Abstract
A comprehensive monitoring strategy is vital for tracking the spread of mosquito-borne Japanese encephalitis virus (JEV), the leading cause of viral encephalitis in Asia. Virus detection consists of passive surveillance of primarily humans and swine, and/or active surveillance in mosquitoes, which may be a valuable proxy in providing insights into ecological processes underlying the spread and persistence of JEV. However, it has not been well characterized whether passive surveillance alone can capture the circulating genetic diversity to make reasonable inferences. Here, we develop phylogenetic models to infer JEV host changes, spatial diffusion patterns, and evolutionary dynamics from data collected through active and passive surveillance. We evaluate the feasibility of using JEV sequence data collected from mosquitoes to estimate the migration histories of genotypes GI and GIII. We show that divergence times estimated from this dataset were comparable to estimates from all available data. Increasing the amount of data collected from active surveillance improved time of most recent common ancestor estimates and reduced uncertainty. Phylogenetic estimates using all available data and only mosquito data from active surveillance produced similar results, showing that GI epidemics were widespread and diffused significantly faster between regions than GIII. In contrast, GIII outbreaks were highly structured and unlinked suggesting localized, unsampled infectious sources. Our results show that active surveillance of mosquitoes can sufficiently capture circulating genetic diversity of JEV to confidently estimate spatial and evolutionary patterns. While surveillance of other hosts could contribute to more detailed disease tracking and evaluation, comprehensive JEV surveillance programs should include systematic surveillance in mosquitoes to infer the most complete patterns for epidemiology, and risk assessment.
Keywords: phylogeography, vector-borne, interspecies transmission, disease spread
1. Introduction
Japanese encephalitis virus (JEV), a mosquito-borne flavivirus, is the leading cause of vaccine-preventable viral encephalitis among humans in Asia (MacKenzie, Gubler, and Peterson 2004). 68,000 new cases in humans are estimated to occur annually and highlight the continued public health threat of JEV (Campbell et al. 2011). JEV is believed to be maintained in an enzootic cycle between mosquitoes, swine, and to some extent, wild aquatic birds (Buescher and Scherer 1959). Mosquitoes, in particular Culex tritaeniorhynchus species, are considered the primary vector for JEV transmission although other mosquito species (i.e. Aedes, Anopheles, etc.) have been shown to carry the virus as well (Buescher and Scherer 1959). Mosquitoes become infected by feeding on viremic vertebrate amplification hosts, of which swine are considered the most important one associated with human outbreaks (MacKenzie, Gubler, and Peterson 2004). Wild aquatic birds are posited to be maintenance reservoirs of the virus (Buescher and Scherer 1959), although scarce JEV surveillance data in this population limit the clarification and elaboration of their role in the disease ecology (Lord, Gurley, and Pulliam 2016). JEV can also infect a wide range of other animals, even though most of them, including humans, are considered incidental or dead-end transmissions (MacKenzie, Gubler, and Peterson 2004). Vector transmission patterns have been shown to vary depending on seasonality, climate, geography, agriculture, and host availability, suggesting that complex interactions underlie JEV disease dynamics (Van den Hurk, Ritchie, and MacKenzie 2009).
Five genotypes (GI–GV) have been identified based on JEV’s envelope (E) gene phylogeny (Chen, Tesh, and Rico-Hesse 1990; Ni and Barret 1995). GII, GIV, and GV are scarcely detected compared with larger prevalences and distributions of GI and GIII. GIII was the major circulating genotype in temperate Asia responsible for human and zoonotic infections from its initial isolation in 1935 until the 1990s, when GI rapidly replaced GIII as the dominant genotype isolated for reasons that remain unclear (Nga et al. 2004). Although it is estimated that this genotype shift occurred during the 1990s, it was not recognized until the early 2000s (Nga et al. 2004).
Disease surveillance to monitor, detect, and isolate JEV among human, animal, and vector hosts has greatly expanded in the last several decades due to the rapid emergence and dominance of GI and the spread of JEV into new geographic regions (MacKenzie, Gubler, and Peterson 2004). JEV outbreaks in humans were first described in Japan during the 1870s and have since spread and become endemic throughout most of Asia by mechanisms that remain elusive but may vary by host, temporal, spatial, or climactic factors (Hanna et al. 1996; Schuh et al. 2013; Lord, Gurley, and Pulliam 2016). As the maintenance hosts of JEV, migratory aquatic birds are believed to contribute to the long-distance movement of the virus into new regions (Buescher and Scherer 1959). Wind-blown infected mosquitoes are also hypothesized to play a role in the emergence of JEV into novel regions such as mainland Australia (Ritchie and Rochester 2001).
Despite greater surveillance efforts, JEV outbreaks continue to occur and spread. Even though information collected from hosts and vectors are indispensable for informing disease prevention and control strategies, discrepancies in host and vector surveillance approaches can contribute to inadequate disease information. Among humans and mammals, viral isolations were conducted largely in response to suspected outbreaks or cases (Hall-Mendelin et al. 2010; Schuh et al. 2013). Although humans can be vaccinated against JEV, swine are less often inoculated despite their role as amplifying hosts (Le Flohic et al. 2013). One reason is that swine are frequently used as sentinel hosts to monitor JEV activity and predict human outbreaks (Borah et al. 2013). In contrast, virus isolation is routinely and actively collected from mosquitoes in many endemic countries with large sample sizes, subjecting it less to sampling biases (Hall-Mendelin et al. 2010; van den Hurk et al. 2012). Vector surveillance may therefore represent a valuable system to monitor cryptic processes such as viral spread and persistence in the absence of large-scale human and animal outbreaks. However, the suitability of gene sequence data collected from active surveillance of mosquitoes for inference of JEV ecological and evolutionary patterns have not been evaluated.
In this study, we use comparative genetic techniques to co-estimate viral phylogeny and migration history. Spatial diffusion patterns and evolutionary dynamics of co-circulating JEV genotypes estimated from gene sequence data collected from active surveillance of mosquitoes and from mixed surveillance of all sampled species are compared. We also model vector and host changes in order to assess the role of interspecies transmission in shaping JEV disease dynamics.
2. Methods
2.1. Dataset
In order to evaluate overall JEV dynamics, a dataset of all available JEV envelope (E) gene sequences of isolates collected between 1935 and 2013 were downloaded from GenBank. Sequences were subject to the following criteria: (1) sequences had known location and isolation year; (2) sequences with the same location, date of isolation, and 100 percent similarity were excluded; and (3) vaccine, derivative, and recombinant sequences were excluded. Since recombinant sequences may invalidate the results of coalescent analyses, sequences in the pruned dataset were screened using the Recombination Detection Program (RDP) version 4.16 under both the default and triplet settings to verify that no recombinants were included (Martin et al. 2010; Schuh et al. 2013). This dataset consisted of 567 taxa (Supplementary Table S1). All JEV genotypes (GI–GV) were represented in our dataset (Supplementary Table S2).
To estimate viral histories, taxa were coded by geographic region of isolation and by animal type. Virus isolation locations were grouped into ten discrete geographic states in East and Southeast Asia based on similar climate and rainfall conditions. Due to its expansive geographic area and climate conditions, Mainland China was subdivided into four states based on annual mean rainfall data (Supplementary Table S2) (Ministry of Environmental Protection 2005). Although we make the assumption that all five genotypes were sourced from a common population in the past, JEV surveillance was limited prior to 1970. We therefore classified isolates collected before this time into an ancestral state (Z) to avoid making any biased distinctions of their geographic origins or spread. GII, GIV, and GV were also assigned to this same ancestral population due to their limited prevalence and distribution. These samples were retained during analyses to inform the phylogenetic tree structure. We do not estimate timing of migration or location of ancient source populations and, therefore, limit all inferences and migration estimates to reflect contemporary patterns of GI and GIII only. Taxa were also classified into one of four animal host categories—mosquito, human/mammals, unidentified/other (non-human or non-mammals), and the previously defined ancestral population. Humans and mammals were combined due to uneven sampling of these hosts observed in GI versus GIII. Preliminary phylogenetic analyses were conducted with the pruned dataset (n = 567), and singletons (n = 15) were excluded to guard against sequencing artifacts (supplementary Table S1). The final dataset consisted of 551 taxa.
2.2. Bayesian phylogenetic and coalescent analysis
To generate a temporal phylogeny of all JEV genotypes using our time-stamped sequence dataset (n = 551 taxa), we applied a relaxed molecular clock in a Bayesian statistical framework implemented in BEAST v.1.8.2 (Drummond et al. 2012). This method allows for nucleotide substitution rate variability between lineages, and incorporates phylogenetic uncertainty by sampling phylogenies and parameter estimates in proportion to their posterior probability. Molecular clock rates were uncorrelated and independently drawn from a lognormal distribution (UCLD) with an initial mean of 0.0033 and a uniform prior ranging from 0 to 0.1. An HKY85 nucleotide substitution model and a Bayesian skyline coalescent prior with twenty coalescent groups were used for all analyses (Drummond et al. 2005).
Four independent chains of one hundred million generations were run, each sampled to produce 10,000 trees per dataset to ensure an adequate sample size of all analysis parameters, including the posterior, prior, nucleotide substitution rates, and likelihoods (effective sample size >200). These analyses were then combined using LogCombiner version 1.8.2 following the removal of 50 percent of the samples. The maximum clade credibility (MCC) phylogenetic tree generated from this analysis showing statistical supports for node age estimates and full taxa names is shown in supporting Supplementary Figure S1.
In order to validate these results, we conducted separate phylogeographic analyses of GI and GIII isolates independent of each other using the same model parameters previously described. Phylogenetic structure and migration rates for each genotype were compared with the results obtained from the joint analysis to evaluate for any sampling biases.
2.3. Estimates of viral geographic migration rates
A reversible discrete phylogeographic method was used to estimate the ancestral state location and estimate migration rates for GI and GIII between Asian regions and within China across all sampled species (Lemey et al. 2009). Isolates were classified into ten discrete geographic states. The ancestral states estimated from these geographical tip-state observations were subsequently mapped onto the internal nodes of phylogenetic trees sampled during the Bayesian analysis to infer migration histories. Given the large number of states, a Bayesian stochastic search variable selection (BSSVS) was employed to reduce the number of parameters to those with significantly non-zero transition rates (Lemey et al. 2009).
To estimate the transition rates of GI and GIII between each geographic state, the analysis was conducted with a truncated Poisson prior where 50 percent prior probability is placed on the minimal rate configuration, comprising eighteen non-zero rates connecting the nineteen locations. We compared these results with those obtained using a uniform prior ranging between the minimum and maximum migration events required to represent all locations on the trees. With this scenario, we have a maximum of 171 non-zero rates connecting the nineteen discrete locations. These locations included one ancestral state (Z) and nine locations for GI and nine locations for GIII. Due to potential sampling biases associated with state Z, migration rates between Z and GI or GIII locations were masked in our model and statistical analyses were restricted to GI and GIII patterns.
The BSSVS also calculates a Bayes factor (BF) allowing the support for individual transitions between location states to be assessed. The number of statistically supported rates using both the Poisson and the uniform prior was similar, although actual BF values did vary where those rates estimated under a uniform prior were generally lower. In particular, strongly supported rates were supported between analyses. Location transition rates with marginal support (BF >3) did vary when using either the Poisson or the uniform prior. The BF is a function of the binary indicator (I) where the number of rates in the migration matrix was augmented. Therefore, when I = 0.5, that rate was estimated in at least 50 percent of the trees simulated under the MCMC. Migration transitions were deemed statistically significant where I > 0.5 and the BF > 6 (Bahl et al. 2013). In this analysis, the minimal cutoffs of 6 ≤ BF ≤ 10, 10 < BF ≤ 30, 30 < BF ≤ 100, and BF > 100 implied substantial, strong, very strong, and decisive statistical support, respectively (Lemey et al. 2009; Bahl et al. 2011).
To determine whether the overall migration of GI and GIII differed, mean, and median migration rates per MCMC step were compared by plotting GI versus GIII rates among: all estimated rates and statistically supported estimated rates (significantly non-zero transition rates). We statistically tested the significance of rate differences (GI > GIII and GIII < GI) with a BF test. Significant differences in migration rates (r) were estimated by the ratio of posterior probabilities (P(r1 > r2 | Data)/P(r2 > r1 | Data)) by prior probabilities (P(r1 > r2)/P(r2 > r1)). For example, the posterior probability of GI mean migration rate (r1) being greater than the probability of GIII mean migration rate (r2) divided by the posterior probability of r2 being greater than r1 multiplied by the inverse estimation for the prior probability of those same rates. The prior probability was estimated with uninformative priors where the odds ratio is ∼1 (Bahl et al. 2009, 2013).
In order to validate these results, we conducted separate phylogeographic analyses of GI and GIII isolates independent of each other using the same model parameters previously described. Phylogenetic structure and migration rates for each genotype were compared with the results obtained from the joint analysis to evaluate for any sampling biases.
2.4. Estimates of viral geographic migration rates in mosquitoes
To describe the JEV phylodynamic patterns among mosquitoes independent of other hosts, a second dataset comprised of 355 isolates sampled from mosquitoes was generated from the final dataset. Analyses were conducted as described above for both GI and GIII from mosquito data. A reversible discrete phylogeographic model analogous to the one previously implemented was used to estimate GI and GIII ancestral state locations and migration rates between Asian regions and within China among mosquitoes. Similarly, this model utilized a truncated Poisson prior where 50 percent prior probability is placed on the minimal rate configuration, comprising seventeen non-zero rates connecting the eighteen locations. In addition, we compared these results with those obtained using a uniform prior ranging between the minimum and maximum migration events required to represent all locations on the trees. With this scenario, we have a maximum of 153 non-zero rates connecting the eighteen discrete locations. These locations included one ancestral state (Z) and eight locations for GI and nine locations for GIII and migrations rates between Z and GI or GIII were masked in the model. Ancestral locations at each node were estimated based on tip classification and mapped onto the MCC tree to infer migration history. The resulting phylogenetic tree structure, demographic histories, and geographic transition patterns were descriptively compared with the corresponding results estimated from all species to assess for similarities and differences in trends.
Mean and median GI versus GIII mosquito migrations rates per MCMC step were plotted to compare differences in migration patterns among: all estimated rates and statistically supported estimated rates. These mosquito plots were overlayed onto those estimated from all sampled species to infer the influence of mosquitoes on overall GI versus GIII migration rates. We also statistically tested the significance of rate differences among mosquitoes (GI > GIII and GIII < GI) with a BF test using an uninformative prior.
2.5. Contributions of host and vector data to time of most recent common ancestor (TMRCA) estimates of JEV
Active disease surveillance in mosquitoes is labor intensive and costly, and sentinel surveillance in swine along with outbreak data from humans and mammals are often relied on as a proxy for monitoring JEV activity (Hall-Mendelin et al. 2010). To assess the contributions of host data on estimates of JEV phylogenetic structure and dates, several datasets were generated with decreasing proportions of mosquito data (Table 1). Mosquito taxa (GI and GIII) were randomly subsampled in graduated proportions relative to the full sample size and removed from the full inclusive dataset. Human/mammal taxa were retained in all the generated datasets as well as any taxa coded as ancestral in order to preserve the overall JEV genotypic structure. Phylogenetic analyses were conducted as described above. TMRCA and Bayesian Confidence Intervals (BCI) ranges were estimated from the resulting MCC trees for each subsampled dataset and compared with those derived from analyses of the full dataset and mosquito-only data.
Table 1.
TMRCA estimates by host contribution.
| Dataset size (n) | Mosquitoa (n/% of dataset) | Human/mammal (n/% of dataset) | TMRCA (95% BCI) |
||||
|---|---|---|---|---|---|---|---|
| JEV overall | GI | GI-a | GI-b | GIII | |||
| 551b | 341/62% | 153/28% | 1879 (1848–1909) | 1935 (1895–1945) | 1960 (1949–1970) | 1962 (1952–1970) | 1914 (1886–1919) |
| 351 | 140 / 40% | 153/44% | 1496 (1062–1817) | 1918 (1881–1944) | 1950 (1932–1966) | 1957 (1942–1971) | 1891 (1860–1913) |
| 301 | 90/30% | 153/51% | 1583 (1156–1859) | 1918 (1888–1944) | 1950 (1932–1965) | 1956 (1938–1970) | 1883 (1860–1912) |
| 265 | 54/20% | 153/58% | 1418 (761–1812) | 1911 (1868–1935) | 1946 (1927–1962) | 1954 (1935–1969) | 1883 (1851–1909) |
| 234 | 23/10% | 153/65% | 1254 (634–1793) | 1902 (1854–1937) | 1937 (1916–1963) | 1919 (1887–1941) | 1875 (1833–1906) |
| 211 | 0/0% | 153/73% | 1198 (530–1649) | 1892 (1864–1941) | 1933 (1909–1959) | 1942 (1918–1966) | 1869 (1831–1903) |
| 355c | 341/96% | 0/0% | 1863 (1788–1925) | 1943 (1922–1957) | 1968 (1958–1975) | 1964 (1956–1971) | 1929 (1911–1944) |
aIncludes GI and GIII mosquito taxa only and does not include mosquito taxa coded to the ancestral state. bFull dataset; cmosquito-only dataset.
3. Results
3.1. Dataset
To reconstruct the phylogenetic and migration history, we downloaded all available JEV isolates (n = 567) from GenBank. A preliminary phylogenetic analysis of the dataset revealed fifteen sequences to be singletons (n = 2 were GI and n = 13 were GIII; Supplementary Table S1), or single tips representing a node. The variance distribution of root-to-tip distances for each tip confirmed these taxa to be outliers, possibly associated with sequencing errors. We therefore excluded these sequences from further analysis. Gross phylogenetic tree topology and divergence time estimates were compared from analyses conducted both with and without these gene sequences. In the final inclusive dataset (n = 551), genotypes GI–GV were represented, with the majority of isolates belonging to GI and GIII (Supplementary Table S2).
Japan was the geographic location with the highest number of both GI and GIII isolates. Mosquitoes were the most represented host in both genotypes (73 percent of GI data; 50 percent of GIII data) and accounted for over 62 percent of the final dataset (Supplementary Table S3). Although humans accounted for 18 percent of isolates identified as GIII, they only comprised about 3 percent of the GI dataset. In contrast, mammals (i.e. swine, boar, horses, and bats) made up a higher proportion of GI’s dataset (23 percent) than GIII’s (13 percent), and may be indicative of differences in either epidemiology or surveillance efforts in detecting these genotypes. For our analyses, we make no assumptions regarding these host discrepancies and therefore combine human and other mammal isolates for GI (n = 80) and GIII (n = 73) in order to avoid sampling bias.
Stratification of the mosquito-only dataset (n = 355) by region indicated some differences in geographic distributions of the samples compared to the inclusive dataset (Supplementary Table S4). GI isolates from mosquitoes were more evenly distributed between regions. GIII isolates from mosquitoes were mostly from Taiwan. All JEV genotypes were represented in the mosquito dataset.
3.2. Phylogenetic reconstruction and relative genetic diversity
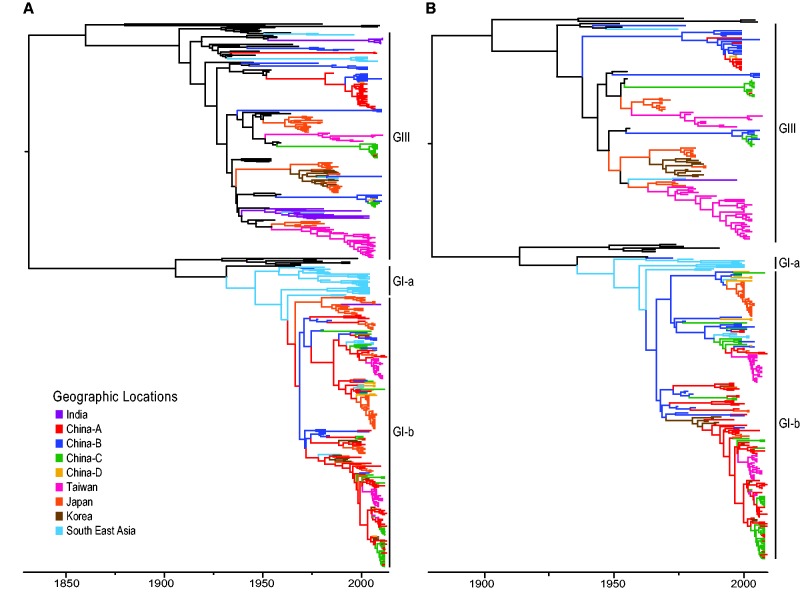
We used a Bayesian relaxed clock phylogenetic approach to account for uncertainty in tree estimation and divergence time estimates. Phylogenies estimated from the full and mosquito-only datasets both indicate the GI ancestor was younger than GIII, and GI was comprised of two major clades (GI-a, GI-b) as described previously (Fig. 1A and B; Supplementary Figs. S1 and S2) (Schuh et al. 2013). While GI-a represented the smaller of the two clades and was confined to Southeast Asia, GI-b represented the largest GI clade and appeared to be genetically homogenous across locations with little obvious geographic structuring of this lineage. In contrast, recent GIII outbreaks exhibited long branches leading to their common ancestor, suggesting missing data. The GIII common ancestor was also significantly older than other circulating genotypes. While GIII isolates were monophyletic, regional outbreaks were paraphyletic and dominated by isolates from few locations. Each analysis showed GI epidemics emerged recently and were widespread; in contrast, GIII outbreaks were older, highly structured, and unlinked to other locations, suggesting localized infectious sources originating from an unsampled ancestral population.
Figure 1.
Phylogenetic tree of the major circulating genotypes in Asia (GI and GIII) inferred from (A) the full dataset of all sampled species, and (B) mosquito-only dataset. Branches are colored by ancestral state locations. GII, GIV, GV viruses, and all samples isolated before 1970 were assigned to the ancestral population from which all JEV genotypes emerged.
A comparison of the projected TMRCA dates for all JEV genotypes, GI, and GIII between the all-species and mosquito-only analyses suggests that the date estimates were close in proximity despite mosquitoes comprising only 62 percent of the full dataset (Table 1; Fig. 1A and B, Supplementary Figs. S1 and S2). Although dates approximated from mosquito-only data were slightly younger than those from the full dataset, the BCI for mosquito results overlapped with those estimates produced by the inclusive dataset with comparable uncertainty ranges. We analyzed the TMRCA projections from subsampled datasets with decreasing proportions of mosquito-only data to assess the role of active surveillance data in JEV tree estimation (Table 1; Supplementary Figs. S3–S7). As the proportion of mosquito data decreased, the resulting TMRCA estimates became older with increased uncertainty for the date estimates. Multiple replicates of the randomly subsampled mosquito dataset produced similar results. A comparison of the dates produced by each dataset suggests the mosquito-only analysis resulted in TMRCA estimates that were most similar to those of the full dataset.
3.3. Ancestral state reconstruction of migration history and rates
In our models of ancestral location history, we simultaneously estimated GI and GIII migration patterns given a particular phylogenetic tree using both datasets. While our analysis assumed a constant migration rate across the phylogeny, the migration rates and locations of GI and GIII were independent. The inclusive dataset was comprised of GI taxa isolated from nine regions, whereas GI isolated from mosquitoes were collected in eight regions. Both datasets consisted of GIII taxa isolated from nine regions since 1970 (Supplementary Tables S2 and S4). We also included all other genotypes and classified viruses isolated before 1970 as part of an ancestral population. These were included in the analysis to maintain phylogenetic tree structure. Therefore, our model estimated 36 migration rates each for GI and GIII among nine locations, and eighteen migration rates from the ancestral population to both GI and GIII’s observed locations. The mosquito model estimated twenty-eight migration rates among eight locations for GI, thirty-six migration rates among nine locations for GIII, and seventeen migration rates from the ancestral population to both GI and GIII’s observed locations. Although our models did allow for transitions between GI and GIII, these were non-overlapping location sets such that given the data, these rates were 0. We used a BSSVS to reduce the number of possible parameters to those with the highest probability and to determine significant migration events (Lemey et al. 2009).
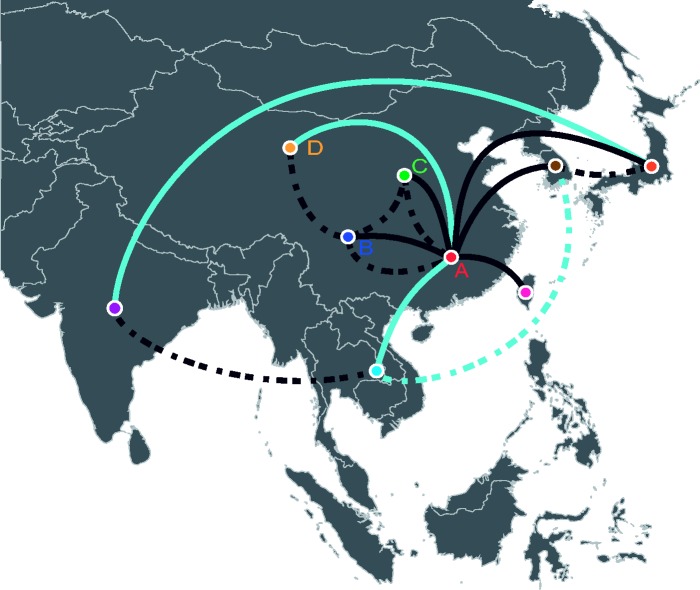
Results based on all sampled species show eight statistically supported migration events for GI and six for GIII (Fig. 2; Tables 2 and 3). An additional four independent emergent events from the ancestral population to GIII were supported. Among mosquitoes, eight mosquito migration events between geographic states were statistically supported for GI, seven for GIII, and three independent transitions between the ancestral population and GIII (Supplementary Tables S5 and S6). The majority of these mosquito transitions were also supported in the analysis of all sampled species. Exceptions were GI transitions between China-B (south mid-China) and other China regions and GIII transitions between Taiwan and Japan that were supported in the mosquito-only analysis but not in the larger analysis (Tables 2 and 3, Supplementary Tables S5 and S6). Several GI transitions were also supported among all sampled species but not in mosquitoes. These include migration events between China-A (south China) and Southeast Asia, China-A and China-B, and India and Japan. The India and Japan transition was characterized by a long tree branch spanning 20 years (Figs. 1 and 2; Tables 2 and 3, and Supplementary Tables S5 and S6).
Figure 2.
Map of discrete geographic states associated with observed location of isolation. Statistically supported migration transitions between geographic states are shown for GI (solid line) and GIII (dashed line). Dark blue lines represent migration transitions supported among all species and among mosquitoes. Light blue lines represent migration transitions supported among all species but not among mosquitoes.
Table 2.
Estimated mean GI migration rates between sampling locations among all sampled species
| India | China-A | China-B | China-C | China D | Taiwan | Japan | Korea | |
|---|---|---|---|---|---|---|---|---|
| SE Asia | 0.20 | 1.08a (0.21, 2.11) | 1.05c | 0.34 | 0.49 | 0.19 | 0.32 | 0.25 |
| India | – | 0.33 | 0.35 | 0.40 | 0.40 | 0.38 | 0.47b (0.02, 1.02) | 0.36 |
| China-A | – | 2.78b,c (0.72, 4.80) | 3.91b,c (1.75, 6.31) | 1.26b (0.24, 2.54) | 0.92b,c (0.16, 1.83) | 2.06b,c (0.77, 3.57) | 1.88b,c (0.65, 3.35) | |
| China-B | – | 1.29c | 0.94c | 0.24 | 0.82 | 0.32 | ||
| China-C | – | 0.55 | 0.24 | 0.24 | 0.33 | |||
| China D | – | 0.25 | 0.28 | 0.31 | ||||
| Taiwan | – | 0.23 | 0.32 | |||||
| Japan | – | 0.56 |
BSSVS statistically supported migration rates in bold with 95% BCI, where the BF was >3 and were observed in at least 50% of the sampled phylogenies. a10 > BF ≥ 30; bBF > 100. cTransitions supported among mosquitoes (see Supplementary Table S5).
Table 3.
Estimated mean GIII migration rates between sampling locations among all sampled species
| SE Asia | India | China-A | China-B | China-C | China D | Taiwan | Japan | Korea | |
|---|---|---|---|---|---|---|---|---|---|
| SE Asia | – | 0.60a,d (0.02, 1.42) | 0.36 | 0.57 | 0.42 | 0.37 | 0.29 | 0.43 | 0.51a (0.01, 1.24) |
| India | – | 0.29 | 0.33 | 0.33 | 0.26 | 0.26 | 0.21 | 0.26 | |
| China-A | – | 1.03c,d (0.09, 2.23) | 0.86c,d (0.06, 1.88) | 0.56d | 0.24 | 0.25 | 0.32 | ||
| China-B | – | 0.61b,d (0.02, 1.42) | 0.84c (0.09, 1.76) | 0.28 | 0.27 | 0.40 | |||
| China-C | – | 0.45 | 0.29 | 0.30 | 0.34 | ||||
| China-D | – | 0.21 | 0.24 | 0.34 | |||||
| Taiwan | – | 0.49d | 0.29 | ||||||
| Japan | – | 1.38c,d (0.33, 2.69) |
BSSV statistically supported migration rates in bold with 95% BCI, where the BF was >3 and were observed in at least 50 percent of the sampled phylogenies. a10 > BF ≥ 30; b30 > BF ≥ 100; cBF > 100. dTransitions supported among mosquitoes (see Supplementary Table S6).
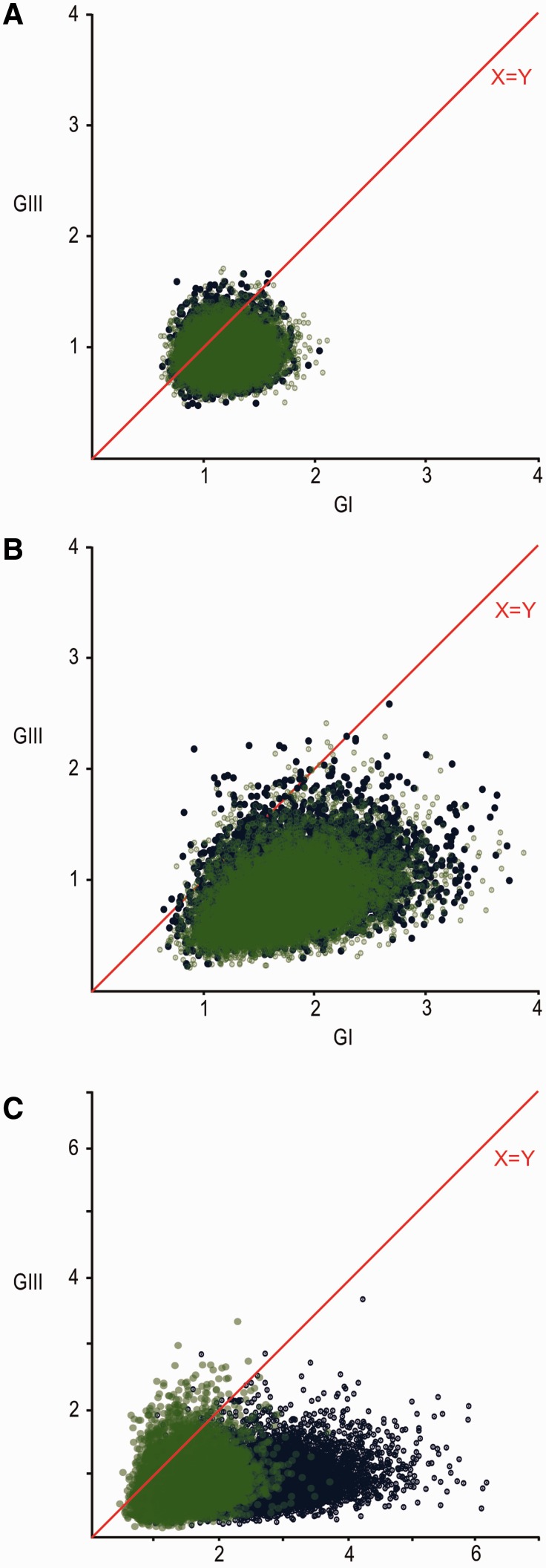
To determine whether the overall migration of GI and GIII differed, mean and median migration rates among all species and among mosquitoes were estimated per MCMC step and compared by plotting GI versus GIII rates for all generated rates and for statistically supported rates (significantly non-zero transition rates determined from the BSSVS). Significant differences between mean migration rates were determined by a BF test. Across all species, the results indicate support for GI migrating faster than GIII based on all estimated rates (Fig. 3A; GI > GIII BF = 4.088; GIII > GI BF = 0.245). Most of these migration rates were close to 0 and not statistically supported. Therefore, the mean GI versus GIII migration rates shown in Fig. 3A show less variance than those supported rates shown in Fig. 3B. Comparison of significantly supported migration rates showed very strong support for mean GI rates being greater than GIII (Fig. 3B; GI > GIII BF = 42.484; GIII > GI BF = 0.024) collectively across all species.
Figure 3.
GI versus GIII mean migration rates per MCMC step: (A) amongst all estimated rates, (B) among statistically supported estimated rates, and (C) among all estimated rates within Mainland China. Blue points represent migration rates estimated across all sampled species; green points represent migration rates estimated from data sampled from mosquitoes only.
Among mosquitoes, GI’s diffusion rate was estimated to be faster than the GIII rate for all estimated mean rates (Fig. 3A; GI > GIII BF = 4.058; GIII > GI BF = 0.246), and significantly non-zero rates (Fig. 3B; GI > GIII BF = 63.008; GIII > GI BF = 0.016) as well. An overlay of the mosquito and all species scatter plots (Fig. 3A–C) shows migration rates estimated from all species data compared with those for mosquitoes-only slightly skewed toward GI.
Since mosquitoes are the primary vector for JEV transmission, we conducted a supplemental ancestral host reconstruction to assess whether the available surveillance information is sufficient to capture the simple vector–host transmission patterns and estimate the mosquito rather than other hosts as the viral ancestor in light of sampling limitations (see Supplementary material for analysis details). The results suggest that most of the GI-b ancestors were estimated to be mosquito vectors (Supplementary Fig. S8). Several GI-b clusters were estimated to be all human/mammal ancestors although these were primarily sampled from the same country and year and may represent outbreaks that did not incorporate mosquito surveillance. GIII consisted of a larger proportion of estimated human/mammal ancestors than GI, and along with the long branches preceding many isolates, suggests surveillance of this genotype has been haphazard.
3.4. GI and GIII viral migration to and from Mainland China
Our ancestral state reconstructions suggest the GI-b lineage emerged several times from China relatively recently and emerging lineages in neighboring regions, such as Southeast Asia, became localized once the population was established. By subdividing Mainland China, we were able to further explore the migration dynamics between the country’s regions using both datasets. In contrast to the localized outbreaks observed in neighboring countries, both analyses indicate the GI lineages within China mixed rapidly, suggesting a large homogenous source population from which outbreaks in these other regions may be sourced (Figs. 1A, B and 2). GI and GIII were also shown to co-circulate in Mainland China. Mean migration rates within China per MCMC step suggest GI migrates faster than GIII across all sampled species (Fig. 3C; BF GI > GIII = 26.878; GIII > GI = 0.037 among all rates in China; GI > GIII BF = 346.870, GIII > GI BF = 0.003 among statistically supported rates only), which was reflected among mosquitoes as well but with lower statistical support (BF GI > GIII = 13.364; GIII > GI = 0.075 among all rates in China; GI > GIII BF = 38.414, GIII > GI BF = 0.026 among statistically supported rates only). Migration events from China into other Asian regions were statistically supported for GI, but no evidence supports GIII migration from China into other regions for either analyses (Tables 2 and 3, Supplementary Tables S5 and S6).
4. Discussion
Recent human outbreaks of JEV in India highlight that a comprehensive surveillance strategy is vital for detecting viral emergence and monitoring its spread before an outbreak occurs (Travasso 2014). In this study, we aim to describe JEV’s spatial diffusion history and host transition patterns from all available data collected through mixed-surveillance methods. We then evaluate the feasibility of using mosquito-only data collected through active surveillance to recover analogous patterns. Our results suggest that evolutionary and spatial diffusion patterns characterizing JEV disease dynamics may be recovered from analyses of mosquito-only data. TMRCA date estimates for all JEV genotypes, GI and GIII inferred from all available JEV data and a subsample of mosquito-only data were comparable with overlapping uncertainty ranges. Both analyses suggest GI is younger than GIII, which supports the observation that GI emerged in Asia recently (Nga et al. 2004). The dates projected from both analyses appear to be more recent than what others have reported, although this may be due to different model settings implemented or dataset construction (Schuh et al. 2013, 2014). Decreasing the proportion of mosquito data from the analyses, and thereby increasing the contribution of human/mammal data, resulted in older root dates with greater uncertainty that were more indicative of previously reported estimates (Soloman et al. 2003; Schuh et al. 2013, 2014). Overall, dates estimated from mosquito data alone were similar to those from the full dataset and illustrates the importance of incorporating this vector information in reconstructing the disease history.
The JEV phylogeny from mosquito data also recovered the population structure patterns observed in the phylogeny estimated from all sampled species. Epidemics belonging to the GI-b clade were genetically homogenous, widespread throughout Asia, and likely emerged recently from a source population in China. In contrast, GIII epidemics appear highly structured and unlinked to other regions, suggesting localized infectious sources originating from an unsampled ancestral population. Long branches leading back to their common ancestor also preceded these outbreaks and may be the result of limited and inadequate surveillance (Schuh et al. 2013). The increased fragmentation observed in GIII populations over time may lead to the emergence of novel GIII variants that diverge from the ancestral population due to limited mixing between circulating populations.
Our study indicates that active surveillance of mosquitoes could recover the majority of JEV GI and GIII migration events between regions that were supported in analyses using all available data. A few transitions were not captured by the mosquito analyses and are likely the result of outbreak surveillance dominated by mammal or human data. Our results further show that GI migrated significantly faster between regions than GIII. Similarly, the rate of mixing in China was faster for GI than GIII. In concurrence with previous studies, our analyses indicate GI may have emerged from Mainland China and seeded local outbreaks in neighboring regions (Gao et al. 2013; Schuh et al. 2014). This suggests that Mainland China may be a transmission hub for GI. In contrast, GIII movement from China into other regions was not supported. Reasons for GI’s faster migration remain enigmatic although it is possible that its rapid spread contributed to GIII’s replacement. Possible mechanisms for this difference include rapid industrialization of livestock production, particularly swine (Oh and Whitley 2011), by natural movement such as wind-blown mosquitoes (Ritchie and Rochester 2001), or in another host such as birds, which have been implicated in the life history of this virus (Buescher and Scherer 1959). In vitro studies have demonstrated higher replication efficiency in mosquito cells of GI compared with GIII (Schuh et al. 2013, 2014). This offers an intriguing mechanism that may explain the disease threat associated with GI, but may also underlie the dynamics of spread within the mosquito population.
The capacity for genomic data from mosquitoes to recapitulate overall disease dynamics may be useful in addressing drawbacks associated with current JEV surveillance strategies. Swine are often deployed as sentinel hosts to monitor for JEV movement and to serve as an early warning indicator for impending human outbreaks (MacKenzie, Gubler, and Peterson 2004; Arai et al. 2008; Hall-Mendelin et al. 2010; Duong et al. 2011; Borah et al. 2013). Although it has the benefit of being cost-effective, sentinel host strategies have disadvantages that compromise its efficacy as a surveillance tool. It requires intensive animal husbandry to maintain a source of immuno-naïve swine, and swine contribute to the transmission cycle as amplifying hosts (Hall-Mendelin et al. 2010). Several studies suggest that swine are not good predictors of JEV risks to humans. High swine seroconversion levels were associated with little evidence of human JEV infection in Sri Lanka (Peiris et al. 1993). Human cases have also appeared in regions that do not have extensive swine keeping (Bi, Zhang, and Parton 2007; Konishi et al. 2009; Nitatpattana et al. 2011; See et al. 2002; Ting et al. 2004), suggesting that other factors, especially unsampled hosts, may contribute to the transmission ecology (Lord, Gurley, and Pulliam 2016).
As a result of these disadvantages, attention has been placed on vector surveillance as a potential predictive system (van den Hurk et al. 2012). Peaks in mosquito infection rates have been shown to precede seroconversion peaks in sentinel swine by 1–2 months, and precede human cases by 2–3 months (Borah et al. 2013). Monitoring for JEV activity among mosquito populations may provide public health officials more time to prepare for impending outbreaks.
To best inform public health efforts, it is essential for surveillance strategies to establish baseline levels of arborovirus activity and routinely monitor for emerging strains. Nevertheless, the appearance of novel strains is often recognized only after human or animal infections, such as the introduction of JEV into northern Australia (Hanna et al. 1996; van den Hurk et al. 2012). Similarly, GI began to supplant GIII as the dominant JEV strain circulating during the 1990s, although this shift was not recognized until the early 2000s (Nga et al. 2004). Since mosquitoes are the primary transmission vectors, viral genomic data from this population are most suited for identifying novel strains (van den Hurk et al. 2012). Mosquito surveillance is also conducted actively with large sample sizes and is a source of robust data that may be less subject to biases.
Overall, our results suggest that data from mosquito surveillance can adequately reflect JEV disease dynamics, although it does have limitations. Low prevalence and elaborate processing steps, such as species sorting and virus extraction, make mosquito surveillance labor intensive. To address these issues, techniques have recently been developed to easily extract viral RNA from mosquito expectorate during feeding (Hall-Mendelin et al. 2010; van den Hurk et al. 2012). Our results also show that while the higher GI migration rate in mosquitoes can account for much of the migration patterns observed, not all disease movement can be attributed to the vector host. Some movement between locations may be attributable to the long-distance migrations of infected livestock (such as domestic poultry that can exhibit high viremia of JEV), wild birds, or unsampled host species that were not captured by our model due to poor surveillance or limited comprehension of their role in the disease ecology (Buescher and Scherer 1959; MacKenzie, Gubler, and Peterson 2004; Le Flohic et al. 2013; Lord, Gurley, and Pulliam 2016). This indicates that while sampling in mosquitoes appears to be sufficient to recover and inform most JEV disease patterns, it should be considered in conjunction with surveillance data from other hosts for the most informative interpretation. Thus, recommendations for comprehensive surveillance programs to protect against and monitor JEV should include systematic surveillance in both mosquitoes and mammalian hosts in regions of high endemicity.
4.1. Data availability
Collection metadata and GenBank accession numbers are provided in the Supplementary material. The BEAST xml file for the phylogeographic reconstruction of ancestral locations used in this study is available at http://dx.doi.org/10.5061/dryad.11c40.
Supplementary data
Supplementary data are available at Virus Evolution online.
Acknowledgements
This study was supported by National Mega Projection Major Drug Development (2009ZX09301-014) and Wuhan Institute of Biological Products Co., Ltd. T.P. and J.B. are supported by funds provided by the University of Texas School of Public Health.
Conflict of interest. None declared.
References
- Arai S., et al. (2008) ‘Japanese Encephalitis: Surveillance and Elimination Effort in Japan From 1982 to 2004’, Japanese Journal of Infectious Diseases, 61: 333–9. [PubMed] [Google Scholar]
- Bahl J., et al. (2009) ‘Gene Flow and Competitive Exclusion of Avian Influenza A Virus in Natural Reservoir Hosts’, Virology, 390/2: 289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl J., et al. (2011) ‘Temporally Structured Metapopulation Dynamics and Persistence of Influenza A H3N2 Virus in Humans’, Proceedings of the National Academy of Sciences, 108/48: 19359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl J., et al. (2013) ‘Influenza A Virus Migration and Persistence in North American Wild Birds’, PloS Pathogens, 9/8, e1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi P., Zhang Y., Parton K. A. (2007) ‘Weather Variables and Japanese Encephalitis in the Metropolitan Area of Jinan City, China’, Journal of Infectious, 55: 551–6. [DOI] [PubMed] [Google Scholar]
- Borah J., Dutta P., Khan S. A., Mahanta J. (2013) ‘Epidemiological Concordance of Japanese Encephalitis Virus Infection Among Mosquito Vectors, Amplifying Hosts and Humans in India’, Epidemiology and Infection, 141: 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher E. L., Scherer W. F. (1959) ‘Ecologic Studies of Japanese Encephalitis Virus in Japan. IX. Epidemiologic Correlations and Conclusions’, American Journal of Tropical Medicine Hygiene, 8: 719–22. [DOI] [PubMed] [Google Scholar]
- Campbell G. L., et al. (2011) ‘Estimated Global Incidence of Japanese Encephalitis: A Systematic Review’, Bulletin of the World Health Organization, 89: 766–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Tesh R. B., Rico-Hesse R. (1990) ‘Genetic Variation of Japanese Encephalitis Virus in Nature’, Journal of General Virology, 71: 2915–22. [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A., Shapiro B., Pybus O. G. (2005) ‘Bayesian Coalescent Inference of Past Population Dynamics From Molecular Sequences’, Molecular Biology and Evolution, 22: 1185–92. [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Dong X., Rambaut A. (2012) ‘Bayesian Phylogenetics With BEAUti and the BEAST 1.7’, Molecular Biology and Evolution 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong V., et al. (2011) ‘Evidence of Japanese Encephalitis Virus Infections in swine Populations in 8 Provinces of Cambodia: Implications for National Japanese Encephalitis Vaccination Policy’, Acta Tropica, 120: 146–50. [DOI] [PubMed] [Google Scholar]
- Gao X., et al. (2013) ‘Southernmost Asia is the Source of Japanese Encephalitis Virus (Genotype 1) Diversity From Which the Viruses Disperse and Evolve Throughout Asia’, PLoS Neglected Tropical Diseases 7/9: e2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Mendelin S., et al. (2010) ‘Exploiting Mosquito Sugar Feeding to Detect Mosquito-Borne Pathogens’, Proceedings of the National Academy of Sciences, 107/25: 11255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J. N., et al. (1996) ‘An Outbreak of Japanese Encephalitis in the Torres Strait, Australia, 1995’, Medical Journal of Australia, 165: 256–60. [DOI] [PubMed] [Google Scholar]
- Konishi E., Sakai Y., Kitai Y., Yamanaka A. (2009) ‘Prevalence of Antibodies to Japanese Encephalitis Virus Among Inhabitants in Java Island, Indonesia, with a Small Pig Population’, American Journal of Tropical Medicine and Hygiene, 80/5: 856–61. [PubMed] [Google Scholar]
- Le Flohic G., Porphyre V., Barbazan P., Gonzalaz J. (2013) ‘Review of Climate, Landscape, and Viral Genetics as Drivers of the Japanese Encephalitis Virus Ecology’, PLoS Neglected Tropical Diseases, 7/9: e2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Drummond A. J., Suchard M. A. (2009) ‘Bayesian Phylogeography Finds Its Roots’, PLoS Computational Biology, 5: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J. S., Gurley E. S., Pulliam J. R. C. (2016) ‘Rethinking Japanese Encephalitis Virus Transmission: A framework for Implicating Host and Vector Species’, PLoS Neglected Tropical Diseases, 9/12: e004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie J. S., Gubler D. J., Petersen L. R. (2004) ‘Emerging Flaviviruses: The Spread and Resurgence of Japanese Encephalitis, West Nile and Dengue Viruses’, Nature Medicine, 10/12 Suppl: S98–S109. [DOI] [PubMed] [Google Scholar]
- Martin D. P., et al. (2010) ‘RDP3: A Flexible and Fast Computer Program for Analyzing Recombination’, Bioinformatics, 26/19: 2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Environmental Protection. (2005) ‘Report on the State of the Environment in China 2004’, Environmental Information Center, Beijing, China. <http://english.mep.gov.cn/SOE/soechina2004/disaster.htm> accessed 19 Feb 2014. [Google Scholar]
- Nga P. T., et al. (2004) ‘Shift in Japanese Encephalitis Virus (JEV) Genotype Circulating in Northern Vietnam: Implications for Frequent Introductions of JEV From Southeast Asia to East Asia’, Journal of General Virology, 85: 1625–1631. [DOI] [PubMed] [Google Scholar]
- Ni H., Barrett A. D. (1995) ‘Nucleotide and Deduced Amino Acid Sequence of the Structural Protein Genes of Japanese Encephalitis Viruses From Different Geographical Locations’, Journal of General Virology, 76: 401–407. [DOI] [PubMed] [Google Scholar]
- Nitatpattana N., et al. (2011) ‘Elevated Japanese Encephalitis Virus Activity Monitored by Domestic Sentinel Piglets in Thailand’, Vector-borne and Zoonotic Diseases, 11/4: 391–394. [DOI] [PubMed] [Google Scholar]
- Oh S. H., Whitley N. C. (2011) ‘Pork Production in China, Japan and South Korea’, Asian-Australian Journal of Animal Science, 24/11: 1629–1636. [Google Scholar]
- Peiris J. S. M., et al. (1993) ‘Japanese Encephalitis in Sri Lanka: Comparison of Vector and Virus Ecology in Different Agro-climatic Areas’, Transactions of the Royal Society of Tropical Medicine & Hygiene, 87/5: 541–548. [DOI] [PubMed] [Google Scholar]
- Ritchie S. A., Rochester W. (2001) ‘Wind-Blown Mosquitoes and Introduction of Japanese Encephalitis Into Australia’, Emerging Infectious Diseases, 7/5: 900–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A. J., Ward M. J., Leigh Brown A. J., Barrett A. D. (2014) ‘Dynamics of the Emergence and Establishment of a Newly Dominant Genotype of Japanese Encephalitis Virus Throughout Asia’, Journal of Virology, 5: 4522–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A. J., Ward M. J., Leigh Brown A. J., Barrett A. D. (2013) ‘Phylogeography of Japanese Encephalitis Virus: Genotype Is Associated With Climate’, PLoS Neglected Tropical Diseases, 7: e2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See E., et al. (2002) ‘Presence of Hemagglutination Inhibition and Neutralization Antibodies to Japanese Encephalitis Virus in Wild Pigs on an Offshore Island in Singapore’, Acta Tropica, 81/3: 233–236. [DOI] [PubMed] [Google Scholar]
- Solomon T., et al. (2003) ‘Origin and Evolution of Japanese Encephalitis Virus in Southeast Asia’, Journal of Virology, 77/5: 3091–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S. H., et al. (2004) ‘Seroepidemiology of Neutralizing Antibodies to Japanese Encephalitis Virus in Singapore: Continued Transmission Despite Abolishment of Pig Farming?’, Acta Tropica, 92: 187–191. [DOI] [PubMed] [Google Scholar]
- Travasso C. (2014) ‘Indian Health Ministry Orders Encephalitis Vaccination in Select Districts After More Than 500 Deaths’, British Medical Journal, 348: g4209. [DOI] [PubMed] [Google Scholar]
- Van den Hurk A. F., Ritchie S. A., MacKenzie J. S. (2009) ‘Ecology and Geographical Expansion of Japanese Encephalitis Virus’, Annual Review of Entomology, 54: 17–35. [DOI] [PubMed] [Google Scholar]
- Van den Hurk A. F., Ritchie S. A., MacKenzie J. S. (2012) ‘Evolutions of Mosquito-Based Arbovirus Surveillance in Australia’, Journal of Biomedicine and Biotechnology, 2012: 325659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.