Abstract
The Hippo pathway is an important regulator of organ size and tumorigenesis. It is unclear, however, how Hippo signaling provides the cellular building blocks required for rapid growth. Here, we demonstrate that transgenic zebrafish expressing an activated form of the Hippo pathway effector Yap1 (also known as YAP) develop enlarged livers and are prone to liver tumor formation. Transcriptomic and metabolomic profiling identify that Yap1 reprograms glutamine metabolism. Yap1 directly enhances glutamine synthetase (glul) expression and activity, elevating steady-state levels of glutamine and enhancing the relative isotopic enrichment of nitrogen during de novo purine and pyrimidine biosynthesis. Genetic or pharmacological inhibition of GLUL diminishes the isotopic enrichment of nitrogen into nucleotides, suppresses hepatomegaly and the growth of liver cancer cells. Consequently, Yap-driven liver growth is susceptible to nucleotide inhibition. Together, our findings demonstrate that Yap1 integrates the anabolic demands of tissue growth during development and tumorigenesis by reprogramming nitrogen metabolism to stimulate nucleotide biosynthesis.
Introduction
Many signaling pathways have been identified to regulate liver growth during organ development and tumor formation. It is unclear, however, how these signals provide energy and cellular building blocks to enable rapid cell division and organ growth. The Hippo signaling pathway has emerged as an evolutionarily conserved master regulator of organ size and tissue growth1. The Hippo kinase cascade regulates the transcriptional co-activator Yap that binds the Tead family of transcription factors to enhance cell proliferation, inhibit cell death and alter cell fate2. The Hippo pathway is frequently perturbed in human cancers3, and many oncogenic pathways feed into the Hippo pathway and sustain growth in a Yap-dependent manner. The molecular mechanisms by which Yap target genes contribute to rapid cell proliferation to increase organ size remain elusive.
Altered cellular metabolism is a hallmark of cancer cells4. Several studies have focused attention on the alternate means by which cancer cells produce ATP, including aerobic glycolysis5 or the anaplerotic use of alternate fuel sources such as glutamine6. While the mTOR pathway can enhance de novo synthesis of building blocks and macromolecules required for cell growth7, it remains unknown whether the Hippo pathway can similarly reprogram cellular metabolism to match the demands for rapid tissue growth.
Here, we investigate zebrafish with hepatocyte-specific transgenic expression of activated yap1S87A that develop hepatomegaly as larvae and are prone to liver tumor formation as adults. Transcriptomic and metabolomic analyses reveal that Yap1 induces glutamine synthetase (glul) to elevate glutamine levels and stimulate de novo nucleotide biosynthesis. Treatment studies in larval and adult zebrafish demonstrate that inhibition of Glul or nucleotide biosynthesis diminishes Yap-induced liver growth. YAP-dependent regulation of GLUL is required for growth of human liver cancer cells. Our studies define a role for Yap in reprogramming cellular metabolism to fulfill the anabolic requirements for tissue growth and illustrate the therapeutic potential of targeting Yap-driven metabolic alterations.
Results
Hepatocyte-specific overexpression of activated Yap causes hepatomegaly and accelerates DMBA-induced liver tumor formation
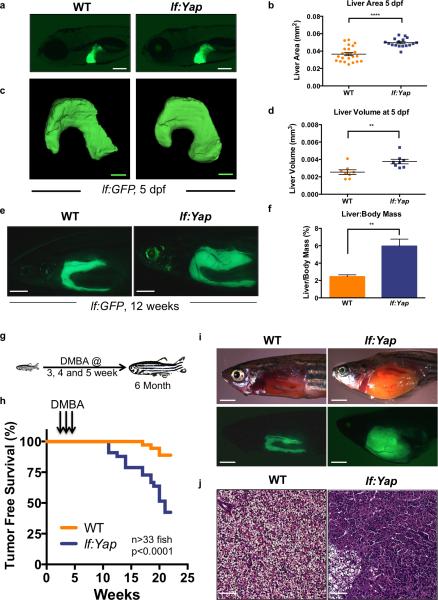
To investigate the impact of Yap activation on liver growth in zebrafish larvae, transgenic zebrafish (Tg(−2.8fabp10a:yap1-1βS87A), referred to as lf:Yap, were generated with hepatocyte-specific expression of constitutively active Yap1, homologous to human YAP1S127A (Fig.S1A). Liver development was visualized using the Tg(−2.8fabp10a:eGFP) (lf:GFP) hepatocyte reporter line8. At 5 days post fertilization (dpf), lf:Yap fish exhibit a 36% increase in liver area (Fig.1A,B) compared to wild-type (WT) siblings. Single-plane illumination confocal microscopy confirmed liver volume enlargement and preservation of gross architectural features of the bi-lobed larval liver in lf:Yap trangenics (Fig.1C,D). Increased liver size was accompanied by hepatocyte hyperproliferation without significantly enlarged cell size, as determined by histological and fluorescence-activated cell sorting (FACS) analyses (Figs.S1B,C). Enhanced liver size in lf:Yap fish persisted into adulthood at 16 weeks post fertilization (wpf), as quantified by fluorescent liver area (Figs.1E, S1D) and a two-fold increase in liver-to-body weight ratio (Fig.1F). Histological analysis of lf:Yap fish revealed liver hyperplasia with increased cellularity, cell proliferation, decreased glycogen, and focally elevated bile duct density (Fig.S1E). These data demonstrate that Yap induces hepatomegaly and promotes liver hyperplasia in zebrafish.
Figure 1. Hepatocyte-specific overexpression of activated Yap causes hepatomegaly and enhances DMBA-induced liver tumor formation.
(a) Fluorescence microscopic analysis of hepatomegaly in lf:GFP;lf:Yap bigenic larvae at 5 dpf. Scale bar: 200μm.
(b) Quantitative analysis of fluorescent liver area in WT and lf:Yap larvae at 5 dpf. n=23 and 17 WT and lf:Yap larvae respectively; see Supplementary Table 4. ****p<0.0001, two-sided Student's t-test, values represent the mean±SEM.
(c) Confocal tomography of liver morphology after lightsheet imaging in WT and lf:Yap larvae at 5 dpf. Scale bar: 100μm.
(d) Quantitative analysis of liver volume at 5 dpf. n=8 larvae; see Supplementary Table 4. **p<0.01, two-sided Student's t-test, values represent the mean±SEM.
(e) Fluorescence microscopic analysis of hepatomegaly in dissected lf:Yap at 12 wpf. Scale bar: 2mm.
(f) Quantitative determination of hepatomegaly in adults as determined by liver:body weight ratio. n=6 adult zebrafish; see Supplementary Table 4. **p<0.01, two-sided Student's t-test, values represent the mean±SEM.
(g) Schematic illustrating the timeline for DMBA exposure.
(h) Kaplan-Meier survival plot showing tumor-free survival of WT and lf:Yap adults exposed to DMBA. n=36 and 33 WT and lf:Yap adult zebrafish respectively; see Supplementary Table 4. p<0.0001; Log rank Mantel-Cox test.
(i) Fluorescence microscopy examination of gross liver morphology of WT and lf:Yap tumors following dissection. Scale bar: 2mm.
(j) H&E stain of liver sections in WT and lf:Yap transgenic fish harboring tumors. Scale bar: 50μm.
To examine whether Yap predisposes to neoplasia, juvenile WT and lf:Yap transgenics were exposed to the carcinogen dimethylbenzanthracene (DMBA)9 (Fig.1G). lf:Yap transgenics formed liver tumors earlier and with greater frequency than WT siblings (Fig.1H), with liver masses occupying the entire abdominal cavity (Fig.1I). Tumor differentiation subtypes ranged from hepatocellular carcinoma (HCC), with occasional foci of sarcomatoid cytology and/or peliosis/spongiosis-hepatis-like change, to cholangiocarcinoma (CCA) (Figs.1J,S1F,G). Ascites was noted in 30% (7/23) of lf:Yap transgenics and 0% (0/5) of WT. These data demonstrate that Yap exerts oncogenic activity enhance chemically induced hepatocarcinogenesis.
Yap alters expression of metabolism-related genes and enhances glul expression
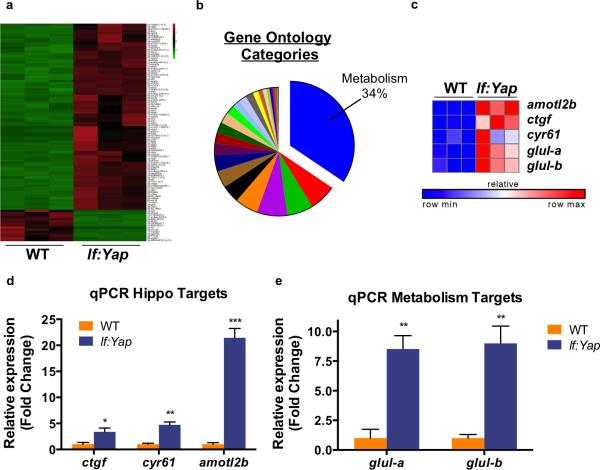
To identify signals responsible for Yap-driven liver hyperplasia, we performed transcriptional profiling in WT and lf:Yap adult livers, revealing a large number of differentially expressed genes (Fig.2A). Gene set enrichment analysis was consistent with a mammalian Hippo signature (Fig.S2A)10. Intriguingly, gene ontology analysis indicated a relationship to metabolism in 34% of annotated categories (Fig.2B), and ~60% of the top 20 biological processes upregulated by Yap were related to metabolism (Fig.S2B). Amongst the most differentially upregulated genes were established Yap targets amotl2, ctgf and cyr61, as well as zebrafish orthologues of glutamine synthetase (GLUL), glula and glulb (Fig.2C). Quantitative PCR (qPCR) verified induction of ctgf, cyr61, amotl2, glula and glulb in lf:Yap livers (Fig.2D,E). GLUL plays a central role in nitrogen assimilation in the liver by catalyzing the formation of glutamine from glutamate and ammonia. While GLUL expression can be regulated by Wnt activity11, Wnt targets cyclinD1, axin2 or cd44 were not induced in lf:Yap transgenics (Fig.S2C,D). yap expression, but not taz, was dramatically increased in lf:Yap livers (Fig.S2E). Collectively, these results suggest that Yap regulates glutamine metabolism.
Figure 2. Yap alters expression of metabolism-related genes and enhances glul expression.
(a) Heatmap of statistically significant differential gene expression between dissected WT and lf:Yap livers. Green indicates decreased expression, while red demonstrates increased expression.
(b) Pie chart of Gene ontology (GO) categories upregulated by Yap. Biological processes related to metabolism are represented in blue.
(c) Heatmap analysis of RNAseq data illustrates the induction of individual Yap target genes (ctfg, amotL2, cyr61) and nitrogen metabolism genes glula and glulb.
(d) qPCR validation of Yap target gene expression in dissected WT and lf:Yap transgenic livers. n=4 WT and lf:Yap adult livers (with respect to ctgf expression). n=3 WT and lf:Yap adult livers (with respect to cyr61 and amotl2 expression); see Supplementary Table 4. *p<0.05, **p<0.01, ***p<0.001, two-sided Student's t-test, values represent the mean±SEM.
(e) qPCR validation of Glul induction in dissected WT and lf:Yap transgenic livers. n=3 WT and lf:Yap adult livers; see Supplementary Table 4. **p<0.01, two-sided Student's t-test, values represent the mean±SEM.
Yap transcriptionally upregulates GLUL in an evolutionarily conserved fashion
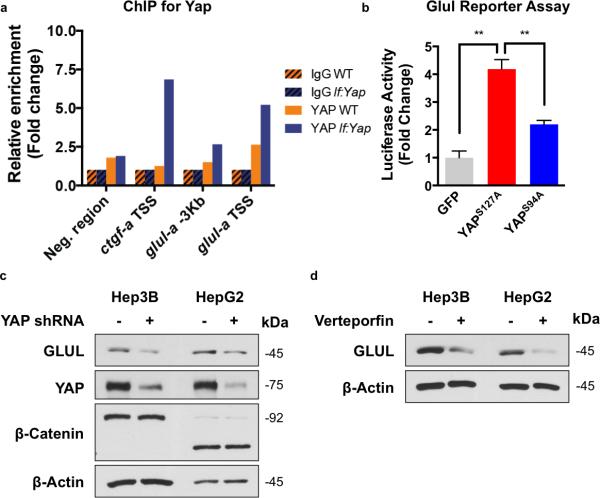
To determine whether Yap directly influences GLUL expression, chromatin immunoprecipitation (ChIP) on isolated adult zebrafish liver lysates was performed. Yap1 occupancy was enriched at the transcriptional start site (TSS) of the glula promoter as avidly as at ctgf (Fig.3A, S3A). Mammalian conservation of Yap-dependent GLUL regulation was demonstrated in, a luciferase reporter assay in HEK-293T cells: constitutively active YAP1S127A enhanced GLUL expression, whereas mutant YAP1 lacking the ability to co-activate TEAD (YAP1S94A) did not equally induce GLUL (Fig.3B). Promoter fragmentation analysis found several regions within the promoter to contribute to the total YAP1-induced activity increase, suggesting several binding sites (Fig.S3B). ChIP analysis for YAP1 in HepG2 cells revealed occupation of the GLUL promoter in the region associated with luciferase activation (Fig.S3C). To further demonstrate the dependency of GLUL expression on Yap activity, genetic and chemical strategies were utilized: shRNA-mediated knockdown of YAP1 and chemical inhibition with verteporfin12 in Hep3B and HepG2 liver cancer cell lines reduced GLUL (Fig.3C,D). Together, these studies demonstrate that direct regulation of GLUL expression by YAP1 is evolutionarily conserved in humans.
Figure 3. Yap transcriptionally upregulates GLUL in an evolutionarily conserved fashion.
(a) Chromatin Immunoprecipitation (ChIP) qPCR analysis of Yap enrichment at glula promoter in WT and lf:Yap adult livers. Shown is the average of 2 WT and lf:Yap adult liver chromatin preps, each derived from a pooled sample of 2 livers; see Supplementary Table 4.
(b) Luciferase GLUL reporter assay in cultured Hek293 cells expressing GFP, YAP1S127A or YAP1S94A. n=3 biologically independent replicates; see Supplementary Table 4. *p<0.05, two-sided Student's t-test, values represent the mean±SEM.
(c) Immunoblot analysis of GLUL, YAP, β-catenin and β-actin expression in HepG2 cells infected with shYAP retrovirus.
(d) Immunoblot analysis of GLUL and β-actin expression in Hep3B and HepG2 cells exposed to verteporfin (VP). Immunoblots are representative of three independent experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Yap reprograms nitrogen metabolism by enhancing GLUL-dependent anabolic assimilation of ammonia for de novo nucleotide biosynthesis
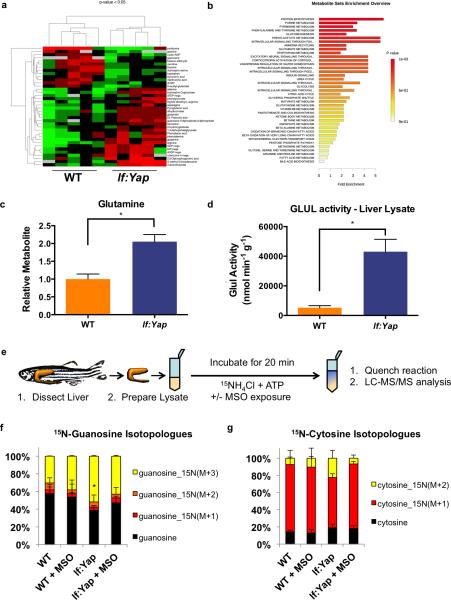
To examine the effect of Yap activation on cellular metabolism, adult WT and lf:Yap livers were subjected to polar steady-state metabolomics analysis by liquid chromatography-based tandem mass spectrometry (LC-MS/MS)13, determining >250 metabolite profiles. Hierarchical clustering based on metabolite abundance revealed significant changes in steady-state levels of many metabolites (Fig.4A, n=5, p<0.05), including enrichment of metabolites associated with protein biosynthesis, purine and pyrimidine metabolism (Fig.4B). Significantly, lf:Yap livers contained twice the amount of glutamine as WT (Fig.4C). Further, GLUL enzyme activity in lf:Yap liver homogenate was increased 10-fold compared to WTs (Fig.4D), and immunohistochemistry demonstrated uniform overexpression of Glul in lf:Yap hepatocytes and heterogeneous expression in Yap-driven liver tumors (Figure S4A). Ammonia excretion rates were decreased when assessed in vivo in lf:Yap fish, consistent with enhanced nitrogen retention due to the catalytic role of GLUL in the formation of glutamine from glutamate and ammonia (Fig.S4B). Similarly, metabolomics analysis detected decreased urea levels in lf:Yap livers (Fig.S4C).
Figure 4. Yap reprograms nitrogen metabolism by enhancing GLUL-dependent anabolic assimilation of ammonia for de novo nucleotide biosynthesis.
(a) Clustergram analysis of polar metabolite abundance from dissected WT and lf:Yap livers as determined by LC-MS/MS via selected reaction monitoring (SRM) analysis. n=5 WT and lf:Yap adult livers; see Supplementary Table 4. p<0.05, two-sided Student's t-test, values represent the mean±SEM.
(b) Metabolite Set Enrichment of polar metabolites from WT and lf:Yap livers as determined by SRM analysis.
(c) Steady state abundance of glutamine in WT and lf:Yap livers as determined by SRM analysis. n=10 WT and lf:Yap adult livers; see Supplementary Table 4. *p<0.01, two-sided Student's t-test, values represent the mean±SEM.
(d) Enzymatic assay of glutamine synthetase activity in dissected WT and lf:Yap transgenic livers. n=8 and 13 WT and lf:Yap adult liver lysates respectively; see Supplementary Table 4. *p<0.01, two-sided Student's t-test, values represent the mean±SEM.
(e) Scheme to monitor the relative isotopic enrichment of 15NH4Cl in the presence or absence of MSO in liver lysates from WT and lf:Yap transgenics.
(f) Percentage of 15N-labelled Guanosine isotopologues in WT and lf:Yap transgenic liver lysates following ammonia assimilation in the presence or absence or MSO. n=5 biologically independent WT and lf:Yap adult liver lysates; see Supplementary Table 4. *p<0.05, two-sided Student's t-test, values represent the mean±SEM.
(g) Percentage of 15N-labelled Cytosine isotopologues in WT and lf:Yap transgenic liver lysates following ammonia assimilation in the presence or absence or MSO. n= 5 biologically independent WT and lf:Yap adult liver lysates; see Supplementary Table 4. Values represent the mean±SEM.
To more precisely determine the GLUL-dependent fate of ammonia, liver lysates were incubated with 15N-labelled NH4Cl and ATP with or without the GLUL inhibitor methionine sulfoximine (MSO) (Fig.4E). Glutamine is essential for de novo nucleotide biosynthesis as it provides nitrogen atoms for both purines and pyrimidines: three of five nitrogen atoms present on the purine guanosine are contributed by glutamine (two in the purine ring, and one in the amino group at C-2). Similarly, two of three nitrogen atoms present on the pyrimidine cytosine are contributed by glutamine. Incorporation of 15N into nucleotide precursors was monitored by LC-MS/MS using selected reaction monitoring (SRM) for potential 15N isotopologues and revealed that 15N-labelled isotopologues of guanosine and cytosine were increased in lf:Yap liver lysates (Figs.4F,G, S4D,E). This response was inhibited by MSO. In contrast, little change was observed in the relative isotopic enrichment of 15N into glutamine or histidine (Fig.S4F,G). These results demonstrate that Yap reprograms nitrogen metabolism by activating GLUL and promoting the incorporation of ammonia-bound nitrogen to form glutamine as a precursor for nucleotide biosynthesis.
GLUL activity and nucleotide biosynthesis contribute to Yap-induced hepatomegaly and the growth of liver cancer cells
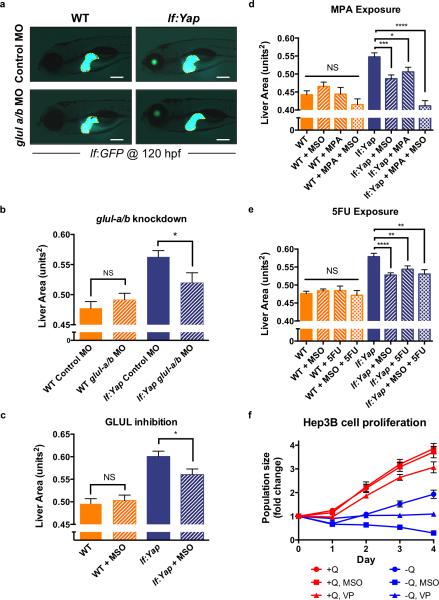
Given the dramatic impact of Yap on GLUL expression and nitrogen metabolism, the requirement of GLUL in mediating Yap-induced rapid liver growth was examined utilizing morpholino (MO)-mediated knockdown of glula and glulb (Fig.S5A). Yap-induced hepatomegaly at 5 dpf was attenuated in glula/glulb morphants (Fig.5A,B). To demonstrate potential therapeutic implications, larvae were exposed to MSO from 3-5 dpf, which effectively inhibited GLUL activity (Fig.S5B) and significantly inhibited Yap-driven hepatomegaly (Fig.5C). Importantly, both glula/glulb knockdown and MSO treatment had no effect on liver size or morphology in WT larvae (Fig.5A-C). Given the increased relative isotopic enrichment of 15N in nucleotide intermediates in lf:Yap transgenics, consistent with stimulation of nucleotide biosynthesis, we investigated whether inhibitors of purine or pyrimidine biosynthesis impacted Yap-driven hepatomegaly: the purine analogue mycophenolic acid (MPA), which inhibits inosine monophosphate dehydrogenase (IMPDH), significantly suppressed Yap-driven hepatomegaly (Fig.5D). Furthermore, the combined treatment of MSO and MPA had an additive effect on liver size (Fig.5D). Similarly, exposure to the pyrimidine analogue 5-Fluorouracil (5FU), which inhibits thymidylate synthase (TS), also suppressed Yap-driven hepatomegaly, but showed no synergy with MSO (Fig.5E). Interestingly, in each case there was no significant impact on WT liver growth (Fig.5D,E). To examine the conserved nature of the Yap-GLUL axis, Hep3B and HepG2 human liver cancer cells were cultured in the presence or absence of glutamine (Q), MSO or VP. VP suppressed proliferation of Hep3B cells in full media and caused cytostasis in glutamine-deficient media (Fig.5F, n=3). Interestingly, MSO exposure had no effect on Hep3B cell proliferation in full media, however, in glutamine-deprived conditions, Hep3B cell number decreased significantly (Fig.5F). Similar effects of VP and MSO were observed in HepG2 cells cultured in glutamine-deprived conditions (Fig.S5C). Together, these studies suggest that GLUL and nucleotide biosynthesis are conditionally required for rapid cell proliferation in the context of Yap-driven hepatomegaly and liver cancer cell growth.
Figure 5. GLUL activity and nucleotide biosynthesis contribute to Yap-induced hepatomegaly and the growth of liver cancer cells.
(a) Morpholino knockdown of glula/b in WT and lf:Yap larvae caused a reduction in yap1-induced hepatomegaly as determined by fluorescence microscopy at 5 dpf. Scale bar: 200μm.
(b) Quantitative analysis of the effect of glula/b morpholino knockdown on liver size. n=45, 42, 46 and 34 WT control morphant, WT glula/b morphant, lf:Yap control morphant and lf:Yap glula/b morphant larvae respectively; see Supplementary Table 4. *p<0.05, two-sided Student's t-test, values represent the mean±SEM.
(c) Quantitative analysis of the effect of MSO treatment from 3-5 dpf on liver size as determined by fluorescence microscopy. n=89, 94, 91 and 104 WT, WT+MSO, lf:Yap and lf:Yap+MSO larvae respectively; see Supplementary Table 4. *p<0.05, two-sided Student's t-test, values represent the mean±SEM.
(d) Quantitative analysis of the effect of MPA treatment from 3-5 dpf on liver size as determined by fluorescence microscopy. n=30, 33, 35, 47, 46, 53, 88 and 64 WT, WT+MSO, WT+MPA, WT+MSO+MPA, lf:Yap, lf:Yap+MSO, lf:Yap+MPA and lf:Yap+MSO+MPA larvae respectively; see Supplementary Table 4. *p<0.05, ***p<0.001, ****p<0.0001, two-sided Student's t-test, values represent the mean±SEM.
(e) Quantitative analysis of the effect of 5FU treatment from 3-5 dpf on liver size as determined by fluorescence microscopy. n=101, 185, 56, 50, 103, 210, 105 and 92 WT, WT+MSO, WT+5FU, WT+MSO+5FU, lf:Yap, lf:Yap+MSO, lf:Yap+5FU and lf:Yap+MSO+5FU larvae respectively; see Supplementary Table 4. **p<0.01, ****p<0.0001, two-sided Student's t-test, values represent the mean±SEM.
(f) Proliferation of Hep3B liver cancer cells over 4 days in the presence or absence of glutamine (Q), MSO or VP. n=3 biologically independent replicates; see Supplementary Table 4. Values represent the mean±SEM.
mTOR pathway is not deregulated by Yap expression or GLUL inhibition, but is required for Yap-induced hepatomegaly
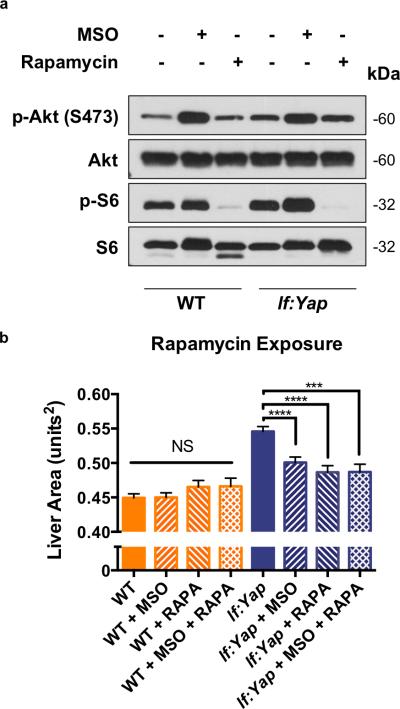
Given the important role of mTOR in stimulating nucleotide biosynthesis to fuel cell growth7, we examined levels of phospho-Akt (p-Akt, S473), an mTORC2 target, as well as phospho-S6 (p-S6), a substrate of S6K: no significant differences were seen in WT and lf:Yap liver homogenates and histological sections (Fig.6A, S6A). Importantly, the mTOR inhibitor Rapamycin (RAPA) completely suppressed p-Akt and p-S6 levels; MSO exposure led to a subtle increase in p-Akt and p-S6 levels (Fig.6A). mTOR pathway activation typically leads to increased cell size7. Hepatocyte size was examined by liver cell counts, liver area and cellularity in liver sections from WT and lf:Yap larvae and was unaffected by Yap expression (Fig.S6B,C,D). To determine whether the mTOR pathway was required for Yap-driven liver growth, WT and lf:Yap larvae were exposed to RAPA with or without MSO, which suppressed Yap-driven liver growth, but did not synergize with MSO (Fig.6B). Together, these experiments demonstrate that Yap does not significantly stimulate the mTOR pathway, however, intact mTOR signaling is necessary for Yap-driven liver growth. Furthermore, the mechanism by which MSO suppresses liver growth is dependent on inhibition of the mTOR pathway.
Figure 6. The mTOR pathway is not deregulated by Yap expression or GLUL inhibition, but it is required for Yap-induced hepatomegaly.
(a) Immunoblot analysis of phospho-Akt (p-Akt, S473), Akt, phospho-S6 (p-S6), and S6 levels in liver tissue isolated from adult WT and lf:Yap transgenic fish 24 hr after exposure to MSO or Rapamycin (RAPA). Immunoblots are representative of three independent experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
(b) Quantitative analysis of the effect of RAPA treatment from 3-5 dpf on liver size as determined by fluorescence microscopy. n=142, 148, 63, 40, 146, 130, 94 and 40 WT, WT+MSO, WT+RAPA, WT+MSO+RAPA, lf:Yap, lf:Yap+MSO, lf:Yap+RAPA and lf:Yap+MSO+RAPA larvae respectively; see Supplementary Table 4. ***p<0.001, ****p<0.0001, two-sided Student's t-test, values represent the mean±SEM.
Yap reprograms the relative isotopic enrichment of nutritional nitrogen into nucleotide biosynthesis in a GLUL-dependent manner to support liver growth
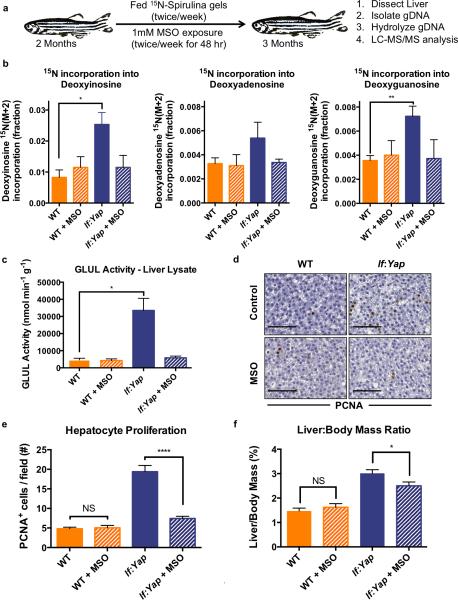
The steady-state metabolomics data indicated that Yap alters nitrogen metabolism, but did not define how nitrogen is utilized in vivo to accelerate tissue growth. To examine the fate of increased glutamine content in lf:Yap transgenics, we developed a feeding methodology for stable isotope labeling in fish to assess relative isotopic enrichment in vivo: 15N-labelled Spirulina gel pellets were prepared to provide a defined and consistent amount of 15N-labelled nutrition. Individual WT and lf:Yap fish were fed twice weekly and maintained in the presence or absence of MSO (twice weekly for 48 hr) for 4 weeks (Fig.7A). Subsequently, liver DNA was extracted and hydrolyzed into nucleotides for analysis (Fig.S7A) to focus exclusively on the fraction of nitrogen incorporated into nucleotides to synthesize DNA and not on other nucleic acid-containing macromolecules, such as RNA. The DNA of lf:Yap transgenic livers contained significantly higher amounts of 15N-labelled nucleotides than WT, with 3 times as much deoxyinosine, twice as much deoxyguanosine and 65% more deoxyadenosine (Fig.7B). Yap-induced increase in nucleotide biosynthesis was mitigated to baseline levels by MSO. The relative isotopic enrichment reveals that Yap reprograms nitrogen metabolism by stimulating anabolic incorporation of glutamine into nucleotides in a GLUL-dependent manner.
Figure 7. Yap reprograms the relative isotopic enrichment of nutritional nitrogen into nucleotide biosynthesis in a Glul-dependent manner to support liver growth.
(a) Scheme describing the long-term 15N-Spirulina feeding studies in adult fish.
(b) Abundance of 15N-labelled nucleotide (Deoxyinosine, Deoxyadenosine and Deoxyguanosine) isotopologues (M+2 fraction) from hydrolyzed genomic DNA of dissected livers from WT and lf:Yap fish, as determined by LC-MS. n=5 WT and lf:Yap adult livers; see Supplementary Table 4. *p<0.05, **p<0.01, two-sided Student's t-test, values represent the mean±SEM.
(c) Glul activity in liver lysates extracted from WT and lf:Yap fish in the presence or absence of MSO. n=12, 12, 18 and 10 biologically independent WT, WT+MSO, lf:Yap and lf:Yap+MSO adult liver lysates respectively; see Supplementary Table 4. *p<0.001, two-sided Student's t-test, values represent the mean±SEM.
(d) Immunohistochemical analysis of PCNA staining in liver sections from WT and lf:Yap transgenic fish 24 hr after exposure to DMSO or MSO. Scale bar: 50μm.
(e) Quantification of hepatocyte proliferation (PCNA+ cells/field) in liver sections from WT and lf:Yap transgenic fish 24 hr after exposure to DMSO or MSO. n=51, 47, 48 and 63 independent WT, WT+MSO, lf:Yap and lf:Yap+MSO immunohistochemically (PCNA) stained fields of view respectively. Fields of view were derived from 4 zebrafish; see Supplementary Table 4. ****p<0.0001, two-sided Student's t-test, values represent the mean±SEM.
(f) Effect of MSO intervention on liver:body weight ratios in WT and lf:Yap adult fish. n=12, 12, 11 and 10 WT, WT+MSO, lf:Yap and lf:Yap+MSO adult zebrafish respectively; see Supplementary Table 4. *p<0.05, two-sided Student's t-test, values represent the mean±SEM.
To evaluate functional consequences and potential therapeutic impact of GLUL inhibition on adult liver hyperplasia, fish were exposed to MSO twice weekly over a 4-week period to efficiently inhibit GLUL activity in vivo (Fig.7D). Short-term exposure to MSO (for 48 hr) suppressed hepatocyte proliferation, as examined by quantification of PCNA+ nuclei in histological sections (Fig.7D,E). Importantly, MSO exposure did not cause cell death as determined by TUNEL staining (Fig.S7B). Prolonged MSO treatment significantly reduced hepatomegaly in lf:Yap transgenics (Fig.7F). MSO exposure had no effect on overall morphology and liver size in WT, suggesting that GLUL is conditionally required for accelerated hepatic growth following induction of Yap1. These data show that Yap elevates GLUL activity and augments glutamine production, which is essential for nucleotide biosynthesis and DNA assembly during rapid liver growth.
Discussion
In this study, transgenic zebrafish expressing activated Yap1 in differentiated hepatocytes demonstrated rapid onset of Yap-dependent larval hepatomegaly that persists into adulthood, along with increased susceptibility to carcinogen-induced hepatocarcinogenesis. Combined transcriptomic and metabolomic analyses revealed that Yap reprograms nitrogen metabolism by inducing GLUL and elevating steady-state levels of glutamine available for biosynthetic growth. The transcriptional regulation of GLUL by YAP and its impact on growth is evolutionarily conserved. Therapeutic prevention studies with the GLUL inhibitor MSO determined that elevated GLUL activity and nucleotide biosynthesis contribute to Yap-driven hepatomegaly. Collectively, our data establish that Yap reprograms glutamine metabolism to provide essential cellular building blocks for hepatic growth. Given that the Hippo pathway is frequently deregulated in cancer, targeting the metabolic alterations driven by Yap activation is a highly attractive and rational therapeutic approach.
Murine studies demonstrated that Yap alters liver cell fate, induce hepatomegaly and promote tumorigenesis14-16. The downstream mechanisms responsible for Yap-driven organ growth, however, have not been fully elucidated. Using zebrafish and unbiased surveys of gene expression and metabolic changes, we identified that Yap reprograms glutamine metabolism through GLUL induction. This results in enhanced de novo nucleotide biosynthesis to meet the anabolic demands of rapid cell proliferation. GLUL is often overexpressed in human hepatoblastoma17, hepatocellular adenoma18 and early stage HCC19. Consistent with these observations, Glul expression and activity are upregulated in premalignant Yap-driven livers. Glul has been previously characterized as a Wnt target in perivenous hepatocytes 11. In contrast, Yap is enriched in a subset of ductal cells during liver homeostasis16, however, Yap levels are elevated in hepatocytes during liver tumorigenesis20. Previous ChIP-seq studies examining YAP and TEAD target genes in MCF7 and embryonic stem cells also showed that Yap binds to the GLUL promoter21, 22. The Hippo and Wnt pathways appear to have a complex relationship, acting synergistically23, 24 and antagonistically25, 26 in different contexts to regulate gene expression. Given the key role of GLUL in nitrogen metabolism, it is not surprising that Met27, Gata328 and Foxo29 also regulate GLUL in other contexts. We speculate that GLUL overexpression provides tumor cells with a competitive advantage by fulfilling the biosynthetic demands for rapid cell proliferation.
One key feature of malignancy is altered cellular metabolism. Tumor cells exploit metabolic pathways to increase nutrient uptake and bioenergetic output. Aerobic glycolysis (“Warburg effect”) and glutaminolysis (glutamine catabolism to generate TCA cycle intermediates) are among the best described metabolic changes in cancer5, 6. A less appreciated aspect of cancer cell metabolism is the need to rewire carbon and nitrogen flux to generate the building blocks required for the anabolic requirements of rapid cell growth30. Growing tissue needs lipids to generate membranes, amino acids for macromolecules and proteins, and nucleotides for DNA and RNA synthesis31. Recent studies examining the metabolic profiles of Kras-induced pancreatic cancer32 and Myc-driven lymphomas33 have identified a common theme in metabolic reprogramming, namely the production of precursors for nucleotide biosynthesis. Here, we discover that Yap reprograms glutamine metabolism to enhance nucleotide biosynthesis and fuel liver growth. Our findings support several recent reports that have elegantly shown that GLUL overexpression can promote cancer cell growth in glutamine-deprived conditions by enhancing nucleotide biosynthesis34, 35. Our studies are also consistent with recent insights by Vander Heiden and colleagues, who showed that Ras-driven lung tumors are less dependent on glutamine catabolism (glutaminolysis) than cultured cells36. Collectively, these studies illustrate the importance of anabolic glutamine utilization for biosynthetic output and cancer cell growth.
Our in vivo relative isotopic enrichment studies enable tracking of nutritional nitrogen in a complex vertebrate organism like very few studies before36, 37. Although our studies focused attention on the anabolic utilization of glutamine for nucleotide biosynthesis, elevated glutamine could also contribute to other biosynthetic outputs including protein synthesis, transamination and the hexosamine biosynthesis pathway. Furthermore, we predict that Yap can alter expression of other metabolic enzymes to enhance utilization of nitrogen and carbon and remodel cellular metabolism. In our work, we highlight the therapeutic potential of targeting anabolic glutamine metabolism with MSO. There is a rich history of targeting the metabolic adaptations of cancer cells as a therapeutic strategy. Many of the first cancer drugs were antimetabolites targeting folate and nucleotide biosynthesis5, 38. Antifolates such as methotrexate and nucleoside analogues including 5FU, gemcitabine and cytarabine remain a lynchpin of modern chemotherapy in many cancers. Indeed, we found that Yap-driven liver growth was sensitive to the nucleoside analogues MPA and 5FU. The ability to convert nutrients into biomass is an essential characteristic of tumors that may provide a therapeutic window for novel antimetabolite strategies.
Functional crosstalk between the mTOR and Hippo pathways during tissue growth is well described39. The mTOR pathway is essential in linking nutrient status with metabolic processes underlying anabolic cell growth7. Intracellular nutrients activate mTOR, altering metabolic flux to generate the lipids, protein and nucleic acids for cell proliferation. For example, the mTOR pathway phosphorylates and thereby activates CAD, which catalyzes the first three steps of pyrimidine biosynthesis40, 41. mTOR also stimulates purine biosynthesis by inducing enzymes associated with the mitochondrial tetrahydrofolate cycle42. Similarly, lysophosphatidic acid, sphingosine 1-phosphate and the mevalonate pathway regulate Yap, thereby establishing Yap as a sensor of nutrient status 43-46. Serum starvation activates Amot130, which binds Yap and facilitates its degradation by recruiting E3 ubiquitin ligase Aip447, 48. In the context of liver growth, bile acids derived from mevalonate precursors activate Yap to promote carcinogenesis49. In addition to serum-borne lipids, glucose deprivation and bioenergetic stress inhibit Yap activation via AMPK and LATS1/250-53. In turn, Yap directly induces the expression of amino acid transporters SLC38A1 and SLC7A5, which are required for cancer cell growth as they stimulate amino acid uptake and activate mTORC154, 55. In light of these studies and the insights provided by our work, we hypothesize that Yap functions to “sense” the nutrient status of the environment and initiate an appropriate growth response. Given the striking similarities between the mTOR and Hippo pathways as networks that integrate metabolite availability to anabolic growth, it will be interesting to further define these links in the context of cancer.
In summary, our study provides compelling evidence that metabolic reprogramming contributes to the accelerated growth phenotype imbued by Yap activation. A greater understanding of the metabolic perturbations specific to malignant cells may provide new pharmacological approaches to combat cancer.
Online Methods
Zebrafish Husbandry
Zebrafish were maintained according to institutional animal care and use committee (IACUC-BIDMC) protocols. Lines used in this study include WT (AB) and Tg(−2.8fabp10a:eGFP)as3, abbreviated lf:GFP56. Tg(−2.8fabp10a:yap1S87A; −0.8cryaa:Venus)s705, abbreviated lf:Yap, were generated using standard techniques57. These zebrafish express a constitutively active form of yap1-1βS87A, a kind gift from Ian Scott, driven by the hepatocyte-specific promoter of fatty acid binding protein (fabp) 10a, otherwise known as liver specific fatty acid binding protein (lfabp or lf)56. This line also contains the A-crystallin (cryaa)-driven selection marker Venus58, to produce green fluorescence in the lens of the eye. Adult zebrafish were used without sex bias (both males and females, in a gender-matched fashion), unless otherwise indicated. For the adult zebrafish studies, at least 3 individuals were used for all assays and the age of the individuals examined was 3-months old (unless otherwise stated in the text).
Chemical Exposure
Zebrafish were exposed to methionine sulfoximine (MSO, 1 mM), mycophenolic acid (MPA, 10 μM), 5-Fluorouracil (5FU, 10 μM) and Rapamycin (RAPA, 0.5 μM) as described. Cultured cells were exposed to Verteporfin (VP, 5 μM) and MSO 1 mM), as described. Where necessary, chemicals were dissolved in DMSO. Chemicals were obtained from Sigma-Aldrich (USA).
Morpholino Injection
Morpholinos (MO) (GeneTools USA) designed against exon2-intron2 splice sites of glula and glulb or mismatched controls glula-MM and glulb-MM (see Supplementary Table 1) were injected into Tg(−2.8fabp10a:eGFP) and Tg(−2.8fabp10a:eGFP); lf:Yap larvae at the one-cell-stage.
Fluorescence determination of liver size
lf:GFP larvae were anesthetized with 0.04 mg/ml tricaine methane sulfonate (MS-222) and imaged by fluorescence microscopy using a Zeiss Discovery V8/Axio Cam MRC. Lightsheet microscopy was performed using a Zeiss Lightsheet Z.1 at the Harvard Center for Biological Imaging (HCBI) core and analyzed using FIJI (NIH) as previously described59.
Histology
Paraformaldehyde (PFA)-fixed larvae, fish were paraffin-embedded, serially sectioned and stained with Hematoxylin and Eosin (H&E), Periodic Acid-Schiff (PAS) or Sirius Red stain using standard protocols60. Analysis of PCNA+ nuclei and cellularity (cells/unit2) was performed using FIJI (NIH) as previously described59.
Immunohistochemistry
Fixed tissue embedded in paraffin was serially sectioned prior to immunohistochemical analysis. Slides were deparaffinized and rehydrated prior to heat-induced antigen retrieval. Antigens were detected using primary antibodies (Supplementary Table 2) in conjunction with an HRP/DAB (ABC) detection kit (Abcam, ab64264) or fluorophore-conjugated secondary antibodies. TUNEL staining was performed according to the manufacturers instructions (R&D Systems).
FACS Analysis
lf:GFP larvae were dissociated at 10 dpf by enzymatic disaggregation in TrypLETM Express (Invitrogen, CO, USA) combined with manual disruption using a pestle. Upon addition of DMEM with 10% FCS, cells were strained through a 35μm nylon mesh and analyzed using a BD FACSAria II flow cytometer (BD Biosciences).
DMBA-induced carcinogenesis
WT and lf:Yap fry were exposed to 5 ppm DMBA for 24 hrs at 3, 4 and 5 weeks9. Fish were monitored closely for tumor development over the subsequent 6 months.
RNA Transcriptomic Analysis
RNA was extracted in Trizol (Life Technologies) from livers isolated from WT and lf:Yap adults and purified using the RNease Mini Kit (Qiagen), according to the manufacturers instructions. RNA quality was checked by Agilent Bioanalyzer Sequencing was performed after library construction on an Illumina HiSeq. polyA sequence data were aligned on to the ZV9 genomic assembly using STAR and differentially affected genes were identified using DeSeq61. Gene ontology (GO) and Gene Set Enrichment analysis (GSEA) of biological processes was determined by GO Slim (Gene Ontology Consortium) and GAGE62.
qRT-PCR
RNA was isolated from pooled zebrafish larvae or isolated livers using Trizol. Following DNAse treatment, cDNA was synthesized using the SuperScript III First-Strand Synthesis kit (Life Technologies). qRT-PCR was performed on biological triplicates using an iCycler with iQ SYBR green (BioRad). Gene expression was analyzed with ef1a as the reference gene (Supplementary Table 1). All of the primers used in this study were purchased from Integrated DNA Technologies (Coralville, IA).
ChIP analysis
Chromatin Immunoprecipitation was performed as previously described63. Cells were cross-linked in 1% formaldehyde for 10min at room temperature after which the reaction was stopped by addition of 0.125M glycine. Cells were lysed and harvested in ChIP buffer (100 mM Tris at pH 8.6, 0.3% SDS, 1.7% Triton X-100, and 5 mM EDTA) and the chromatin disrupted by sonication using a Diagenode Bioruptor sonicator UCD-200 to obtain fragments of average 200-500 bp in size. Suitable amounts of chromatin were incubated with specific antibodies overnight (Supplementary Table 2). Immunoprecipitated complexes were recovered on Protein-A/G agarose beads (Pierce) and, after extensive washes; DNA was recovered by reverse crosslinking and purification using QIAquick PCR purification kit (QIAGEN).
Cell lines
No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. Cell lines were purchased from ATCC just prior to commencing the experiments. The cell lines were not authenticated and were not tested for mycoplasma contamination.
Luciferase assay
Hek293T cells were obtained from American Type Culture Collection (ATCC) and maintained in Dulbecco's Modified Eagle Medium (DMEM; Cellgro) supplemented with 10% FBS (Gibco). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. HEK293T cells were co-transfected with glutamine synthetase reporter constructs29, the Renilla plasmid, and mutant isoforms of yap64, 65, including pCMV-Yap1-2α-S127A (Addgene, #17794), or pCMV-Yap1-2α-S94A (Addgene, #33102). pCMV-EGFP (Addgene, #11153) was used as control. Cells were harvested 48 hrs after transfection using the Dual-Glo Luciferase Assay System (Promega) and assayed according to the manufacturer's directions.
Yap knockdown in liver cancer cell lines
HepG2 and Hep3B cells were obtained from ATCC and maintained in Minimum Essential Medium (MEM; Cellgro) with Earle's salts and L-glutamine supplemented with 10% fetal bovine serum. Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. For shRNA silencing of Yap a single-stranded oligonucleotide encoding Yap target shRNA, and its complement, were synthesized (Supplementary Table 1). The oligonucleotide sense and antisense pair was annealed and inserted into the pLKO.1 backbone. To produce lentiviral supernatents, HEK293T cells were cotransfected with control or shRNA-containing pLKO.1 vectors, VSVG and psPAX2 for 48 hours. HepG2 and Hep3B cells at approximately 40% confluence were infected with lentiviral particles for 24 hrs followed by selection in 2 μg/mL puromycin for 48 hrs prior to cell lysis.
Liver cancer cell line growth assay
HepG2 and Hep3B cells were seeded in 96 well plates in MEM containing Earle's salts and L-glutamine supplemented with 10% fetal bovine serum. Twenty-four hours after seeding, cells were washed with PBS and incubated in MEM media containing 10% dialyzed fetal bovine serum (Gibco), with or without L-glutamine. Where appropriate media was supplemented with 1 mM MSO or 10 μM verteporfin. Cell proliferation was monitored using the Sulforhodamine B (SRB) assay. Adherent cells were fixed by addition of 12.5% (w/v) trichloroacetic acid and incubation at 4°C for 1 hour. Wells were washed with water and cells were stained by addition of WRB solution (0.5% (w/v) SRB in 1% acetic acid). Wells were washed twice with 1% acetic acid and allowed to dry at room temperature. SRB was solubilized with 10 mM Tris, pH 10.5 and absorbance at 510 nm was measured.
Immunoblot analysis
Cell lysates were prepared using RIPA buffer, resolved by SDS-PAGE and electrophoretically transferred onto nitrocellulose. Membranes were probed with the primary antibody overnight and detected with secondary antibody conjugated with HRP (Supplementary Table 2). Antibody complexes were visualized by enhanced chemiluminescence using X-ray film.
Steady-state Metabolomics Analysis
Livers were isolated from adult WT and lf:Yap transgenic adults and a methanol extraction was performed. Polar metabolites were isolated and enriched using the methodology outlined by Yuan et al13. Metabolite fractions were collected and analyzed by targeted LC-MS/MS via selected reaction monitoring (SRM) with positive/negative ion polarity switching using a 5500 QTRAP hybrid triple quadrupole mass spectrometer (A/B SCIEX). Metabolic pathway enrichment analysis was performed using MetaboAnalyst66.
GLUL activity assay
Glul activity was assayed using the γ-glutamyltransferase assay as previously described67. Briefly, WT and lf:Yap transgenic larvae or adult livers were lysed in 50 mM imidazole and samples were purified using micro bio-spin gel p-6 columns (BioRad). Lysates were incubated in activity buffer (50 mM imidazole, 50 mM L-glutamine, 25 mM hydroxylamine, 25 mM sodium arsenate, 2mM manganese chloride, 0.16 mM adenosine diphosphate) at 27 °C for 30 min. The reactions were quenched in stop solution (2.42% iron chloride and 1.45% trichloroacetic acid in 1.82 N hydrochloric acid). Absorbance was read at 520 nm and a calibration curve of glutamyl-γ-hydroxamate (Sigma) was used to calculate nmol of product formed.
Determination of ammonia excretion rate
The rate of ammonia excretion was determined by placing individual fish into small water containers and monitoring ammonia buildup over time using methods previously established68 and an ammonia detection kit (Biovision Inc.) according to the manufacturers instructions.
Analysis of the relative isotopic enrichment in liver lysates following 15NH4Cl exposure
Ammonia incorporation into nucleotide precursors was measured using adaptations to previously established methods69. Briefly, liver homogenates isolated from WT and lf:Yap adults were prepared in lysis buffer (100mM NaCl, 25mM Imidazole, 25mM HEPES, 1mM MgCl2, 1mM MnCl2, 0.1mM KCl and 1% NP-40 pH 7.4 supplemented with Complete protease inhibitors (Roche)). Liver homogenates (90 μl) containing the same amount of protein were incubated a reaction cocktail (10 μl) containing 10 mM 15NH4Cl (Cambridge Isotopes Laboratories, Inc.), 10 mM ATP and 10 mM sodium glutamate and incubated at 37 °C for 20 min before the reaction was quenched in 80% methanol (900μl). Polar metabolites were enriched using the methodology outlined by Yuan et al13 and quantified using SRM on a 5500 QTRAP mass spectrometer using a protocol to detect 15N-labelled isotopologues of nucleotide precursors (Supplementary Table 3).
15N-Spirulina feeding protocol
15N-labelled Spirulina (98%) (Cambridge Isotopes Laboratories, Inc.) gels containing 50 mg/ml 15N-labelled Spirulina, 5 mg/ml Gemma Micro 300 (Skretting) and 1% agar were aliquoted into 30 μl pellets. Individual WT and Tg(fabp10:yap1S87A) transgenic adults were given a 15N-labelled Spirulina gel pellet twice a week over a 4 week period.
Analysis of the relative isotopic enrichment of 15N in genomic DNA
Livers were isolated from adult WT and lf:Yap transgenic adults and genomic DNA was isolated using Quick-gDNA™ MiniPrep (Zymo Research) according to the manufacturers instructions and re-purified by sodium acetate precipitation. DNA from 15N-labelled spirulina and unlabelled spirulina was extracted as previously described70. Purified DNA was quantified with PicoGreen (Life Technologies), and 100 ng of DNA was treated with DNA Degradase Plus (Zymo Research) for 24 hrs. Completion of DNAse digestion was measured by PicoGreen quantification. Hydrolyzed nucleotides were enriched using the methodology outlined by Yuan et al13 and quantified using SRM on a 5500 QTRAP mass spectrometer using a protocol to detect 15N-labelled isotopologues of nucleotides (Supplementary Table 3).
Statistics and Reproducibility
Sample sizes and reproducibility for each figure are denoted in the figure legends. Unless otherwise noted, all data including immunoblots and microscopy images are representative of at least three biologically independent experiments. For zebrafish experiments, no statistical method was used to predetermine sample size. Furthermore, the experiments were not randomized and the Investigators were not blinded to allocation during experiments and outcome assessment. Statistical significance between conditions was assessed by two-tailed Student's t-tests. All error bars represent s.e.m., and significance between conditions is denoted as *p<0.05; **p<0.01; ***p<0.001 and ****p<0.0001. Raw data from independent replicate experiments can be found in the Statistics Source (Supplementary Table 4). Statistical data for each quantitative experiment, including mean, s.e.m., and exact p values can be found in the Statistics Source Data.
Supplementary Material
Acknowledgements
This work was supported by an Irwin Arias Postdoctoral Fellowship (AGC) and Liver Scholar Award (AGC) from the American Liver Foundation, HDDC Pilot Feasibility Grant from the Harvard Digestive Disease Center, P30 DK034854 (AGC, DY), NIH NIGMS T32GM007753 (KLH), NIH NCI 5K08CA172288 (KJE), NIH NIDDK R01DK60322 (DYRS), NIH NIDDK R01DK090311 (WG), NIH K08DK105351 (DY), R01AR064036 (FDC) and R01DK099559 (FDC), and the Packard Foundation (DYRS). JMA is partially supported by NIH NCI 5P01CA120964 and 5P30CA006516. GGG is supported by an American-Italian Cancer Foundation postdoctoral research fellowship. KJE was a Robert Black Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-109-10). DY is a Gilead Sciences Scholar in Liver Disease. WG is supported by the Claudia Adams Barr Program for Innovative Cancer Research, and is a Pew Scholar in the Biomedical Sciences.
Footnotes
Author Contributions:
A.G.C. and W.G. conceived the study, reviewed results and wrote the manuscript. K.L.H. contributed to experimental design. A.G.C. and K.L.H. performed the majority of the experiments and data analysis. K.K.B. performed cell culture experiments and immunoblotting. K.E. and D.Y.S. generated lf:Yap fish and K. E. performed pathological analysis of liver tumors. S.B., K.O., A.T. and S.N. assisted in zebrafish experiments. M.Y., E.C.L. and J.M.A. developed methods and analyzed metabolomics samples. G.G.G. performed CHIP experiments. S.C. and Y.H. analyzed RNAseq datasets. D.Y., A.M., D.E.C, F.D.C., J.A., Y.H., and D.Y.S. provided overall input. All authors reviewed the manuscript.
Data Availability
The RNA sequencing data that support the findings of this study have been deposited in the GEO database: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82246. Source data for all of the figures in the entire have been provided as Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Piccolo S, Dupont S, Cordenonsi M. The Biology of YAP/TAZ: Hippo Signaling and Beyond. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 2.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Her GM, Chiang CC, Chen WY, Wu JL. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 2003;538:125–133. doi: 10.1016/s0014-5793(03)00157-1. [DOI] [PubMed] [Google Scholar]
- 9.Spitsbergen JM, et al. Neoplasia in zebrafish (Danio rerio) treated with 7,12- dimethylbenz[a]anthracene by two exposure routes at different developmental stages. Toxicol Pathol. 2000;28:705–715. doi: 10.1177/019262330002800511. [DOI] [PubMed] [Google Scholar]
- 10.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cellrelated traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 11.Benhamouche S, et al. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev Cell. 2006;10:759–770. doi: 10.1016/j.devcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEADYAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ionswitching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 16.Yimlamai D, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt A, et al. Differential expression of glutamine synthetase and cytochrome P450 isoforms in human hepatoblastoma. Toxicology. 2011;281:7–14. doi: 10.1016/j.tox.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Bioulac-Sage P, et al. Hepatocellular adenoma subtype classification using molecular markers and immunohistochemistry. Hepatology. 2007;46:740–748. doi: 10.1002/hep.21743. [DOI] [PubMed] [Google Scholar]
- 19.Nault JC, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology. 2014 doi: 10.1002/hep.27372. [DOI] [PubMed] [Google Scholar]
- 20.Li H, et al. Deregulation of Hippo kinase signalling in human hepatic malignancies. Liver Int. 2012;32:38–47. doi: 10.1111/j.1478-3231.2011.02646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lian I, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbluh J, et al. beta-Catenin-Driven Cancers Require a YAP1 Transcriptional Complex for Survival and Tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao J, et al. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varelas X, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Barry ER, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuneva MO, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 2011;7:e1002229. doi: 10.1371/journal.pgen.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Vos KE, et al. Modulation of glutamine metabolism by the PI(3)KPKB- FOXO network regulates autophagy. Nat Cell Biol. 2012;14:829–837. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- 30.Howell JJ, Ricoult SJ, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc Trans. 2013;41:906–912. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- 31.Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem Sci. 2015;40:130–140. doi: 10.1016/j.tibs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157:1088–1103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bott AJ, et al. Oncogenic Myc Induces Expression of Glutamine Synthetase through Promoter Demethylation. Cell Metab. 2015;22:1068–1077. doi: 10.1016/j.cmet.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tardito S, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson SM, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016 doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014;20:1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 39.Tumaneng K, et al. YAP mediates crosstalk between the Hippo and PI(3)KTOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robitaille AM, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Sahra I, Hoxhaj G, Ricoult SJ, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller E, et al. Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem Biol. 2012;19:955–962. doi: 10.1016/j.chembiol.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Sorrentino G, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, et al. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc Natl Acad Sci U S A. 2014;111:E89–98. doi: 10.1073/pnas.1319190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adler JJ, et al. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci U S A. 2013;110:17368–17373. doi: 10.1073/pnas.1308236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adler JJ, et al. Amot130 adapts atrophin-1 interacting protein 4 to inhibit yes-associated protein signaling and cell growth. J Biol Chem. 2013;288:15181–15193. doi: 10.1074/jbc.M112.446534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anakk S, et al. Bile acids activate YAP to promote liver carcinogenesis. Cell Rep. 2013;5:1060–1069. doi: 10.1016/j.celrep.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeRan M, et al. Energy Stress Regulates Hippo-YAP Signaling Involving AMPK-Mediated Regulation of Angiomotin-like 1 Protein. Cell Rep. 2014;9:495–503. doi: 10.1016/j.celrep.2014.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W, et al. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015 doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mo JS, et al. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat Cell Biol. 2015 doi: 10.1038/ncb3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Enzo E, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015 doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park YY, et al. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology. 2016;63:159–172. doi: 10.1002/hep.28223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansen CG, Ng YL, Lam WL, Plouffe SW, Guan KL. The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res. 2015;25:1299–1313. doi: 10.1038/cr.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Her GM, Yeh YH, Wu JL. 435-bp liver regulatory sequence in the liver fatty acid binding protein (L-FABP) gene is sufficient to modulate liver regional expression in transgenic zebrafish. Dev Dyn. 2003;227:347–356. doi: 10.1002/dvdy.10324. [DOI] [PubMed] [Google Scholar]
- 57.Thermes V, et al. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 58.Kurita R, et al. Suppression of lens growth by alphaA-crystallin promoterdriven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish. Dev Biol. 2003;255:113–127. doi: 10.1016/s0012-1606(02)00079-9. [DOI] [PubMed] [Google Scholar]
- 59.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goessling W, et al. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 61.Collins JE, White S, Searle SM, Stemple DL. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res. 2012;22:2067–2078. doi: 10.1101/gr.137901.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galli GG, et al. Prdm5 regulates collagen gene transcription by association with RNA polymerase II in developing bone. PLoS Genet. 2012;8:e1002711. doi: 10.1371/journal.pgen.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 65.Gaffney CJ, et al. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene. 2012;509:215–222. doi: 10.1016/j.gene.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deuel TF, Louie M, Lerner A. Glutamine synthetase from rat liver. Purification, properties, and preparation of specific antisera. J Biol Chem. 1978;253:6111–6118. [PubMed] [Google Scholar]
- 68.Bucking C, Lemoine CM, Walsh PJ. Waste nitrogen metabolism and excretion in zebrafish embryos: effects of light, ammonia, and nicotinamide. J Exp Zool A Ecol Genet Physiol. 2013;319:391–403. doi: 10.1002/jez.1802. [DOI] [PubMed] [Google Scholar]
- 69.Skaper SD, O'Brien WE, Schafer IA. The influence of ammonia on purine and pyrimidine nucleotide biosynthesis in rat liver and brain in vitro. Biochem J. 1978;172:457–464. doi: 10.1042/bj1720457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morin N, Vallaeys T, Hendrickx L, Natalie L, Wilmotte A. An efficient DNA isolation protocol for filamentous cyanobacteria of the genus Arthrospira. J Microbiol Methods. 2010;80:148–154. doi: 10.1016/j.mimet.2009.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.