ABSTRACT
In mammals, the time period that follows fertilization is characterized by extensive chromatin remodeling, which enables epigenetic reprogramming of the gametes. Major changes in chromatin structure persist until the time of implantation, when the embryo develops into a blastocyst, which comprises the inner cell mass and the trophectoderm. Changes in DNA methylation, histone variant incorporation, and covalent modifications of the histones tails have been intensively studied during pre-implantation development. However, modifications within the core of the nucleosomes have not been systematically analyzed. Here, we report the first characterization and temporal analysis of 3 key acetylated residues in the core of the histone H3: H3K64ac, H3K122ac, and H3K56ac, all located at structurally important positions close to the DNA. We found that all 3 acetylations occur during pre-implantation development, but with different temporal kinetics. Globally, H3K64ac and H3K56ac were detected throughout cleavage stages, while H3K122ac was only weakly detectable during this time. Our work contributes to the understanding of the contribution of histone modifications in the core of the nucleosome to the “marking” of the newly established embryonic chromatin and unveils new modification pathways potentially involved in epigenetic reprogramming.
Keywords: Epigenetics, histone acetylation, lateral surface, mammalian embryo, nucleosome core
Introduction
Histones can be extensively modified covalently. Currently, the most studied modifications are the ones at the N-terminal tails of histones, which protrude from the nucleosome.1 It is now clear that histone modifications can affect practically all DNA-dependent processes, ranging from cell division to gene regulation and DNA damage response, thereby playing a key role in defining cellular identity. Posttranslational modifications of histones are multiple and include phosphorylation, acetylation, ubiquitylation, and methylation, which are the most studied ones. The best characterized mode of action of histone modifications is indirect, since histone tail modifications can be bound by specific proteins (referred to as ‘readers’), which subsequently effect cellular functions through chromatin remodeling and/or transcriptional regulation.2 One example of such a reader is TFIID, a multiprotein complex of the basal transcriptional machinery. One of the TFIID subunits, TAF1, harbors a double chromodomain that recognizes methylated H3K4.3 The recognition of H3K4me3 by TAF1 promotes the binding of TFIID to transcription start sites and the subsequent assembly of the preinitiation complex, a required step for RNA Polymerase II transcription.3 A similar scenario has been proposed for H1 acetylation, specifically for the H1.4 isoform, whereby acetylation of H1.4K34 serves as a binding platform for TAF1, thereby facilitating transcriptional activation.4
However, the above-cited model implies that modified residues need to be accessible for recognition by their binders. This is the case for all N-terminal histone tails and for some of the C-terminal tails, which protrude outwards of the nucleosome core particle.5 However, histones are also modified within the core of the nucleosome, in positions that are in close contact to the DNA and that may not be readily accessible to protein binders.6-8 In particular, H3 can be modified through acetylation of K56, K64, K122, and K115,9-12 which are globular residues located within a structurally important position. Lysine acetylation at H3K122 and H3K64 has been correlated with active genomic regions and both modifications have been shown to have a direct and causative function in stimulating transcription. Indeed, the addition of an acetyl group results in charge neutralization of the lysine residue and could promote nucleosome destabilization due to a decrease in the affinity between histones and DNA when acetylation occurs on a lysine situated close to the DNA.9,11 H3K56ac is the best characterized lateral surface modification, but most work has been performed in yeast, and its precise function in mammalian cells is not fully understood. H3K56ac is enriched at some transcriptional regulatory regions,12 and is linked to the core transcriptional network in human embryonic stem cells.13 H3K56ac has been linked to replication in S. cerevisiae14 and was identified as a modification occurring predominantly on newly synthesized H3, promoting binding to the CAF-1 assembly factor.15 In vitro, H3K56ac causes enhanced DNA unwrapping and an increase in nucleosome disassembly rate, thereby facilitating DNA accessibility.16,17 The location of these 3 residues within the nucleosome core particle at positions that can potentially directly affect the binding and wrapping of the DNA around the histone octamers makes them interesting candidates to mediate large scale epigenetic reprogramming.
After fertilization, the 2 parental genomes undergo epigenetic reprogramming, which is thought to mediate the reversal from a highly differentiated state in the gametes to totipotency, necessary to start a new developmental program.18,19 To address the potential contribution of changes in modifications of the nucleosome core residues to reprogramming after fertilization, we determined the dynamics of 3 key modifications in the nucleosome core during mouse pre-implantation development. We systematically analyzed the localization patterns of H3K56ac, H3K64ac, and H3K122ac. We found that all 3 analyzed core nucleosome acetylation sites have different temporal patterns during pre-implantation development. In particular, acetylation of H3K64 opposes the pattern of trimethylation of the same residue (H3K64me3). H3K64ac is detected exclusively in the male pronucleus (PN) from pronuclear stage 3 (PN3) and in both pronuclei from PN4. Subsequently, H3K64ac is present throughout pre-implantation development. H3K122ac is present in the growing and fully mature oocyte like H3K56ac and H3K122ac, but then, H3K122ac is only present at extremely weak levels upon fertilization and until the blastocyst stage. Additionally, we found that H3K56ac was abundant throughout pre-implantation development, but displayed no correlation with replication foci. None of the 3 analyzed modifications was enriched in the inner cell mass or the trophectoderm at the early blastocyst stage. Our results suggest that acetylation in the nucleosome core at histone H3 is dynamic during pre-implantation development, and should therefore be considered as a potential regulator of chromatin remodeling after fertilization, during the transitions of cellular plasticity at the beginning of development.
Results
H3K64ac has been linked to transcriptional activation and increased nucleosome dynamics.9 We analyzed, for the first time, the kinetics of this histone modification during early mouse development. Immunostaining with a previously characterized specific H3K64ac antibody9 revealed a strong staining in the mature, germinal vesicle stage (GV) oocyte that recapitulated the DAPI localization, suggesting that H3K64ac is widely distributed in the mature oocyte (Fig. 1A). During meiotic resumption, at the metaphase plate of the meiosis I (MI), H3K64ac was undetectable on the maternal chromatin, and remained so in both pronuclei after fertilization in early pronuclear stages in the zygote (PN2, Fig. 1A). We first detected H3K64ac at the mid-pronuclear stages (PN3), which is the time at which replication is known to start in zygotes.20,21 Interestingly, at this stage H3K64ac was readily detected on the paternal chromatin, but not in the maternal chromatin, (Fig. 1A). This parental asymmetry in H3K64ac localization was only transient, as a few hours later, at pronuclear stage 4 (PN4), H3K64ac was present in both pronuclei at similar levels. The pattern of localization of H3K64ac was largely euchromatic, with a discrete ‘dotty’ pattern throughout the nucleoplasm (Fig. 1A). After the first mitotic division, in early 2-cell stage embryos, H3K64ac levels dropped and, although detectable, were lower than in zygotes at PN3 and PN4 stages (Fig. 1A). This kinetics of changes in acetylation levels in 2-cell embryos are similar to other acetylation sites, such as H3K27ac, which also decreases following the first mitosis,22 but differ to those of other acetylation sites, such as H3K9, which remain abundant in zygotes and 2-cell embryos.23
Figure 1.
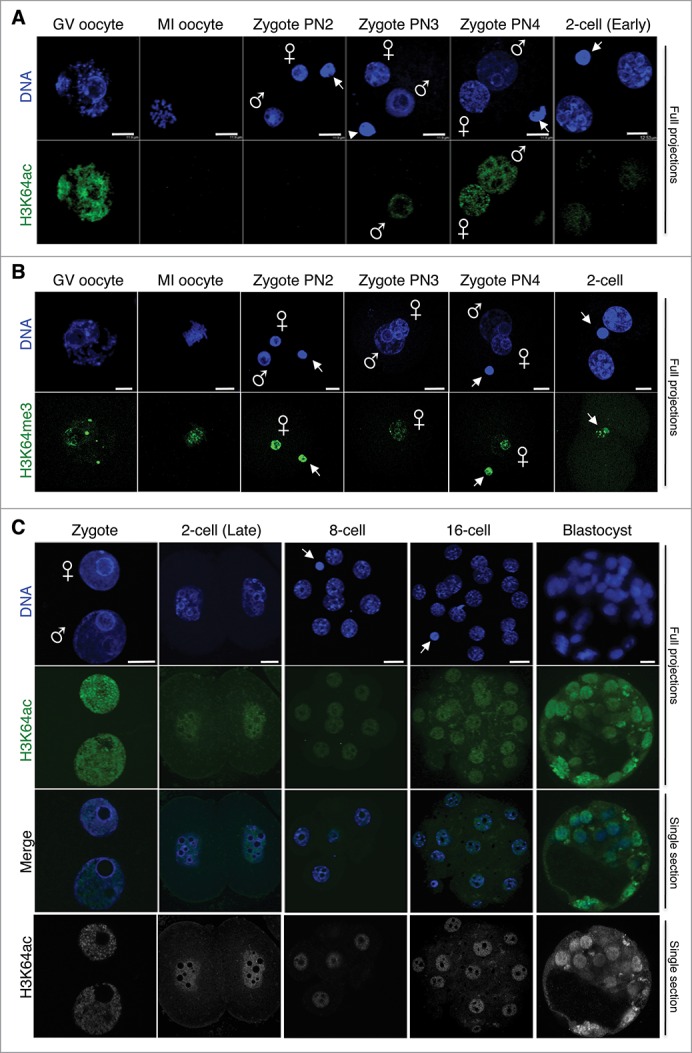
Dynamics of H3K64ac during mouse pre-implantation development. Freshly collected embryos were fixed and stained with an H3K64ac antibody (green). DNA is shown in blue. A. Maximal projections of Z-sections taken every 1 μm on a confocal microscope of oocytes, zygotes at different pronuclear (PN) stages and early 2-cell stage embryos. The arrowheads point to the polar body; male and female pronuclei are indicated. Images were acquired using the same confocal parameters and therefore the fluorescence levels are directly comparable. B. H3K64me3 opposes the temporal kinetics of H3K64ac. Oocytes at the germinal vesicle (GV) or MI stage, zygotes at different PN stages and early 2-cell stage embryos were immunostained with an H3K64me3 antibody (green). The DNA was stained with DAPI (blue). Shown are maximal intensity projections of confocal Z-sections acquired as in A. Male and female pronuclei are indicated, and the polar body is demarcated by an arrow. Note that H3K64me3 is absent from the paternal chromatin throughout. C. H3K64ac levels at different developmental stages as indicated. Top and middle parts show full projections of Z-sections taken every 1 μm (cleavage stages) or 2 μm (blastocyst) on a confocal microscope of DNA (blue) and H3K64ac (green). The ‘Merge’ panels show a merge image of the corresponding middle sections for the blue (DNA) and green (H3K64ac) channels. The same confocal section corresponding to the H3K64ac staining is shown in gray scale at the bottom. From A through C, between 12 and 24 embryos per stage were analyzed in at least 3 independent experiments. Scale bar is 12 μm.
We previously documented that H3K64 can also be trimethylated.24 Notably, H3K64me3 was also enriched in the chromatin of GV oocytes, but contrary to H3K64ac, it remained present on the metaphase plate of MI oocytes and was detected on the maternal pronucleus throughout zygotic development (Fig. 1B). While H3K64ac was still detected in nuclei of early 2-cell stage embryos, H3K64me3 became largely undetectable in early 2-cell stage embryos and was only visible in the polar body, as reminiscent of the maternal chromatin extruded after completion of meiosis (Fig. 1B), similarly to other heterochromatic marks, such as H4K20me3.25 Thus, while H3K64me3 was present at the earliest developmental stages after fertilization, inherited exclusively from the maternal germline, H3K64 became acetylated only after pronuclear stage 3 and was first detected on the paternal chromatin only. The two modifications of the same amino acid have therefore an opposing pattern in terms of their temporal dynamics after fertilization, suggesting that they define distinct chromatin states.
We next examined the levels of H3K64ac throughout the subsequent developmental stages, up to blastocyst stage. H3K64ac was readily detected from late 2-cell stage throughout all analyzed stages: 8-cell, 16-cell, and early blastocyst (Fig. 1C). The distribution of H3K64ac was euchromatic, excluded from the big nucleoli, and clearly visible from the 8-cell stage. In the blastocyst stage, H3K64ac was detected at similar levels in both the ICM and the trophectoderm. The overall pattern of H3K64ac contrasts with that of H3K64me3, which remains undetectable from the 2-cell stage to implantation.24
Acetylation of H3K122 is catalyzed by CBP/p300,11 which are known to be expressed in pre-implantation mouse embryos.26 We therefore assessed for the first time the distribution of H3K122ac in early embryos, initially focusing on the detailed dynamics from the mature oocyte and throughout zygotic stages after fertilization. As shown in Fig. 2A, H3K122ac was detected in the fully grown GV oocyte, where it was distributed throughout the nucleus and excluded from the DAPI-ring regions surrounding the nucleolar-like bodies (NLBs), indicating a largely euchromatic localization, as expected from its known function related to active enhancers and active genomic elements.11 The presence of H3K122ac in the oocyte might potentially reflect remodeling taking place during oocyte growth and maturation. Upon meiotic resumption, in MI oocytes, H3K122ac became undetectable on the metaphase chromosomes (Fig. 2A). Immediately after fertilization, at PN0, at the time when the decondensing maternal chromosomes can be observed, there was no H3K122ac visible on the maternal chromatin (Fig. 2A). A faint antibody signal could be observed at the position of the forming second polar body, identified by a clear protrusion, which did not colocalized with the condensed maternal chromosomes (Fig. 2A, arrowhead, the dashed line demarcates the plasma membrane). This accumulation of H3K122ac signal could potentially reflect residual histones, which have been suggested to be ‘disposed of’ upon polar body extrusion.27 At this time, the sperm nucleus can be observed, which showed no accumulation of H3K122 (Fig. 2A, PN0, the position of the paternal PN is indicated). H3K122ac becomes detectable again at the PN2 stage, and its levels increased slightly toward the end of the first cell cycle at stages PN3/PN4 (Fig. 2A). Similarly to H3K64ac, the abundance of H3K122ac dropped in early 2-cell stage embryos (Fig. 2A). Compared to late zygotes, the levels of H3K122ac remained very low throughout cleavage stages from the 2-cell to the 8-cell stage (Fig. 2B). However, a robust staining was observed at the blastocyst stage, where H3K122ac was readily distributed in both the ICM and the trophectoderm, with a nuclear localization that was excluded from the DAPI-rich regions (Fig. 2B). Thus, cleavage stages are characterized by low levels of H3K122ac globally.
Figure 2.
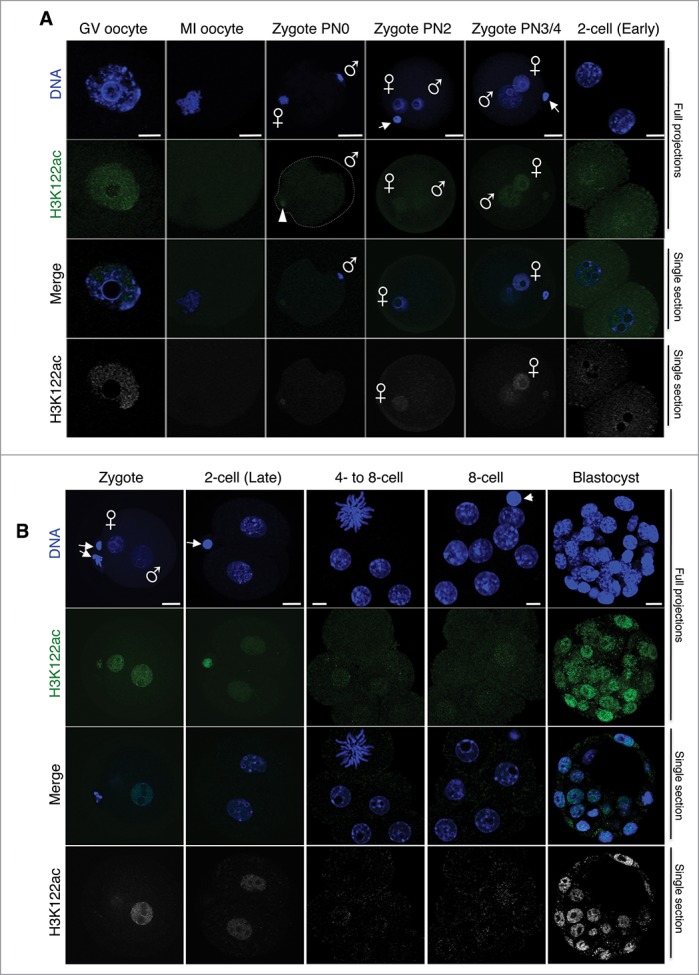
H3K122ac is present at low levels in cleavage stage embryos. (A) H3K122ac was analyzed in oocytes and zygotes immediately after fertilization, freshly collected from natural matings. The top 2 panels show maximal projections of confocal Z-sections for H3K122ac (green) or DAPI (blue) of representative embryos of at least 16 analyzed per stage. The bottom panel shows a middle confocal section where the gray and blue channels are merged. When present, the polar body is indicated by an arrowhead. The male and female pronuclei are indicated. Scale bar is 12 μm. (B) Embryos at the indicated stages were freshly collected, fixed, stained with an H3K122ac (green) antibody, and analyzed using confocal microscopy. DNA is shown in blue. Shown are full projections of Z-sections taken every 1 μm (cleavage) or 2 μm (blastocyst). The bottom panel is the same channel for the merge shown on the third row, but showing the H3K122ac channel in gray scale. When present, the polar body is indicated by an arrowhead. The male and female pronuclei are indicated. At least 10 embryos from independent experiments were analyzed per stage. Embryos shown were imaged under identical conditions, thus fluorescence levels are directly comparable. Scale bar is 12 μm.
Finally, we also studied a third acetylation site within the nucleosome core, H3K56ac. While in yeast H3K56ac has been shown to facilitate incorporation of newly deposited histones during S-phase,15 the role of H3K56ac in higher eukaryotes remains unclear. We detected H3K56ac in the GV oocyte, which was distributed throughout the nucleoplasm, but excluded from the pericentromeric chromatin domains, identified as bright DAPI-rich foci (Fig. 3A). H3K56ac was undetectable on the metaphase plate of MI oocytes, and remained so upon meiotic resumption after fertilization, with both maternal and paternal chromatin showing no detectable levels of H3K56ac in early zygotes at PN1 stage (Fig. 3A). H3K56ac was first detected after fertilization at the mid-zygotic stages, around the PN2-PN3 stages, and was equally distributed in the 2 pronuclei (Fig. 3A). At late zygotic stages, H3K56ac remained detectable in both pronuclei. We found no enrichment in the DAPI-rich regions surrounding the NLBs, and H3K56ac showed a dotty pattern throughout both pronuclei (Fig. 3A, Zygote PN4–5). In early 2-cell embryos, H3K56ac was detected throughout the whole nucleus, indicating that both paternal and maternal chromatin retained high levels of H3K56ac (Fig. 2A and B). A systematic, side-by-side comparison of H3K56ac staining from zygotes through cleavage stages revealed that H3K56ac was present at all analyzed developmental stages, without exception, and showed a dotty-like pattern in embryonic nuclei, with the clearest pattern at the zygote stage (Fig. 3B). Thus, H3K56ac showed similar temporal kinetics in maternal and paternal pronuclei and was first detected in mid-zygotes after fertilization, with high levels persisting throughout pre-implantation development.
Figure 3.
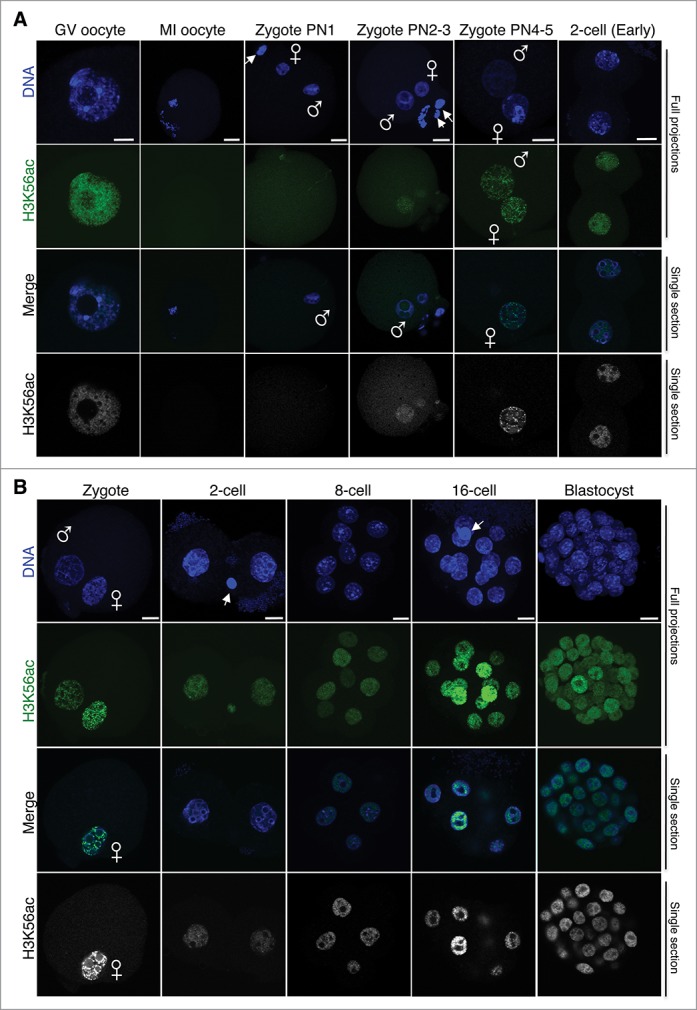
H3K56ac is first detected at the mid-zygote stage after fertilization and persists throughout pre-implantation development. (A) Analysis of H3K56ac immediately after fertilization and until the early 2-cell stage. Freshly collected mouse embryos from natural matings at the indicated stages were stained with an H3K56ac antibody (green) and with DAPI (blue). Full projections of Z-sections acquired every 1 μm (top 2 parts) and middle, merge section (third part) are shown. The gray scale view of the green (H3K56ac) channel of the merge image confocal section is shown at the bottom. Male and female pronuclei are indicated, and the polar body, where visible, is demarcated by an arrow. The PN classification was done as described by Adenot and colleagues.20 At least 10 embryos were analyzed per stage, performed in 2 independent experiments. Scale bar is 12 μm. (B) Dynamics of H3K56ac throughout cleave stages. Embryos were collected at the indicated stages, fixed, and processed for immunostaining using an H3K56ac. As in A, top and second panels are maximal projections of confocal Z-sections acquired every 1 μm (cleavage stages) or 2 μm (blastocyst) of DAPI (blue) or H3K56ac (green) channels. The merge image shows a middle confocal section of the same embryos and the bottom panel is the gray scale of the green (H3K56ac) channel. At least 14 embryos were analyzed per stage, performed in 2 independent experiments. Scale bar is 12 μm.
We next investigated the dotty pattern observed for H3K56ac (Fig. 3) and H3K64ac (Fig. 1A) in more detail. A closer, higher magnification analysis using immunostaining for the 2 modifications in the zygote, 2-cell, 8-cell, and 16-cell stage embryos confirmed a persistent dotty pattern throughout cleavage stages, for which we found no obvious correlation with specific nuclear localization or coincidence with DAPI-rich regions (Fig. 4A). The observation of H3K64ac and H3K56ac dots being particularly sharp in zygotes prompted us to investigate whether these dots were related to replication sites at this stage. To address this, we performed short BrdU pulses of 30 min in zygotes at early-mid S-phase or in late S-phase and performed co-immunostaining for BrdU and H3K64ac or H3K56ac. In zygotes at early-mid S-phase, H3K64ac was distributed widely throughout the nucleoplasm, as described above. At this time point, BrdU signal was also found throughout the nucleus (Fig. 4B). Some regions in the nucleus were stained with the BrdU antibody as well as the H3K64ac antibody (Fig. 4B). However, a large part of the BrdU and the H3K64ac signals clearly did not colocalized. Thus, given the broad distribution of the H3K64ac staining, we conclude that there does not appear to be a specific colocalization between H3K64ac and replication. At later stages of S-phase, BrdU is usually detected exclusively in the regions surrounding the NLBs and close to the nuclear periphery (Fig. 4B), reflecting a late replication pattern of genomic regions localized in these parts of the nucleus.21 Despite of a global distribution in the nucleus of H3K64ac, we observed no colocalization with BrdU at this stage, suggesting that replication in the zygote does not entail H3K64ac (Fig. 4B). We performed similar analysis with H3K56ac, which revealed that there was no strict correlation between acetylation of H3K56 and incorporation of BrdU (Fig. 4B).
Figure 4.
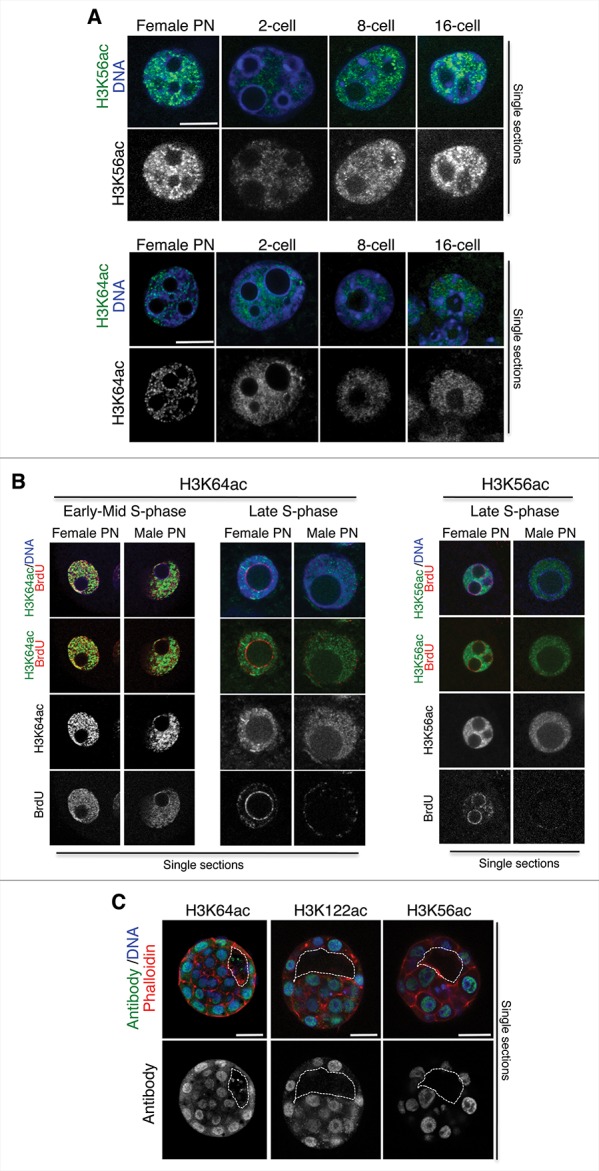
Relationship between H3K56ac or H3K64ac and S-phase progression. (A) Higher magnification of a representative female pronucleus or nuclei from 2-, 8-, or 16-cell stage embryos stained with the H3K56ac or the H3K64ac antibody. Shown are merge images of single confocal sections. Scale bar is 12 μm. (B) Zygotes were subjected to a BrdU pulse for 30 min during early/mid or late S-phase and processed for immunostaining with an anti-BrdU and an anti-H3K64ac (left) or an anti-H3K56ac (right) antibody. Representative zygotes of 10 analyzed at different stages of S-phase derived from 3 (H3K56ac) or 2 (H3K64ac) independent experiments are shown. Note, that the late replication pattern is characterized by BrdU incorporation at the satellite sequences around the NLBs and remaining BrdU at the peripheral nuclear regions. Shown are single sections of Z-stack sections taken every 0.5 μm where the diameter of the male pronucleus is maximal. Male and female pronuclei are shown. Scale bar is 12 μm. (C) Middle section of an early blastocyst stained with the H3K64ac, the H3K122ac, or the H3K56ac antibody. Shown are the merge images of the green (antibody), blue (DAPI), or red (cortical actin stained with phalloidin) channels. The white line delineates the blastocyst cavity. Scale bar is 20 μm. Representative embryos from 8, 9, or 12 embryos for the H3K64ac, H3K122ac, and H3K56ac antibodies are shown.
Lastly, we asked whether any of the analyzed modifications preferentially localized to each of the 2 lineages of the early blastocyst. Indeed, some histone modifications, such as H3K27me3 and DNA methylation are enriched in either the ICM or the trophectoderm.28,29 We therefore performed immunostaining of early blastocysts using antibodies to detect H3K64ac, H3K56ac, or H3K122ac. To firmly distinguish between inner and outer cells, we additionally co-stained for cortical actin using phalloidin. As shown in Fig. 4C, none of the 3 analyzed modifications was detectably enriched in the ICM or the trophectoderm, but were rather equally abundant in both lineages. These results suggest that, globally, chromatin dynamics related to these core nucleosome modifications might be involved in the epigenetic reprogramming, but not in setting up lineage specification.
Discussion
After fertilization, the early mammalian embryo is characterized by large-scale chromatin remodeling, including changes in the incorporation of histone variants, global changes in DNA, and histone tail modification and alterations in nuclear organization.18,19 Here, we have characterized the temporal dynamic patterns of the 3 key acetylation sites of histone H3 occurring in the globular domain at the core of the nucleosome, on the lateral surface. We focused on these residues given their potential role to regulate DNA-histone interactions. Our data suggest that acetylation of lateral surface residues occurs throughout pre-implantation development, which contrasts to the absence of methylation of the other lateral surface or core domain residues that have been so far investigated, H3K64 and H3K79, respectively, during this developmental time window.24,30
All three analyzed core nucleosome acetylation sites were present throughout pre-implantation development; however, they displayed different temporal patterns. H3K56ac and H3K64ac were abundant throughout pre-implantation, while H3K122ac, despite being also present in the growing and fully mature oocyte, was only detected at very low levels up to before implantation. Considering H3K122ac implication in enhancer regulation,11 the low levels of this modification throughout cleavage stages might suggest a less frequent enhancer usage or regulation through an alternative histone modification signature, perhaps H3K64 itself. However, it should be noted that H3K27ac, another modification typically associated with active enhancers,31 is undetectable in 2-cell stage embryos.22 The usage of enhancer has been thoroughly studied in zygotes and 2-cell stage embryos using reporter constructs upon microinjections.32,33 However, it is unknown whether endogenous enhancers are required for gene expression in the early embryo and, if so, at what extent and for which genes. It also awaits investigation if H2K122ac indeed marks enhancers during these developmental phases.
A transcriptionally repressive stage builds up at the 2-cell stage, and has been associated with the acquisition of a chromatin configuration that can be partially released via HDAC treatment32,34,35 and may be associated with high histone mobility.36 This suggests that histone acetylation is, not surprisingly, linked to the initial activation of embryonic transcription. Histone hyperacetylation on the tail of histone H4 has been documented to precede embryonic transcription in the mouse,20 and H3K9ac was also readily detected from the PN3 stages,23 which mark the onset of embryonic transcription and replication.21,37 Our results show that lateral surface modifications were also present at these developmental phases. H3K64ac was detected in the male PN from approximately PN3 and in both pronuclei from PN4 onwards, suggesting that acetylation of H3K64 may contribute to activation of the first wave of activation during ZGA. Indeed, H3K64ac has been shown to induce transcriptional activation.9 The transient asymmetry in H3K64ac, whereby the paternal PN becomes acetylated prior to the maternal one, is in line with previous observations documenting epigenetic asymmetry of additional acetylation sites occurring on the histone tails, such as H3K27ac, H4K12ac, and H4K16.22,38 Functionally, it is known that the extent of the release of transcriptional activity after sodium butyrate treatment is lower in the male as compared to the female pronucleus 39and, therefore, higher acetylation levels of H3K64ac in the male versus the female pronucleus support this observation.
The temporal dynamics of H3K64ac after fertilization opposes the pattern of H3K64me3 at the earliest (PN0-PN3) zygotic stages, suggesting that modifications on H3K64 help creating ‘opposing’ chromatin states between the male and female PN globally: while the female PN is typically enriched with heterochromatic marks, the male pronucleus is mostly devoid of them.
The role of H3K56ac in higher eukaryotes is still unclear. Here, we analyzed it for the first time through early mammalian development. We find that H3K56ac, known to affect unwrapping of the DNA,16 is dynamic during periods of epigenetic reprogramming. In human ES cells, H3K56ac was described to be enriched on the promoter of pluripotent genes as well as on histone genes.13 It is possible that H3K56ac may contribute to the great deal of chromatin architecture changes occurring during this developmental window, as well as to the transcription of histone (and pluripotency) genes, which are needed to ensure the cell divisions and changes in chromatin assembly pathways occurring in the embryo.
Histone modifications occurring on the lateral surface of the nucleosome core have remained largely under investigated. To the best of our knowledge, the only other modification on the core of the nucleosome that has been investigated in pre-implantation embryos to date is methylation of H3K79.30 Like H3K64me3, H3K79me3 localizes to the maternal pericentromeric heterochromatin regions at fertilization, but becomes rapidly undetectable at early pronuclear stages and remains undetectable throughout pre-implantation development.30 A similar pattern was observed for H3K79me2, which was found to be largely absent in interphase nuclei of cleavage stages. Thus, it would seem that while acetylation of lysines situated at the nucleosome core, specifically at the lateral surface, is persistent in early mouse embryos, methylation is not.
There is still debate as to whether histone modifications are a cause or a consequence of the transcriptional process, in particular those occurring at the tail. However, growing evidence suggest that modifications of histone residues in close contact to the DNA affect directly the stability of the nucleosome and DNA accessibility, and are therefore causative to chromatin disruption.6,7,40,41 Future experiments will have to address whether these marks have also a causative function during epigenetic reprogramming. Our work contributes to the molecular characterization of the embryonic chromatin at the earliest developmental stages, and will help integrating our understanding of the large-scale chromatin remodeling processes that takes place during this important developmental time window.
Materials and methods
Collection of mouse embryos
Mouse embryos were collected from natural matings of CD1 crosses bred in a 12 h light cycle as previously described.42 Zygotes, 2-cell stage, 4-cell stage embryos, and morulas were obtained by puncturing the oviduct approximately 12, 34, 46 and 58 h post-coitum (hpc), respectively. Blastocysts were collected by flushing the uterus with M2 medium (Sigma) 72 hpc. All embryos were fixed immediately after collection, excepting for the BrdU labeling experiments, where embryos were shortly subjected to a BrdU pulse as indicated in the corresponding figure legends. Experiments with animals were carried out according to valid legislation in France and under the approval of the appropriate ethical committee (COMETH).
Immunostaining
Embryos were fixed immediately after collection following the removal of the zona pellucida with acid Tyrode's solution (Sigma) and processed as described.43 After permeabilization embryos were washed 3 times in PBS-T (0.1% Tween in PBS), blocked and incubated with the primary antibodies for ˜12 h at 4°C, followed by 2 washes in PBS-T, blocking for 30 min and incubation for 2 h at 25°C with the corresponding secondary antibodies: A488-conjugated goat anti rabbit IgG, (Life Technologies A11070) or Cy3-conjugated goat anti mouse IgG, Jackson ImmunoResearch (115–165–146) used at a 1:500 dilution. After washing, embryos were mounted in Vectashield (Vector Laboratories) containing 4′-6-Diamidino-2-phenylindole (DAPI). For visualizing cell-cell boundaries, blastocysts were stained with Alexa Fluor 635 phalloidin (Molecular Probes). The antibodies used were: H3K64ac (9; 1:200 dilution); H3K122ac (ref. 11; 1:150 dilution), H3K56ac (Abcam ab76307; 1:250 dilution) and anti-BrU (SIGMA-Aldrich B8434-clone BU-33; 1:250 dilution)
BrdU labeling
Zygotes were subjected to a BrdU pulse of 30 min during mid and late S-phase and processed for immunostaining with anti-BrdU and the indicated antibodies. The immunostaining protocol was as above, except that an HCl denaturation step and a subsequent neutralization step were performed after fertilization (0.1 N HCl for 30 min at RT followed by a 10 min 100 mM Tris-HCl pH8.8). S-phase patterns were staged according to.21
Confocal analysis
Confocal microscopy was performed using a 63x oil objective in a Leica SP2 UV inverted microscope. For imaging of blastocysts a 40x oil objective in a Leica SP2 AOBS MP confocal was used. Sections were taken every 0.5 or 2 μm for cleavage stage embryos or blastocysts, respectively. All the stainings were repeated independently at least 2 times with at least 10 embryos analyzed per stage.
Funding Statement
M.E.T.-P acknowledges funding by EpiGeneSys NoE, ERC-Stg ‘NuclearPotency’, the EMBO YIP and the Fondation Schlumberger pour l'Education et la Recherche. C. Z-B is supported through the Université de Strasbourg. Work in R.S. laboratory is supported by the Fondation pour la Recherche Médicale (FRM), by the Agence Nationale de Recherche (ANR, CoreAc), La Ligue National Contre Le Cancer (Equipe Labellisée) and INSERM Plan Cancer (Epigénétique et Cancer).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693-705; PMID:17320507; http://dx.doi.org/ 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000; 403:41-5; PMID:10638745; http://dx.doi.org/ 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 3.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007; 131:58-69; PMID:17884155; http://dx.doi.org/ 10.1016/j.cell.2007.08.016 [DOI] [PubMed] [Google Scholar]
- 4.Kamieniarz K, Izzo A, Dundr M, Tropberger P, Ozretic L, Kirfel J, Scheer E, Tropel P, Wisniewski JR, Tora L, et al.. A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation. Genes Dev 2011; 26:797-802; http://dx.doi.org/ 10.1101/gad.182014.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997; 389:251-60; PMID:9305837; http://dx.doi.org/ 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol 2004; 11:1037-43; PMID:15523479; http://dx.doi.org/ 10.1038/nsmb851 [DOI] [PubMed] [Google Scholar]
- 7.Tropberger P, Schneider R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat Struct Mol Biol 2013; 20:657-61; PMID:23739170; http://dx.doi.org/ 10.1038/nsmb.2581 [DOI] [PubMed] [Google Scholar]
- 8.Tessarz P, Santos-Rosa H, Robson SC, Sylvestersen KB, Nelson CJ, Nielsen ML, Kouzarides T. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 2014; 505:564-8; PMID:24352239; http://dx.doi.org/ 10.1038/nature12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Cerbo V, Mohn F, Ryan DP, Montellier E, Kacem S, Tropberger P, Kallis E, Holzner M, Hoerner L, Feldmann A, et al.. Acetylation of histone H3 at lysine 64 regulates nucleosome dynamics and facilitates transcription. Elife 2014; 3, e01632; PMID:24668167; http://dx.doi.org/ 10.7554/eLife.01632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci U S A 2011; 108:12711-6; PMID:21768347; http://dx.doi.org/ 10.1073/pnas.1106264108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Bühler M, et al.. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 2013; 152:859-72; PMID:23415232; http://dx.doi.org/ 10.1016/j.cell.2013.01.032 [DOI] [PubMed] [Google Scholar]
- 12.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 2005; 121:375-85; PMID:15882620; http://dx.doi.org/ 10.1016/j.cell.2005.03.011 [DOI] [PubMed] [Google Scholar]
- 13.Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, et al.. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 2009; 33:417-27; PMID:19250903; http://dx.doi.org/ 10.1016/j.molcel.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 2008; 134:244-55; PMID:18662540; http://dx.doi.org/ 10.1016/j.cell.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 2005; 436:294-8; PMID:16015338; http://dx.doi.org/ 10.1038/nature03714 [DOI] [PubMed] [Google Scholar]
- 16.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 2009; 36:153-63; PMID:19818718; http://dx.doi.org/ 10.1016/j.molcel.2009.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.North JA, Shimko JC, Javaid S, Mooney AM, Shoffner MA, Rose SD, Bundschuh R, Fishel R, Ottesen JJ, Poirier MG. Regulation of the nucleosome unwrapping rate controls DNA accessibility. Nucleic Acids Res 2012; 40:10215-27; PMID:22965129; http://dx.doi.org/ 10.1093/nar/gks747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton A, Torres-Padilla ME. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 2014; 15:723-34; PMID:25303116; http://dx.doi.org/ 10.1038/nrm3885 [DOI] [PubMed] [Google Scholar]
- 19.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol 2009; 10:526-37; PMID:19603040; http://dx.doi.org/ 10.1038/nrm2727 [DOI] [PubMed] [Google Scholar]
- 20.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 1997; 124:4615-25; PMID:9409678 [DOI] [PubMed] [Google Scholar]
- 21.Bouniol-Baly C, Nguyen E, Besombes D, Debey P. Dynamic organization of DNA replication in one-cell mouse embryos: relationship to transcriptional activation. Exp Cell Res 1997; 236:201-11; PMID:9344600; http://dx.doi.org/ 10.1006/excr.1997.3708 [DOI] [PubMed] [Google Scholar]
- 22.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 2010; 12:853-62; PMID:20676102; http://dx.doi.org/ 10.1038/ncb2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boskovic A, Bender A, Gall L, Ziegler-Birling C, Beaujean N, Torres-Padilla ME. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics 2012; 7:747-57; PMID:22647320; http://dx.doi.org/ 10.4161/epi.20584 [DOI] [PubMed] [Google Scholar]
- 24.Daujat S, Weiss T, Mohn F, Lange UC, Ziegler-Birling C, Zeissler U, Lappe M, Schübeler D, Torres-Padilla ME, Schneider R. H3K64 trimethylation marks heterochromatin and is dynamically remodeled during developmental reprogramming. Nat Struct Mol Biol 2009; 16:777-81; PMID:19561610; http://dx.doi.org/ 10.1038/nsmb.1629 [DOI] [PubMed] [Google Scholar]
- 25.Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, et al.. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci 2004; 117:2491-501; PMID:15128874; http://dx.doi.org/ 10.1242/jcs.01238 [DOI] [PubMed] [Google Scholar]
- 26.Kwok RP, Liu XT, Smith GD. Distribution of co-activators CBP and p300 during mouse oocyte and embryo development. Mol Reprod Dev 2006; 73:885-94; PMID:16596650; http://dx.doi.org/ 10.1002/mrd.20440 [DOI] [PubMed] [Google Scholar]
- 27.Wen D, Banaszynski LA, Rosenwaks Z, Allis CD, Rafii S. H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos. Nucleus 2014; 5:369-75; PMID:25482190; http://dx.doi.org/ 10.4161/nucl.36231 [DOI] [PubMed] [Google Scholar]
- 28.Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 2003; 130:4235-48; PMID:12900441; http://dx.doi.org/ 10.1242/dev.00625 [DOI] [PubMed] [Google Scholar]
- 29.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241:172-82; PMID:11784103; http://dx.doi.org/ 10.1006/dbio.2001.0501 [DOI] [PubMed] [Google Scholar]
- 30.Ooga M, Inoue A, Kageyama S, Akiyama T, Nagata M, Aoki F. Changes in H3K79 methylation during preimplantation development in mice. Biol Reprod 2008; 78:413-24; PMID:18003948; http://dx.doi.org/ 10.1095/biolreprod.107.063453 [DOI] [PubMed] [Google Scholar]
- 31.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al.. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010; 107:21931-6; PMID:21106759; http://dx.doi.org/ 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiekowski M, Miranda M, DePamphilis ML. Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol 1991; 147:403-14; PMID:1916016; http://dx.doi.org/ 10.1016/0012-1606(91)90298-H [DOI] [PubMed] [Google Scholar]
- 33.Bevilacqua A, Fiorenza MT, Mangia F. Developmental activation of an episomic hsp70 gene promoter in two-cell mouse embryos by transcription factor Sp1. Nucleic Acids Res 1997; 25:1333-8; PMID:9060426; http://dx.doi.org/ 10.1093/nar/25.7.1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latham KE, Solter D, Schultz RM. Acquisition of a transcriptionally permissive state during the 1-cell stage of mouse embryogenesis. Dev Biol 1992; 149:457-62; PMID:1309712; http://dx.doi.org/ 10.1016/0012-1606(92)90300-6 [DOI] [PubMed] [Google Scholar]
- 35.Schultz RM, Worrad DM. Role of chromatin structure in zygotic gene activation in the mammalian embryo. Semin Cell Biol 1995; 6:201-8; PMID:8562912; http://dx.doi.org/ 10.1006/scel.1995.0028 [DOI] [PubMed] [Google Scholar]
- 36.Boskovic A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EV, Meshorer E, Torres-Padilla ME. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev 2014; 28:1042-7; PMID:24831699; http://dx.doi.org/ 10.1101/gad.238881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res 1995; 218:57-62; PMID:7537698; http://dx.doi.org/ 10.1006/excr.1995.1130 [DOI] [PubMed] [Google Scholar]
- 38.Adenot PG, Campion E, Legouy E, Allis CD, Dimitrov S, Renard J, Thompson EM. Somatic linker histone H1 is present throughout mouse embryogenesis and is not replaced by variant H1 degrees. J Cell Sci 2000; 113 (Pt 16):2897-907; PMID:10910774 [DOI] [PubMed] [Google Scholar]
- 39.Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol 1993; 159:366-78; PMID:8365573; http://dx.doi.org/ 10.1006/dbio.1993.1248 [DOI] [PubMed] [Google Scholar]
- 40.Brehove M, Wang T, North J, Luo Y, Dreher SJ, Shimko JC, Ottesen JJ, Luger K, Poirier MG. Histone Core Phosphorylation Regulates DNA Accessibility. J Biol Chem 2015; 290(37):22612-21; PMID:26175159; http://dx.doi.org/ 10.1074/jbc.M115.661363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J Biol Chem 2009; 284:23312-21; PMID:19520870; http://dx.doi.org/ 10.1074/jbc.M109.003202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hogan BL, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. 2nd. edn, (Cold Spring Harbor Laboratory Press, 1994). [Google Scholar]
- 43.Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol 2006; 50:455-61; PMID:16586346; http://dx.doi.org/ 10.1387/ijdb.052073mt [DOI] [PubMed] [Google Scholar]


