Abstract
DNA damage response is finely tuned, with several pathways including those for DNA repair, chromatin remodeling and cell cycle checkpoint, although most studies to date have focused on single pathways. Genetic diseases characterized by genome instability have provided novel insights into the underlying mechanisms of DNA damage response. NBS1, a protein responsible for the radiation-sensitive autosomal recessive disorder Nijmegen breakage syndrome, is one of the first factors to accumulate at sites of DNA double-strand breaks (DSBs). NBS1 binds to at least five key proteins, including ATM, RPA, MRE11, RAD18 and RNF20, in the conserved regions within a limited span of the C terminus, functioning in the regulation of chromatin remodeling, cell cycle checkpoint and DNA repair in response to DSBs. In this article, we reviewed the functions of these binding proteins and their comprehensive association with NBS1.
Keywords: Nijmegen breakage syndrome, NBS1, RNF20, MRE11, ATM, RPA, RAD18
INTRODUCTION
The human genome is constantly challenged by a plethora of insults, and its integrity is maintained by so-called DNA damage responses including cell cycle checkpoint, chromatin remodeling and DNA repair [1, 2]. These pathways in DNA damage response must be closely associated in order to facilitate DNA repair with sequential access of the repair proteins and the appropriate duration of repair time. DNA double-strand breaks (DSBs) are the most deleterious form of DNA damage and can be generated by extrinsic stress, such as exposure to ionizing radiation, and intrinsic accidents during DNA replication. A DSB generated in a cell, if not correctly rejoined, potentially leads to cellular lethality or development of malignancy. To cope with DSBs, vertebrates have evolved two major repair pathways: homologous recombination repair (HRR) and nonhomologous end joining (NHEJ). HRR produces 3′ single-strand DNA (ssDNA) at DSB ends by resection of 5′ DNA to facilitate strand invasion into the homologous region of a sister chromatid, which in turn serves as a template for reconstruction of the damaged DNA sequences [3]. NHEJ, which is a dominant process in higher eukaryotes, enables cells to rejoin DSBs directly, or after processing the DNA ends, at an appropriate chromosomal end. These repairs must be coordinated with the cell cycle checkpoint to arrest cell growth until the repair is completed. At least two checkpoints, G1 and G2, are involved in DSB repair. Normal progression through G1 is promoted through activity of the cyclin-dependent protein kinases 2 (CDK2), which is inhibited by p21. In the G1 checkpoint, the amount of p53 in the damaged cells is increased and it mediates the expression of p21 through enhanced transcription [1, 2]. Similarly, the G2 checkpoint enables cells to arrest the cell cycle in the G2 phase via Chk2 activation, which in turn inactivates CDK1, a kinase required for passage from the G2 to the M phase, via CDC2 phosphorylation, a binding partner of CDK1 [1, 2]. Because DNA in eukaryotes is packaged into condensed chromatin, chromatin remodeling to relax the tight chromatin structure is an essential step in introducing the repair proteins to the sites of DSBs [4]. DNA damage response must be orchestrated through functional interplay among the checkpoint, chromatin remodeling and DNA repair (rejoining), while the spatiotemporal regulatory mechanism is yet to be elucidated. NBS1, the protein responsible for Nijmegen breakage syndrome (NBS), is unique in physically binding to key proteins of the DNA damage response within a limited region (approximately 100 amino acids in length) of NBS1 at the C terminus, and regulates the respective pathways [5]. Thus, NBS1 disruption results in dysfunctional pathways of DNA damage response; consequently, patients with NBS are characterized by high frequencies of malignancy and high sensitivity to ionizing radiation.
NIJMEGEN BREAKAGE SYNDROME
NBS is a rare autosomal recessive genetic disorder characterized by microcephaly and a predisposition to malignancy. In 1981, two brothers affected with NBS were first described by C. Weemaes at the University of Nijmegen [6]. After the first recognition of the disorder, the disease appeared to be prevalent in the Eastern and Central European populations, particularly in Poland and the Czech Republic. To date, 150 patients have been reported in many other European countries, North and South America, Morocco, New Zealand and Japan [7]. Microcephaly is a hallmark symptom of NBS, which is detected in 100% of patients after the age of 1 month. By the age of 20 years, over 40% of NBS patients develop a malignant disease, predominantly of lymphoid origin, and the incidence is one of the highest among repair-deficiency diseases. NBS patients are particularly prone to recurrent infections, such as those of the respiratory and urinary tracts. The humoral immunodeficiency in NBS patients is agammaglobulinemia, particularly IgG and IgA deficiency; however, 10% of patients show a normal Ig status [7]. NBS patients show chromosomal instability in peripheral lymphocytes in the form of inversions and translocations involving chromosomes 7 and 14 at the sites of IgH or TCR genes. Malignancy and recurrent infections are the major cause of death in these individuals. Freckles are the most frequently reported skin pigmentation abnormality, as observed in patients with the UV-sensitivity disease xeroderma pigmentosum (XP) [6]. Half of the patients show minor skeletal anomalies, such as encountered in patients with the DNA crosslinking agent-sensitivity disease Fanconi anemia (FA). The frequency of heterozygous carriers in the Eastern and Central European population is estimated to be one case per 177 newborns [7].
Cells from an NBS patient show high sensitivity to ionizing radiation (IR) and abnormal cell cycle checkpoints, which are signs similar to those found in ataxia–telangiectasia (A-T) [1, 2]. For these reasons, NBS has been considered to be a variant of A-T and is further divided into two groups: AT variant 1 and AT variant 2 or Berlin breakage syndrome. This subdivision of the variants was proposed on the basis of complementation studies of cell hybrids of AT variants 1 and 2 using DNA synthesis as an endpoint, although the inadequacy of this endpoint in AT hybrids has been indicated [8]. The putative gene responsible for NBS was mapped to 8q21–q24 [9, 10], and we and others independently cloned NBS1, the underlying gene, from the candidate region in 1998 [11–14]. It is noted that two putative variants were caused by the mutation of a single NBS1 gene.
The NBS1 gene consists of 16 exons over 50 kb, and it encodes a 754-amino acid NBS1 protein (85 kDa). All but one patient in the Eastern and Central European populations were homozygous for a 5-base pair (bp) NBS1 deletion, 657del5, revealing the presence of a common founder [11]. All 11 mutations identified to date are located between exons 6 and 10, producing either a 73- or a 52-kDa fragment of NBS1 with a low level of expression [15, 16]. Two further mutations have been reported in exons 4 and 10 in French siblings, and a homozygous mutation in exon 5 in a Japanese child; however, no clinical sign of NBS was presented in these individuals [17, 18]. All efforts to develop an Nbs1-null mouse have failed because of embryonic lethality of the Nbs1-null phenotype. It is noteworthy that all NBS cell lines derived from human fibroblast cells and mouse MEF cells express the C-terminal fragment of NBS1 at a low level, although the expression levels and the truncated species are different among cell lines and mutants [19].
NBS1 PROTEIN
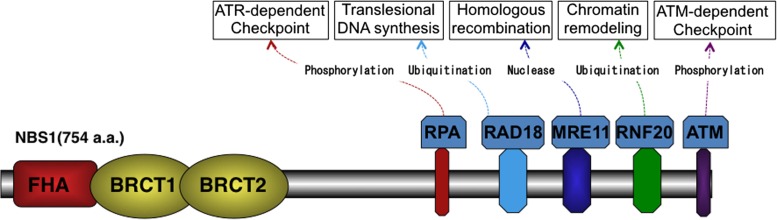
The NBS1 protein contains two functional regions at the N terminus (1–196 amino acids) and the C terminus (665–693 amino acids). Weak homology to yeast Xrs2 protein (29%) was first recognized at the N-terminal sequence [1, 2], which consists of a forkhead-associated (FHA) domain (20–108 amino acids) and a BRCA1 C terminus (BRCT1: 111–197 amino acids; BRCT2: 219–327 amino acids) domain (Fig. 1). The FHA domain is required for binding of NBS1 to CtIP (a homolog of yeast Sae2), which resects DSB ends to produce 3′ ssDNA for HRR. Similarly, both FHA and BRCT1/2 interact with the phosphoserine of MDC1 to accumulate at DSB sites, which are visualized as radiation-induced nuclear foci using immunofluorescence. The C terminus of NBS1 contains at least five regions conserved in vertebrates: an RPA-binding region (549–563 amino acids), an RAD18-binding region (650–665 amino acids), an MRE11-binding region (682–693 amino acids), an RNF20-binding region (704–708 amino acids) and an ATM-binding region (734–754 amino acids). The MRE11-binding region, identified using yeast two-hybrid experiments, is essential for the MRE11/RAD50 complex accumulation at the sites of DSBs and the subsequent HRR pathway [20, 21]. This MRE11-binding region is well conserved in eukaryotes, including yeast (Saccharomyces cerevisiae). Similarly, the RNF20-binding region, recognized by yeast two-hybrid assay, is essential for chromatin remodeling, although the sequence is not conserved in yeast [22]. The ATM- and RPA-binding regions, and the RAD18-binding region, which are independently identified, are also required for the regulation of the cell cycle checkpoint and translesion DNA synthesis, respectively; however, no clear sequence homology in their binding regions was detected in yeast [19, 23, 24].
Fig. 1.
Structure of NBS1 protein. NBS1 consists of FHA and BRCT1/2 regions at the N terminus and multiple protein-binding regions at the C terminus. FHA and BRCT1/2 regions are involved in the IR-induced nuclear foci via interaction with phosphoproteins. Protein-binding regions at the C terminus contain ATM-, RNF20-, MRE11-, RAD18- and RPA-binding regions, which regulate cell cycle checkpoint, DNA repair and chromatin remodeling.
MRE11 AND HOMOLOGOUS RECOMBINATION
MRE11 has a nuclease activity that generates 3′ ssDNA tails during the S and G2 phases, which invade homologous strands of the sister chromatid and exploit it as a template for DNA synthesis of the damaged DNA sequences. NBS1 has a role in the recruitment of MRE11 nuclease to the sites of DSBs, physically interacting with it at the C terminus (Fig. 1), so the accumulation of MRE11 nuclease has been shown to be abolished in NBS1 clones lacking the MRE11-binding region [20]. Thus, NBS1 is essential for HRR in vertebrates, such as in human and chicken cells. Conversely, the disruption of NBS1 or cells from NBS patients severely reduced HRR ability as measured by HRR reporter assay using DR-GFP and SCneo [3, 21]. In contrast, the A-T cells or ATM-depleted cells had normal HRR activity as measured by the reporter assay, although the precise mechanism was not understood [3]. NBS1 activity is modified during the S and G2 phases through phosphorylation at the S432 site with CDK [25]. Indeed, NBS1 mutation at the CDK-mediated phosphorylation site significantly reduces the generation of ssDNA tails during the S and G2 phases. Thus, NBS1 plays a critical role in HRR, although HRR is considered to play a minor role in mammalian DSB repair when DSB is induced through IR exposure. However, NBS cells are highly sensitive to IR, suggesting a dysfunction of NHEJ. NHEJ is further categorized into two subgroups: Ku protein-dependent NHEJ (or canonical NHEJ) and microhomology-mediated end-joining (MMEJ or alternative NHEJ). Our NBS1-deficient chicken DT40 cells showed normal Ku-dependent NHEJ, but not MMEJ, when measured using the end-joining assay with the microhomology DNAs [21]. This finding is consistent with the observation that NBS1 disruption causes MRE11 dysfunction, which is required for MMEJ. Thus, the MRE11-binding domain of NBS1 at the C terminus predominantly determines IR sensitivity by both pathways of HRR and MMEJ in mammalian and chicken cells. This is functionally and structurally similar to yeast DSB repair, because Xrs2 shows sequence homology with the mammalian MRE11-binding region and the disruption of Xrs2 impairs both HRR and NHEJ [26].
RNF20 AND CHROMATIN REMODELING
Chromatin remodeling is usually initiated by histone modification at DSB sites and is followed by the accumulation of a protein, termed as a chromatin remodeling factor that relaxes the chromatin structure in an ATP-dependent fashion. NBS1 and ATM physically interact with RNF20, a ubiquitin ligase of histone H2B, after IR exposure [22, 27] (Fig. 1), suggesting that RNF20 is involved in NBS1- and ATM-associated DSB repair. Indeed, RNF20-depleted cells compromise the accumulation of HRR proteins such as RAD51 and BRCA1, and NHEJ proteins such as XRCC4, which inhibits DSB repair. RNF20-mediated H2B ubiquitylation is directly involved in DSB repair, because overexpression of the K120R H2B mutant, which lacks a lysine to be ubiquitylated, significantly attenuates the accumulation of repair proteins at DSB sites. The role of RNF20 in DSB repair is associated with chromatin remodeling, because the impaired accumulation of the repair protein at the DSB sites after RNF20 depletion was restored via treatment with several agents that induce chromatin relaxation [22]. This is further supported by the observation that the concurrent depletion of RNF20 and SNF2H (Fig. 2a), a chromatin remodeling factor, resulted in no more reduction of HRR ability than that of a single depletion, indicating epistatic function in the common pathway [19]. Moreover, the RNF20–SNF2 h pathway is controlled by FACT, a histone chaperone, as it physically binds to RNF20 and that mutation of FACT at the binding sites leads to failure of RNF20 accumulation at the DSB sites [28]. Consequently, accumulation of SNF2 h at DSB sites is compromised by the depletion of either RNF20 or FACT.
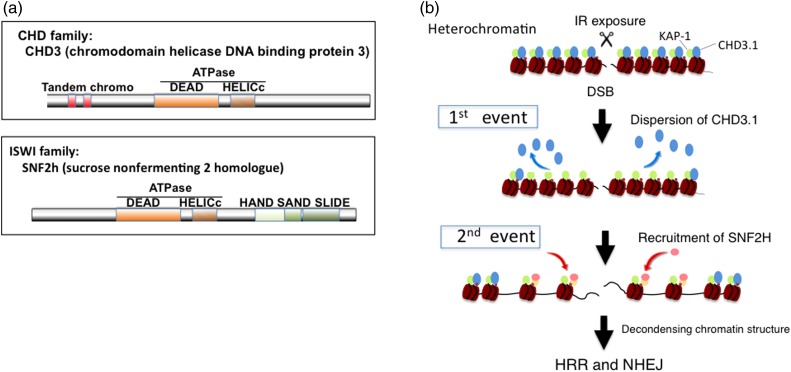
Fig. 2.
Two-event model of IR-induced chromatin remodeling with CHD3.1 and SNF2 h. (a) The chromatin remodeling factors CHD3.1 and SNF2 h are family proteins containing ATPase domains (DEAD and HELICs). CHD3.1 belongs to the CHD3 (chromodomain helicase DNA binding proteins 3) family and participates in maintenance of transcriptional repression at heterochromatic nucleosomes. SNF2 h (sucrose nonfermenting 2 homolog) belongs to the ISWI family, which relaxes the compact structure of heterochromatic nucleosomes. (b) IR-induced phosphorylation of KAP-1 initiates the dispersion of CHD3.1 from histone and thereafter SNF2 h is accumulated via H2B ubiquitination by RNF20.
CHD3.1, another chromatin remodeling factor (Fig. 2a), detached from heterochromatin after IR exposure, and this detachment is a prerequisite for chromatin remodeling before processing of DSB repair. The radiation-induced dispersion of CHD3.1 requires phosphorylation of KAP-1 at S824, which causes dissociation between KAP-1 and CHD3.1, so that CHD3.1 detaches from the histone at the DSB sites [29]. Interestingly, CHD3.1 functions in the same pathway as that of H2AX phosphorylation, although this modification of H2AX is not necessary for the RNF20–SNF2 h pathway. It may be noted that the phosphorylation of H2AX at damage sites is very rapid, whereas the ubiquitylation of H2B is much slower. Recently, Klement et al. [30] proposed a model in which the relaxation of heterochromatin at DSB sites requires two events: CHD3.1 initially detaches from histone via KAP-1 phosphorylation, but this detachment is not sufficient for chromatin decondensation, particularly in heterochromatin. An additional recruitment of SNF2 h and substitution with CHD3.1 are required for the completion of chromatin remodeling at the DSB site (Fig. 2b).
RAD18 AND TRANSLESION DNA SYNTHESIS
In the original paper by C. Weemases, NBS was classified as a UV-sensitivity disease, similar to XP [6]. XP is a UV-sensitivity recessive genetic disease resulting from a defect in nucleotide excision repair (NER). However, our experiments using enzyme linked immunosolvent assay with antibodies to cyclobutane pyrimidine dimer and 6-4 photoproduct showed normal activity of NER in NBS1-deficient cells, although they were significantly UV-sensitive [19]. In addition, a defect in translesion DNA synthesis (TLS) is the underlying mechanism of another type of XP, known as XP variant. TLS is initiated by ubiquitination of PCNA, which causes a DNA polymerase switch with translesion DNA polymerase, Pol eta, to perform DNA synthesis across the damaged DNA. The ubiquitin-conjugating enzyme RAD6 and ubiquitin ligase RAD18 are recruited to sites of stalled replication after UV exposure and mediate monoubiquitination of PCNA. We analyzed TLS in NBS1-deficient cells using UV-induced Pol eta focus formation as a marker of TLS. The results indicated that NBS1 deficiency severely impairs both Pol eta focus formation and monoubiquitination of PCNA. These defects are caused by the impaired recruitment of RAD18, because UV-induced RAD18 foci were not formed in NBS1-defective cells. Upon UV exposure, NBS1 binds directly to RAD18 via the RAD18-binding region at the C terminus and recruits RAD18 to the damage sites (Fig. 1) [19]. This sequence of events has been further confirmed through the development of a knock-in (KI) mouse that lacks the RAD18-binding region of NBS1. The NBS1-KI mouse, which is viable, is UV sensitive, and cells of the mouse show normal UV-induced NBS1 but not RAD18 focus formation, indicating that NBS1 controls RAD18 recruitment after UV exposure [31].
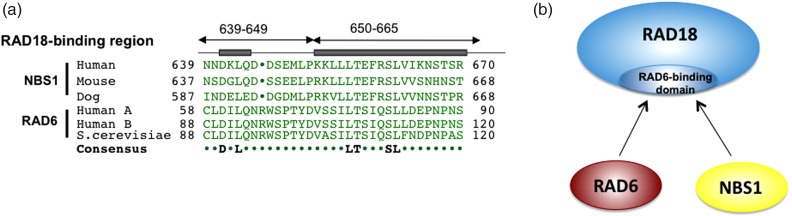
The Rad18-binding region of NBS1 shows high sequence homology with that of RAD6, suggesting that RAD6 and NBS1 compete to bind to the same surface of RAD18 (Fig. 3a, 3b). However, it is puzzling that RAD18 can be recruited by NBS1 and can function with RAD6 if bindings to NBS1 and RAD6 are mutually exclusive [19]. This question was resolved through our immune-precipitation experiment with an antibody to RAD18, showing homodimer formation of RAD18 in cells, which was later confirmed by another group using X-ray crystallography [32]. Thus, NBS1 plays a key role in the initiation of translesion DNA synthesis, although its functional role in the response to DSBs remains a mystery.
Fig. 3.
Structural and functional similarity of NBS1 and RAD6. (a) NBS1 has sequence homology with RAD6 in their RAD18-binding regions. (b) NBS1 and RAD6 compete for the same binding surface of RAD18.
ATM/RPA AND CELL CYCLE CHECKPOINT
In response to DSBs, ATM undergoes spatial relocalization and catalytic activation, resulting in growth arrest via regulation of the cell cycle checkpoint. The G1 checkpoint is initiated through phosphorylation of p53, MDM2 and Chk2 kinase by ATM activation, and similarly the G2 checkpoint with phosphorylation of CDC2 by activation of both ATM and Chk2 kinases [1, 2, 33]. In undamaged cells, the dormant ATM protein is present as a homodimer, which dissociates into an active monomer upon generation of DSBs. This model was originally associated with autophosphorylation of human ATM at Ser1981 for monomerization of ATM dimers [34]. However, surprisingly, mutation of Ser1981 at the autophosphorylation sites in mouse ATM did not affect ATM activation and subsequent signal transduction. Further studies suggested that dispersion of PP2A and PP5 phosphatases from ATM is more important for ATM activation. Another hypothesis is that autophosphorylation is required only for the retention of activated ATM at the DSB sites, rather than for the initial recruitment of ATM. Thus, ATM activation is still being debated, but it is considered that NBS1 plays a central role in ATM recruitment at DSB sites. NBS1 binds to ATM at the extreme C terminus (Fig. 1) and the mutation of ATM-binding sites abrogates ATM accumulation at DSB sites. Similarly, a mutant lacking the binding sites showed a severely affected G2 checkpoint, indicating that the interaction of ATM and NBS1 via the ATM-binding domain is required for the induction of IR-induced checkpoints [34].
ATR, a so-called ATM- and Rad3-related kinase, is a member of the ATM kinase family, and both ATR and ATM are known as master regulators of the DNA damage response. DSBs rapidly induce ATM activation, which in turn produces ssDNAs at the DSB end for initiation of HRR. Since ssDNA is the DNA structure to trigger for ATR activation, ATR kinase is progressively activated in a TopBP1-dependent manner and then phosphorylates Chk1 kinase at Ser345, leading to phosphorylation of CDC2 and following inactivation of CDK1. The ssDNA is usually coated with RPA immediately after production, and can directly interact with NBS1 at the RPA-binding domain, leading to the recruitment of TopBP1 to the ssDNA sites and resulting in ATR activation [35, 36]. According to this model, mutation of NBS1 at the RPA-binding region affects the G2 checkpoint but does not affect ATM activation. ATR and ATM appear to have a distinct role in cell cycle checkpoints: the NBS1–ATM pathway induces the checkpoint immediately after DSB generation and the NBS1–ATR pathway likely maintains the checkpoint during HRR.
NBS1 IN ICL REPAIR AND CENTROSOME MAINTENANCE
One NBS patient, designated as EUFA1020, has been misdiagnosed as having FA, an autosomal recessive genetic disease [37]. FA is characterized by aplastic anemia, skeletal anomalies and occasionally microcephaly. Cells in FA exhibit defective repair of inter-crosslinking DNA (ICL) damage, which is caused by anti-tumor drugs such as mitomycin C and cisplatin. Some of these clinical symptoms and cellular features of FA are similar to those of NBS, as NBS occasionally displays skeletal anomalies and cells of patients are highly sensitive to mitomycin C. Repair of ICL damage consists of sequential steps including: (1) detection of ICL damage by FA protein complex; (2) nucleolytic incisions in the strand by structure-specific nucleases for unhooking the ICL; (3) TLS-mediated DNA synthesis of the strand opposite to that flanking the ICLs; (4) removal of remaining ICL adducts and gap filling; and (5) rejoining DSBs generated when the replication fork stalls at ICL [38]. Because NBS1 is a HRR protein, it may be involved in step 5 of DSB repair. However, an unexpected role of NBS1 was shown by our removal assay of ICL damage using a dot blot with psoralen–polyethylene oxide–biotin linked to ICL [39]. NBS cells show a defect in ICL removal at step 4, indicating a defect in either step 2 or step 3 but not step 5. Because NBS1 plays a critical role in TLS, NBS1 may be involved in step 3 of ICL repair. This explanation is further supported by a recent observation that high cellular sensitivity to cisplatin was observed after the depletion of TLS polymerase, Pol eta, which is recruited to the damaged sites by NBS1 [19, 40].
Microcephaly is a hallmark symptom of NBS. Genetic disorders characterized by microcephaly, such as primary recessive microcephaly (MCPH) and ATR–Seckel, are defective in centrosome maintenance [41, 42]. Similarly, NBS1 is required for proper centrosome duplication, and the depletion of NBS1 causes the accumulation of excess centrosomes [43]. The excess centrosomes in these syndromes cause defects in cell division and the resulting depletion of the neural progenitor pool, consequently leading to small brain size. This process is also observed in IR-induced microcephaly, as observed in atomic bomb survivors in Hiroshima and Nagasaki who were exposed in utero and have a high risk of microcephaly (56% incidence at 1 Sv exposure) [44]. IR exposure induces excess centrosomes in cultured human and mouse cells, similar to those in NBS1-deficient cells and ATR-deficient cells [45, 46].
CONCLUSION
NBS patients show several clinical symptoms and cellular features, which are associated with the multiple functions of NBS1, including high IR sensitivity and immunodeficiency associated with both defects in chromatin remodeling and DNA repair (HRR and NHEJ), microcephaly associated with a defect in centrosome maintenance, sun sensitivity associated with a defect in TLS and predisposition to malignancy associated with abnormal cell cycle checkpoints. Upon DSB generation, NBS1 coordinately regulates these pathways by binding at a unique C-terminal region to five key proteins: RNF20 (chromatin remodeling), RAD18 (TLS), ATM and RPA (cell cycle checkpoints), and MRE11 (HRR and NHEJ). Further study of the structural modification of NBS1 and the time course of binding selectivity could broaden our understanding of their precise regulatory mechanisms in response to DSBs.
FUNDING
Funding to pay the Open Access publication charges for this article was provided by the Research Accomplishment Report Committee of the 15th International Congress of Radiation Research.
ACKNOWLEDGMENT
The author wishes to thank the laboratory members and staff who have assisted in this NBS1 project.
REFERENCES
- 1.Tauchi H, Matsuura S, Kobayashi J et al. . Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene 2002;21:8967–80. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi J, Antoccia A, Tauchi H et al. . NBS1 and its functional role in the DNA damage response. DNA Repair (Amst) 2004;3:855–61. [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto S, Iijima K, Mochizuki D et al. . Homologous recombination repair is regulated by domains at the N- and C-terminus of NBS1 and is dissociated with ATM functions. Oncogene 2007;26:6002–9. [DOI] [PubMed] [Google Scholar]
- 4.Kato A, Komatsu K. RNF20-SNF2H pathway of chromatin relaxation in DNA double-strand break repair. Genes (Basel) 2015;6:592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito Y, Komatsu K. Functional role of NBS1 in radiation damage response and translesion DNA Synthesis. Biomolecules 2015;5:1990–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weemaes CM, Hustinx TW, Scheres JM et al. . A new chromosomal instability disorder: the Nijmegen breakage syndrome. Acta Paediatr Scand 1981;70:557–64. [DOI] [PubMed] [Google Scholar]
- 7.Antoccia A, Kobayashi J, Tauchi H et al. . Nijmegen breakage syndrome and functions of the responsible protein, NBS1. Genome Dyn 2006;1:191–205. [DOI] [PubMed] [Google Scholar]
- 8.Komatsu K, Okumura Y, Kodama S et al. . Lack of correlation between radiosensitivity and inhibition of DNA synthesis in hybrids (A-T x HeLa). Int J Radiat Biol 1989;56:863–7. [DOI] [PubMed] [Google Scholar]
- 9.Komatsu K, Matsuura S, Tauchi H et al. . The gene for Nijmegen breakage syndrome (V2) is not located on chromosome 11. Am J Hum Genet 1996;58:885–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuura S, Weemaes C, Smeets D et al. . Genetic mapping using microcell-mediated chromosome transfer suggests a locus for Nijmegen breakage syndrome at chromosome 8q21-24. Am J Hum Genet 1997;60:1487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuura S, Tauchi H, Nakamura A et al. . Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet 1998;19:179–81. [DOI] [PubMed] [Google Scholar]
- 12.Tauchi H, Matsuura S, Isomura M. Sequence analysis of an 800-kb genomic DNA region on chromosome 8q21 that contains the Nijmegen breakage syndrome gene, NBS1. Genomics 1999;55:242–7. [DOI] [PubMed] [Google Scholar]
- 13.Varon R, Vissinga C, Platzer M et al. . Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 1998;93:467–76. [DOI] [PubMed] [Google Scholar]
- 14.Carney JP, Maser RS, Olivares H et al. . The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 1998;93:477–86. [DOI] [PubMed] [Google Scholar]
- 15.Varon R, Dutrannoy V, Weikert G et al. . Mild Nijmegen breakage syndrome phenotype due to alternative splicing. Hum Mol Genet 2006;15:679–89. [DOI] [PubMed] [Google Scholar]
- 16.Salewsky B, Hildebrand G, Rothe S et al. . Directed alternative splicing in Nijmegen breakage syndrome: proof of principle concerning its therapeutical application. Mol Ther 2016;24:117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warcoin M, Lespinasse J, Despouy G et al. . Fertility defects revealing germline biallelic nonsense NBN mutations. Hum Mutat 2009;30:424–30. [DOI] [PubMed] [Google Scholar]
- 18.Shimada H, Shimizu K, Mimaki S et al. . First case of aplastic anemia in a Japanese child with a homozygous missense mutation in the NBS1 gene (I171 V) associated with genomic instability. Hum Genet 2004;115:372–6. [DOI] [PubMed] [Google Scholar]
- 19.Yanagihara H, Kobayashi J, Tateishi S et al. . NBS1 recruits RAD18 via a RAD6-like domain and regulates Pol η-dependent translesion DNA synthesis. Mol Cell 2011;43:788–97. [DOI] [PubMed] [Google Scholar]
- 20.Tauchi H, Kobayashi J, Morishima K et al. . The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50/hMRE11/NBS1 complex DNA repair activity. J Biol Chem 2001;27:12–15. [DOI] [PubMed] [Google Scholar]
- 21.Tauchi H, Kobayashi J, Morishima K et al. . Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 2002;420:93–8. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura K, Kato A, Kobayashi J et al. . Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell 2011;41:515–28. [DOI] [PubMed] [Google Scholar]
- 23.Shiotani B, Nguyen HD, Håkansson P et al. . Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep 2013;3:1651–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005;434:605–11. [DOI] [PubMed] [Google Scholar]
- 25.Falck J, Forment JV, Coates J et al. . CDK targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination. EMBO Rep 2012;13:561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haber JE. The many interfaces of Mre11. Cell 1998;95:583–6. [DOI] [PubMed] [Google Scholar]
- 27.Moyal L, Lerenthal Y, Gana-Weisz M et al. . Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell 2011;41:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira DV, Kato A, Nakamura K et al. . Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J Cell Sci 2014;127:763–72. [DOI] [PubMed] [Google Scholar]
- 29.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 2011;18:831–9. [DOI] [PubMed] [Google Scholar]
- 30.Klement K, Luijsterburg MS, Pinder JB et al. . Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol 2014;207:717–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komatsu K. presentation at the 15th International Congress of Radiation Research from 25–29 May 2015, Kyoto, Japan. [Google Scholar]
- 32.Huang A, Hibbert RG, de Jong RN et al. . Symmetry and asymmetry of the RING-RING dimer of Rad18 . J Mol Biol 2011;410:424–35. [DOI] [PubMed] [Google Scholar]
- 33.Buscemi G, Savio C, Zannini L et al. . Chk2 activation dependence on Nbs1 after DNA damage. Mol Cell Biol 2001;21:5214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 2013;14:197–210. [PubMed] [Google Scholar]
- 35.Morishima K, Sakamoto S, Kobayashi J et al. . TopBP1 associates with NBS1 and is involved in homologous recombination repair. Biochem Biophys Res Commun 2007;362:872–9. [DOI] [PubMed] [Google Scholar]
- 36.Hirokawa T, Shiotani B, Shimada M et al. . CBP-93872 inhibits NBS1-mediated ATR activation, abrogating maintenance of the DNA double-strand break-specific G2 checkpoint. Cancer Res 2014;74:3880–9. [DOI] [PubMed] [Google Scholar]
- 37.Nakanishi K, Taniguchi T, Ranganathan V et al. . Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol 2002;4:913–20. [DOI] [PubMed] [Google Scholar]
- 38.Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 2012;26:1393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchida K, Komatsu K. Impaired removal of DNA interstrand cross-link in Nijmegen breakage syndrome and Fanconi anemia, but not in BRCA-defective group. Cancer Sci 2008;99:2238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee YS, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci USA 2014;111:2954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome. Trends Genet 2009; 25:501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmood S, Ahmad W, Hassan MJ. Autosomal Recessive Primary Microcephaly (MCPH): clinical manifestations, genetic heterogeneity and mutation continuum. Orphanet J Rare Dis 2011; 6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimada M, Sagae R, Kobayashi J et al. . Inactivation of the Nijmegen breakage syndrome gene leads to excess centrosome duplication via the ATR/BRCA1 pathway. Cancer Res 2009;69:1768–75. [DOI] [PubMed] [Google Scholar]
- 44.Otake M, Schull WJ, Yoshimaru H. A review of forty-five years study of Hiroshima and Nagasaki atomic bomb survivors. Brain damage among the prenatally exposed. J Radiat Res 1991;32(Suppl):249–64. [DOI] [PubMed] [Google Scholar]
- 45.Shimada M, Kobayashi J, Hirayama R. Differential role of repair proteins, BRCA1/NBS1 and Ku70/DNA-PKcs, in radiation-induced centrosome overduplication. Cancer Sci 2010;101:2531–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimada M, Hirayama R, Komatsu K. High LET radiation amplifies centrosome overduplication through a pathway of γ-tubulin monoubiquitination. Int J Radiat Oncol Biol Phys 2013;86:358–65. [DOI] [PubMed] [Google Scholar]