ABSTRACT
There is accumulating evidence that the histone methyltransferase enhancer of zeste homolog 2 (EZH2), the main component of the polycomb-repressive complex 2 (PRC2), is involved in melanoma progression and metastasis. Novel drugs that target and reverse such epigenetic changes may find a way into the management of patients with advanced melanoma. We provide a comprehensive up-to-date review of the role and biology of EZH2 on gene transcription, senescence/apoptosis, melanoma microenvironment, melanocyte stem cells, the immune system, and micro RNA. Furthermore, we discuss EZH2 inhibitors as potential anti-cancer therapy.
KEYWORDS: Chromatin, EZH2, epigenetic changes, H3K27 trimethylation, histone, melanoma, polycomb-repressive complex
Introduction
Melanoma is currently the 5th and 7th most common cancer in American men and women, respectively.1 When diagnosed early, melanoma is easily treatable with surgery alone. Once the cancer metastasizes, however, the prognosis becomes increasingly bleak, treatment becomes more difficult, and the survival rate drops significantly. Prior to 2011, the prognosis for patients with metastatic melanoma was dismal, with an overall 5-year mortality rate exceeding 95%.2 Recently, immunotherapy with checkpoint inhibitors has revolutionized the management of metastatic melanoma.2 The combination of the monoclonal antibodies ipilimumab (the cytotoxic T-lymphocyte-associated antigen 4 inhibitor) and nivolumab (the programmed cell-death protein 1 inhibitor) was associated with a response rate of 57.6%, compared to 43.7% for nivolumab and 19% for ipilimumab alone.3 In addition, the median progression-free survival (PFS) was 11.5 months in the combination group, compared to 6.9 months in the nivolumab group and 2.9 months in the ipilimumab group,3 suggesting that synergistic treatments are superior to monotherapy. Despite these impressive results, approximately 40% of patients with metastatic melanoma do not respond to immunotherapy. Identifying biomarkers that predict response to immunotherapy is still an unmet need.4 Moreover, identifying oncogenic or epigenetic alterations that may contribute to immunotherapy resistantance will be important to find candidate drug targets and ultimately enhance the efficacy of immunotherapy.
There is accumulating evidence that epigenetic alterations including DNA methylation patterns, post-translational modification of histones and chromatin remodeling are common in cancer and play a large role in its progression and metastasis.5,6 EZH2 is a histone modifier that functions as the catalytic component of the Polycomb Repressive Complex 2 (PRC2).7 It is a lysine methyltransferase and promotes addition of the repressive mark histone H3K27me3 to target chromatin, thereby inducing chromatin compaction and transcriptional repression by restricting access to transcriptional regulators like RNA Polymerase II and other transcription associated factors.8 Silencing of tumor suppressor genes by H3K27me3 has been implicated in the initiation and advancement of melanoma.7-9
Overexpression of EZH2 is associated with thicker primary melanoma, higher Clark's level of invasion, loss of p16 protein staining, and strong expression of cyclin D1.10 Our group analyzed a panel of melanoma cell lines by immunoblot and showed higher levels of H3K27me3 and EZH2 protein in the metastatic melanoma cell line (WM266-4) relative to the primary melanoma cell line (WM115) which are both derived from the same patient.8 We also revealed that the WM266-4 cell line overexpresses EZH2 and is more proliferative and invasive than the WM115 cell line.8 EZH2 prepares the landscape and provides the proper microenvironment for melanoma to survive.9,11 It contributes to the transcriptional silencing of tumor suppressor and differentiation genes, promoting uncontrolled cell proliferation and cancer progression.10,12,13
Other specific oncogenic or epigenetic alterations may play a role in immune evasion and resistance to immunotherapy. Activation of the WNT/β-catenin pathway has been shown to result in defective production of the chemokine CCL4 which in turn fails to recruit CD103+ dendritic cells, required for T cell priming, to the melanoma microenvironment.14 Hence, activation of the WNT/β-catenin pathway may contribute to immune evasion and resistant to immunotherapy in metastatic melanoma.14 Likewise, EZH2 mediated H3K27 hypermethylation has been shown to repress the tumor production of T helper1- type chemokines CXCL9 and CXCL10 resulting in low tumor infiltrating CD8+ cells and poor patient outcome in ovarian cancer.15 The latter data has not been investigated in melanoma yet. Perhaps EZH2 overexpression may silence important immune-protective genes and hence allow cancer cells to evade immune surveillance.15 There is a growing interest in epigenetic alterations and its role in the formation of resistance to either BRAF inhibitors or current immune therapy in melanoma.16 Future studies should look into combining EZH2 inhibitor with either BRAF inhibitors or immune therapy in melanoma tumor models to overcome a range of resistant mechanisms and enhance the clinical efficacy of these drugs.16
It is not surprising that EZH2 has emerged as a potential target of interest. A number of small molecule-EZH2 inhibitors have been developed 17-19 and have already entered phase I/II trials in lymphoma.20 We have shown that treatment of WM115, WM115EZ (WM115 melanoma cells transfected with a plasmid expressing EZH2), and WM226-4 cells treated with the EZH2 inhibitor GSK126 resulted in depletion of H3K27me3 in all three cell lines with a significant restoration of RUNX3 and E-cadherin (tumor suppressor genes) expression in the EZH2 overexpression WMT115EZ and WM266-4 cells.8 Furthermore, depletion of EZH2 via siRNA knockdown enhanced RUNX3 and E-Cadherin expression in WM266-4 cells.8 Chromatin immunoprecipitation (ChIP) performed in the matched melanoma cell lines WM115 and WM266-4 along with the engineered cell line WM115EZ, showed significantly higher enrichment of H3K27me3 at the RUNX3 and E-cadherin promoter in aggressive melanoma cell lines (WM115EZ and WM226-4) compared to less aggressive melanoma cell line WM115. The H3K27me3 enrichment in both promoters was reduced by GSK126 treatment in WM115EZ and WM266-4 cells.8 Our data along with others emphasize the importance of silencing of tumor suppressor genes by H3K27me3 in melanoma progression.7-9
The purpose of this article is to provide a comprehensive up-to-date review of the role and biology of EZH2, the main component of PRC2, in melanoma. We review in detail the effect of EZH2 overexpression on gene transcription, senescence/apoptosis, melanoma microenvironment, melanocyte stem cells, the immune system, and micro RNA. Also, we review the potential targeted therapy of EZH2 in cancer.
Epigenetic gene regulation
The human genome, composed of three billion base pairs, is tightly compacted into chromatin inside the cell nucleus. If the chromatin were highly compacted at all times, gene transcription would not be possible, but chromatin compaction is very dynamic, allowing genes to be turned on and off.17 Epigenetic regulations refer to heritable changes in gene expression that occur in the absence of alterations to DNA.16 Epigenetic modifications are defined as changes in the DNA or associated proteins that can alter chromatin compaction, thereby allowing or denying the transcriptional machinery access to gene promoters.5,6 There are two major groups of epigenetic modifications: 1) DNA methylation and 2) histone post-translational modifications (histone-PTMs).5,6,9 DNA methylation leads to transcriptional repression while histone PTMs leads to either transcriptional activation or repression.5,9 DNA methylation results from the addition of methyl groups to DNA by DNA methyltransferases (DNMT). Histone PTMs represent the addition of chemical groups to certain histone amino acid residues.9 There are over 100 different sites known to experience histone PTMs through any of the numerous types of modifications including lysine methylation, acetylation, ubiquitination, arginine methylation and serine phosphorylation.5,9 Histone methylation can cause transcriptional repression or activation while histone acetylation causes transcriptional activation. Methylation of lysines 27 (H3K27 methylation) and 9 (H3K9 methylation) of Histone 3 is correlated with transcriptional repression while methylation of lysine 4 of histone 3 (H3K4 methylation) is correlated with transcriptional activation.5,9
One particular set of epigenetic modifying enzymes of focus in this review are the Polycomb group proteins (PcG) initially described in Drosophila.21,22 These proteins play important roles in cellular proliferation, differentiation, and maintenance of stem cell identity.23,24 PcG proteins form two main complexes: Polycomb-repressive complex 1 and 2 (PRC1, PRC2).25,26 PRC1 is formed by BMI1, RING1A/B, CBX, and PHC subunits.27 RING1A/B are ubiquitin E3 ligases that catalyze the monoubiquitination of histone H2A at lysine 119 (H2AK119ub1) causing transcriptional silencing.7,9 The PRC2 complex includes five subunits: EZH2, EED, SUZ12, RbAp46/48, and AEBP2 (Fig. 1). EZH2 lacks enzymatic function on its own; however, it gains robust histone lysine methyltransferase activity when it complexes with two other noncatalytic subunits of the PRC2 complex: the zinc-finger containing SUZ12 and the EED.13 PRC2 without the RbAp46/48 subunit can still maintain substantial enzymatic activity. The AEBP2 subunit acts as a cofactor that interacts with the other four subunits and helps stabilize the overall architecture of PRC2. It also enables the PRC2 complex to target to specific DNA sites and enhances its methyltransferase activity.28-30 PRC2 is responsible for adding up to three methyls to the 27th amino acid of histone 3 (H3K27me3).31 H3K27me3 is a marker for chromatin condensation and gene silencing (Fig. 1).32,33 PRC2-induced H3K27 trimethylation recruits PRC1 by binding the chromodomain of the PHC subunits.9,34 This classical sequential or hierarchical model postulates that once PCR2 induces H3K27 trimethylation it recruits PCR1 by binding the chromodomain of the PHC subunits.9,34 Once recruited, PRC1 induces transcriptional repression of the target gene by catalyzing the ubiquitination of lysine 119 of histone H2 or by an H2AK119ub1-independent mechanism.35,36 However, the relationship between PRC1 and PRC2 is more complex than described in this classic model. Several genome-wide profiling studies showed that PRC1 and PRC2 do not always bind the same regions, suggesting alternative mechanisms for the establishment of polycomb-mediated regulation of transcription.37-39 In addition, ubiquitination of H2AK2119 can be mediated by CUL4B, and loss of CUL4B results in down-regulation of H3K27me3.40
Figure 1.
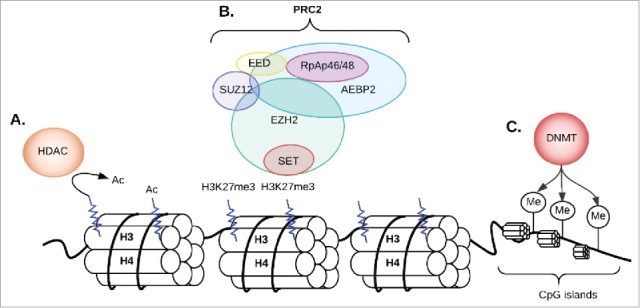
The PRC2 complex and histone methylation; (A) If the lysine amino acid 27 on histone 3 is pre-acetylated, the histone deacetylase (HDAC) enzyme will remove the acetyl group from an N-acetyl lysine amino acid first; (B) EZH2 (one of the 5 subunits of PCR2 complex) adds methyl groups to Histone 3 at Lysine 27. Triple methylation of H3K27 (H3K273m) leads to transcription repression and silencing of genes; (C) DNA methyltransferases (DNMT) add a methyl group to cytosines within CG dinucleotides or CNG trinucleotides (N can be C, A, G or T) in CpG islands leading to transcriptional repression. Ac: acetylation; M: methylation; CG: cytosine-guanine.
PRC2 works in harmony with other epigenetic silencing enzymes such as histone deacetylases (HDACs) and DNMTs to establish a global and stable state of gene silencing.41,42 In addition to its role as a gene repressor, EZH2 may also function as a gene activator. (43–45) EZH2 activates NF-κB target genes through the formation of a ternary complex with the NF-κB components RelA and RelB that does not require other PRC2 subunits.43 EZH2 overexpression can also lead to its interaction with Wnt signaling components and subsequent activation of the c-myc and cyclin D1 genes independent of its methyltransferase activity.44 Phosphorylation of EZH2 at Ser21, mediated directly or indirectly by the PI3K-Akt pathway, can alter its function from a polycomb repressor to a transcriptional co-activator.45 Many of the EZH2-activated genes are downregulated upon EZH2 knockdown, suggesting that EZH2 can activate genes independent of its methyltransferase activity.45
Ezh2 overexpression can drive melanoma progression
Significantly increased EZH2 expression has been observed in melanoma cells when compared to nevus cells.46 It has been proposed that EZH2 plays a role in facilitating the progression from benign nevi to metastatic melanoma epigenetically by silencing important tumor suppressor genes. The BRAF mutation, often seen in benign nevi,47 is not capable alone of driving melanoma progression because most nevi become senescent.48 EZH2 upregulation has been shown to silence the cell cycle inhibitor p21 and bypass the senescent state, driving cancer progression.49 We speculate that EZH2 overexpression may be an early event in melanomagenesis.
Most reports are consistent in showing that expression of EZH2 and H3K27me3 is significantly higher in metastatic melanoma as compared to primary melanoma,50-53 although one report revealed a significant overexpression of H3K27me3, but not EZH2, in primary melanoma compared to metastatic melanoma.46 EZH2 highly expressing melanoma cells significantly correlated with Ki67-expression, a marker indicative of cell cycle engagement and proliferative activity, thus implicating EZH2 in the regulation of proliferation of melanoma at primary and metastatic sites.54 In a study of nearly 700 archival patient tissue samples, encompassing different cancer types including melanoma, breast, endometrial, and prostate cancer, there was a significant association between EZH2 expression and tumor cell proliferation.10 EZH2 overexpression is also associated with features of aggressive clinical behavior and poor survival.10 EZH2 high-melanoma patient group, based on RNAseq and clinical data from the Cancer Genome Atlas, showed a significantly shorter survival as compared to EZH2 low group.54 Moreover, primary melanoma and lymph node metastases patients of EZH2 high group developed distant metastases significantly faster than patients of low EZH2 group.54 Cell migration, invasion, and proliferation are inhibited after EZH2 knockdown in both cutaneous and uveal melanoma cell lines.12,52,55,56
Somatic heterozygous mutations of Y646 in EZH2 are found in ∼2–3% of melanoma samples.57,58 Based on the Cancer Genome Atlas and two other dataset, cutaneous melanoma is the only solid cancer, besides lymphomas, with non-synonymous gain of function mutations affecting tyrosine 646 (Y646*).54 These mutations increase H3K27me3 at the promoters of EZH2 target genes.17 Perhaps unsurprisingly, melanoma cell lines with EZH2 gain-of-function mutations form larger tumors compared to control cells in a xenograft mouse model.59-61
The biology of EZH2 and how it affects melanoma progression
EZH2, enriched at the promoters of tumor suppressor genes, is linked to the proliferation and survival of melanoma cells by its disruption of tumor-suppressive pathways and activation of tumor-promoting pathways.53,62-64 Fig. 2 illustrates a working hypothesis of EZH2 biology in driving melanoma progression and metastasis.
Figure. 2.
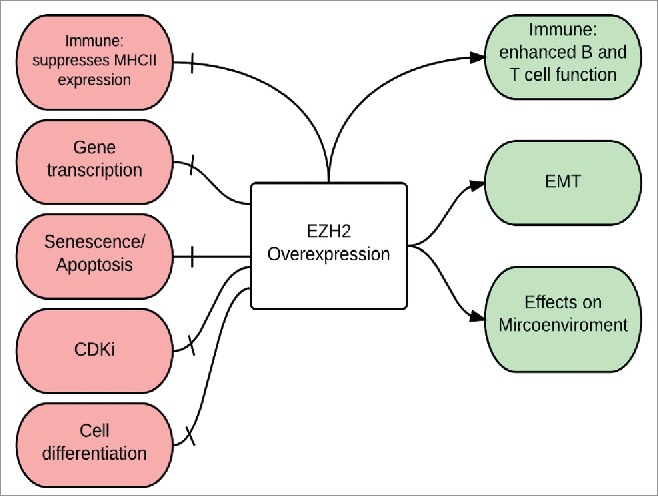
Biology of EZH2 overexpression in melanoma The Immune system: EZH2 overexpression has a positive effect on the immune system by enhancing the function of both B and T cells, however, it has a negative effect by suppressing MHC II expression and hence help melanoma cells escape immune surveillance; Gene transcription: EZH2 silences tumor suppressor genes; Senescence and apoptosis: EZH2 silences apoptotic genes and helps melanoma cells escape senescence; CdKI: EZH2-mediated silencing of cyclin-dependent kinase inhibitors (CdkI), also called INK4 proteins, results in inhibition of apoptosis and senescence and stimulation of melanoma cell proliferation; Cell differentiation: EZH2 enhance the process of de-differentiation to neural-like stem cells; Epithelial-Mesenchymal Transition (EMT) and Microenvironment: EZH2 regulates the EMT and causes morphologic changes in melanocytes (increased motility, branching, aggressive growth, and cell migration) that help shape the interactions between melanoma cells and the microenvironment.
EZH2 regulates gene transcription
EZH2 overexpression strongly contributes to the transcriptional silencing of tumor suppressor and differentiation genes, promoting uncontrolled cell proliferation and cancer progression (Table 1).13 The axonal guidance genes, NRCAM, CEACAM1, SORCS1, ADAM23, and MME are downregulated by exogenous EZH2 gain-of-function mutations in melanoma cells in vitro.61 EZH2 epigenetically silences the INK4b-ARF-INK4a locus, which encodes the p15INK4b, p16INK4a and p14ARF proteins that protect organisms from inappropriate growth signals and promote senescence and apoptosis.65 The INK4 proteins are cyclin-dependent kinase inhibitor (CDKi) that block CdK-induced phosphorylation of EZH2 (preventing EZH2 from binding to other proteins in the PCR2 complex) and pRb (inhibiting E2F activation).66-68 EZH2 and HDAC1 compete for interaction with pRB2/p130, indicating that PRC2 participates in the regulation of pRB/E2F activity. This results in EZH2-induced cyclin A activation and cell cycle progression.69,70
Table 1.
Genes and pathways regulated by EZH2 overexpression in melanoma.
| Gene Name | Gene function | Regulation status |
|---|---|---|
| CIITA | Encodes for the class II major histocompatibility complex (MHC II) | |
| DAB2IP | Ras GTPase-activating protein that promotes apoptosis | |
| E-Cadherin | Enables cell-cell adhesion | |
| KIF2C | Encodes for kinesin-like protein KIF2C****Regulates microtubule dynamics ****Required for chromosome segregation****Promotes melanoma cell motility, invasion, and lung colonization | |
| KIF22 | Encodes for kinesin-like protein KIF22****Essential role in metaphase chromosome alignment and maintenance****Promotes melanoma cell motility, invasion, and lung colonization | |
| CEACAM1 | Plays a role in the differentiation and arrangement of 3D tissue structure, angiogenesis, apoptosis, tumor suppression, metastasis, and the modulation of innate and adaptive immune responses ****May be regulated by MITF in melanocytes | |
| ADAM23 | Endodes for the enzyme disintegrin and metalloproteinase domain-containing protein 23****ADAM proteins negatively modulate integrin-mediated cell proliferation, adhesion, and migration | |
| AMD1 | A tumor suppressor gene that suppresses EMT and metastatic spread of melanoma | |
| MME | Associated with the generation of angiostatin, which inhibits angiogenesis | |
| MITF | Encodes a transcription factor that regulates the differentiation and development of melanocytes | |
| NRCAM | Neuronal cell adhesion molecule | |
| Notch | Active Notch signaling is associated with increased cell motility and lung colonization in melanoma | |
| Oca2 | Encodes for the P protein that is involved in small molecule transport, specifically tyrosine, which is a precursor of melanin | |
| PAX3 | Encodes for the transcription factor PAX3 ****Regulates melanocyte differentiation from neural crest cells during development | |
| P15 | Encodes for a CdKI that inhibits the cell cycle at G1 phase | |
| P16 | Encodes for a CdKI that inhibits the cell cycle at G1 phase | |
| P21 | Encodes for a CdKI that inhibits the cell cycle at G1 phase | |
| PTEN | A tumor suppressor gene that negatively regulates the AKT/PKB signaling pathway | |
| SRF | Active SRF signaling is associated with increased cell motility and lung colonization in melanoma | |
| SOX 10 | Encodes for the transcription factor SOX 10 that is involved in the regulation of embryonic development and in the determination of cell fate | |
| TGF-β | Controls proliferation and differentiation | |
| Wnt signaling pathways | A group of signal transduction pathways made up of proteins that pass signals from outside to inside of the cell. ****Implicated in oncogenesis and in several developmental processes, including regulation of cell fate |
CdKI: cyclin-dependent kinase inhibitor; PTEN: phosphatase and tensin homolog; Oca2: oculocutaneous albinism II (OCA2) gene; MITF: microphthalmia-associated transcription factor; MME: mouse macrophage metalloelastase; CEACAM1: carcinoembryonic antigen-related cell adhesion molecule 1.
EZH2 overexpression is significantly associated with a loss of the cell cycle suppressor protein p16 and increased expression of cyclin D1 in melanoma.71 Although EZH2 is predominantly a repressor of transcription, it can positively regulate genes that promote invasion such as KIF2C, KIF22 and TOP2A.21,72 KIF2C and KIF22 genes promote melanoma cell motility, invasion, and lung colonization.72 Hence, throughout melanoma progression, EZH2 might dynamically repress diverse tumor suppressor genes to achieve either growth or invasion.54
EZH2-dependent suppression of senescence/apoptosis in melanoma cells
EZH2 overexpression helps melanoma cells escape death and maintain resistance to senescence by downregulating genes involved in apoptosis such as CDKN1A and the phosphatase and tensin homolog (PTEN).49,73,74 Oncogenic stimuli in melanocytes provoke an oncogene-induced senescence program, resulting in formation of the melanocytic nevus,48 which is a benign proliferation of melanocytes. Melanocytes with EZH2 overexpression escape senescence and progress to melanoma through EZH2- mediated inhibition of p21.49 EZH2 overexpression induces silencing of DAB2IP, a Ras GTPase-activating protein that promotes apoptosis through the tumor necrosis factor-mediated JNK signaling pathway.75,76 EZH2 depletion activates p21 and induces both senescence and apoptosis in melanoma.74-77
EZH2 promotes the microenvironment for melanoma survival and progression
During development the cells of the neural crest adopt stem cell features and a remarkable migratory capacity, which allows the cells to disseminate through the embryonic tissue and to colonize distant sites, including the skin where they give rise to melanocytes.54 Mesenchymal-like cell motility is utilized by many cell-types that undergo epithelial-mesenchymal transition (EMT) and migrate as single cells throughout the embryo using a protrusion-based mesenchymal mode of motility.78 Melanoma cells revert to highly motile neural crest-like cells during metastasis.79 EMT involves the loss of cell-cell adhesions through downregulation of E-cadherin, the acquisition of a motile phenotype, and acquisition of a gene-expression signature reminiscent of mesenchymal cells. Long-term exposure to signals such as TGF-β may not only induce EMT, but may also lead to the acquisition of stem-like properties.80 The high level of TGF- β found in melanoma likely increases invasion of melanoma cells, promotes tumor immunosuppression and angiogenesis, and thereby promotes melanoma progression.81
EZH2 overexpression causes downregulation of E-cadherin in melanoma.12,31,82 E-cadherin is upregulated again after treating these cells with GSK126, an EZH2 inhibitor. EZH2 thereby regulates EMT and helps shape the interactions between melanoma cells and their microenvironment.12,51,61,82,83 Moreover, during melanomagenesis EZH2 epigenetically repress AMD1 which is a tumor suppressor gene that suppresses EMT and metastatic spread of melanoma.54
EZH2 regulates the transcriptional program of melanoma cells with high Notch and SRF activity.72 Active Notch and SRF signaling is associated with increased cell motility and lung metastasis in melanoma.84-87 Activation of SRF signaling is associated with nuclear translocation of the myocardin-related transcription factor (MRTF-A; also called MKL) which leads to increased SRF-dependent transcription.88,89 Loss of MRTF-A in B16 melanoma cells leads to decreased experimental lung metastasis.89 Activation of SRF was not sufficient to drive cell invasion when EZH2 function was blocked, hence, EZH2 is required for efficient lung colonization by metastatic melanoma.74
Melanoma cell lines expressing exogenous EZH2 gain-of-function mutations exhibit dramatic changes in 3D culture morphology, including prominent branching, disruption of spheroid formation, enhanced motility, aggressive growth morphology, a decrease in cell contractility, and an increase in collective cell migration. These changes are attenuated by treatment with GSK126, the EZH2 inhibitor.61 EZH2 suppresses the pigment production in invasive melanoma cells by repressing Oca2 levels, and melanoma with the highest levels of EZH2 expression had the lowest levels of pigmentation.72,74,90 EZH2 also regulates the amelanotic phenotype of motile cells in vivo by suppressing Oca2.74 It has been shown that motile melanoma cells have lower pigment levels and higher Brn2 promoter activity compared with non-motile cells.90
EZH2 gain-of-function mutations have been shown to occur concurrently with BRAF mutations in melanoma.55 Knockdown of BRAFV600E in vitro caused downregulation of EZH2 levels; 11 hence, EZH2 may complement the function of BRAF. As BRAF signaling drives cell division, EZH2 prepares the landscape and proper microenvironment for melanoma to survive.9,11
Melanoma cells switch between mutually exclusive invasive and proliferative states under the surveillance of EZH2.91 The proliferative state is characterized by high expression of the melanocyte transcription factor MITF and low Brn2 expression, while the opposite is true in the invasive state.92,93
EZH2 determines melanoma cellular differentiation
A potential explanation for the aggressive nature of melanoma is melanocyte natural history. While mature melanocytes are stationary, during development they are derived from the highly motile neural crest cells that migrate extensively throughout the embryo to form neurons, glia, melanocytes, smooth muscle cells, and cells of the craniofacial connective tissue.94,95 MITF, SOX-10, Wnt signaling, and Pax-3 all play an important role in the differentiation process of neural crest cells to melanocytes.96,97 Pax-3 activates MITF expression but prevents its binding to the promoter of Dct. Upon activation of Wnt signaling, Pax-3 is displaced from the Dct promoter, and MITF drives Dct transcription and (hence) pigment production.98 Pax-3 acts to determine both cell fate and maintain cells in a less differentiated state. EZH2 plays an important role in maintaining cells in a progenitor-stem cell-like state through silencing genes associated with differentiation.38,99,100 Genome-wide analysis of melanoma cell lines proposes that melanoma cells can exist in two states, proliferative or invasive, and that a “phenotype switch” between the two may drive invasion and metastasis.101 Perhaps EZH2 helps melanoma cells switch between these different states. Nevi also are found to express a number of neural crest related motility factors suggesting that melanoma may retain characteristics of the less differentiated neural crest cells.79 Melanoma cells may revert to a neural crest-like state that promotes motility and metastasis. A large number of differentiation-related factors such as Sox, Fox, Pax, components of Wnt signaling, and TGF-β are silenced by EZH2.38,102-104 By silencing differentiation related genes, EZH2 helps melanoma cells de-differentiate to their stem cell origin. This function of EZH2 is highlighted in the normal epidermis, where EZH2 levels are high in keratinocyte progenitors but decrease with differentiation.83 Loss of EZH2 in the developing skin increases differentiation.83
Recent research has shown that the tumor invasion front (tumor cells scattered in the stroma at the invasive margin of the tumor) contains tumor cells with stem cell-like properties that might promote metastasis.62,63 Intratumoral heterogeneity in EZH2 and H3K27me3 expression is a feature of melanoma, but it appears to be highly expressed at the invasive tumor margin.46 EZH2, H3K4me2, and H3K27me3 may function as putative markers for melanoma cells with stem cell properties at the melanoma invasive front.46
Notch signaling is also implicated in melanoma progression.105,106 It can promote EMT and motility in numerous cancer types 105,107,108 and is capable of maintaining melanoma cells in a less differentiated stem cell-like state.75,76 Genome-wide analysis identified an overlapping set of genes that was associated with high Notch and SRF activity and regulated by EZH2.74 EZH2 mediates transcriptional activation and repression of numerous target genes involved in cellular differentiation ultimately determining cell fate.23,109
EZH2 role in immune surveillance of melanoma
EZH2 is expressed at high levels in germinal center (GC) B cells, which are involved in the generation of antibodies with high affinities to their antigen.110 An EZH2 deficiency in GC B cells may reduce the efficacy of the long-lasting humoral responses.111 EZH2 deficiency also has a negative effect on T-cell immunity by preventing expansion of early T cell precursors in the thymus 112 and reducing T cell antigen receptor (TCR)-driven proliferation of T cells.113 Other research noted that EZH2 and H3K27me3 overexpression is associated with significant lymphocytic infiltration in melanoma tissues.46 These findings suggest that EZH2 plays an important role in boosting both T and B cell immune function which is supposed to provide an environment that is hostile to melanoma proliferation and invasion.113
H3K4me3 and acetylated histones H3 and H4 114,115 have been implicated in gene activation while histone H3 dimethylation at Lys 9 (H3K9me2) or H3K27me3 have been implicated in gene silencing of immune cells.115-117 EZH2 is involved in transcriptional downregulation of IFN-γ-induced expression of CIITA (essential for the transcriptional activation of MHC-II genes) in uveal melanoma.118,119 Downregulation of MHC expression is frequently noted on cancer cells; the lack of MHC expression impairs cellular immune recognition allowing cancer cells to escape efficient T cell-mediated tumor eradication. Increased expression of EZH2 and H3K27 is noted around the CIITA-Promoter IV (CIITA-PIV) in uveal melanoma cells. EZH2 knockdown results in incremental CIITA expression levels after IFN-γ induction.120
Currently, stimulation of the host's immune system represents an emerging treatment option in the management of metastatic melanoma. We speculate that EZH2 down-regulates expression of immune response and antigen presentation genes, which allows tumor cells to evade the immune response. Indeed, the role of EZH2 in immune dysregulation deserves further study.
EZH2-induced downregulation of microRNA (miRNA) promote melanoma progression
The initiation and progression of melanoma have been associated with a number of signaling pathways, including Ras/Raf/MEK/ERK, WNT/ß-catenin, and PTEN/AKT. Recently microRNAs (miRNAs), small noncoding RNAs that regulate gene expression post transcriptionally and function as tumor suppressor genes, have been implicated as key regulators in melanoma.121-124 miRNA are capable of regulating EZH2 transcription, and EZH2 can epigenetically suppress some miRNAs.123,125-127 Low expression of miR-137 is correlated with poor survival in stage IV melanoma patients, and miR-137 overexpression inhibits invasion, migration, and proliferation of melanoma cells.55 In uveal melanoma, miR-137 suppresses cell proliferation through downregulation of MITF and cyclin-dependent kinase 6 55,128-130 and by targeting BCL2, c-Met, YB1, and EZH2.131 Other miRNAs, including miR-34a and miR-182, regulate uveal melanoma development.55,132,133 YB1, a transcriptional and translational factor is upregulated in melanoma and promotes proliferation, survival, invasion, and chemosensitivity of melanoma cells.131,134 Likewise, miR-214, reported in cutaneous and uveal melanoma 135 modulates EZH2 expression.136 miR-101 suppresses invasion and proliferation in melanoma by targeting MITF and EZH2 expression.131 Interestingly, genomic loss of miRNA-101 leads to overexpression of EZH2.52
Introducing miR-124a into uveal melanoma cells resulted in inhibition of cell growth, migration, and invasion both in vivo and in vitro by downregulating CDK4, CDK6, cyclin D2, and EZH2. miR-124a expression was found to be regulated via epigenetic mechanisms, with its expression restored when cells were treated with a DNA hypomethylating agent, 5-aza-2′-deoxycytidine, and a histone deacetylase inhibitor, trichostatin A.133 Ectopic overexpression of miR-31 in various melanoma cell lines inhibited cell migration and invasion by targeting oncogenic kinases such as SRC, MET, NIK (MAP3K14) and the melanoma specific oncogene RAB27a.137 miR-31 overexpression resulted in downregulation of EZH2 and a de-repression of its target gene rap1GAP.137 A significant induction of miR-31 expression occurred upon depletion of EZH2 by RNA interference or by treatment with either DZNep or the DNA methylation inhibitor, 5′aza-dC.137
Regulation of EZH2 expression
EZH2 expression is regulated by molecular factors (Fig. 3) that control cell proliferation and self-renewal, such as E2F and c-myc, whereas its repression is induced by differentiation-promoting factors, such as pRb and p16INK4b.138,139 c-myc, a key regulator of embryonic cell pluripotency, is directly involved in transcriptional upregulation of all components of PRC2 including EZH2.139 c-myc binds PRC2 subunits promoters and induces the acetylation of histones H3 and H4, leading to transcriptional activation.139 Ectopic expression of pRb and p16INK4b results in E2F target gene repression, including repression of EZH2 in melanoma cell lines.32 MEK-ERK pathway upregulation, common in melanoma, is found to mediate a transcriptional activation of EZH2.140 miRNAs, as discussed earlier, are also capable of downregulating the expression of EZH2 in melanoma.126,127,137
Figure 3.
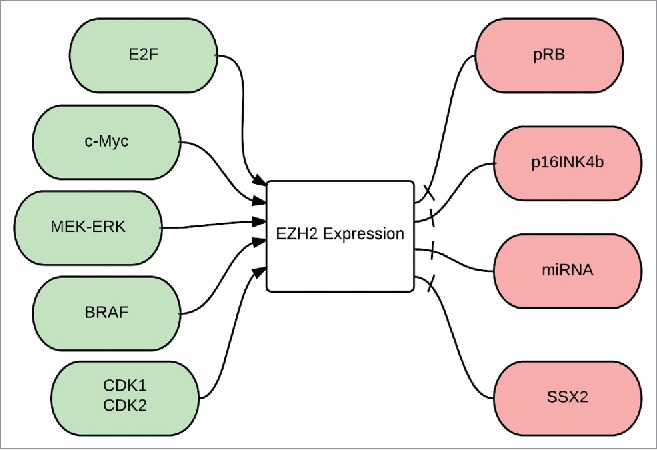
Regulation of EZH2 expression: E2F: is a group of genes that codifies a family of transcriptional factors in higher eukaryotes, cMyc is a regulatory gene that codes for a transcriptional factor, MEK-ERK is a pathway also known as the Ras-Raf-MEK-ERK pathway, BRAF is a human gene that makes a protein called B-Raf, CdK1 and 2: Cyclin dependent kinases 1 and 2, pRB: phosphorylated retinoblastoma gene, p16INK4B is a tumor suppressor gene, miRNA: micro RNA, SSX2: synovial sarcoma X gene.
There is a strong link between BRAFV600E signaling with EZH2 expression in melanoma. Upregulation of the DNMT1 and EZH2 genes is likely an important mechanism in the hypermethylation of tumor suppressor genes driven by the BRAFV600E signaling, as knockdown of BRAFV600E caused a dramatic decrease in the expression of both genes in melanoma.84 Expression of most of these BRAFV600E-target genes is identical in melanoma and papillary thyroid cancer, suggesting that these genes are commonly regulated by BRAFV600E through epigenetic mechanisms in human cancers.141
SSX proteins, expressed in about 30% of melanomas 142 were first discovered as part of the fusion oncogene SYT-SSX that plays an important role in the progression of synovial sarcoma.143,144 SSX2, a germ line-chromatin associated specific protein antagonizes BMI1 and EZH2 and negatively regulates the levels of H3K27me3 in melanoma cells (Fig. 3).145 SSX2 binds directly to dsDNA in vivo leading to a change in the chromatin structure, reducing the binding of PcG complexes and presence of H3K27me3 at promoters of PcG target genes. SSX2 knockdown in melanoma cells increases genome-wide levels of H3K27me3 and EZH2.145
In order to bind to the PRC2 complex and exert its molecular function, EZH2 must be phosphorylated in several specific sites.146 EZH2 is phosphorylated by Cdk1 and Cdk2 during cell cycle progression.147-149 EZH2 expression is inhibited by hypo-phosphorylated pRb inhibiting E2F during the G1 phase of the cell cycle; however during the transition from G1 to S phase, E2F is activated by phosphorylated pRb, and that results in overexpression of EZH2. Phosphorylation of EZH2 is also modulated by environmental signals that induce Akt activation which in turn phosphorylate EZH2 at Serine 21 (Ser21). Akt-dependent phosphorylation (i.e. pS21 EZH2) reduces the affinity of EZH2 for histone H3, which results in a decrease of the H3K27 methylation and transcriptional activation.150
EZH2 inhibitors as potential anti-cancer therapy
EZH2 is frequently overexpressed in melanoma and contributes to tumorigenesis by altering cell fate decisions and regulating pathways involved in proliferation, differentiation, and cell migration. These observations have prompted researchers to investigate the role of EZH2 catalytic activity in tumor models. Pharmacological inhibition of EZH2 by GSK503 in melanoma-bearing mice reduced melanoma progression and doubled survival times.54 Moreover, genetic ablation of EZH2 in mouse effectively prevented metastasis formation.54 Targeting EZH2 by GSK 126 has potent effects on the growth of both wild-type and EZH2 mutant human melanoma in vitro particularly in cell lines harboring the EZH2Y646 activating mutation. This was associated with cell cycle arrest, reduced proliferative capacity in both 2D and 3D culture systems, and induction of apoptosis.151
EZH2 has emerged as attractive drug target through the discovery of several catalytic small-molecule inhibitors of EZH2.17-19 GSK126 is a highly selective, S-adenosylmethionine-competitive, small molecule inhibitor of EZH2 methyltransferase activity; it decreases global H3K27me3, and thereby reactivates PRC2 target genes. (54) Studies have shown that GSK126 is very effective at inhibiting proliferation in DLBCL cells.17,20 In a phase I study, tazemetostat (EPZ-6438), a selective small molecule inhibitor of EZH2, was administered orally twice daily to 21 patients with B cell Non-Hodgkin's lymphoma (B cell NHL) and solid tumors. Patients were treated with tazemetostat at dose levels of 100, 200, 400, 800, and 1,600 mg BID. Preliminary results from the phase 1 trial were presented at the International Congress on Malignant Lymphoma (ICML) on June 20, 2015.152 Tazemetostat as a monotherapy produced a 60% overall response rate in heavily pre-treated patients with relapsed or refractory NHL, with an acceptable safety and tolerability profile.152 Tazemetostat has also shown efficacy in patients with INI1 negative and SMARCA4 negative malignant solid tumors (malignant rhabdoid tumors and epitheloid sarcomas). INI1 and SMARCA4 are subunits of the SWIF/SNF complexes, a family of multi-subunit complexes that use the energy of adenosine triphosphate (ATP) hydrolysis to remodel nucleosomes. Subsequently, genome-wide sequencing has identified mutations in genes encoding different subunits of the SWI/SNF complexes in a large number of tumors including sarcomas and malignant rhabdoid tumors.153 Updated data from patients with advanced solid tumors in the phase 1 study was presented at ESMO's European Cancer Congress in Vienna, Austria on September 26, 2015. Results from the study showed tazemetostat as a monotherapy resulted in a 55% disease control rate in nine patients with INI1-negative or SMARCA4-negative tumors who were treated at or above the recommended phase 2 dose of 800mg twice daily.152 The preclinical data as well as the promising data from EZH2 inhibitors in lymphoma supports the value of clinical development of EZH2 inhibitors in the treatment of metastatic melanoma. Future research should investigate whether inhibition of EZH2 may enhance the efficacy of immunotherapy and overcome resistance.
Conclusions
There is strong evidence for a significant role of EZH2 in melanoma. EZH2 expression is significantly higher in metastatic melanoma as compared to primary melanoma. EZH2, by disruption of tumor-suppressive pathways and activation of tumor-promoting pathways, promotes melanoma progression and invasion. EZH2 overexpression helps melanoma cells escape death and maintain resistance to senescence by downregulating genes involved in apoptosis. In Fig. 2 we provided a working hypothesis of EZH2 biology in driving melanoma progression and metastasis. Gain of function somatic mutations of EZH2 are reported in 2–3% of melanoma cases. However, the attractiveness of EZH2 is not limited to these rare mutations. Small molecule inhibitors of EZH2 are active against both wild-type and mutant forms of the protein. Novel drugs that target EZH2 may find their way into the management of patients with advanced melanoma. We speculate that EZH2 is even involved in melanomagenesis and hence targeting EZH2 may be of interest in chemoprevention. Future studies should further investigate the role EZH2 might play in the immune surveillance and whether EZH2 inhibitors might overcome mechanisms of resistant to both immune therapy and targeted therapy in metastatic melanoma.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Mrs. Dorothy Graves for the preparation of this manuscript.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016. January; 66(1):7-30; PMID:26742998; http://dx.doi.org/ 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Atrash S, Makhoul I, Mizell JS, Hutchins L, Mahmoud F. Response of metastatic mucosal melanoma to immunotherapy: It can get worse before it gets better. J Oncol Pharm Pract 2016. January 24; PMID:26811403 [DOI] [PubMed] [Google Scholar]
- 3.Larkin J, Hodi FS, Wolchok JD. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015. September 24; 373(13):1270-1; PMID:26398076; http://dx.doi.org/ 10.1056/NEJMc1509660 [DOI] [PubMed] [Google Scholar]
- 4.Buchbinder EI, Hodi FS. Melanoma in 2015: Immune-checkpoint blockade - durable cancer control. Nat Rev Clin Oncol 2016. February; 13(2):77-8; PMID:26787285; http://dx.doi.org/ 10.1038/nrclinonc.2015.237 [DOI] [PubMed] [Google Scholar]
- 5.Berger SL. The complex language of chromatin regulation during transcription. Nature 2007. May 24; 447(7143):407-12; PMID:17522673; http://dx.doi.org/ 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- 6.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004. February; 4(2):143-53; PMID:14732866; http://dx.doi.org/ 10.1038/nrc1279 [DOI] [PubMed] [Google Scholar]
- 7.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011. January 20; 469(7330):343-9; PMID:21248841; http://dx.doi.org/ 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verma SK, Tian X, LaFrance LV, Duquennet C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, et al.. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2; ACS Med Chem Lett. 2012 Oct 19; 3(12):1091-6; doi: 10.1021/ml3003346. eCollection 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchesi Irene BL. Role of Enhancer of Zeste Homolog 2 Polycomb Protein and Its Significance in Tumor Progression and Cell Differentiation. In: Danuta Radzioch, editor. Chromatin Remodelling Intech; 2013. [Google Scholar]
- 10.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP, Akslen LA. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol 2006. January 10; 24(2):268-73; PMID:16330673; http://dx.doi.org/ 10.1200/JCO.2005.01.5180 [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Tu SW, Hsieh JT. Downregulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem 2005. June 10; 280(23):22437-44; PMID:15817459; http://dx.doi.org/ 10.1074/jbc.M501379200 [DOI] [PubMed] [Google Scholar]
- 12.Avaritt NF, Mahmoud F, Sengupta D, Makhoul I, Hutchins L, Tackett A. Role of the EZH2 histone methyltransferase in melanoma. Pigment Cell Melanoma Res 2014. November; 27(6):1003-241; PMID:25346049; http://dx.doi.org/ 10.1111/pcmr.12321 [DOI] [PubMed] [Google Scholar]
- 13.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 2008. December 1; 647(1–2):21-9; PMID:18723033; http://dx.doi.org/ 10.1016/j.mrfmmm.2008.07.010 [DOI] [PubMed] [Google Scholar]
- 14.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015. July 9; 523(7559):231-5; PMID:25970248; http://dx.doi.org/ 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 15.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, Sun Y, Zhao E, Vatan L, Szeliga W, et al.. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015. November 12; 527(7577):249-53; PMID:26503055; http://dx.doi.org/ 10.1038/nature15520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher SJ, Tiffen JC, Hersey P. Histone Modifications, Modifiers and Readers in Melanoma Resistance to Targeted and Immune Therapy. Cancers (Basel) 2015. September 25; 7(4):1959-82; PMID:26426052; http://dx.doi.org/ 10.3390/cancers7040870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al.. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci U S A. 2012. February 21; 109(8):2989-94; PMID:22323599; http://dx.doi.org/ 10.1073/pnas.1116418109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, et al.. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett 2012. October 19; 3(12):1091-6; PMID:24900432; http://dx.doi.org/ 10.1021/ml3003346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G, Kumar N, et al.. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther 2014. April; 13(4):842-54; PMID:24563539; http://dx.doi.org/ 10.1158/1535-7163.MCT-13-0773 [DOI] [PubMed] [Google Scholar]
- 20.Ribrag V, Soria JC, Reyderman L, Chen R, Salazar P, Kumar N, et al.. O7.2Phase 1 first-in-human study of the enhancer of zeste-homolog 2 (EZH2) histone methyl transferase inhibitor E7438. Ann Oncol 2015. March; 26(suppl 2):ii10; http://dx.doi.org/ 10.1093/annonc/mdv085.2 [DOI] [Google Scholar]
- 21.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010. June; 35(6):323-32; PMID:20346678; http://dx.doi.org/ 10.1016/j.tibs.2010.02.009 [DOI] [PubMed] [Google Scholar]
- 22.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 2009. November; 136(21):3531-42; PMID:19820181; http://dx.doi.org/ 10.1242/dev.033902 [DOI] [PubMed] [Google Scholar]
- 23.Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol 2008. April; 20(2):201-7; PMID:18291635; http://dx.doi.org/ 10.1016/j.ceb.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 24.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 2004; 38:413-43; PMID:15568982; http://dx.doi.org/ 10.1146/annurev.genet.38.072902.091907 [DOI] [PubMed] [Google Scholar]
- 25.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002. October 18; 111(2):197-208; PMID:12408864; http://dx.doi.org/ 10.1016/S0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- 26.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 1999. July 9; 98(1):37-46; PMID:10412979; http://dx.doi.org/ 10.1016/S0092-8674(00)80604-2 [DOI] [PubMed] [Google Scholar]
- 27.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 2010. September 3; 7(3):288-98; PMID:20804966; http://dx.doi.org/ 10.1016/j.stem.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 28.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 2005. May 24; 15(10):942-7; PMID:15916951; http://dx.doi.org/ 10.1016/j.cub.2005.04.051 [DOI] [PubMed] [Google Scholar]
- 29.Montgomery ND, Yee D, Montgomery SA, Magnuson T. Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. J Mol Biol 2007. December 14; 374(5):1145-57; PMID:17997413; http://dx.doi.org/ 10.1016/j.jmb.2007.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004. October 13; 23(20):4061-71; PMID:15385962; http://dx.doi.org/ 10.1038/sj.emboj.7600402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008. March; 28(5):1862-72; PMID:18086877; http://dx.doi.org/ 10.1128/MCB.01589-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003. October 15; 22(20):5323-35; PMID:14532106; http://dx.doi.org/ 10.1093/emboj/cdg542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003. April 4; 300(5616):131-5; PMID:12649488; http://dx.doi.org/ 10.1126/science.1084274 [DOI] [PubMed] [Google Scholar]
- 34.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 2002. November 15; 16(22):2893-905; PMID:12435631; http://dx.doi.org/ 10.1101/gad.1035902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al.. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 2010. May 14; 38(3):452-64; PMID:20471950; http://dx.doi.org/ 10.1016/j.molcel.2010.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 2004. June 4; 14(5):637-46; PMID:15175158; http://dx.doi.org/ 10.1016/j.molcel.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 37.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al.. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014. June 5; 157(6):1445-59; PMID:24856970; http://dx.doi.org/ 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al.. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006. May 18; 441(7091):349-53; PMID:16625203; http://dx.doi.org/ 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- 39.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al.. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 2008. October; 4(10):e1000242; PMID:18974828; http://dx.doi.org/ 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu H, Yang Y, Ji Q, Zhao W, Jiang B, Liu R, Yuan J, Liu Q, Li X, Zou Y, et al.. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell 2012. December 11; 22(6):781-95; PMID:23238014; http://dx.doi.org/ 10.1016/j.ccr.2012.10.024 [DOI] [PubMed] [Google Scholar]
- 41.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al.. Epigenetic stem cell signature in cancer. Nat Genet 2007. February; 39(2):157-8; PMID:17200673; http://dx.doi.org/ 10.1038/ng1941 [DOI] [PubMed] [Google Scholar]
- 42.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al.. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006. February 16; 439(7078):871-4; PMID:16357870; http://dx.doi.org/ 10.1038/nature04431 [DOI] [PubMed] [Google Scholar]
- 43.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, Liou YC, Yu Q. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell 2011. September 2; 43(5):798-810; PMID:21884980; http://dx.doi.org/ 10.1016/j.molcel.2011.08.011 [DOI] [PubMed] [Google Scholar]
- 44.Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, Sun L, Zhang Y, Chen Y, Li R, et al.. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol 2007. July; 27(14):5105-19; PMID:17502350; http://dx.doi.org/ 10.1128/MCB.00162-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al.. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012. December 14; 338(6113):1465-9; PMID:23239736; http://dx.doi.org/ 10.1126/science.1227604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kampilafkos P, Melachrinou M, Kefalopoulou Z, Lakoumentas J, Sotiropoulou-Bonikou G. Epigenetic Modifications in Cutaneous Malignant Melanoma: EZH2, H3K4me2, and H3K27me3 Immunohistochemical Expression is Enhanced at the Invasion Front of the Tumor. Am J Dermatopathol 2015. February; 37(2):138-44; PMID:25614949; http://dx.doi.org/ 10.1097/DAD.0b013e31828a2d54 [DOI] [PubMed] [Google Scholar]
- 47.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, et al.. High frequency of BRAF mutations in nevi. Nat Genet 2003. January; 33(1):19-20; PMID:12447372; http://dx.doi.org/ 10.1038/ng1054 [DOI] [PubMed] [Google Scholar]
- 48.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 2005. August 4; 436(7051):720-4; PMID:16079850; http://dx.doi.org/ 10.1038/nature03890 [DOI] [PubMed] [Google Scholar]
- 49.Fan T, Jiang S, Chung N, Alikhan A, Ni C, Lee CC, Hornyak TJ. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res 2011. April; 9(4):418-29; PMID:21383005; http://dx.doi.org/ 10.1158/1541-7786.MCR-10-0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al.. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A 2003. September 30; 100(20):11606-11; PMID:14500907; http://dx.doi.org/ 10.1073/pnas.1933744100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. J Cutan Pathol 2007. August; 34(8):597-600; PMID:17640228; http://dx.doi.org/ 10.1111/j.1600-0560.2006.00678.x [DOI] [PubMed] [Google Scholar]
- 52.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al.. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008. December 12; 322(5908):1695-9; PMID:19008416; http://dx.doi.org/ 10.1126/science.1165395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamaguchi H, Hung MC. Regulation and Role of EZH2 in Cancer. Cancer Res Treat 2014. July; 46(3):209-22; PMID:25038756; http://dx.doi.org/ 10.4143/crt.2014.46.3.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, Arenas-Ramirez N, Haeusel J, Zhang Y, Bonalli M, et al.. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015. January 22; 6:6051; PMID:25609585; http://dx.doi.org/ 10.1038/ncomms7051 [DOI] [PubMed] [Google Scholar]
- 55.Chen X, Wang J, Shen H, Lu J, Li C, Hu DN, Dong XD, Yan D, Tu L, et al.. Epigenetics, microRNAs, and carcinogenesis: functional role of microRNA-137 in uveal melanoma. Invest Ophthalmol Vis Sci 2011. March 2; 52(3):1193-9; PMID:21051724; http://dx.doi.org/ 10.1167/iovs.10-5272 [DOI] [PubMed] [Google Scholar]
- 56.Bryant RJ, Cross NA, Eaton CL, Hamdy FC, Cunliffe VT. EZH2 promotes proliferation and invasiveness of prostate cancer cells. Prostate 2007. April 1; 67(5):547-56; PMID:17252556; http://dx.doi.org/ 10.1002/pros.20550 [DOI] [PubMed] [Google Scholar]
- 57.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al.. A landscape of driver mutations in melanoma. Cell 2012. July 20; 150(2):251-63; PMID:22817889; http://dx.doi.org/ 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al.. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012. September; 44(9):1006-14; PMID:22842228; http://dx.doi.org/ 10.1038/ng.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwabe M, Lubbert M. Epigenetic lesions in malignant melanoma. Curr Pharm Biotechnol 2007. December; 8(6):382-7; PMID:18289047; http://dx.doi.org/ 10.2174/138920107783018372 [DOI] [PubMed] [Google Scholar]
- 60.Sigalotti L, Covre A, Fratta E, Parisi G, Colizzi F, Rizzo A, Danielli R, Nicolay HJ, Coral S, Maio M, et al.. Epigenetics of human cutaneous melanoma: setting the stage for new therapeutic strategies. J Transl Med 2010. June 11; 8:56, 5876-8-56; PMID:20540720; http://dx.doi.org/ 10.1186/1479-5876-8-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barsotti AM, Ryskin M, Zhong W, Zhang WG, Giannakou A, Loreth C, Diesl V, Follettie M, Golas J, Lee M, et al.. Epigenetic reprogramming by tumor-derived EZH2 gain-of-function mutations promotes aggressive 3D cell morphologies and enhances melanoma tumor growth. Oncotarget 2015. February 20; 6(5):2928-38; PMID:25671303; http://dx.doi.org/ 10.18632/oncotarget.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012. July 6; 150(1):12-27; PMID:22770212; http://dx.doi.org/ 10.1016/j.cell.2012.06.013 [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011. March; 17(3):330-9; PMID:21386836; http://dx.doi.org/ 10.1038/nm.2305 [DOI] [PubMed] [Google Scholar]
- 64.Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta 2011. January; 1815(1):75-89; http://dx.doi.org/ 10.1016/j.bbcan.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 65.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 2006. September; 7(9):667-77; PMID:16921403; http://dx.doi.org/ 10.1038/nrm1987 [DOI] [PubMed] [Google Scholar]
- 66.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol 2008. May; 28(10):3457-64; PMID:18332116; http://dx.doi.org/ 10.1128/MCB.02019-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Mönch K, Minucci S, Porse BT, Marine JC, et al.. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 2007. March 1; 21(5):525-30; PMID:17344414; http://dx.doi.org/ 10.1101/gad.415507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999. January 14; 397(6715):164-8; PMID:9923679; http://dx.doi.org/ 10.1038/16476 [DOI] [PubMed] [Google Scholar]
- 69.Tonini T, Bagella L, D'Andrilli G, Claudio PP, Giordano A. Ezh2 reduces the ability of HDAC1-dependent pRb2/p130 transcriptional repression of cyclin A. Oncogene. 2004. June 17; 23(28):4930-7; PMID:15077161; http://dx.doi.org/ 10.1038/sj.onc.1207608 [DOI] [PubMed] [Google Scholar]
- 70.Tonini T, D'Andrilli G, Fucito A, Gaspa L, Bagella L. Importance of Ezh2 polycomb protein in tumorigenesis process interfering with the pathway of growth suppressive key elements. J Cell Physiol. 2008. February; 214(2):295-300; PMID:17786943; http://dx.doi.org/ 10.1002/jcp.21241 [DOI] [PubMed] [Google Scholar]
- 71.Bachmann IM, Straume O, Akslen LA. Altered expression of cell cycle regulators Cyclin D1, p14, p16, CDK4 and Rb in nodular melanomas. Int J Oncol 2004. December; 25(6):1559-65; PMID:15547691 [PubMed] [Google Scholar]
- 72.Wu ZL, Zheng SS, Li ZM, Qiao YY, Aau MY, Yu Q. Polycomb protein EZH2 regulates E2F1-dependent apoptosis through epigenetically modulating Bim expression. Cell Death Differ 2010. May; 17(5):801-10; PMID:19893569; http://dx.doi.org/ 10.1038/cdd.2009.162 [DOI] [PubMed] [Google Scholar]
- 73.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 2009. March 20; 136(6):1122-35; PMID:19303854; http://dx.doi.org/ 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manning CS, Hooper S, Sahai EA. Intravital imaging of SRF and Notch signalling identifies a key role for EZH2 in invasive melanoma cells. Oncogene 2014. November 10; 34(33):4320-32; PMID:25381824; http://dx.doi.org/ 10.1038/onc.2014.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moriyama M, Osawa M, Mak SS, Ohtsuka T, Yamamoto N, Han H, Delmas V, Kageyama R, Beermann F, Larue L, et al.. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J Cell Biol 2006. May 8; 173(3):333-9; PMID:16651378; http://dx.doi.org/ 10.1083/jcb.200509084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zabierowski SE, Baubet V, Himes B, Li L, Fukunaga-Kalabis M, Patel S, McDaid R, Guerra M, Gimotty P, Dahmane N, et al.. Direct reprogramming of melanocytes to neural crest stem-like cells by one defined factor. Stem Cells 2011. November; 29(11):1752-62; PMID:21948558; http://dx.doi.org/ 10.1002/stem.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balasubramanian S, Adhikary G, Eckert RL. The Bmi-1 polycomb protein antagonizes the (−)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival. Carcinogenesis 2010. March; 31(3):496-503; PMID:20015867; http://dx.doi.org/ 10.1093/carcin/bgp314 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 78.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009. November 25; 139(5):871-90; PMID:19945376; http://dx.doi.org/ 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 79.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet 2005. October; 37(10):1047-54; PMID:16142232; http://dx.doi.org/ 10.1038/ng1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al.. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008. May 16; 133(4):704-15; PMID:18485877; http://dx.doi.org/ 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Javelaud D, Alexaki VI, Mauviel A. Transforming growth factor-β in cutaneous melanoma. Pigment Cell Melanoma Res 2008. April; 21(2):123-32; PMID:18426405; http://dx.doi.org/ 10.1111/j.1755-148X.2008.00450.x [DOI] [PubMed] [Google Scholar]
- 82.Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J, Hutchinson P, Tse G, et al.. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 2013. October 14; 24(4):466-80; PMID:24075834; http://dx.doi.org/ 10.1016/j.ccr.2013.08.018 [DOI] [PubMed] [Google Scholar]
- 83.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schübeler D, van Nimwegen E, Christofori G. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 2013. June 10; 23(6):768-83; PMID:23764001; http://dx.doi.org/ 10.1016/j.ccr.2013.04.020 [DOI] [PubMed] [Google Scholar]
- 84.Hou P, Liu D, Dong J, Xing M. The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells. Cell Cycle 2012. January 15; 11(2):286-95; PMID:22189819; http://dx.doi.org/ 10.4161/cc.11.2.18707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M, Liu ZJ. Activation of Notch1 signaling is required for β-catenin-mediated human primary melanoma progression. J Clin Invest 2005. November; 115(11):3166-76; PMID:16239965; http://dx.doi.org/ 10.1172/JCI25001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pinner S, Jordan P, Sharrock K, Bazley L, Collinson L, Marais R, Bonvin E, Goding C, Sahai E. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res 2009. October 15; 69(20):7969-77; PMID:19826052; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 2010. May; 11(5):353-65; PMID:20414257; http://dx.doi.org/ 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 2000. August 3; 406(6795):532-5; PMID:10952316; http://dx.doi.org/ 10.1038/35020106 [DOI] [PubMed] [Google Scholar]
- 89.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol 2009. March; 11(3):257-68; PMID:19198601; http://dx.doi.org/ 10.1038/ncb1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tiffen J, Gallagher SJ, Hersey P. EZH2: an emerging role in melanoma biology and strategies for targeted therapy. Pigment Cell Melanoma Res 2015. January; 28(1):21-30; PMID:24912396; http://dx.doi.org/ 10.1111/pcmr.12280 [DOI] [PubMed] [Google Scholar]
- 91.Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res 2008. February 1; 68(3):650-6; PMID:18245463; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-2491 [DOI] [PubMed] [Google Scholar]
- 92.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 2006. December 15; 20(24):3426-39; PMID:17182868; http://dx.doi.org/ 10.1101/gad.406406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goodall J, Carreira S, Denat L, Kobi D, Davidson I, Nuciforo P, Sturm RA, Larue L, Goding CR. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor-negative melanoma cells. Cancer Res 2008. October 1; 68(19):7788-94; PMID:18829533; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-1053 [DOI] [PubMed] [Google Scholar]
- 94.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol 2008. July; 9(7):557-68; PMID:18523435; http://dx.doi.org/ 10.1038/nrm2428 [DOI] [PubMed] [Google Scholar]
- 95.Sauka-Spengler T, Bronner-Fraser M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis 2008. November; 46(11):673-82; PMID:19003930; http://dx.doi.org/ 10.1002/dvg.20436 [DOI] [PubMed] [Google Scholar]
- 96.Hornyak TJ, Hayes DJ, Chiu LY, Ziff EB. Transcription factors in melanocyte development: distinct roles for Pax-3 and Mitf. Mech Dev 2001. March; 101(1–2):47-59; PMID:11231058; http://dx.doi.org/ 10.1016/S0925-4773(00)00569-4 [DOI] [PubMed] [Google Scholar]
- 97.Tachibana M, Takeda K, Nobukuni Y, Urabe K, Long JE, Meyers KA, Aaronson SA, Miki T. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet 1996. September; 14(1):50-4; PMID:8782819; http://dx.doi.org/ 10.1038/ng0996-50 [DOI] [PubMed] [Google Scholar]
- 98.Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 2005. February 24; 433(7028):884-7; PMID:15729346; http://dx.doi.org/ 10.1038/nature03292 [DOI] [PubMed] [Google Scholar]
- 99.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 2009. March 20; 136(6):1122-35; PMID:19303854; http://dx.doi.org/ 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007. May 1; 21(9):1050-63; PMID:17437993; http://dx.doi.org/ 10.1101/gad.1524107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res 2010. December; 23(6):746-59; PMID:20726948; http://dx.doi.org/ 10.1111/j.1755-148X.2010.00757.x [DOI] [PubMed] [Google Scholar]
- 102.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006. May 1; 20(9):1123-36; PMID:16618801; http://dx.doi.org/ 10.1101/gad.381706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 2004. July 1; 18(13):1592-605; PMID:15231737; http://dx.doi.org/ 10.1101/gad.1200204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al.. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006. April 21; 125(2):301-13; PMID:16630818; http://dx.doi.org/ 10.1016/j.cell.2006.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pinnix CC, Lee JT, Liu ZJ, McDaid R, Balint K, Beverly LJ, Brafford PA, Xiao M, Himes B, Zabierowski SE, et al.. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res 2009. July 1; 69(13):5312-20; PMID:19549918; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, et al.. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res 2004. August 1; 64(15):5270-82; PMID:15289333; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-0731 [DOI] [PubMed] [Google Scholar]
- 107.Liu ZJ, Xiao M, Balint K, Smalley KS, Brafford P, Qiu R, Pinnix CC, Li X, Herlyn M. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res 2006. April 15; 66(8):4182-90; PMID:16618740; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-3589 [DOI] [PubMed] [Google Scholar]
- 108.Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci U S A 2008. April 29; 105(17):6392-7; PMID:18427106; http://dx.doi.org/ 10.1073/pnas.0802047105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007. May 24; 447(7143):425-32; PMID:17522676; http://dx.doi.org/ 10.1038/nature05918 [DOI] [PubMed] [Google Scholar]
- 110.Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, Elemento O. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 2010. December 9; 116(24):5247-55; PMID:20736451; http://dx.doi.org/ 10.1182/blood-2010-04-280149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 2003. February; 4(2):124-31; PMID:12496962; http://dx.doi.org/ 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- 112.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wülfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell 2005. May 6; 121(3):425-36; PMID:15882624; http://dx.doi.org/ 10.1016/j.cell.2005.02.029 [DOI] [PubMed] [Google Scholar]
- 113.He S, Wang J, Kato K, Xie F, Varambally S, Mineishi S, Kuick R, Mochizuki K, Liu Y, Nieves E, et al.. Inhibition of histone methylation arrests ongoing graft-versus-host disease in mice by selectively inducing apoptosis of alloreactive effector T cells. Blood 2012. February 2; 119(5):1274-82; PMID:22117046; http://dx.doi.org/ 10.1182/blood-2011-06-364422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cuddapah S, Barski A, Zhao K. Epigenomics of T cell activation, differentiation, and memory. Curr Opin Immunol 2010. June; 22(3):341-7; PMID:20226645; http://dx.doi.org/ 10.1016/j.coi.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al.. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009. January 16; 30(1):155-67; PMID:19144320; http://dx.doi.org/ 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al.. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012. August 16; 488(7411):404-8; PMID:22842901; http://dx.doi.org/ 10.1038/nature11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007. September 21; 130(6):1083-94; PMID:17825402; http://dx.doi.org/ 10.1016/j.cell.2007.08.019 [DOI] [PubMed] [Google Scholar]
- 118.Gobin SJ, Peijnenburg A, Keijsers V, van den Elsen PJ. Site α is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity 1997. May; 6(5):601-11; PMID:9175838; http://dx.doi.org/ 10.1016/S1074-7613(00)80348-9 [DOI] [PubMed] [Google Scholar]
- 119.Morris AC, Beresford GW, Mooney MR, Boss JM. Kinetics of a gamma interferon response: expression and assembly of CIITA promoter IV and inhibition by methylation. Mol Cell Biol 2002. July; 22(13):4781-91; PMID:12052885; http://dx.doi.org/ 10.1128/MCB.22.13.4781-4791.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Holling TM, Bergevoet MW, Wilson L, Van Eggermond MC, Schooten E, Steenbergen RD, Snijders PJ, Jager MJ, Van den Elsen PJ. A role for EZH2 in silencing of IFN-gamma inducible MHC2TA transcription in uveal melanoma. J Immunol 2007. October 15; 179(8):5317-25; PMID:17911618; http://dx.doi.org/ 10.4049/jimmunol.179.8.5317 [DOI] [PubMed] [Google Scholar]
- 121.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature 2007. February 22; 445(7130):851-7; PMID:17314971; http://dx.doi.org/ 10.1038/nature05661 [DOI] [PubMed] [Google Scholar]
- 122.Mueller DW, Bosserhoff AK. The evolving concept of ‘melano-miRs’-microRNAs in melanomagenesis. Pigment Cell Melanoma Res 2010. October; 23(5):620-6; PMID:20557479; http://dx.doi.org/ 10.1111/j.1755-148X.2010.00734.x [DOI] [PubMed] [Google Scholar]
- 123.Mueller DW, Bosserhoff AK. Role of miRNAs in the progression of malignant melanoma. Br J Cancer 2009. August 18; 101(4):551-6; PMID:19638982; http://dx.doi.org/ 10.1038/sj.bjc.6605204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bell RE, Levy C. The three M's: melanoma, microphthalmia-associated transcription factor and microRNA. Pigment Cell Melanoma Res 2011. December; 24(6):1088-106; PMID:22004179; http://dx.doi.org/ 10.1111/j.1755-148X.2011.00931.x [DOI] [PubMed] [Google Scholar]
- 125.Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I, Kreis S. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res 2010. May 15; 70(10):4163-73; PMID:20442294; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4512 [DOI] [PubMed] [Google Scholar]
- 126.Benetatos L, Voulgaris E, Vartholomatos G, Hatzimichael E. Non-coding RNAs and EZH2 interactions in cancer: long and short tales from the transcriptome. Int J Cancer 2013. July 15; 133(2):267-74; PMID:23001607; http://dx.doi.org/ 10.1002/ijc.27859 [DOI] [PubMed] [Google Scholar]
- 127.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol 2009. January; 41(1):87-95; PMID:18834952; http://dx.doi.org/ 10.1016/j.biocel.2008.09.005 [DOI] [PubMed] [Google Scholar]
- 128.Dynek JN, Chan SM, Liu J, Zha J, Fairbrother WJ, Vucic D. Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res 2008. May 1; 68(9):3124-32; PMID:18451137; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-6622 [DOI] [PubMed] [Google Scholar]
- 129.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al.. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005. July 7; 436(7047):117-22; PMID:16001072; http://dx.doi.org/ 10.1038/nature03664 [DOI] [PubMed] [Google Scholar]
- 130.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 2006. September; 12(9):406-14; PMID:16899407; http://dx.doi.org/ 10.1016/j.molmed.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 131.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, et al.. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol 2013. March; 133(3):768-75; PMID:23151846; http://dx.doi.org/ 10.1038/jid.2012.357 [DOI] [PubMed] [Google Scholar]
- 132.Yan D, Dong XD, Chen X, Yao S, Wang L, Wang J, Wang C, Hu DN, Qu J, Tu L. Role of microRNA-182 in posterior uveal melanoma: regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS One 2012; 7(7):e40967; PMID:22848417; http://dx.doi.org/ 10.1371/journal.pone.0040967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen X, He D, Dong XD, Dong F, Wang J, Wang L, Tang J, Hu DN, Yan D, Tu L. MicroRNA-124a is epigenetically regulated and acts as a tumor suppressor by controlling multiple targets in uveal melanoma. Invest Ophthalmol Vis Sci 2013. March 1; 54(3):2248-56; PMID:23404119; http://dx.doi.org/ 10.1167/iovs.12-10977 [DOI] [PubMed] [Google Scholar]
- 134.Kosnopfel C, Sinnberg T, Schittek B. Y-box binding protein 1–a prognostic marker and target in tumour therapy. Eur J Cell Biol 2014. Jan-Feb; 93(1–2):61-70; PMID:24461929; http://dx.doi.org/ 10.1016/j.ejcb.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 135.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 2008. May; 18(5):549-57; PMID:18379589; http://dx.doi.org/ 10.1038/cr.2008.45 [DOI] [PubMed] [Google Scholar]
- 136.Penna E, Orso F, Taverna D. miR-214 as a Key Hub that Controls Cancer Networks: Small Player, Multiple Functions. J Invest Dermatol 2015 Apr; 135(4):960-9; PMID:25501033; http://dx.doi.org/22948084 10.1038/jid.2014.479 [DOI] [PubMed] [Google Scholar]
- 137.Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, Fullen DR, Johnson TM, Giordano TJ, Palanisamy N, et al.. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget 2012. September; 3(9):1011-25; PMID:22948084; http://dx.doi.org/ 10.18632/oncotarget.622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol 2001. October; 21(20):6820-32; PMID:11564866; http://dx.doi.org/ 10.1128/MCB.21.20.6820-6832.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Neri F, Zippo A, Krepelova A, Cherubini A, Rocchigiani M, Oliviero S. Myc regulates the transcription of the PRC2 gene to control the expression of developmental genes in embryonic stem cells. Mol Cell Biol 2012. February; 32(4):840-51; PMID:22184065; http://dx.doi.org/ 10.1128/MCB.06148-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fujii S, Fukamachi K, Tsuda H, Ito K, Ito Y, Ochiai A. RAS oncogenic signal upregulates EZH2 in pancreatic cancer. Biochem Biophys Res Commun 2012. January 20; 417(3):1074-9; PMID:22222375; http://dx.doi.org/ 10.1016/j.bbrc.2011.12.099 [DOI] [PubMed] [Google Scholar]
- 141.Liu S, Zhang B, Zhao Y, Chen P, Ji M, Hou P, Shi B. Association of BRAFV600E mutation with clinicopathological features of papillary thyroid carcinoma: a study on a Chinese population. Int J Clin Exp Pathol 2014. September 15; 7(10):6922-8; PMID:25400776 [PMC free article] [PubMed] [Google Scholar]
- 142.dos Santos NR, Torensma R, de Vries TJ, Schreurs MW, de Bruijn DR, Kater-Baats E, Ruiter DJ, Adema GJ, van Muijen GN, van Kessel AG. Heterogeneous expression of the SSX cancer/testis antigens in human melanoma lesions and cell lines. Cancer Res 2000. March 15; 60(6):1654-62; PMID:10749136 [PubMed] [Google Scholar]
- 143.Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM, Gusterson BA, Cooper CS. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994. August; 7(4):502-8; PMID:7951320; http://dx.doi.org/ 10.1038/ng0894-502 [DOI] [PubMed] [Google Scholar]
- 144.Garcia CB, Shaffer CM, Alfaro MP, Smith AL, Sun J, Zhao Z, Young PP, VanSaun MN, Eid JE. Reprogramming of mesenchymal stem cells by the synovial sarcoma-associated oncogene SYT-SSX2. Oncogene 2012. May 3; 31(18):2323-34; PMID:21996728; http://dx.doi.org/ 10.1038/onc.2011.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gjerstorff MF, Relster MM, Greve KB, Moeller JB, Elias D, Lindgreen JN, Schmidt S, Mollenhauer J, Voldborg B, Pedersen CB, et al.. SSX2 is a novel DNA-binding protein that antagonizes polycomb group body formation and gene repression. Nucleic Acids Res 2014. October; 42(18):11433-46; PMID:25249625; http://dx.doi.org/ 10.1093/nar/gku852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Caretti G, Palacios D, Sartorelli V, Puri PL. Phosphoryl-EZH-ion. Cell Stem Cell 2011. March 4; 8(3):262-5; PMID:21362566; http://dx.doi.org/ 10.1016/j.stem.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, Bagchi A, Simon JA, Huang H. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol 2010. November; 12(11):1108-14; PMID:20935635; http://dx.doi.org/ 10.1038/ncb2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 2010. December 1; 24(23):2615-20; PMID:21123648; http://dx.doi.org/ 10.1101/gad.1983810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, et al.. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 2011. January; 13(1):87-94; PMID:21131960; http://dx.doi.org/ 10.1038/ncb2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 2005. October 14; 310(5746):306-10; PMID:16224021; http://dx.doi.org/ 10.1126/science.1118947 [DOI] [PubMed] [Google Scholar]
- 151.Tiffen JC, Gunatilake D, Gallagher SJ, Gowrishankar K, Heinemann A, Cullinane C, Dutton-Regester K, Pupo GM, Strbenac D, Yang JY, et al.. Targeting activating mutations of EZH2 leads to potent cell growth inhibition in human melanoma by derepression of tumor suppressor genes. Oncotarget 2015. September 29; 6(29):27023-36; PMID:26304929; http://dx.doi.org/ 10.18632/oncotarget.4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.ZH2 Inhibitor — EPZ-6438 for Non-Hodgkin Lymphoma and INI1-Deficient Tumors [Internet]. [2016].Available from: http://www.epizyme.com/programs/tazemetostat/ [Google Scholar]
- 153.Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol 2015; 10:145-71; PMID:25387058; http://dx.doi.org/ 10.1146/annurev-pathol-012414-040445 [DOI] [PubMed] [Google Scholar]


