Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules that have key regulatory roles in cancer, acting as both oncogenes and tumor suppressors. Due to the potential roles of miRNAs in improving cancer prognostic, predictive, diagnostic and therapeutic approaches, they have become an area of intense research focus in recent years. MiRNAs harbor attractive features allowing for translation to the clinical world, such as relatively simple extraction methods, resistance to molecular degradation, and ability to be quantified. Numerous prognostic, predictive and diagnostic miRNA signatures have been developed. To date however, miRNA analysis has not been adopted for routine clinical use. The objectives of this article are to provide an overview of miRNA research and review a selection of miRNA studies in breast cancer, cervical cancer, sarcoma, and nasopharyngeal carcinoma to highlight advances and challenges in miRNA cancer research.
Keywords: microRNA, breast cancer, cervical cancer, sarcoma, nasopharyngeal carcinoma
INTRODUCTION
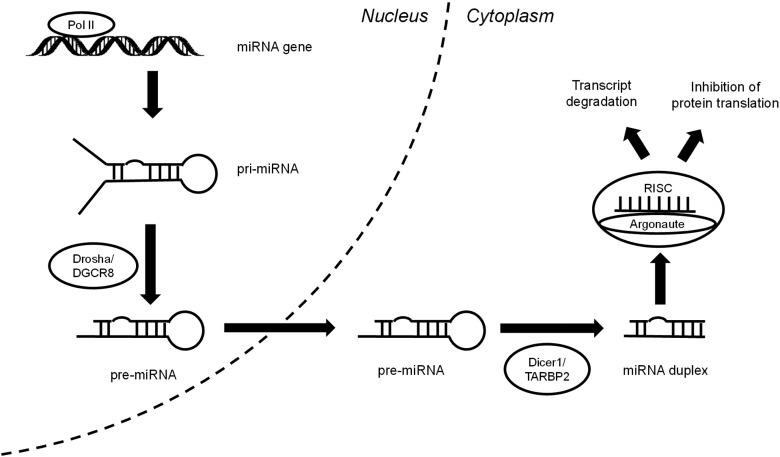
MicroRNAs (miRNAs) are non-coding RNAs composed of 18–25 nucleotides [1], which were first described in C. elegans by Lee et al. in 1993 [2]. Since their discovery, miRNAs have been demonstrated to have a key role in gene regulation in many different systems. There are several important steps involved in the synthesis of miRNAs (Fig. 1) [1]. Beginning in the nucleus, RNA polymerase II produces pri-miRNA transcripts. These are structurally similar to protein-coding gene transcripts with an additional stem-loop structure. This stem-loop interacts with the ribonuclease Drosha and double-stranded RNA binding protein, DiGeorge syndrome critical region gene 8 (DGCR8), to generate a precursor miRNA (pre-miRNA). Upon transport to the cytoplasm, the ribonuclease Dicer1 and transactivation-responsive RNA-binding protein 2 (TARBP2) convert the pre-miRNA into a double-stranded miRNA duplex. Following strand separation, the mature miRNA strand combines with Argonaute and forms the RNA-induced silencing complex (RISC). Gene regulation then occurs through either binding to target mRNA transcripts with perfect complementarity, resulting in transcript degradation, or more frequently, binding with imperfect complementarity, leading to inhibition of protein translation.
Fig. 1.
MicroRNA biosynthesis.
MiRNAs are dysregulated in almost all human cancers [3] and can function as either oncogenes or tumour suppressors, depending upon their target transcripts. For example, miRNA-21 (miR-21), one of the most overexpressed miRNAs in human epithelial malignancies, downregulates a myriad of tumour suppressors such as phosphatase and tensin homolog (PTEN) [4], ras homolog gene family member B (RhoB) [5], tropomyosin 1 (TPM1) [6], and programmed cell death 4 (PDCD4) [7], thereby resulting in increased tumour cell proliferation, invasion, and metastasis [4–7]. MiRNA levels are also correlated with response to ionizing radiation. Downregulating proteins in its biosynthesis pathway (e.g. Drosha and Dicer) increases radioresistance via a decreased DNA-damage response [8].
MiRNAs harbor attractive features allowing for translation to clinical practice, such as simple extraction, resistance to molecular degradation, and accurate, reproducible quantification using quantitative real-time reverse transcription PCR (qRT-PCR) and other analytical methods [9]. Combined with their potential to improve cancer diagnosis, classification, prognosis and therapies, miRNAs have garnered significant research attention in recent years. For example, with the identification of upregulated miR-141 in prostate cancer patients [10], circulating serum miRNAs were speculated as a possible non-invasive diagnostic tool. MiRNA profiling may also facilitate better classification of tumors into treatment subgroups [11], especially for poorly differentiated tumors that are difficult to define by histology. A number of prognostic miRNA signatures have been generated and validated in various cancers [12, 13]. Additionally, miRNAs have been investigated for drug therapy (e.g. miR-122 antagonists for decreasing risk of hepatocellular carcinoma [14]). Predictive signatures have also been proposed based on select miRNAs (e.g. miR-16, -29b, -150, -1254 and let-7e [15]) associated with radiosensitivity and radioresistance.
The general approach to miRNA profiling (Fig. 2) involves: (i) specimen selection (cells, organisms, fresh tissue, fixed tissue, body fluids); (ii) RNA extraction; (iii) sample quality control; (iv) profiling miRNA with qRT-PCR, microarray, or RNA-sequencing; and (v) data analysis [16]. qRT-PCR is the gold standard because it provides the best absolute miRNA quantification with the greatest sensitivity and dynamic range compared with the other methods [16], although several other platforms are commonly used in miRNA research.
Fig. 2.
MicroRNA profiling workflow.
Global miRNA profiling of cancers has yielded important research findings. In particular, profiling of formalin-fixed, paraffin-embedded (FFPE) samples from human tumors, which are correlated with clinical details, has significantly improved our understanding of miRNAs in this disease. The purpose of this article is to provide an overview of miRNA research and highlight a selection of miRNA studies in breast cancer, cervical cancer, sarcoma, and nasopharyngeal carcinoma (NPC) conducted by our group to underscore recent advances and challenges in miRNA cancer research.
BREAST CANCER
Breast cancer is the second most commonly diagnosed cancer, and fifth most common cause of cancer mortality worldwide [17]. MiRNA dysregulation has been identified as a frequent phenomenon in breast cancer. Four main mechanisms of dysregulation have been proposed, including epigenetic modification, genetic alteration, changes in miRNA biogenesis, and transcriptional repression [18]. Some validated dysregulated miRNAs are highlighted in Table 1 [19–25]. As shown in this Table, global miRNA profiling in breast cancer has yielded a number of potential biomarkers and therapeutic targets. We will focus specifically on two studies of miRNA profiling in breast cancer from FFPE specimens to highlight its use [19, 26].
Table 1.
Dysregulated miRNAs in breast cancer
| Upregulated miRNAs [19–21] | Downregulated miRNAs [19, 21–25] |
|---|---|
| miR-9, miR10b, miR-21, miR-27a, miR-29a, miR-96, miR-146a, miR-155, miR-181, miR-191, miR-196a, miR-221/222, miR-373, miR-375, miR-520c, and miR589 | miR-30a, miR-31, miR-34a, miR-125, miR-126, miR-146a, miR-146b, miR-195, miR-200, miR-205, miR-206, miR-221, and let-7 |
Prior to 2008, the majority of global miRNA profiling in cancer utilized frozen tissues; however, formalin fixation is a common practice for specimen preservation. Thus, Hui et al. [19] examined the feasibility of miRNA analysis on FFPE tissues. They performed miRNA expression profiling on 40 archived FFPE breast lumpectomy specimens using Taqman Low-Density Arrays (TLDAs). The expression of 365 miRNAs in 34 invasive ductal cancers and 6 reduction mammoplasty normal epithelial breast tissues was assessed and confirmed using the gold standard single-well qRT-PCR. The results from both methods were highly correlated and confirmed the dysregulation of miR-21, miR-155, miR-191, miR-196a, miR-125b and miR-221, which were previously reported. Thus, this study provided support for performing miRNA profiling on FFPE tissues.
Clinical trial materials are a rich source of uniformly collected and processed archival FFPE tumor tissues for evaluation and are ideal for addressing primary or secondary research aims involving miRNA profiling [26, 27]. For example, 71 FFPE blocks from participants of a Phase III clinical trial comparing Tamoxifen alone vs Tamoxifen plus breast radiotherapy in women with node-negative breast cancer [28] served as the cohort for a subsequent miRNA study by Shi et al. [26]. Through global miRNA profiling with qRT-PCR, they identified a 6.7-fold higher expression of miR-301 in tumours as compared with normal breast tissue [26]. Overexpression of this miRNA was also associated with increased incidence of nodal and distant relapse [26]. However, the prognostic value of miR-301 required further validation in a larger patient cohort.
CERVICAL CANCER
Cervical cancer is the fourth most common cancer in females and accounts for 7.5% of all female cancer deaths [17]. Identifying prognostic and predictive biomarkers are important aims of miRNA research in cervical cancers; however, cervical tissues have presented several challenges in miRNA profiling.
In 2010, Pereira et al. [29] studied miRNA expression in human cervical tissues using a 381-probe microarray platform on a mixed set of 19 normal, 14 pre-cancerous, and four cancerous cervical tissues derived from 25 patients. During their investigation, they noted highly variable miRNA expression among the normal samples. This variability was confirmed to be non-technical and related to the inter-patient biological differences, such as human papillomavirus (HPV) status and age. This study emphasized the importance of considering interpatient variations that may influence miRNA expression profiles, including natural genetic variation, age, viral infections, and non-neoplastic diseases, when interpreting miRNA profiling results in cervical tissues [29].
A subsequent study in cervical cancers further underscored additional considerations in miRNA analyses. In 2015, How et al. [30] described their investigation of developing a nine-miRNA signature for cervical cancers. They formulated a candidate prognostic signature for disease-free survival (DFS), which included well-characterized miRNAs involved in neoplasia, based on a training cohort of frozen tissues. However, there was difficulty validating this signature in an independent cohort comprised of FFPE tissue samples. They identified several reasons contributing to this challenge including intratumor heterogeneity, poor correlation between miRNAs in frozen and FFPE samples, and poor reliability of profiling platforms (Table 2).
Table 2.
Sources of expression heterogeneity in cervical tissues
Despite difficulties with profiling in cervical tissues, positive findings have been reported in the literature as well. For example, based on the global miRNA profiling of 79 cervical cancer fresh frozen punch biopsies and 11 normal samples, Kogo et al. [31] identified a difference in miR-218 expression between malignant cervical and normal cervical epithelial tissues. Downregulation of miR-218 was prognostic for reduced DFS, overall survival and lymph node recurrence. Further investigations of miR-218 biology determined that it was targeting the survivin axis to modulate cancer proliferation, migration and invasion. In this process, YM155 was identified as a promising small molecule inhibitor of survivin, which could reduce nodal metastasis in cervical cancer xenograft models [31].
SARCOMA
Sarcomas are a heterogeneous group of rare tumors that provide diagnostic and prognostic challenges. Its 5-year overall survival is ∼60–80%, with distant metastasis (DM) as the main contributor to mortality [32]. Tumor size and grade have demonstrated prognostic significance, but variability in clinical outcome suggests the need for more robust biomarkers. Few prognostic signatures for soft tissue sarcomas have been validated. Wong et al. [33] were the first to validate a miRNA signature for distant metastasis–free survival (DMFS) in undifferentiated pleomorphic sarcomas (UPS), the most common and aggressive subtype of adult soft-tissue sarcomas. Using TaqMan Human Micro-RNA Array-A, 110 fresh frozen UPS samples were analyzed to identify and validate a six-miRNA signature for DMFS. This signature, comprised of miR-132, miR-138, miR-143, miR-221, miR-224 and miR-491–5p, was independently prognostic of known clinical factors. Functional studies demonstrated that these miRNAs were targeting the Rho adhesion pathway in UPS metastasis.
NASOPHARYNGEAL CARCINOMA
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy of the head and neck that has a preponderance in Southeast Asia. Patients with early stage NPC have a favorable outcome when treated with radiotherapy, with 5-year local control rates exceeding 90% [34]. However, late stage disease (Stage III–IVB) has a worse 5-year overall survival rate of only ∼68% or less [34]. Furthermore, DM continues to occur and is the major factor contributing to mortality in this population. Improved prognostic markers associated with DM are therefore clearly required. Bruce et al. [35] profiled 734 miRNAs on a training set comprised of 125 NPC FFPE biopsy samples, and a four-miRNA expression signature for DM was validated on an independent cohort of 121 FFPE biopsies. This signature demonstrated prognostic value in addition to tumor node metastasis staging, the mainstay of clinical classification in NPC. In addition, this four-miRNA signature was observed to possess potential predictive value in identifying a low-risk group of locally advanced NPC patients who could be treated with radiotherapy alone, thereby sparing the additional toxicity of chemotherapy. Other signatures comprised of different sets of miRNAs have also been reported for NPC, including a five-miRNA signature by Liu et al. [36]. Bruce et al. [35] attempted to compare the four-miRNA with the five-miRNA signature to determine if superiority could be established; however, due to differences in technical platforms, patient population, and availability of clinical data, no definitive conclusions could be drawn. With an accumulating number of miRNA signatures reported in the literature, prioritization of miRNA signatures to translate for clinical use could become problematic without the ability to compare signatures derived from different methodologies.
LESSONS LEARNED
In recent years, research in miRNA profiling has garnered significant interest. Dysregulation of miRNAs in a variety of different cancers have been reported, and associated miRNA signatures have been developed. Along the way, a number of challenges in miRNA research have also emerged. Based on the research reviewed in this article, there are some lessons that can be learned.
Three main problems have been identified in the miRNA profiling workflow; they involve tissue preparation, profiling platform selection, as well as data normalization and analysis. The first issue speaks to the need for more careful consideration of differences between specimen types. Tissue preservation technique appears to affect miRNA expression results, as described in How et al.'s study [30], which demonstrated the difficulty in comparing miRNA profiling of frozen and FFPE tissues. Thus, when defining training and validation cohorts for future miRNA signature testing, it would be prudent to extract RNA from tissues prepared in a similar manner to maximize comparability.
Technical platform variability has also proven to be an important consideration in investigating miRNAs. Several platforms have been used to quantify miRNAs; however, these techniques have significant methodological differences. For example, TLDAs use multiplex reverse transcription followed by TaqMan PCR, while NanoString uses optical quantification of fluorescently tagged miRNAs. Consistency between profiling platforms has been called into question by studies such as that of How et al. [30], which demonstrated that profiling of FFPE RNA samples by TLDA and NanoString had poor correlation in regards to miRNA abundances and tumor-to-normal tissue expression differences [37]. This, therefore, increases the difficulty of interpreting results across different platforms. In addition, for PCR-based techniques, the quantification may not distinguish between abundances of miRNA at different levels of maturation. Specifically, pri- and pre-miRNA levels cannot be discriminated if the amplicon is shared by both [38]. Thus, it is important to understand the limitations of each methodology.
As well, the study of miRNAs has created some new challenges that have arisen since past research with mRNA transcripts. Normalization of data is required for decreasing technical bias; however, data normalization methods for miRNA analysis remain varied and unreliable in some cases. For example, methods that require a reliable estimate of the distribution of miRNA expression values have been noted to lack applicability in miRNA datasets compared with mRNA data, since the majority of global miRNA profiling array platforms test much fewer transcripts than mRNA platforms [37, 39]. This yields a smaller set of miRNAs with stable expression. Thus, the use and selection of controls become paramount in miRNA studies. It has been suggested that a greater number of normalization controls should be used and controls should be validated for stability before use [39]. In general, adapting mRNA methodologies to miRNA studies has its limitations, and novel approaches for miRNA-specific research have been explored. In this era with little consensus on miRNA data normalization methods, it is imperative for researchers to ensure that signatures generated are not normalization method–specific to ensure robustness and generalizability.
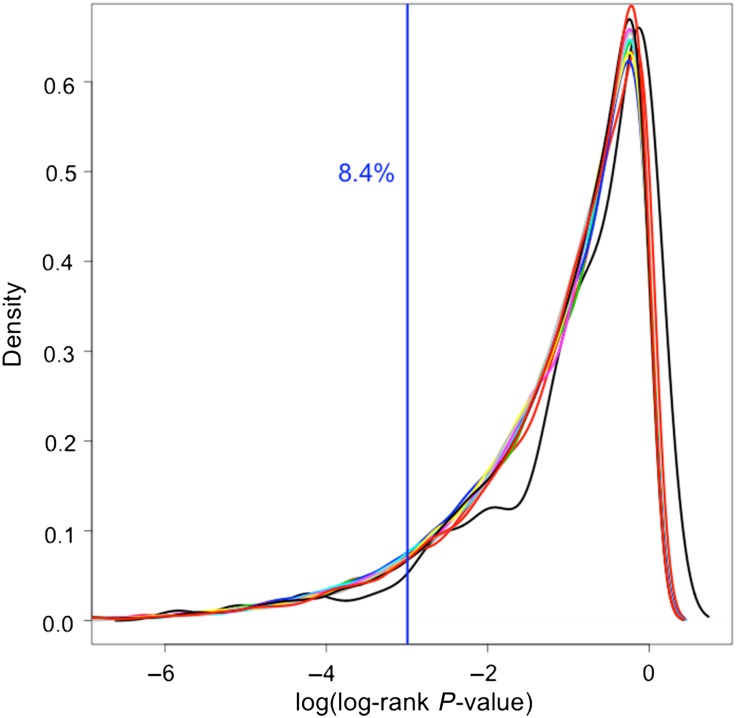
Lastly, the biological validity of some miRNA signatures has been called into question [35, 40]. MiRNA signatures are currently developed from algorithms based on finding associations between expression and disease outcome. Hence, causality is often not taken into account, and algorithms may generate signatures that are not biologically significant, despite their statistical significance [40]. For example, based on our experience, we were able to validate 8.4% of 90 000 randomly generated miRNA signatures, comprised of 2–10 miRNAs each, using the Bruce et al. training and validation sample set [35] (Fig. 3). Thus, it is important to consider the limitations of algorithms for miRNA signature generation and how selection of algorithms may impact results. Utilization of further methods to evaluate signatures of likely biological significance, such as the Bruce et al.'s approach of prioritizing highly significant signatures for further molecular target identification and pathway analysis, is needed.
Fig. 3.
Validation of randomly generated miRNA signatures. Density plot is shown with different color plots denoting different signature sizes (2–10 miRNAs). Vertical line denotes log-rank P < 0.05 in our validation set.
Currently, a variety of methods exist for generation of miRNA signatures. As a result, numerous signatures have been developed. However, for the majority of cancers, miRNA signatures are far from being adopted for routine clinical use. In future years, the focus needs to shift towards facilitating the translation of these miRNA findings into the clinical environment. Several steps are likely required. As described in this review, variable profiling methodologies have created challenges in interpreting data and comparing results between studies. Thus, careful optimization of the workflow of miRNA profiling is needed, including the adoption of standardized protocols (e.g. uniform sample preparation, quantification, normalization, and analysis). In addition, validation of candidate diagnostic, prognostic, and predictive signatures in large patient cohorts will be needed to confirm clinical significance of findings.
In conclusion, much remains to be learned regarding miRNA biology in human cancers. Their value and role as prognostic or predictive biomarkers warrant further investigations. Lessons learned from the work thus far underscore the importance of uniformity in tissue types, consistency in platforms, rigour in data normalization and analyses, and the use of sizeable cohorts for validation.
FUNDING
This work was supported by the Canadian Institutes of Health Research, the Ontario Institute for Cancer Research, the Canadian Breast Cancer Foundation and the Canadian Breast Cancer Research Alliance. Funding to pay the Open Access publication charges for this article was provided by the Canadian Institutes of Health Research.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Tara Spence for her advice in the preparation of this manuscript. Results from this study were presented at the 15th International Congress of Radiation Research (ICRR 2015), Kyoto, Japan, 25–29 May 2015.
REFERENCES
- 1.Lujambio A, Lowe SW. The microcosmos of cancer. Nature 2012;482:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–54. [DOI] [PubMed] [Google Scholar]
- 3.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang JG, Wang JJ, Zhao F et al. . MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 2010;411:846–52. [DOI] [PubMed] [Google Scholar]
- 5.Liu M, Tang Q, Qiu M et al. . miR-21 targets the tumor suppressor RhoB and regulates proliferation, invasion and apoptosis in colorectal cancer cells. FEBS Lett 2011;585:2998–3005. [DOI] [PubMed] [Google Scholar]
- 6.Zhu S, Wu H, Wu F et al. . MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008;18:350–9. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Xin S, He Z et al. . MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell Physiol Biochem 2014;33:1631–42. [DOI] [PubMed] [Google Scholar]
- 8.Francia S, Michelini F, Saxena A et al. . Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012;488:231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell PS, Parkin RK, Kroh EM et al. . Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai X, Chen A, Bai Z. Integrative investigation on breast cancer in ER, PR and HER2-defined subgroups using mRNA and miRNA expression profiling. Sci Rep 2014;4:6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling H, Krassnig L, Bullock M et al. . MicroRNAs in testicular cancer diagnosis and prognosis. Urol Clin North Am 2016;43:127–34. [DOI] [PubMed] [Google Scholar]
- 13.Iorio M, Croce C. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 2009;27:5848–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanford RE, Hildebrandt-Eriksen ES, Petri A et al. . Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 20120;327:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, Boohaker R, Jiang C et al. . A radiosensitivity MiRNA signature validated by the TCGA database for head and neck squamous cell carcinomas. Oncotarget 2015;6:34649–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferlay J, Soerjomataram I, Ervik M et al. . GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. 2013: France. http://globocan.iarc.fr/ (24 August 2015, date last accessed). [Google Scholar]
- 18.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015;5:1122–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hui AB, Shi W, Boutros PC et al. . Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest 2009;89:597–606. [DOI] [PubMed] [Google Scholar]
- 20.Piva R, Spandidos DA, Gambari R. From microRNA functions to microRNA therapeutics: novel targets and novel drugs in breast cancer research and treatment. Int J Oncol 2013;43:985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christodoulatos GS, Dalamaga M. Micro-RNAs as clinical biomarkers and therapeutic targets in breast cancer: Quo vadis? World J Clin Oncol 2014;5:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SJ, Shin JY, Lee KD et al. . MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res 2012;14:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang K, Ahn H, Sim J et al. . Loss of microRNA-200a expression correlates with tumor progression in breast cancer. Transl Res 2014;163:242–51. [DOI] [PubMed] [Google Scholar]
- 24.Zhao FL, Dou YC, Wang XF et al. . Serum microRNA-195 is down-regulated in breast cancer: a potential marker for the diagnosis of breast cancer. Mol Biol Rep 2014;41:5913–22. [DOI] [PubMed] [Google Scholar]
- 25.Hu J, Xu J, Wu Y et al. . Identification of microRNA-93 as a functional dysregulated miRNA in triple-negative breast cancer. Tumour Biol 2015;36:251–8. [DOI] [PubMed] [Google Scholar]
- 26.Shi W, Gerster K, Alajez NM et al. . MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res 2011;71:2926–37. [DOI] [PubMed] [Google Scholar]
- 27.Simon RM, Paik S, Hayers DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009;101:1446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fyles AW, McCready DR, Manchul LA et al. . Tamoxifen with or without breast irradiation in women 50 years of age or older with early breast cancer. N Engl J Med 2004;351:963–70. [DOI] [PubMed] [Google Scholar]
- 29.Pereira PM, Margues JP, Soares AR et al. . MicroRNA expression variability in human cervical tissues. PLoS One 2010;5:e11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.How C, Pintilie M, Bruce JP et al. . Developing a prognostic micro-RNA signature for human cervical carcinoma. PLoS One 2015;10:e0123946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kogo R, How C, Chaudary N et al. . The microRNA-218∼Survivin axis regulates migration, invasion, and lymph node metastasis in cervical cancer. Oncotarget 2015;6:1090–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendenhall WM, Indelicato DJ, Scarborough MT et al. . The management of adult soft tissue sarcomas. Am J Clin Oncol 2009;36:436–42. [DOI] [PubMed] [Google Scholar]
- 33.Wong P, Hui A, Su J et al. . A potential therapeutic target in undifferentiated pleomorphic sarcomas. Oncotarget 2015;6:39127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee AW, Ng WT, Chan YH et al. . The battle against nasopharyngeal cancer. Radiother Oncol 2012;104:272–8. [DOI] [PubMed] [Google Scholar]
- 35.Bruce JP, Hui AB, Shi W et al. . Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget 2015;6:4537–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N, Chen NY, Cui RX et al. . Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol 2012;13:633–41. [DOI] [PubMed] [Google Scholar]
- 37.How C. Characterization of altered microRNA expression in cervical cancer. Ph.D. Thesis. The University of Toronto Graduate Department of Medical Biophysics 2013. [Google Scholar]
- 38.Schmittgen T, Lee E, Jiang J et al. . Real-time PCR quantification of precursor and mature microRNA. Methods 2008;44:31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B, Wang XF, Howell P et al. . A personalized microRNA microarray normalization method using a logistic regression model. Bioinformatics 2010;26:228–34. [DOI] [PubMed] [Google Scholar]
- 40.Boutros PC, Lau SK, Pintilie M et al. . Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A 2009;106:2824–8. [DOI] [PMC free article] [PubMed] [Google Scholar]