Abstract
Current methods for engineering enzymes modify enzymes themselves and require a detailed mechanistic understanding or a high-throughput assay. Here, we describe a new approach where catalytic properties are modulated with synthetic binding proteins, termed monobodies, directed to an unmodified enzyme. Using the example of a β-galactosidase from Bacillus circulans, we efficiently identified monobodies that restricted its substrates for its transgalactosylation reaction and selectively enhanced the production of small oligosaccharide prebiotics.
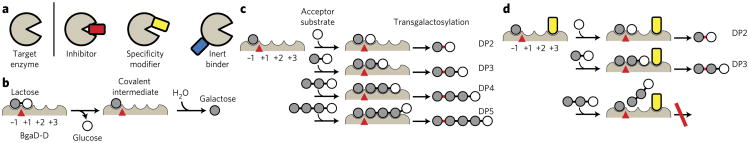
Engineering enzymes with desirable catalytic properties remains a major goal of biotechnology. Essentially all standard enzyme-engineering approaches, such as structure-guided design and directed evolution1–5, modify the enzyme itself. However, rational and directed evolution efforts can be hampered if the structure and catalytic mechanism of the target enzyme or a close homolog are unknown, or if the protein is poorly suited to expression in heterologous hosts or high-throughput screening. De novo enzyme design, still in its early stages, also requires detailed understanding of the reaction mechanism6. We wondered whether an alternative approach might be possible in which the target enzyme is unchanged; we hypothesized that, instead, synthetic binding proteins that bind to the target enzyme and modify its activity could be found through high-throughput protein design technologies. Among various classes of binding proteins defined in terms of their effect on enzyme and their respective utilities (Fig. 1a), we focused the present study on altering the substrate specificity, a generally challenging task in protein engineering.
Figure 1. Modulation of enzyme catalytic properties with synthetic binding proteins.
(a) Schematic drawing of different classes of binding proteins. (b–d) Schematic representations of the hydrolysis reaction (b) and of the transgalactosylation reaction of BgaD-D in the absence (c) or the presence (d) of a specificity modifier (yellow). Hypothesized subsites are labeled −1, +1, +2 and +3. The catalytic site where hydrolysis and transgalactosylation reactions occur is shown as a red triangle. In c and d, the initial step of lactose cleavage is omitted, and the reactions from the covalent intermediate are shown for brevity. A sugar linkage newly formed by the transgalactosylation reaction is marked in red. DP2, DP3, DP4 and DP5 denote di-, tri-, tetra- and pentasaccharides, respectively. The specificity modifier restricts the access of larger oligosaccharides to the subsites (d). The inhibition of DP4 production naturally leads to the inhibition of the production of DP5 and larger oligosaccharides (not shown).
We tested our approach on a β-galactosidase from B. circulans ATCC 31382. This enzyme catalyzes lactose hydrolysis and transgalactosylation reactions (Fig. 1b,c, respectively)7. Because its transgalactosylation reactions yield galacto-oligosaccharides (GOSs) that are recognized as beneficial prebiotics8, the enzyme is used for industrial production of GOSs under the trademark Biolacta. However, because GOSs of any length can be the substrate of the transgalactosylation reaction (Fig. 1c), the enzyme produces a wide range of these compounds, leading to low yields of GOSs of a specific size. Such broad substrate specificity is observed for many enzyme families that form and/or break biopolymers, including proteases, glycosidases and lipases. Tailoring the substrate specificity of these classes of enzymes is often desired but challenging.
We envisioned enhancing the production of short GOSs by engineering the β-galactosidase in a way that prevented the enzyme from using large GOS species as acceptor substrates. Cloning and functional expression of this enzyme in Escherichia coli have recently been reported7, and four residues that constitute the putative active site have been identified on the basis of inactivating mutations9. It is a large protein, and its shortest active fragment used in this work was ~100 kDa (termed BgaD-D; see Online Methods)10. Its three-dimensional structure in the apo form has very recently been reported11 after the completion of the experiments described here. It has low homology, even at positions surrounding the active site, to well-characterized members—such as E. coli LacZ (25% sequence identity)—of the GH2 glycosidase family to which BgaD belongs7. Moreover, unlike LacZ, BgaD does not require Mg or Na ions for catalysis7. Although BgaD probably utilizes the same catalytic mechanism as other members of the GH2 family, the molecular details of its substrate recognition remain to be elucidated. Furthermore, no high-throughput assays for profiling GOSs are available. Thus, it has been challenging to alter catalytic properties of BgaD-D using conventional approaches. In fact, we have generated more than 1,000 point mutations at positions surrounding the putative active site predicted from homology modeling of the BgaD-D structure (saturation mutagenesis at a total of 54 positions and additional combinations; see Supplementary Results, Supplementary Fig. 1), but we have identified only mutations that affect the catalytic efficiency and none that alter the profile of oligo-saccharide products.
We designed our strategy according to the general active site architecture of glycoside hydrolases and transferases12–15. We hypothesized that BgaD-D has subsites (−1, +1, +2, +3 and so on) that each recognize a glycosyl residue, with hydrolysis and transgalactosylation occurring using the same catalytic site located between −1 and +1 (Fig. 1b,c). We further hypothesized that blocking subsite +3 diminishes the production of tetrasaccharides and larger oligosaccharides (DP4, DP5 and so on) while minimally affecting lactose hydrolysis and trisaccharide (DP3) production (Fig. 1d). Structure-guided mutagenesis directed to subsites distant from the catalytic residues has been used successfully to alter the substrate specificities of other glycosyltransferases and other enzyme types16–18, supporting our strategy. However, we emphasize that we have no knowledge of the presence and precise locations of such subsites in BgaD-D. These hypotheses, in turn, posit that binding proteins with desired specificity-enhancing characteristics should bind to the close vicinity of the catalytic center but should not inhibit lactose hydrolysis.
We generated a series of monobodies, synthetic binding proteins based on the tenth human fibronectin type III (FN3) domain (Supplementary Fig. 2a), directed to BgaD-D by performing combinatorial library selection using phage and yeast display technologies as previously established19,20. The monobody system has generated numerous potent and specific binding proteins21. Monobodies have a strong tendency to bind to a functional site in the target protein21–23, and we envisioned that their small size might be beneficial in precisely targeting a specific subsite. The initial unbiased selection yielded a total of 68 monobodies (Supplementary Table 1). On the basis of phylogenetic analysis of these monobodies (Supplementary Fig. 3a), we produced 11 clones representing each ‘family’ of these monobodies as purified proteins. We identified monobodies, represented by Mb(BgaD_L02), Mb(BgaD_S09) and Mb(BgaD_S10), that bound strongly to three distinct epitopes of BgaD-D in the absence of substrates, as determined in competitive binding experiments using purified monobody samples (Supplementary Fig. 4). (For brevity, hereafter we use abbreviated names for monobodies from which “BgaD_” is omitted.) However, none showed significant effects on BgaD-D catalytic activity (Supplementary Fig. 4), suggesting that they are inert binders binding to locations distant from the active site (Fig. 1a and Supplementary Fig. 2b). The absence of inhibitors in the initial pool of monobodies was surprising given our previous experience, but this result could be rationalized by the fact that BgaD-D is a large protein containing sugar-binding domains in addition to the catalytic domain. In parallel, we attempted to select for monobodies whose binding was displaced by oligosaccharides, but were unsuccessful, probably because of the low affinity and hence incomplete blockage of the active site by oligosaccharides.
To obtain monobodies that bind in the close vicinity of the active site, we performed a second selection campaign in which we used the three monobodies to mask the dominant, nonfunctional epitopes (Supplementary Fig. 2b). This selection yielded six different monobodies (Supplementary Table 1), and by characterizing three of them we identified Mb(L14), which bound tightly to BgaD-D (Kd = 31 ± 2.3 nM; Supplementary Fig. 4b) and potently inhibited BgaD-D as assayed by o-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolysis, a proxy assay for the disaccharide hydrolysis activity (Supplementary Fig. 4d). These results suggest that Mb(L14) may bind to regions critical to catalysis, such a subsites −1 and/or +1 (Fig. 1b).
We then hypothesized that desired specificity enhancers should bind to the close vicinity of the active site pocket and that their epitopes may overlap with the epitope of the inhibitory monobody, Mb(L14). Accordingly, we performed the third selection campaign, in which we recovered 11 monobodies that are competed with by Mb(L14) (Supplementary Table 1 and Supplementary Fig. 2b). By characterizing seven of these in the ONPG hydrolysis assay, we obtained Mb(L19), which was competitively inhibited by Mb(L14) (Supplementary Fig. 4c) but minimally affected ONPG hydrolysis (Supplementary Fig. 4d). Because it bound weakly to BgaD-D (Kd = 600 ± 170 nM as assayed using yeast surface display; Supplementary Fig. 4a), we then improved the affinity of Mb(L19) by error-prone PCR and directed evolution. A new monobody, Mb(L23), had higher affinity (Kd = 12 ± 3 nM as assayed using yeast surface display) and maintained the desired binding profile of Mb(L19) (Supplementary Fig. 4). Neither Mb(L19) nor Mb(L23) competed with the three inert monobodies, as expected (Supplementary Fig. 4c).
Next, we characterized these monobodies to determine whether they alter the substrate specificity of the enzyme as we hypothesized (Fig. 1d). Their effects are shown in plots of the amount of each oligosaccharide versus reaction time and also the amount of consumed lactose (Fig. 2a; Supplementary Figs. 5 and 6). The latter is a common presentation format for comparing oligosaccharide productivity that compensates for different catalytic rates (Fig. 2a, right column)10,24. Mb(L23) showed little effect on lactose hydrolysis and the production of mono-, di- and trisaccharides (Fig. 2a and Supplementary Fig. 6). By contrast, it greatly diminished the production of tetraoligosaccharides and larger species (DP4+) (Fig. 2a). Figure 2b compares the product profiles at the time point when the total amount of oligosaccharides reaches the maximum, a comparison that reflects the actual use of the enzyme (because of the galactosidase activity of the enzyme, longer reaction times beyond this point lead to GOS hydrolysis). Notably, the BgaD-D–Mb(L23) complex produced the highest amount of trisaccharide (DP3) and produced DP4 and DP5+ species in amounts 4.5- and 30-fold lower, respectively, than were seen with BgaD-D alone or with the other monobodies (Fig. 2b). This pattern of restricted GOS production is consistent with our model (Fig. 1d).
Figure 2. Monobodies modulate catalytic properties of BgaD-D.
(a) Effects of monobodies on GOS production by BgaD-D (see Supplementary Figs. 5 and 6 for the complete data sets). The amounts of indicated sugars relative to the total amount of sugar (wt/wt) are plotted as a function of reaction time at left; the degree of lactose consumption is shown at right. (b) Relative amounts of oligosaccharides in each of the reactions shown in a, measured at the time point when the total GOS amount reaches the maximum. The degree of lactose consumption is also shown. (c) Effects of oligosaccharides on monobody–BgaD-D interaction (see also Supplementary Fig. 7b). In c, monobody names are abbreviated for brevity. In all panels, error bars indicate s.d. (n = 3), and where none are visible, the errors are within the size of the markers.
In contrast to Mb(L23), although Mb(L14) potently inhibited the hydrolysis activity of BgaD-D toward lactose and similarly reduced the rates of transgalactosylation reactions, it did not affect the oligosaccharide production profile. These results are consistent with a model in which the inhibitor monobody occupies the enzyme active site (for example, -1 and/or +1 in Fig. 1b), and only the free enzyme is catalytically active for either lactose hydrolysis or transgalactosylation. The inert monobody, Mb(L02), likewise showed no effects, as expected (Fig. 2a).
To characterize the monobodies' mechanisms of action, we tested the effects of mutations at a putative active site residue, E447 (ref. 9). Mutating this residue to Gln, Arg or Lys did not affect binding of the inert monobodies, as expected (Supplementary Fig. 7a). By contrast, these mutations all abolished binding of Mb(L14). The sensitivity of Mb(L14) to the subtle perturbation by the E447Q mutation suggests that it binds directly to the active site. The specificity-enhancing monobodies, Mb(L19) and Mb(L23), were moderately affected by the mutations, more strongly so by E447K and E447R than E447Q (Supplementary Fig. 7a), suggesting that they bind to a region near E447. Together with the binding competition results (Supplementary Fig. 4c), these results further support our model (Supplementary Fig. 2b) whereby Mb(L14) binds directly to the active site and Mb(L19) and Mb(L23) bind near the active site.
To clarify whether the specificity-modifying monobodies achieve their function by limiting access of larger substrates to the enzyme, we then tested the effects of oligosaccharides on the monobody–BgaD-D interactions. These oligosaccharides should bind to the enzyme in the mode for the products of transglycosylation by spanning from the -1 site to the positively numbered sites (Fig. 1c). Whereas oligosaccharides of different sizes all blocked the binding of the inhibitor monobody, Mb(L14), only the longer oligosaccharides, DP4 and DP5—but not DP3—inhibited the specificity-enhancing monobodies (Fig. 2c and Supplementary Fig. 7b). DP4 and DP5 are precisely the oligosaccharides whose production was reduced by the specificity-enhancing monobodies (Fig. 2b). The results strongly support the proposed mechanism of the specificity-enhancing monobodies (Fig. 1d).
Our success in altering enzyme specificity using monobodies illustrates that synthetic binding proteins can precisely modulate enzyme properties without modifying the enzyme itself. We accomplish this goal with limited knowledge of the structure-function relationship of the enzyme, in particular the recognition of large substrates, and without a high-throughput enzyme assay. Instead, we started this project with a general reaction mechanism for this class of enzymes (Fig. 1) and a back-of-the-envelope design for monobody action (Fig. 1d). The ability to define a desired profile for monobodies in terms of target binding was effective in identifying candidate monobodies and in minimizing the number of monobodies requiring detailed characterization, an important factor with our GOS profiling assay, which takes a week to complete. We successfully identified a desired specificity enhancer by performing detailed enzyme assays with only 20 monobodies. These monobodies will be powerful tools for investigating the substrate-recognition mechanism, which will help recapitulate the specificity modifying effect of the monobody with mutations within the enzyme. Although we focused this study on modulating the substrate specificity, the other two classes of monobodies (inhibitors and inert binders; Fig. 1a) will be useful tools for different types of applications. Many enzymes, including esterases and proteases, recognize substrates in an extended conformation using subsites in a manner conceptually similar to glycosyltransferases, and these types represents the majority of industrial enzymes currently in use25. We anticipate that our strategy would be applicable to restricting the substrates of these enzymes to shorter species. Therefore, this work substantially expands enzyme-engineering technologies as well as the utility of synthetic binding proteins.
Online Methods
Protein expression and purification
The commercially available Biolacta preparation (Amano Enzyme) contains four isozymes, termed BgaD-A (residues 36–1737), BgaD-B (residues 36–1422), BgaD-C (36–1249) and BgaD-D (residues 36–847), and GOSs are produced mostly by the three isozymes other than BgaD-A10. We used the smallest, BgaD-D, for all experiments in this study. BgaD-D and its active site mutants (E447Q, E447K and E447R) were prepared as a fusion protein C-terminal to a biotin-acceptor tag (Avi-tag) and His6 tag using pCold II vector (Takara). The proteins were produced as previously described7, except that the protein was produced in BL21(DE3) containing the pBirAcm plasmid (Avidity) in the presence of 50 μM d-biotin for in vivo biotinylation. Monobodies were prepared as His10-tagged proteins using the pHFT2 vector or as His6-tagged and biotinylated proteins using the pHBT vector as previously described22,26. All proteins were purified using Ni-Sepharose columns (GE Healthcare) and further purified with a Superdex size-exclusion column (Superdex200 for BgaD-D and Superdex75 for monobodies, GE Healthcare). Representative SDS-PAGE of purified samples is shown in Supplementary Figure 8. For enzyme assay experiments, we used BgaD-D without Avi-tag7 and monobodies from which the affinity tag had been removed.
Phage display and yeast-surface display
The monobody libraries used and general selection methods have been described previously20,21. The buffers used for binding reaction and washing were BSS (50 mM Tris-HCl, pH 7.4, containing 150 mM NaCl and 1 mg/mL bovine serum albumin) and BSST (BSS and 0.1% Tween 20), respectively, for both phage display and yeast surface display selection experiments. In the initial selection campaign, apo BgaD-D was used as a target and the selection was performed in an unbiased manner (Supplementary Fig. 2b). The BgaD-D concentrations used for rounds 1, 2 and 3 of phage display selection were 300 nM, 200 nM and 100 nM, respectively. Monobody-displayed phages were captured onto biotinylated target enzyme immobilized to streptavidin-coated magnetic beads (Z5481/2, Promega) and then eluted in 0.1 M Gly-HCl, pH 2.1. After gene shuffling among phage clones within each enriched population and transfer of the resulting gene pool to a yeast surface display vector, we performed library sorting using the target enzyme concentration of 100 nM, as described previously20. Phylogenetic tree analysis was performed using the program Phylogeny (EMBL-EBI)27,28 In the second selection campaign, library sorting experiments were performed as described above except that 2 μM of Mb(L02), 40 μM of Mb(S09) and 3 μM of Mb(S10) were added to the biotinylated target enzyme before mixing the target and a monobody library (Supplementary Fig. 2b). We alternated library sorting with the enzyme-monobody complex and sorting with the enzyme only to ensure that selected monobodies bound to the enzyme, not to the competitor monobodies. In the third campaign, we performed library sorting in an unbiased manner as described above and then subjected the recovered population to negative selection using 2 μM of Mb(L14) as a competitor.
Affinity of monobodies to BgaD-D was initially determined using yeast surface display, as described previously18. Yeast cells displaying a monobody were incubated with varying concentrations of BgaD-D, washed with the buffer and stained with appropriate fluorescently labeled secondary detection reagents, before analysis on a flow cytometer (Guava EasyCyte 6/L, Millipore). Kd values were determined from plots of the mean fluorescent intensity against BgaD-D concentration by fitting the 1:1 binding model using SigmaPlot software (Systat Software).
Affinity maturation using error prone mutagenesis
To generate an error prone PCR library from the Mb(L19) parent sequence, we performed PCR in the presence of 0.3 mM MnCl2, 7 mM MgCl2, 0.2 mM each of dATP, dGTP, dCTP and dTTP, and 2.5 units of Taq DNA polymerase, and constructed the library using yeast homologous recombination, as described previously20,29. The resultant yeast surface display library was subjected to selections using yeast surface display as described above.
Affinity measurements using purified proteins
Affinity of purified monobodies were determined using a bead-based assay30. Streptavidin-coated Dynabeads M280 (Invitrogen) at 20 μg/mL were incubated with an appropriate concentration of biotinylated monobody (10-30 nM) at 4 °C for 30 min with rotation, and then blocked with 10 μM d-biotin for 15 min. The monobody-immobilized beads were washed and resuspended in BSS. 10 μL of the beads solution was then transferred to a well of a 96-well filter plate (MultiScreenHTS HV, 0.45 μm pore size, Millipore) and drained by vacuum. 20 μL of biotinylated BgaD-D or its active site mutant at various concentrations (0–5,000 nM) in BSS was added to the wells of the filter plate containing the monobody-immobilized beads, and the plate was incubated at 25 °C with shaking for 20 min. The wells were drained and washed twice with BSST. After draining, 20 μL of 10 μg/mL of DyLight650-conjugated to streptavidin (Thermo) in BSS was added to each of the wells. After incubation at 4 °C with shaking for 30 min, the wells were drained and washed twice with BSST. The beads were resuspended in 180 μL of BSS, and the fluorescence emission in the far-red channel was analyzed on Guava EasyCyte 6/L. Kd values were determined from plots of the median fluorescent intensity against BgaD-D concentration by fitting the 1:1 binding model using SigmaPlot software (Systat Software). Kd values obtained using purified monobodies generally agreed with those determined using yeast surface display (Supplementary Fig. 2a and b), consistent with our previous observations20.
Preparation of purified GOS
Ten percent (wt/vol) Vivinal GOS solution (FrieslandCampina) was separated using a Bio-gel P-2 extra fine size-exclusion column (26 mm × 100 cm; Bio-Rad Laboratories) in H2O at a flow rate of 0.4 mL/min, and fractions were analyzed with MALDI-TOF mass spectroscopy as described previously10. The fractions containing disaccharides (DP2), trisaccharides (DP3), tetrasaccharides (DP4), pentasaccharides (DP5) and larger oligosaccharides were respectively pooled, lyophilized and kept at −20 °C until use.
Competition binding assay
Competition binding assay was performed using the bead-based assay with purified proteins as described above. The monobody-competition binding assay for testing the specificity of monobodies was carried out as described above except that 20 μL of an appropriate concentration of biotinylated BgaD-D pre-incubated with or without a competitor monobody was added at 200 times the Kd value for the monobody-BgaD-D interaction in BSS. The concentration of biotinylated BgaD-D used was 10 nM for Mb(L02) and Mb(S10), 30 nM for Mb(L14) and Mb(L23), 40 nM for Mb(L19) or 180 nM for Mb(S09) to account for different affinity. For the oligosaccharide-competition binding assay, the biotinylated BgaD-D was pre-incubated with or without 20% (wt/vol) of oligosaccharide (lactose, DP3, DP4 or DP5) in BSS for 5 min on ice, and then 20 μL of the mixture was transferred to the wells where monobody binding took place. Experiments were performed in triplicate.
Hydrolysis activity assay
Hydrolysis activity was assayed using 4 mM o-nitrophenyl β-d-galactopyranoside (ONPG) as a substrate in the assay buffer (50 mM sodium phosphate buffer, pH 7.4, containing 150 mM NaCl and 0.01% Triton X-100) at 25 °C. All reagents were pre-incubated at 25 °C for 10 min, and the reaction was initiated by mixing 50 μL of substrate solution containing 8 mM ONPG and 50 μL of protein solution containing 62 nM BgaD-D and/or 10 μM monobody. After 10 min incubation at 25 °C, the reactions were terminated by the addition of 250 μL of 2% (wt/vol) Na2CO3, and then the absorbance at 405 nm was measured using a SpectraMax 340PC plate reader (Molecular Devices). The absorbance for the assay solution containing the substrate but no proteins was subtracted as the background from the other reaction solutions to determine the catalytic activity. Experiments were performed in triplicate.
Quantification of GOS production
The production of GOSs was measured using 5% (wt/vol) lactose as a substrate in the assay buffer (20 mM sodium phosphate buffer, pH 7.0, containing 150 mM NaCl) at 25 °C. The reactions were initiated by mixing 125 μL of substrate solution containing 20% (wt/vol) lactose and 375 μL of protein solution containing 0.4 μM BgaD-D and/or an appropriate concentration of monobody (133–266 μM). Samples were withdrawn periodically and boiled for 10 min to terminate the reaction. The amounts of monosaccharides, disaccharides and GOSs were determined using a CK04S column (Mitsubishi Chemical) on an HPLC (LC-30AD, Shimadzu) equipped with an evaporative light scattering detector (ELSD-LTII, Shimadzu). The assay samples were eluted from the column using H2O at a flow rate of 0.4 mL/min at 80 °C. For separation and determination of lactose and other disaccharides (DP2), an Asahipak NH2P-40 3E column (Shodex) was used with a gradient of H2O (solvent A) and acetonitrile (solvent B) at a flow rate of 0.3 mL/min at 30 °C. Sugar concentrations were determined from peak areas. Glucose, galactose, lactose and 4′-galactosyllactose purchased from Wako Chemicals, and tetrasaccharides and larger oligosaccharides prepared as described above, were used as reference compounds for producing standard curves for these assays. Experiments were performed in triplicate.
Supplementary Material
Acknowledgments
We acknowledge the use of the University of Chicago Genomics Core, which is supported by US National Institutes of Health grant P30CA014599 to the University of Chicago Comprehensive Cancer Center.
Footnotes
Accession codes. The DNA sequences of the monobodies have been deposited in the GenBank Nucleotide database with accession codes KT153227–KT153250.
Author Contributions: S.T. and S. Koide designed the research. S.T. and A.K. generated and characterized the monobodies. S.T. and T.T. analyzed effects of the monobodies on enzyme reactions. T.T. and S.I. conducted mutagenesis analysis of BgaD. S. Koikeda and S. Koide supervised the research. S.T. and S. Koide wrote the manuscript and the other authors commented on it.
Competing Financial Interests: The authors declare competing financial interests: details accompany the online version of the paper.
Additional Information: Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Wilks HM, et al. Science. 1988;242:1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]
- 2.Chen KQ, Arnold FH. BioTechnology. 1991;9:1073–1077. doi: 10.1038/nbt1191-1073. [DOI] [PubMed] [Google Scholar]
- 3.Bornscheuer UT, et al. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 4.Davids T, Schmidt M, Bottcher D, Bornscheuer UT. Curr Opin Chem Biol. 2013;17:215–220. doi: 10.1016/j.cbpa.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Goldsmith M, Tawfk DS. Curr Opin Struct Biol. 2012;22:406–412. doi: 10.1016/j.sbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Mak WS, Siegel JB. Curr Opin Struct Biol. 2014;27:87–94. doi: 10.1016/j.sbi.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Song J, et al. Biosci Biotechnol Biochem. 2011;75:1194–1197. doi: 10.1271/bbb.110014. [DOI] [PubMed] [Google Scholar]
- 8.Barile D, Rastall RA. Curr Opin Biotechnol. 2013;24:214–219. doi: 10.1016/j.copbio.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Bultema JB, Kuipers BJ, Dijkhuizen L. FEBS Open Bio. 2014;4:1015–1020. doi: 10.1016/j.fob.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song J, et al. Biosci Biotechnol Biochem. 2011;75:268–278. doi: 10.1271/bbb.100574. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa K, et al. FEBS J. 2015;282:2540–2552. doi: 10.1111/febs.13298. [DOI] [PubMed] [Google Scholar]
- 12.Strokopytov B, et al. Biochemistry. 1995;34:2234–2240. doi: 10.1021/bi00007a018. [DOI] [PubMed] [Google Scholar]
- 13.Vocadlo DJ, Davies GJ, Laine R, Withers SG. Nature. 2001;412:835–838. doi: 10.1038/35090602. [DOI] [PubMed] [Google Scholar]
- 14.Blake CC, et al. Proc R Soc Lond B Biol Sci. 1967;167:378–388. doi: 10.1098/rspb.1967.0035. [DOI] [PubMed] [Google Scholar]
- 15.Davies GJ, Wilson KS, Henrissat B. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Veen BA, et al. J Mol Biol. 2000;296:1027–1038. doi: 10.1006/jmbi.2000.3528. [DOI] [PubMed] [Google Scholar]
- 17.Chen-Goodspeed M, Sogorb MA, Wu F, Raushel FM. Biochemistry. 2001;40:1332–1339. doi: 10.1021/bi001549d. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt J, Brocca S, Schmid RD, Pleiss J. Protein Eng. 2002;15:595–601. doi: 10.1093/protein/15.7.595. [DOI] [PubMed] [Google Scholar]
- 19.Koide A, Bailey CW, Huang X, Koide S. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 20.Koide A, Wojcik J, Gilbreth RN, Hoey RJ, Koide S. J Mol Biol. 2012;415:393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojcik J, et al. Nat Struct Mol Biol. 2010;17:519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbreth RN, et al. Proc Natl Acad Sci USA. 2011;108:7751–7756. doi: 10.1073/pnas.1102294108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sha F, et al. Proc Natl Acad Sci USA. 2013;110:14924–14929. doi: 10.1073/pnas.1303640110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albayrak N, Yang ST. Biotechnol Bioeng. 2002;77:8–19. doi: 10.1002/bit.1195. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Yang X, Yang S, Zhu M, Wang X. Comput Struct Biotechnol J. 2012;2:e201209017. doi: 10.5936/csbj.201209017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. Proc Natl Acad Sci USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, et al. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Goujon M, et al. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koide S, Koide A, Lipovsek D. Methods Enzymol. 2012;503:135–156. doi: 10.1016/B978-0-12-396962-0.00006-9. [DOI] [PubMed] [Google Scholar]
- 30.Nishikori S, et al. J Mol Biol. 2012;424:391–399. doi: 10.1016/j.jmb.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.