ weights were inferred for branches on the path between antigen–antisera pairs in the tree, in accordance with the directionality of the measurements, from an antigen to the root of the tree (up) or from the root to an antiserum (down) (see Fig. 2 from Steinbrück and McHardy (2012)). To assess the antigenic weight of a particular amino acid position, we used the average of all available antigenic ‘up’ or ‘down’ weights of the branches with an amino acid change at this position. In contrast to our earlier study, only internal branches were considered, as we observed a tendency towards systematic bias and that highly variable antigenic weights were assigned to the terminal branches, caused by single isolate variations. To avoid assigning large weights to positions based on little data and to penalize the estimated weight for a lack of data, we divided the weight of each branch with a reconstructed change for this position by the total number of amino acid changes on the branch. If three or fewer branches contributed antigenic weights to an amino acid position, we considered the estimate for this site to be less reliable.
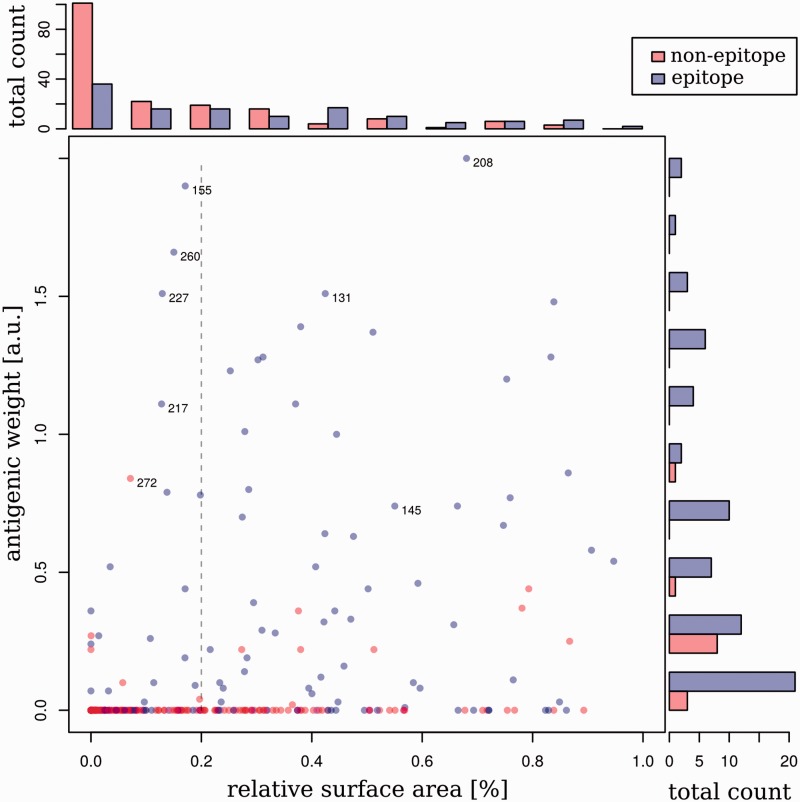
Figure 2.
Distribution of antigenicity-altering effects. Antigenic weights are plotted versus the relative available surface area (RSA) for residues within the epitope regions (blue) and outside the epitope regions (red) on HA of human influenza A/H3N2 viruses. Residues with an RSA of more than 5 per cent lie to the right of the dashed gray line. On the side of the plot are histograms showing the number of sites with particular antigenic weights (right) or RSA values (top). Note that the histogram on the right-hand side of this figure compares only the antigenic weights between non-epitopes and epitope sites, whereas the histogram at the top of this figure shows the comparison of RSAs between non-epitope and epitope residues.
2.2 Spatial coordinates and surface accessibility
The protein structure model (PDB identifier 3HMG) of the HA of human influenza A/H3N2 viruses was obtained from the RCSB database (Berman et al. 2000). The coordinates of the atoms were used to represent the spatial coordinates of the corresponding amino acid residues. To classify residues as exposed or buried, the relative solvent accessibility (RSA was computed by estimating the accessible surface area (ASA) with CCP4 (Lee and Richards 1971; Winn et al. 2011) and normalization with the respective maximum surface area. A measurement for normalization was initially proposed by Chothia (1976) and recently improved by Tien et al. (2013) as previous maximum surface area measurements underestimated the largest allowed ASA (Tien et al. 2013). Residues with an RSA of 5 per cent or more were defined as exposed, following Tien et al. (2013). To determine the influence of the protein structure on our results, we repeated our complete analysis with an influenza structure based on a more recent viral strain (PDB identifier 2YP7), a structure of an HA trimer in connection with a neutralizing antibody (1QFU) and a structure predicted from the consensus of all 258 sequences used in our antigenic tree inference (prediction was performed with the Phyre 2 webserver (Kelley and Sternberg 2009)). The identified patches were identical for the different structures. We also found that the root mean square deviation between these structures and the 3HMG model was very small: 0.794 Å for 2YP7, 0.427 Å for 1QFU, and 0.526 Å for the consensus structure, as determined on the , N, and O atoms of the protein head with the ‘super’ command in Pymol v1.5 (Schrodinger 2010), without additional refinement cycles. For the graph-cut clustering, residue coordinates were normalized so that the largest dimension of the protein was of length one, to ensure the normalized variance of input variables in the optimization.
2.3 Clustering and visualization
We used the antigenic weights of individual sites and the spatial coordinates from the protein structure model as input for a graph-cut-based clustering to infer dense patches of sites with a large antigenic impact. As described in Tusche, Steinbrück, and McHardy (2012), this divides the set of all analyzed sites into relevant (Pos) and irrelevant (Neg) subsets. The Neg set thus included sites for which the analyzed datasets did not provide sufficient evidence for antigenic impact; however, this does not preclude that changes at some of these sites might be of antigenic relevance, which would be observed only when analyzing different datasets. The graph-cut approach was applied to a graph where each node n represented a protein site. Two additional nodes P and N represented the Pos and the Neg sets. Each residue n was connected with an edge to both P and N, and to all its neighboring residues on the protein structure within a distance of δ. Edges to P and N were weighted with and , respectively, and edges between residues m and n are weighted with the proximity exp(−dist(m,n)), where dist(m,n) measures the Euclidean distance between the Cα atoms of the amino acid residues. For a residue n, we set to be equal to the antigenic weight of that residue and , with being the largest antigenic weight in the data. The measure exp(−dist(m,n)) was large if the residues m and n were close to each other. The graph-cut divided the set of all residues by placing the nodes into the positive class that 1, had a large antigenic weight , 2, were close to other nodes in the positive class and 3, were far away from nodes in the negative class. This was achieved by searching for the set of edges with minimal costs which, when removed, cut all paths between P and N:
where corresponds to the neighboring residues within a distance of δ to n, and β determines the size of the positive and negative classes. Based on the value of β, the result gradually changes between two extreme (and undesirable) cases: It either assigns all sites to one of the two classes (β = 0, Neg in our implementation) or distributes sites between Pos and Neg (large β), so that (the right part of the cost equation) is minimal. The latter case ignores spatial distances and we refer to it as ‘saturation’. To determine the optimal value for β, we performed a parameter search. Since the value of β required for saturation is dependent on the size of the protein, the total number of residues and their antigenic weights, we iteratively increased β by one and defined as the first β for which we did not observe updates in the assignments for 1,000 iterations (our implementation returns a warning and an empty result when β reaches 5,000). We then set the final value for (i.e. we set β to the value returning a compromise between the two extreme cases as a result). We set δ for to 22 Å, which is half the diameter of the largest epitope in the HA of subtype H3 (epitope D has a maximum diameter of ∼44 Å). Subsequently, the selected residues were grouped into patches if their pairwise distances were less than δ. Any remaining single residues were discarded. The resulting patches were visualized on the original protein structure with PyMOL v1.4 (Schrodinger 2010).
2.4 HA phylogeny used for evaluation
For further evaluation of the relevance of the identified antigenic patches for the evolution of human influenza A/H3N2 viruses, we inferred a phylogeny from 7,127 HA sequences of seasonal human influenza A/H3N2 isolates, sampled between 1968 and 2014. The sequences were downloaded from NCBI’s Influenza Virus Resource database (Bao et al. 2008). A multiple sequence alignment was calculated with MUSCLE (Edgar 2004) and used to infer a phylogenetic tree with FastTree under the GTR model (Price, Dehal, and Arkin 2009). To root the tree, A/Hong Kong/1-1/1968 was used. Ancestral states for the amino acid sequences were reconstructed for all internal nodes of the tree using our implementation of the Fitch algorithm (Fitch 1971). Amino acid changes between parent and child node sequences were then mapped to the branches of the tree, and the tree was visualized using Cytoscape v3.1.1 (Shannon et al. 2003).
3. Results
3.1 Distribution of antigenic weights
We used antigenic tree inference (Steinbrück and McHardy 2012) to map the antigenic distances derived from HI titers between viral strains and reference antisera onto a phylogenetic tree. The phylogeny was constructed from the corresponding sequences of the HA1 subunit of HA from human influenza A/H3N2 viruses sampled between 1968 and 2002 (Section Materials and Methods). Subsequently, amino acid changes were reconstructed for the branches of the tree using ancestral character state reconstruction with PAML v4.5 (Yang 2007). Antigenic weights for individual sites were inferred from these data as the mean of all antigenic weights of the branches with a mutation at this position (Materials and Methods section). As we had previously observed a tendency towards systematic bias and noise for the antigenic weights of terminal branches (Steinbrück and McHardy 2012), caused by single isolate variations, we only considered internal branches.
To characterize the sites with antigenic weights, we first visualized their distribution on the protein structure of HA of influenza subtype H3 and determined the RSA for each residue (Fig. 1). There were six buried sites with antigenic weights in the epitopes (RSA < 5%). These sites might not be directly involved in antibody interactions but could contribute to antigenic evolution by compensating for stability or any fitness disadvantages caused by nearby antigenic changes. Epitope sites had more surface exposure than sites outside the epitopes (, Kolmogorov–Smirnov test; : epitope values have a larger RSA value) and a smaller portion of buried sites (52 out of 131 epitope sites versus 123 out of 180 non-epitope sites; Fig. 2). However, as there was no significant correlation between surface exposure and the antigenic weight (r = 0.139 with Pearson’s correlation; P = 0.215) for sites with non-zero antigenic weights on HA1, we included both surface and buried residues in our subsequent antigenic patch inference, as buried sites might be located close to key antigenic regions on the protein surface and changes here might indirectly affect antigenicity.
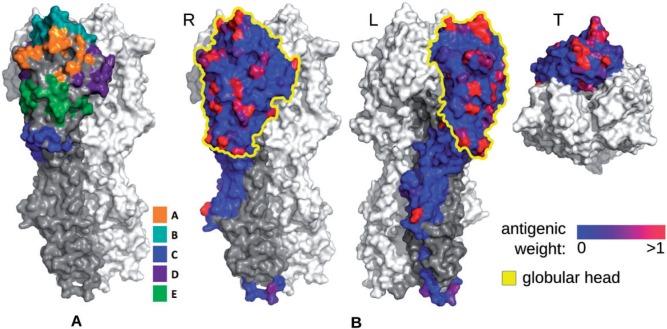
Figure 1.
Antigenicity-altering effects of protein sites on the influenza A/H3N2 HA surface. (A) Location of the five epitope regions A–E, colored according to the legend. (B) Inferred antigenic weights for residues on the surface of the HA1 chain of HA protein for human influenza A/H3N2 viruses. The value of the weight is shown with a blue–red gradient for one subunit of the HA homotrimer in three different orientations. The globular head is indicated in yellow; HA2 is colored dark gray. It is evident that not all epitope sites have large antigenic weights and that sites with large antigenic weights are mostly found as spatial clusters within the epitope sites and not outside the epitopes. Sites with antigenic weights outside of the head are not supported by the known biology of the antibody–HA interactions and any likely antigenic hitchhikers found in genotype–phenotype inference.
Not surprisingly, sites in the epitopes had larger antigenic weights than sites outside these regions (, Kolmogorov–Smirnov test; : epitope sites have larger antigenic weights). Still, 93 out of the 131 epitope sites had no antigenic impact assigned and thus had no discernible relevance for immune evasion in the past. Within the epitope sites, a tendency for sites with weights to cluster was evident (Fig. 1b), and many sites were found in the vicinity of the RBS in the globular head of the protein. As expected, the sites outside the epitope regions were mostly assigned low antigenic weights: 167 out of 180 had no antigenic weight, including forty-nine of the fifty-seven exposed sites. The remaining thirteen sites with antigenic weights were mostly found at disconnected positions outside the head region or within the stem. Even though there are sites in the stem recognized by broadly neutralizing antibodies, alterations at these sites do not affect receptor recognition but affect viral entry into the cell, and would not be recognized by an HI assay (Laursen and Wilson 2013; DiLillo et al. 2014). Thus these thirteen sites are likely to be antigenic hitchhikers that have been falsely identified as being relevant by antigenic tree inference, due to their co-occurrence with antigenicity-altering changes on the branches of the tree.
Taken together, our findings indicated that 1, some sites with antigenic weights seemed to cluster on the protein structure, particularly in epitope regions where this would be expected according to viral interactions with the host’s immune system and 2, that antigenic tree inference returned in addition to such plausible assignments (see also Steinbrück und McHardy (2012)) a likely false set of antigenic weight assignments for some sites, which was particularly evident for sites located outside of the head region of HA1. We therefore devised a method named AntiPatch to define the antigenically most relevant patches of sites on the protein structure of HA1 by taking additional spatial information into consideration.
3.2 Inference of antigenic patches
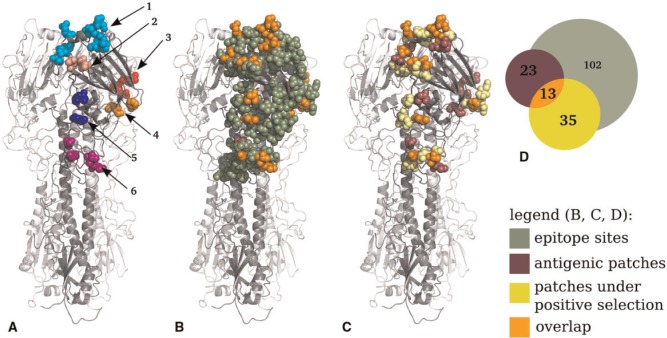
We determined spatial clusters of sites with large antigenic weights on the three-dimensional structure of HA using a graph-cut algorithm and residue clustering (Tusche, Steinbrück, and McHardy 2012) with a newly derived parameter optimization procedure (Section Materials and Methods). We identified six antigenic patches of twenty-three residues overall in the HA1 subunit of HA (Fig. 3, Table 1) associated with the altered behavior of a viral isolate in HI assays. In terms of the identified patch sites, the results of the patch inference method were robust to variations in the method and to variation of the structural information used and to the user-defined input parameter delta (see below). One large patch (patch 1) includes ten residues, and five patches have four or fewer residues each. With the exception of residue 272, all patch sites are part of the epitopes. Patch 1 is located on top of the protein head and includes residues 189, 196, and 227 of the RBS (Skehel 2009). Patch 2 is located within epitope A and overlaps with the 130-loop of the RBS. Patch 2 surrounds and includes residue 145, which has repeatedly been reported as being under positive selection (Bush et al. 1999; Kosakovsky Pond et al. 2008). Changes at this site have very large antigenic weights (Smith et al. 2004; Lee et al. 2007; Liao et al. 2008; Huang, King, and Yang 2009; Koel et al. 2013; Liao, Lin, and Lin 2013). The other four patches are located within epitopes C–E (Table 1).
Figure 3.
Location of the inferred antigenic patches on influenza A/H3N2 HA. (A) Antigenic patches on the HA homotrimer, numbered according to Table 1. (B) Location of epitope sites (dark green), antigenic patch sites (purple), and sites included in both sets (orange). Note that only residue 272 is not included in the overlap. (C) Comparison of antigenic patch sites (purple) to patches of sites under positive selection (Tusche, Steinbrück, and McHardy 2012) (yellow) and to sites included in both sets (orange). (D) Venn diagram showing the overlap of antigenic patches, patches under selection, and epitope sites (Supplementary Fig. S1). Note that there is no coherence between circle size and site counts.
Table 1.
Identified antigenic patches, patch sites, and their placement within the five epitope regions.
| Patch no. | Sites | Epitope |
|---|---|---|
| 1 | 131, 156, 158, 159, 189, 196, | ABBBBB |
| 155, 186, 217, 227 | BBDD | |
| 2 | 137, 144, 145 | AAA |
| 3 | 50, 276, 272, 278 | CC-C |
| 4 | 62, 75 | EE |
| 5 | 207, 208 | DD |
| 6 | 174, 260 | DE |
Sites are enumerated according to the H3 numbering convention (Nobusawa, Aoyama, and Kato 1991).
To identify the influence of the size of the neighborhood in patch inference, we assessed the effect of varying δ throughout the range from 1 to 65 Å. After increasing the neighborhood stepwise starting at δ = 22, we determined patches of successively larger sizes, while the number of patches decreased down to a single patch at δ = 35. The sites that were assigned to the individual patches did not change drastically. Only two sites (site 80 (at δ = 25) and site 262 (at δ = 45) with antigenic weights of 0.8 and 0.72, respectively) were additionally assigned to the patches (Supplementary Table S2). When we used smaller values (δ < 22), this resulted in smaller patches and an increase in the number of patches, as the patches were split up into smaller ones (a decrease from δ = 20 to δ = 15). For smaller neighborhoods (δ = 10), the number of patches decreased further and for δ < 8, we did not find any patches. Only one additional site (site 262) was added to a patch (at δ = 20) and subsequently removed (at δ = 10). In summary, these experiments showed that the sites placed in patches were robust to variations in the size of the neighborhood parameter throughout a large range of tested values.
3.3 Comparison to antigenic ‘clusters’
With the exception of positions 159, 186, and 227, all residues of patch 1 had changed in past transitions between consecutive antigenic clusters of influenza A/H3N2 strains over the studied time period (Smith et al. 2004). Of the twenty-three sites included in the six antigenicity-altering patches, eighteen are part of forty-five sites that have changed in antigenic cluster transitions (Smith et al. 2004; Supplementary Fig. S1). Koel et al. (2013) used site-directed mutagenesis to confirm that of these forty-five sites, positions 145, 155, 156, 158, 159, 189, and 193 had the strongest antigenic impact on previous antigenic evolution (Koel et al. 2013). Position 145 is included in patch 2; all other sites, except for position 193, are part of patch 1, which indicates the importance of these particular two patches for antigenic evolution. Site 193 had the lowest antigenic impact of the seven changes in one transition between predominating antigenic types and had no antigenic impact in another, suggesting that it was less relevant than the other six sites, which is in line with our other findings. Large antigenic weights were also inferred for the remaining five sites of patch 1, namely residues 159, 186, 208, 227, and 272 (0.86, 0.79, 2.0, 1.5, and 0.8 antigenic units, respectively), indicating the relevance of these sites for antigenic evolution. In fact, residue 208 was assigned the largest antigenic weight of all sites (Fig. 2). As the five sites were not part of the antigenic cluster transitions, these changes are likely to reflect antigenic variations between subsets of the strains that were never fixed in a new predominating antigenic variant. As evolution is a stochastic process, this does not preclude their relevance for antigenic evolution.
3.4 The role of antigenic patches in the evolution of human influenza A/H3N2 viruses
To gain more detailed insight into the relevance of individual patches, we studied their appearance in the ten antigenic cluster transitions between 1968 and 2002 (Smith et al. 2004). Two transitions (SI87–BE89, BE92–WU95) were accompanied by a single change at position 145 of patch 2; four had several changes in patch 1 (TX77–BA79, BA79–SI87), or in patch 1 and additional patches (WU95–SY97, SY97–FU02). The remaining four transitions showed changes in patches 1 and 2, and other patches (Supplementary Table S1). Thus changes in either patch 1 or 2 consistently seem to accompany antigenic cluster transitions, whereas changes in the other patches occur more sporadically.
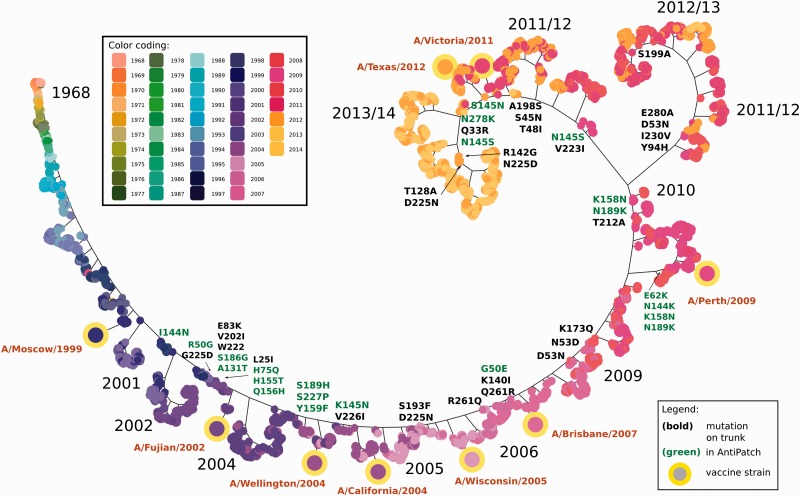
We then studied the genetic evolution of the influenza A HA for the subtype H3N2 after 2002 and inferred a second phylogeny from 7,127 HA sequences sampled from 1968 to February 2014 (Materials and Methods section: HA phylogeny used for evaluation). This shows the typical ‘cactus-like’ structure (McHardy and Adams 2009), with a single surviving lineage connecting the early isolates from 1968, which are close to the root of the tree, to the sequences from 2014. In addition to that, we derived a larger subtree with a diverging lineage that includes some of the circulating strains sampled between 2011 and 2013 (Fig. 4). Using a maximum parsimony reconstruction of the ancestral amino acid sequences for the internal nodes of the tree, we inferred the amino acid changes for the branches. Changes providing a selective advantage (e.g. by altering antigenicity of the virus) will be enriched on the trunk of the phylogenetic tree, which corresponds to the changes that became fixed in the evolution of the influenza A/H3N2 population (Fitch et al. 1997; Steinbrück and McHardy 2012).
Figure 4.
Phylogeny showing the genetic evolution of influenza A HA sequences of subtype H3 since 1968. A single trunk line (and a major branch around 2011) shows the evolution of HA since 1968. Amino acid changes occurring on the trunk and the major branch line are indicated for the subtree containing strains that occurred after 2002. Changes on patch sites are colored in green. Note that branches are scaled for visualization purposes only and do not represent molecular distances. Leaf nodes in the phylogenetic tree are colored based on the year in which the corresponding virus was isolated.
After 2002, five antigenically distinct viral variants successively became predominant in the human population, named after the respective vaccine strains used, A/California/7/2004, A/Wisconsin/67/2005, A/Brisbane/10/2007, A/Perth/16/2009, A/Victoria/361/2011 (Fig. 4) (Who 2005a,b, 2006, 2007a,b, 2008a,b, 2009, 2014). Amino acid changes by which the predominant variants differed from the preceding ones are evident from the trunk of the HA tree and from the backbone of a larger subtree including A/Perth/16/2009. All variants except A/Wisconsin/67/2005 differed by one or more amino acid changes in the antigenic patches from the previously circulating strains: A/California/7/2004-like viruses differed most notably by the N145K change in patch 2, which has been experimentally confirmed to result in an antigenic distance of 2.6–4 antigenic units on its own (Smith et al. 2004). In the subsequent A/Wisconsin/67/2005 variant, S193F and D225N were introduced. The D225N change was shown to drastically reduce the HA receptor binding avidity of the strain (Lin et al. 2012) and infections with these viral strains caused less illness. Notably, differences in receptor binding avidity can also substantially influence HI assays (Lin et al. 2012). Therefore, differences in the HI assays of A/Wisconsin/67/2005-like viruses relative to the preceding variant may have also been due to the lower receptor avidity, instead of an alteration in antigenicity. This would be in line with the absence of changes in the antigenic patches that distinguished A/Wisconsin/67/2005-like viruses from their precursors. The subsequent A/Brisbane/10/2007 variant had amino acid changes that were most notable in patch 3 (G50E) and a site in the neighborhood of residues 137, 144, and 145 of patch 2 (K140I).
Subsequently, A/Perth/16/2009-like strains circulated that had changes in patches 1 (K158N, N189K), 2 (N144K), and 4 (E62K). The subsequent A/Victoria/361/2011-like strains had two of the same changes as A/Perth/16/2009 in patch 1 (K158N, N189K) and, in addition, a change in patch 2 (N145S) at a site that is known to alter antigenicity drastically, all located on the trunk of the tree. The vaccine strain A/Victoria/361, however, included additional changes (H156Q, G186V) in antigenic patch 1 and a change (S219Y) that is not found in a patch. In our evaluation phylogeny, all changes were located on a terminal branch, indicative of either egg-adaptation changes or infrequent variants that are not likely to rise to predominance. An altered antigenicity, due to egg-adaptation, was confirmed for A/Victoria/361 by the WHO (Who 2013) and (Skowronski et al. 2014), unlike other circulating strains. A/Victoria/361/2011 was therefore replaced one year later by the WHO with A/Texas/50, which represented a better match to circulating viruses (Who 2013; Barr 2014). It seems advisable that isolates with changes in antigenic patches found on terminal branches should be excluded as candidates for a vaccine strain update, because of their likely antigenic mismatch to other circulating strains. Such isolates should be considered as vaccine strain updates only if the changes are located on the root of a subtree, including multiple isolates already sequenced within a particular season, particularly if these also demonstrate a substantial rise in prevalence relative to previous seasons (Steinbrück and McHardy 2011). Viruses with changes at site 145 in patch 2 (S145N then N145S) and with a change in patch 3 (N278K) were circulating throughout 2013. These recent viral isolates thus again had an S at position 145, and were antigenically similar to one another and to the A/Victoria/361/2011-like vaccine strain A/Texas/50/2012. Therefore, another vaccine strain update was considered to be unnecessary (Who 2014).
In summary, antigenically novel variants that were becoming abundant were detectable based on alterations in antigenic patches, which were located on larger branches or the trunk in the respective evaluation phylogeny, suggesting that this analysis could aid in selecting vaccine strains. Isolates with alterations in antigenic patches at terminal branches, such as A/Victoria/361/2011, should not be considered as vaccine candidates, as it is likely that such changes will not be abundant in circulating viruses.
3.5 Comparison to patches under selection
We investigated the effect of the antigenic patch inference algorithm by comparing sites with antigenic weights before and after the application of the algorithm to sites previously determined to be in patches under positive selection (Tusche, Steinbrück, and McHardy 2012). Before clustering, twenty-five sites (out of eighty-two sites) with an antigenic weight were located in patches, out of a total of thirty-five sites under positive selection. After spatial clustering, this was reduced to thirteen of twenty-three sites being located in antigenic patches (Fig. 3d, Supplementary Fig. S1). Thus, the percentage of overlap between sites in patches under selection and antigenic sites was substantially increased from 30 to 57 per cent by antigenic patch inference, supporting the notion that false-positive antigenic sites determined by antigenic tree inference alone were removed by the patch inference. There was a significant enrichment of sites under selection in antigenic patches in comparison to the entire head region (hypergeometric distribution; N = 230, K = 35, n = 23, k = 13; : sites under positive selection are sampled from antigenic patches and the overall head region of HA at the same rates; ). However, still, a striking discrepancy was evident between sites in patches under positive selection and antigenic patches (43 per cent; ten of twenty-three antigenic patch sites were not part of patches under selection), indicating that changes in antigenicity and measurements thereof using currently available data do not allow us to determine all the molecular changes that are relevant for the adaptive evolution of HA of human influenza A/H3N2 viruses. The antigenic weights in antigenic patches were significantly different from those in the patches under positive selection (two-sided Kolmogorov–Smirnov-test;: both samples were derived from the same distribution; P = 0.002). This indicates that alterations of the antigenicity do not explain all the sites in patches under evolutionary pressure, in line with (Meyer and Wilke 2015). Possibly, sites that do not alter their antigenicity but directly affect their binding avidity to sialic acid residues on the host cell surfaces are under selection, which has recently been described as affecting influenza A/H1N1 evolution in mouse models (Hensley et al. 2009). In line with this notion, six sites (192, 193, 197, 198, 199, and 229) that are found only in patches under positive selection but not in the antigenic patches are located in the RBS.
4. Discussion
Knowledge of the sites of influenza A/H3N2 viruses that alter viral antigenicity is of substantial importance for viral surveillance and vaccine design (Gershoni et al. 2007; Lees, Moss, and Shepherd 2011). Here, we combined genetic, evolutionary, phenotypic, and structural information to detect regions on the protein structure of HA of human influenza A/H3N2 viruses where alterations had a large antigenic impact. We used the HI titer measurements to determine the antigenic impact of branch-associated amino acid changes in HA1 by inference of an antigenic tree and, from this tree, the relevance of particular amino acid changes and sites. A subsequent clustering of sites based on their antigenic weights and spatial proximity revealed that six areas of the HA1 subunit have been most relevant for changing viral antigenicity over a 35-year period of viral evolution. The six identified patches are located in the protein head region, mostly close to the RBS and within the known epitope regions (Fig. 3). They include many previously described antigenicity-altering sites and sites that have changed during antigenic cluster transitions of human influenza A/H3N2 viruses. The ten antigenic cluster transitions during the study period all included amino acid changes in either the first or second antigenic patch, or both. Amino acid changes in these two patches were also preferentially fixed and found on the trunk of the HA phylogeny of viral strains since 2002. This is a simpler model for the antigenic evolution of influenza A/H3N2 viruses than the previous hypothesis, which stated that changes in epitopes A and B are required for an antigenic cluster transition (Wilson and Cox 1990; Huang and Yang 2011).
Antibodies interact with influenza HA in three ways: they disrupt viral attachment to sialic acids on the host cell surface, they prevent the release of virions and they block viral fusion with the host cell (Laursen and Wilson 2013; DiLillo et al. 2014). Only the first interaction is associated with a hemagglutination effect and can be quantified with an HI assay. Amino acid changes in regions outside of the protein head region are unlikely to show a signal in an HI assay, and we therefore restricted our study to the HA1 subunit. HI titers are commonly used to estimate antigenic characteristics of circulating viral strains within the global surveillance network of the WHO (Russell et al. 2008). However, titers may be imprecise or show variable results (Who 2011; Steinbrück and McHardy 2012), and measurements might be influenced by the effects of egg-adaptations (Lin et al. 2012), NA activity (Sandbulte et al. 2011) or alterations of receptor binding avidity (Li et al. 2013). To avoid bias caused by egg-adaptation, we excluded terminal branches from our antigenic tree analysis and thus considered only amino acid changes that were supported by two or more viral strains (Materials and Methods section). For the identification of further antigenically relevant regions in the influenza A NA protein, it should be straightforward to include measurements characterizing NA alterations. Our method could easily be adapted to similar phenotypic measures, such as data from a recently described neutralization assay (Terletskaia-Ladwig, Meier, and Enders 2013), if a suitable distance matrix comparing the viral strains and a structural model could be made available.
Previously, computational methods that identified antigenic sites based on their correlation with antigenic distances or location on the protein structure have been suggested (Lee et al. 2007; Liao et al. 2008; Huang, King, and Yang 2009; Lees, Moss, and Shepherd 2011). This is the first attempt to reconstruct and redefine antigenic areas on the protein structure as a replacement for the broadly defined epitope regions via joint consideration of spatial, antigenic, evolutionary, and genetic information. A refined definition of the antigenically relevant sites as antigenic patches is further supported by Meyer and Wilke (2015), indicating that the epitope sites themselves rely on a historically imprecise definition and are rather broadly grouped to epitope regions (Meyer and Wilke 2015). By integrating the available data on structure, evolution, and antigenicity, we identified the antigenically relevant areas of HA, including relevant sites with lower antigenic weights if these were in the vicinity of a cluster of relevant sites on the protein structure. This agrees with the underlying model of molecular interactions between the viral surface proteins and host antibodies. Lees, Moss, and Shepherd (2011) mapped sphere-shaped clusters of amino acid substitutions between predominant strains onto a grid of the HA protein structure to predict antigenic distances (Lees, Moss, and Shepherd 2011), which indicated a large number (76) of potentially relevant sites. Here, we clustered all HA residues together, based on their antigenic weights and location on the protein structure without restricting our attention to a particular set of substitutions or a specific cluster shape. Sun et al. (2013) used ridge regression to infer antigenic weights based on genetic and antigenic profiles, identifying thirty-nine antigenicity-associated sites on the protein surface, but did not consider their spatial distances. Of the total twenty-three AntiPatch sites reported here, fourteen were found also by Sun and colleagues. Of the other nine sites, five were involved in antigenic cluster transitions and were assigned above-average antigenic weights (an average of 1.2 antigenic units for the nine sites; the average of all sites in HA1 is 0.13 antigenic units). Huang, King, and Yang (2009) used a decision tree to detect the ‘antigenic critical positions’ that were relevant for classifying antigenic clusters, without consideration of the protein structure (Huang, King, and Yang 2009). They described eleven sites, including positions 137, 145, 156, 158, and 189 in patches 1 and 2, and positions 62, 260, and 278 in patches 3, 4, and 6. Residues 155 and 156 have been confirmed as being responsible for the antigenic cluster transition to A/Fujian-like viruses in 2003 (Jin et al. 2005). In our previous study, in which we described antigenic tree inference (Steinbrück and McHardy 2012), we used a strict criterion to define relevance and described seven sites with antigenic weights that were larger than one antigenic unit (positions 112, 137, 144, 155, 156, 189, and 208). Here, by removing all sites with antigenic weights that were located far from spatial clusters of antigenic sites on HA, we eliminated changes that are likely to have no real effects on HI assay measurements (such as changes in the stem region of HA) and have potentially been identified due to their co-evolution with antigenicity-altering changes in the antigenic tree inference. Six of the seven sites we described in Steinbrück and McHardy (2012) are located in the patches; five of them are in patch 1 or 2, which again stresses the importance of these regions for antigenic evolution.
We observed a notably small overlap between sites in the antigenic patches (thirteen of twenty-three patch sites) and patches in the sites under selection (Tusche, Steinbrück, and McHardy 2012). In comparison to other studies, four out of the eighteen sites found by (Bush et al. 1999) were included in the antigenic patches (sites 145, 156, 158, and 186). Patch sites 137, 155, 196, and 276 were also reported to be under positive selection elsewhere (Fitch et al. 1997; Kosakovsky Pond et al. 2008) (Supplementary Fig. S1). Earlier, Smith et al. (2004) noted that the sites involved in antigenic cluster transitions and the sites under selection (Bush et al. 1999) seem to be distinct; however, the authors compared sites under positive selection in a different period from the one studied here using antigenic measurements. Here, we directly compared antigenic patches with patches under selection identified for the same period of time and found that this was the case. The lack of overlap was also apparent when comparing other studies reporting sites under positive selection (Bush et al. 1999; Kosakovsky Pond et al. 2008; Murrell et al. 2012) with studies reporting sites of antigenic impact (Koel et al. 2013). These findings raise questions on the nature of the selective advantage provided by changes at sites that are under positive selection but do not seem to influence antigenicity. Possibly, such sites directly affect receptor binding avidity instead of altering antigenicity, or alter the binding behavior to negatively charged cell surface structures. It is also possible that additional antigenicity-altering sites might be missed, due to limitations in the panels of ferret antisera used in HI assays and their lack of similarity to the human immune response (a recent article indicated such effects for current H1N1 viruses (Linderman et al. 2014)), or their inability to detect antibody–HA interactions for antibodies binding outside of the head region. Indeed, observing changes in the sites under positive selection but which are distinct from the antigenic patches could indicate these effects. The practical relevance of the changes at these additional sites for shaping the antigenic evolution of human H3N2 viruses remains to be determined, as the observed antigenic patch changes allowed us to explain antigenic evolution over the examined time period, and also became abundant or even fixed within circulating human viruses. Taken together, however, the results indicate that more refined models that include multiple factors shaping the evolution of human influenza A viruses should be considered. Future work could include modeling of avidity patches (based on their distance to the RBS) in the patch inference step, thus allowing HI assay data tentatively to be dissected into the effects of altered antibody–antigen interactions or of altered avidity, similarly to the BMDS model of Bedford et al. (2014). Determining the sites or patches of sites that are associated with phenotypes, such as the host receptor’s binding avidity, protein stability, or binding to the negatively charged phospholipids on the cell surface, could provide further insight into the mechanisms shaping the evolution of human influenza A/H3N2 viruses.
Software
A C++ program for the residue clustering and all datasets used in this publication can be accessed at https://github.com/hzi-bifo/AntiPatch/wiki.
Supplementary data
Supplementary data are available at Virus Evolution online.
Acknowledgements
This work was funded by Heinrich Heine University and Helmholtz Center for Infection Research. We thank John McCauley, the head of the WHO influenza vaccine selection committee, for providing insightful comments and discussing the manuscript. We also thank Marc Daxer for creating the evaluation HA phylogeny.
Conflict of interest: None declared.
References
- Bao Y., et al. (2008) ‘The Influenza Virus Resource at the National Center for Biotechnology Information’, Journal of Virology, 82: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow D. J., Edwards M. S., Thornton J. M. (1986) ‘Continuous and Discontinuous Protein Antigenic Determinants’, Nature, 322: 747–8. [DOI] [PubMed] [Google Scholar]
- Barr I. G., et al. (2014) ‘WHO recommendations for the viruses used in the 2013–2014 Northern Hemisphere influenza vaccine: Epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from October 2012 to January 2013’, Vaccine, 32: 4713–25. [DOI] [PubMed] [Google Scholar]
- Bedford T., et al. (2014) ‘Integrating Influenza Antigenic Dynamics with Molecular Evolution’, Elife, 3: e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., et al. (2000) ‘The Protein Data Bank’, Nucleic Acids Research, 28: 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S., Holmes E. C., Pybus O. G. (2011) ‘The Genomic Rate of Molecular Adaptation of the Human Influenza A Virus’, Mol Biol Evol, 28: 2443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner P., et al. (2006) ‘A Global Initiative on Sharing Avian Flu Data’, Nature, 442: 981. [Google Scholar]
- Bush R. M., et al. (1999) ‘Positive Selection on the H3 Hemagglutinin Gene of Human Influenza Virus A’, Molecular Biology and Evolution, 16: 1457–65. [DOI] [PubMed] [Google Scholar]
- Cattoli G., et al. (2011) ‘Antigenic Drift in H5N1 Avian Influenza Virus in Poultry Is Driven by Mutations in Major Antigenic Sites of the Hemagglutinin Molecule Analogous to Those for Human Influenza Virus’, Journal of Virology, 85: 8718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. (1976) ‘The Nature of the Accessible and Buried Surfaces in Proteins’, Journal of Molecular Biology, 105: 1–12. [DOI] [PubMed] [Google Scholar]
- Darriba D., et al. (2011) ‘ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution’, Bioinformatics, 27: 1164–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K., et al. (2010) ‘Structures of Influenza A Proteins and Insights into Antiviral Drug Targets’, Nature Structural and Molecular Biology, 17: 530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D. J., et al. (2014) ‘Broadly Neutralizing Hemagglutinin Stalk-Specific Antibodies Require FcgammaR Interactions for Protection Against Influenza Virus in vivo’, Nature Medicine, 20: 143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer P. R., et al. (2011) ‘Influenza Vaccine Immunology’, Immunological Reviews, 239: 167–77. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M. (1971) ‘Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology’, Systematic Zoology, 20: 406–16. [Google Scholar]
- Fitch W. M., et al. (1997) ‘Long Term Trends in the Evolution of H(3) HA1 Human Influenza Type A’, Proceedings of the National Academy of Sciences of the United States of America, 94: 7712–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten R. J., et al. (2009) ‘Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans’, Science, 325: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni J. M., et al. (2007) ‘Epitope Mapping: The First Step in Developing Epitope-Based Vaccines’, BioDrugs, 21: 145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003) ‘A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood’, Systematic Biology, 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hensley S. E., et al. (2009) ‘Hemagglutinin Receptor Binding Avidity Drives Influenza A Virus Antigenic Drift’, Science, 326: 734–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst G. K. (1943) ‘Studies of Antigenic Differences Among Strains of Influenza A by Means of Red Cell Agglutination’, Journal of Experimental Medicine, 78: 407–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Honda W. (2006) ‘CED: A Conformational Epitope Database’, BMC Immunology, 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. W., Yang J.-M. (2011) ‘Changed Epitopes Drive the Antigenic Drift for Influenza A (H3N2) Viruses’, BMC Bioinformatics, 12 (Suppl 1): S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. W., King C.-C., Yang J.-M. (2009) ‘Co-Evolution Positions and Rules for Antigenic Variants of Human Influenza A/H3N2 Viruses’, BMC Bioinformatics, 10 (Suppl 1): S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger B. W., et al. (2012) ‘An Overlapping Protein-Coding Region in Influenza A Virus Segment 3 Modulates the Host Response’, Science, 337: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., et al. (2005) ‘Two Residues in the Hemagglutinin of A/Fujian/411/02-Like Influenza Viruses Are Responsible for Antigenic Drift from A/Panama/2007/99’, Virology, 336: 113–9. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992) ‘The Rapid Generation of Mutation Data Matrices from Protein Sequences’, Computer Applications in the Biosciences, 8: 275–82. [DOI] [PubMed] [Google Scholar]
- Kelley L. A., Sternberg M. J. (2009) ‘Protein Structure Prediction on the Web: A Case Study Using the Phyre Server’, Nature Protocols, 4: 363–71. [DOI] [PubMed] [Google Scholar]
- Koel B. F., et al. (2013) Substitutions Near the Receptor Binding Site Determine Major Antigenic Change During Influenza Virus Evolution’, Science, 342: 976–9. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., et al. (2008) A Maximum Likelihood Method for Detecting Directional Evolution in Protein Sequences and Its Application to Influenza A Virus’, Molecular Biology and Evolution, 25: 1809–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., et al. (2011) ‘A Random Effects Branch-Site Model for Detecting Episodic Diversifying Selection’, Molecular Biology and Evolution, 28: 3033–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S., et al. (2011) ‘Prevalence of Epistasis in the Evolution of Influenza A Surface Proteins’, PLoS Genetics, 7: e1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapedes A., Farber R. (2001) ‘The Geometry of Shape Space: Application to Influenza’, The Journal of Theoretical Biology, 212: 57–69. [DOI] [PubMed] [Google Scholar]
- Laursen N. S., Wilson I. A. (2013) ‘Broadly Neutralizing Antibodies Against Influenza Viruses’, Antiviral Research, 98: 476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Richards F. M. (1971) ‘The Interpretation of Protein Structures: Estimation of Static Accessibility’, Journal of Molecular Biology, 55: 379–400. [DOI] [PubMed] [Google Scholar]
- Lee M. S., et al. (2007) ‘Identifying Potential Immunodominant Positions and Predicting Antigenic Variants of Influenza A/H3N2 Viruses’, Vaccine, 25: 8133–9. [DOI] [PubMed] [Google Scholar]
- Lees W. D., Moss D. S., Shepherd A. J. (2011) ‘Analysis of Antigenically Important Residues in Human Influenza A Virus in Terms of B-Cell Epitopes’, Journal of Virology, 85: 8548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., et al. (2013) ‘Single Hemagglutinin Mutations That Alter Both Antigenicity and Receptor Binding Avidity Influence Influenza Virus Antigenic Clustering’, Journal of Virology, 87: 9904–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. C., et al. (2008) ‘Bioinformatics Models for Predicting Antigenic Variants of Influenza A/H3N2 Virus’, Bioinformatics, 24: 505–12. [DOI] [PubMed] [Google Scholar]
- Liao Y. C., Lin H. H., Lin C. H. (2013) ‘Monitoring the Antigenic Evolution of Human Influenza A Viruses to Understand How and When Viruses Escape from Existing Immunity’, BMC Research Notes, 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. P., et al. (2012) ‘Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin’, Proceedings of the National Academy of Sciences of the United States of America, 109: 21474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman S. L., et al. (2014) ‘Potential Antigenic Explanation for Atypical H1N1 Infections Among Middle-Aged Adults During the 2013–2014 Influenza Season’, Proceedings of the National Academy of Sciences of the United States of America, 111: 15798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy A. C., Adams B. (2009) ‘The Role of Genomics in Tracking the Evolution of Influenza A Virus’, PLoS Pathogens, 5: e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R., García-Sastre A. (2011) ‘Influenza A Viruses: New Research Developments’, Nature Reviews Microbiology, 9: 590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. G., Wilke C. O. (2012) ‘Integrating Sequence Variation and Protein Structure to Identify Sites Under Selection’, Molecular Biology and Evolution,: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. G., Wilke C. O. (2015) ‘Geometric Constraints Dominate the Antigenic Evolution of Influenza H3N2 Hemagglutinin’, PLoS Pathogens, 11: e1004940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari N.-A. M., et al. (2007) ‘The Annual Impact of Seasonal Influenza in the US: Measuring Disease Burden and Costs’, Vaccine, 25: 5086–96. [DOI] [PubMed] [Google Scholar]
- Murrell B., et al. (2012) ‘Detecting Individual Sites Subject to Episodic Diversifying Selection’, PLoS Genetics, 8: e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. (2005) ‘Selectionism and Neutralism in Molecular Evolution’, Molecular Biology and Evolution, 22: 2318–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Yang Z. (1998) ‘Likelihood Models for Detecting Positively Selected Amino Acid Sites and Applications to the HIV-1 Envelope Gene’, Genetics, 148: 929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusawa E., Aoyama T., Kato H. (1991) ‘Comparison of Complete Amino Acid Sequences and Receptor-Binding Properties Among 13 Serotypes of Hemagglutinins of Influenza A Viruses’, Journal of Virology, 182: 475–85. [DOI] [PubMed] [Google Scholar]
- Nozawa M., Suzuki Y., Nei M. (2009) ‘Reliabilities of Identifying Positive Selection by the Branch-Site and the Site-Prediction Methods’, Proceedings of the National Academy of Sciences of the United States of America, 106: 6700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S. L. K., Frost S. D. W., Muse S. V. (2005) ‘HyPhy: Hypothesis Testing Using Phylogenies’, Bioinformatics, 21: 676–9. [DOI] [PubMed] [Google Scholar]
- Posada D. (2008) ‘jModelTest: Phylogenetic Model Averaging’, Molecular Biology and Evolution, 25: 1253–6. [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2009) ‘FastTree: Computing Large Minimum Evolution Trees With Profiles Instead of a Distance Matrix’, Molecular Biology and Evolution, 26: 1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C. A., et al. (2008) ‘Influenza Vaccine Strain Selection and Recent Studies on the Global Migration of Seasonal Influenza Viruses’, Vaccine, 26 (Suppl 4): D31–4. [DOI] [PubMed] [Google Scholar]
- Sandbulte M. R., et al. (2011) ‘Discordant Antigenic Drift of Neuraminidase and Hemagglutinin in H1N1 and H3N2 Influenza Viruses’, Proceedings of the National Academy of Sciences of the United States of America, 108: 20748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. L. C. (2010) The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC. [Google Scholar]
- Shannon P., et al. (2003) ‘Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks’, Genome Research, 13: 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A. C.-C., et al. (2007) ‘Simultaneous Amino Acid Substitutions at Antigenic Sites Drive Influenza A Hemagglutinin Evolution’, Proceedings of the National Academy of Sciences of the United States of America, 104: 6283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J. (2009) ‘An Overview of Influenza Haemagglutinin and Neuraminidase’, Biologicals: Journal of the International Association of Biological Standardization, 37: 177–8. [DOI] [PubMed] [Google Scholar]
- Skowronski D. M., et al. (2014) ‘Low 2012–13 Influenza Vaccine Effectiveness Associated with Mutation in the Egg-Adapted H3n2 Vaccine Strain Not Antigenic Drift in Circulating Viruses’, PLoS One, 9: e92153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. J., et al. (2004) ‘Mapping the Antigenic and Genetic Evolution of Influenza Virus’, Science, 305: 371–6. [DOI] [PubMed] [Google Scholar]
- Steinbrück L., McHardy A. C. (2011) ‘Allele Dynamics Plots for the Study of Evolutionary Dynamics in Viral Populations’, Nucleic Acids Research, 39: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrück L., McHardy A. C. (2012) ‘Inference of Genotype–Phenotype Relationships in the Antigenic Evolution of Human Influenza A (H3N2) Viruses’, PLOS Computational Biology, 8: e1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., et al. (2013) ‘Using Sequence Data to Infer the Antigenicity of Influenza Virus’, mBio, 4: e00230–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. (2004) ‘New Methods for Detecting Positive Selection at Single Amino Acid Sites’, Journal of Molecular Evolution, 59: 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. (2006) ‘Natural Selection on the Influenza Virus Genome’, Molecular Biology and Evolution, 23: 1902–11. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Gojobori T. (1999) ‘A Method for Detecting Positive Selection at Single Amino Acid Sites’, Molecular Biology and Evolution, 16: 1315–28. [DOI] [PubMed] [Google Scholar]
- Terletskaia-Ladwig E., Meier S., Enders M. (2013) ‘Improved High-Throughput Virus Neutralisation Assay for Antibody Estimation Against Pandemic and Seasonal Influenza Strains from 2009 to 2011’, Journal of Virological Methods, 189: 341–7. [DOI] [PubMed] [Google Scholar]
- Tien M. Z., et al. (2013) ‘Maximum Allowed Solvent Accessibilites of Residues in Proteins’, PLoS One, 8: e80635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., et al. (2012) ‘A Distinct Lineage of Influenza A Virus from Bats’, Proceedings of the National Academy of Sciences of the United States of America,: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., et al. (2013) ‘New World Bats Harbor Diverse Influenza A Viruses’, PLoS Pathogens, 9: e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusche C., Steinbrück L., McHardy A. C. (2012) ‘Detecting Patches of Protein Sites of Influenza A Viruses Under Positive Selection’, Molecular Biology and Evolution, 29: 2063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., et al. (1992) ‘Evolution and Ecology of Influenza A Viruses’, Microbiological Reviews, 56: 152–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Who (2005a) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2005–2006 Influenza Season’, WHO Weekly Epidemiological Record, 80: 66–71.15771206 [Google Scholar]
- Who (2005b) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2006 Influenza Season’, WHO Weekly Epidemiological Record, 80: 342–7. [PubMed] [Google Scholar]
- Who (2006) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2007 Influenza Season’, WHO Weekly Epidemiological Record, 81: 390–5. [PubMed] [Google Scholar]
- Who (2007a) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2007–2008 Influenza Season’, WHO Weekly Epidemiological Record, 82: 69–74. [Google Scholar]
- Who (2007b) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2008 Influenza Season’, WHO Weekly Epidemiological Record, 82: 351–6. [PubMed] [Google Scholar]
- Who (2008a) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2008–2009 Influenza Season’, WHO Weekly Epidemiological Record, 83: 81–7. [Google Scholar]
- Who (2008b) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2009 Southern Hemisphere Influenza Season’, WHO Weekly Epidemiological Record, 83: 366–72. [PubMed] [Google Scholar]
- Who (2009) ‘Recommended Composition of Influenza Virus Vaccines for Use in 2009–2010 Influenza Season (Northern Hemisphere Winter)’, WHO Weekly Epidemiological Record, 84: 65–72. [PubMed] [Google Scholar]
- Who (2011) Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. Geneva: World Health Organization. [Google Scholar]
- Who (2013) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2013–2014 Northern Hemisphere Influenza Season’, WHO Weekly Epidemiological Record, 88: 101–14. [PubMed] [Google Scholar]
- Who (2014) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2014–2015 Northern Hemisphere Influenza Season’, WHO Weekly Epidemiological Record, 89: 93–104. [PubMed] [Google Scholar]
- World Health Organisation (WHO) (2009) Fact sheet no. 211. In. [Google Scholar]
- Wiley D. C., Skehel J. J. (1987) ‘The Structure and Function of the Hemagglutinin Membrane Glycoprotein of Influenza Virus’, The Annual Review of Biochemistry, 56: 365–94. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. (1981) ‘Structural Identification of the Antibody-Binding Sites of Hong Kong Influenza Haemagglutinin and Their Involvement in Antigenic Variation’, Nature, 289: 373. [DOI] [PubMed] [Google Scholar]
- Wilson I., Cox N. J. (1990) ‘Structural Basis of Immune Recognition of Influenza Virus Hemagglutinin’, The Annual Review of Immunology, 8: 737–71. [DOI] [PubMed] [Google Scholar]
- Winn M. D., et al. (2011) ‘Overview of the CCP4 Suite and Current Developments’, Acta Crystallographica Section D: Biological Crystallography, 67: 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise H. M., et al. (2009) ‘A Complicated Message: Identification of a Novel PB1-Related Protein Translated from Influenza A Virus Segment 2 Mrna’, Journal of Virology, 83: 8021–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise H. M., et al. (2012) ‘Identification of a Novel Splice Variant Form of the Influenza A Virus M2 Ion Channel with an Antigenically Distinct Ectodomain’, PLoS Pathogens, 8: e1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2000) ‘Maximum Likelihood Estimation on Large Phylogenies and Analysis of Adaptive Evolution in Human Influenza Virus A’, Journal of Molecular Evolution, 51: 423–32. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007) ‘PAML 4: Phylogenetic Analysis by Maximum Likelihood’, Molecular Biology and Evolution, 24: 1586–91. [DOI] [PubMed] [Google Scholar]
- Yang Z., dos Reis M. (2011) ‘Statistical Properties of the Branch-Site Test of Positive Selection’, Molecular Biology and Evolution, 28: 1217–28. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. (2002) ‘Codon-Substitution Models for Detecting Molecular Adaptation at Individual Sites Along Specific Lineages’, Molecular Biology and Evolution, 19: 908–17. [DOI] [PubMed] [Google Scholar]
- Zwickl D. J. (2006) ‘Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion’, Ph.D. dissertation, The University of Texas at Austin. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.