Abstract
OBJECTIVE
Individual MRI characteristics (e.g., volume) are routinely used to identify survival-associated phenotypes for glioblastoma (GBM). This study investigated whether combinations of MRI features can also stratify survival. Furthermore, the molecular differences between phenotype-induced groups were investigated.
METHODS
Ninety-two patients with imaging, molecular, and survival data from the TCGA (The Cancer Genome Atlas)-GBM collection were included in this study. For combinatorial phenotype analysis, hierarchical clustering was used. Groups were defined based on a cutpoint obtained via tree-based partitioning. Furthermore, differential expression analysis of microRNA (miRNA) and mRNA expression data was performed using GenePattern Suite. Functional analysis of the resulting genes and miRNAs was performed using Ingenuity Pathway Analysis. Pathway analysis was performed using Gene Set Enrichment Analysis.
RESULTS
Clustering analysis reveals that image-based grouping of the patients is driven by 3 features: volume-class, hemorrhage, and T1/FLAIR-envelope ratio. A combination of these features stratifies survival in a statistically significant manner. A cutpoint analysis yields a significant survival difference in the training set (median survival difference: 12 months, p = 0.004) as well as a validation set (p = 0.0001). Specifically, a low value for any of these 3 features indicates favorable survival characteristics. Differential expression analysis between cutpoint-induced groups suggests that several immune-associated (natural killer cell activity, T-cell lymphocyte differentiation) and metabolism-associated (mitochondrial activity, oxidative phosphorylation) pathways underlie the transition of this phenotype. Integrating data for mRNA and miRNA suggests the roles of several genes regulating proliferation and invasion.
CONCLUSIONS
A 3-way combination of MRI phenotypes may be capable of stratifying survival in GBM. Examination of molecular processes associated with groups created by this combinatorial phenotype suggests the role of biological processes associated with growth and invasion characteristics.
Keywords: imaging genomics, clustering, differential expression analysis, combinatorial phenotype analysis, glioblastoma, radiogenomics, oncology
Despite aggressive treatment, the prognosis for patients diagnosed with glioblastoma (GBM) remains poor, with a median survival time of 12–15 months.32 Surgery never completely removes the tumor due to widespread infiltration of tumor cells, leading to recurrence. Tumor samples are used to establish diagnosis, but the rest of the tumor is unavailable for pathologic or genomic interrogation. The nonuniformity of GBM may explain why it is so difficult to control with conventional therapies.
Imaging provides the only nondestructive means to evaluate the morphology of the entire tumor. Imaging features potentially represent a phenotypic characterization of the genetics and biology of the tumor.4 This study seeks combinations of imaging features that are predictive of tumor behavior and therapeutic response.
Identifying noninvasive surrogates of clinical outcome and associated molecular differences might suggest the etiology of the underlying disease state.4,29 Several studies13,24,38 indicate that MRI phenotypes might potentially serve as noninvasive proxies of cancer-associated molecular processes. Although the standard of care for treating GBM is relatively nonspecific (i.e., resection, radiotherapy, and temozolomide33), personalized treatment may have different efficacies among GBM subtypes.25 Better patient stratification using clinical imaging methodology may offer a more patient-specific treatment. Molecular information concerning the GBM is only available from surgical specimens. Thus, a noninvasive radiographic method to determine molecular and clinical characteristics may provide prognostic data to help guide the frequency of surveillance imaging for tumor progression or recurrence, possibly leading to earlier and more effective therapy.
The Cancer Genome Atlas (TCGA) contains comprehensive genomic data (e.g., expression, copy number, methylation, and microRNA [miRNA]). The Cancer Imaging Archive (TCIA) is a publicly available repository of patient-derived images across multiple modalities (SPECT, CT, and MRI) and tumor types. The TCIA images are from patients who have corresponding molecular data in TCGA. Using genomic and imaging data from the TCGA-GBM archive, we performed a computational analysis to identify radiological feature combinations that might stratify survival. Two phenotype groups were defined based on these feature combinations. Differential analysis of mRNA and miRNA expression was performed to identify molecular differences between these groups.
We showed that a combinatorial phenotype comprising 3 radiological characteristics (volume-class, T1/FLAIR ratio, and hemorrhage) can stratify survival. The median survival difference between the less aggressive phenotype and the more aggressive phenotype was 8 months. Molecular examination of these 2 groups suggests that genes and miRNAs related to growth, invasion, and proliferation underlie this phenotypic difference.
Methods
Patients
Patients were included in the study based on the availability of MRI data from TCIA and were selected to ensure consistency in glioma history and treatment profile. Specifically, patients with no prior glioma and those who received standard of care (concurrent chemotherapy and radiotherapy after resection) were selected, for a total of 92 cases. The clinical data for these untreated, primary GBM cases were obtained from the cBioPortal for Cancer Genomics (Memorial Sloan Kettering Cancer Center; http://www.cbioportal.org/public-portal/). To assess stratification of survival by the combinatorial phenotype, we divided data into a training set (44 patients; 14 females, 30 males) and a validation set (48 patients; 18 females, 30 males), based on median survival and vital (alive vs dead) status. We also verified that patients in these 2 sets had similar demographic and functional (i.e., Karnofsky Performance Scale score) characteristics.
Imaging Features
Radiology annotations for the 92 patients with GBM and their preoperative images (from TCIA) were obtained from the TCGA Glioma Phenotype Research Group (https://wiki.cancerimagingarchive.net/display/Public/TCGA+Glioma+Phenotype+Research+Group) based on the VASARI (Visually AcceSAble Rembrandt Images) standardized feature set.9 This contains annotations produced via consensus reads by a team of radiologists affiliated with the TCGA Glioma Phenotype Research Group. See details at https://wiki.nci.nih.gov/display/CIP/VASARI and https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project. Gutman et al.9 discuss details of specific imaging protocols and the semantic scoring system used by the radiologists.
VASARI characteristics studied were volume-class, enhancement quality, proportion contrast-enhancing tumor, proportion noncontrast-enhancing tumor, proportion necrosis, proportion edema, T1/FLAIR-envelope ratio, enhancing margin thickness, distribution, and hemorrhage. This subset corresponded to the imaging-specific rather than anatomical (location) subset of the VASARI features. Several of these imaging characteristics were described as independently prognostic in previous studies10,18,26 and most are qualitative (categorical data), whereas some (i.e., major and minor axis lengths) are quantitative (continuous-valued). For consistency, all variables were made categorical.
Instead of using the length variables separately, we computed an “approximate” 3D volume using the formula (4π*a2*b/3)/8, with “a” being the major axis length and “b” being the minor axis length. These measurements were derived from the region of T2 signal abnormality. For consistency (i.e., to make all variables categorical), we converted the volume into an ordinal scale based on its magnitude with respect to the median volume across all cases (1 = ≤ median, 2 = > median). This dichotomized version of the volume is referred to as “volume-class.”
The T1/FLAIR ratio attribute compares the relative size of the abnormality in the T1-weighted images to the size in the T2-FLAIR images. It can have 3 categories: expansive (size of precontrast T1 abnormality region approximately equal to FLAIR envelope), mixed (T1 abnormality less than FLAIR abnormality), and infiltrative (T1 region significantly smaller than FLAIR region). Hemorrhage represents intrinsic hemorrhage in the tumor matrix (observed in T1-weighted images or T2-weighted images).
Genomic Features
Level 3 expression data for both mRNA (Affymetrix U133Av2 BI Platform: Affymetrix HT HG U133A) and miRNA (UNC Agilent Human miRNA 8 × 15 K) were downloaded from the TCGA data portal. These data are normalized and were used for differential expression analysis.
Survival Data
Patient-specific survival data came from the cBioPortal for Cancer Genomics. Clinical data are available at http://www.cbioportal.org/public-portal/study.do?cancer_study_id=gbm_tcga. We extracted “Overall Survival (months)” and “Overall Survival Status” for each patient. Further demographic information (sex, age, prior glioma status, and treatment history) was also obtained from the cBioPortal (or from the TCGA portal [http://cancergenome.nih.gov/]).
Clustering Analysis
To examine whether these image features stratify the cases in any clinically relevant manner, we hierarchically clustered them (using R package “cluster”) using Gower’s similarity metric with complete linkage. Gower’s distance is applicable for mixed (combination of discrete, interval-scaled, and categorical) data7 and is defined as
where dijk is the distance between individuals i and j based on variable k. wijk is a weight given to the ijk comparison, taking values of “0” for an invalid comparison and “1” for a valid comparison.
Viewing the cluster plot allowed identification of imaging variables with a range consistent within a cluster that also changed across the cluster boundary. This led to “cluster blocks,” within which the variable is approximately constant and between which there is a change. This allowed creation of a combinatorial phenotype via element-wise multiplication of the individual imaging variables. For example, a case with “high” volume (Category 2), “low” T1/FLAIR ratio (Category 1), and “no” hemorrhage (Category 1) will take the value 2 (2*1*1) for its combinatorial phenotype.
Tree-Based Partitioning Analysis to Find Cutpoint on the Combinatorial Phenotype
Using survival data from the 92 patients, we used a k-adaptive partitioning scheme (R package “kaps”) to estimate a cutpoint on the combinatorial phenotype that induces a statistically significant difference in survival between the 2 groups. This cutpoint is initially estimated on the training set (44 cases) and validated on a validation set (remaining 48 cases absent from the training set). The survival difference between the 2 cutpoint-induced groups was estimated using a log-rank test. The split value obtained in the training set was also used to compare differences in progression-free survival (PFS) via the log-rank test. The performance of the combinatorial phenotype relative to individual variables in the combination is assessed via area under the survival receiver operating characteristic (ROC) curve. Area under the curve (AUC) can take a value between 0 (poor) and 1 (perfect discrimination). An AUC > 0.5 suggests predictive ability better than random chance.
Differential Expression Analysis Between Cutpoint-Induced Phenotype Classes
Cutpoint analysis (described above) identified a split value on the combinatorial radiophenotype that partitioned the data into 2 distinct survival groups. Next, we looked for molecular differences (differential expression of mRNA and miRNA) between these 2 groups, using the Comparative Marker Selection module within the GenePattern Suite (Broad Institute).
Normalized Level 3 expression data for both mRNA and miRNA were downloaded from the TCGA data portal. These data were used for differential expression analysis via the Comparative Marker Selection module within the publicly available GenePattern platform (http://genepattern.broadinstitute.org/). The algorithm uses a 2-sided t-test to identify genes/miRNAs differentially expressed between the 2 phenotype classes. These classes are induced by the split value (“2”) obtained from the partitioning algorithm used above. Cases with “volume-class:T1/FLAIR:hemorrhage” combination values greater than “2” are designated Group 1. The rest are designated Group 0.
Functional Analysis and Integrative Analysis
Functional analysis (i.e., pathway analysis) of the differentially expressed genes and miRNAs was done using Gene Set Enrichment Analysis (GSEA; Broad Institute). We used the GSEA desktop version (http://www.broadinstitute.org/gsea/) with the settings “t-test” metric and “equalized and balanced” randomization during 1000 permutations. The t-test is used for consistency with the metric used for differential expression analysis (from GenePattern). Multiple testing correction for significance was done using false discovery rate (FDR) computation.1
For relating differentially expressed miRNAs with differentially expressed mRNAs, the miRNA target filter feature was used within Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems) to find miRNAs targeting the differentially expressed genes. This integrative miRNA:mRNA analysis looks for target relationships between miRNAs and the genes derived from differential expression analysis. Also, the miRNA target filter in IPA looks for concordant changes in expression (i.e., anticorrelated expression changes in miRNA:mRNA abundance).
Integrative Network Analysis of miRNA and mRNA Entities via Ingenuity Pathway Network Analysis
Core analysis and functional analysis were performed on the gene and miRNA lists. We explored “Direct Interactions” using “Experimentally Observed or High Confidence Predictions” in the IPA Knowledge Base (“Genes only”).
Results
Clustering Analysis Suggests Unsupervised Clustering Structure Is Driven by a Combination of 3 Imaging Variables
Study of the hierarchical clustering plot (based on Gower’s distance and complete linkage; Fig. 1) suggested 3 categorical variables were influencing clustering structure: volume-class, T1/FLAIR ratio, and hemorrhage. Next, the combinatorial phenotype was derived as an element-wise product of the categorical values taken by each of the 3 variables (volume-class, T1/FLAIR ratio, and hemorrhage). For example, if volume-class is Category 1, T1/FLAIR ratio is Category 2, and hemorrhage is Category 2, then the combined phenotype takes a value equal to 1*2*2 (i.e., 4).
FIG. 1.
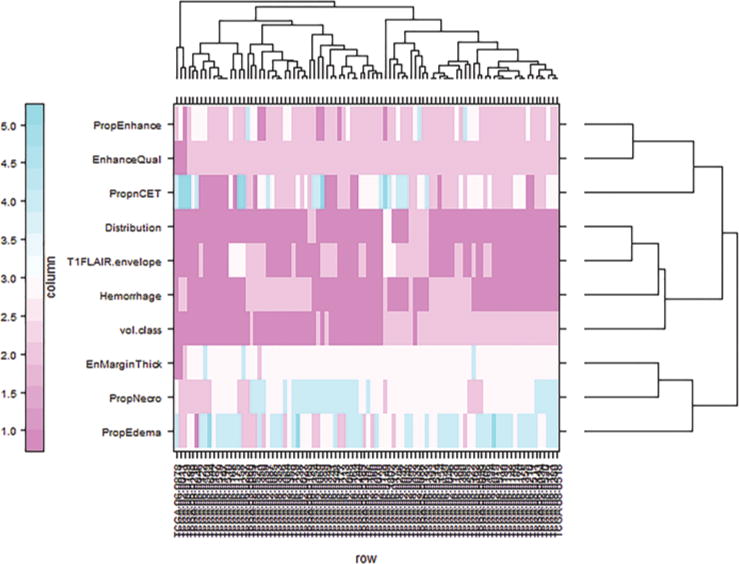
Cluster plot obtained using Gower’s distance metric and complete linkage method. The plot represents the 10 VASARI features along rows and the 92 cases along columns. EnhanceQual = enhancement quality; EnMarginThick = enhancing margin thickness; PropEdema = proportion edema; PropEnhance = proportion contrast-enhancing; PropnCET = proportion noncontrast-enhancing tumor; PropNecro = proportion necrosis; T1FLAIR.envelope = T1/FLAIR-envelope ratio; vol.class = volume-class. Figure is available in color online only.
The Combinatorial Radiophenotype Comprising Volume-Class, T1/FLAIR Ratio, and Hemorrhage Significantly Stratifies Survival
The association of this combinatorial phenotype with survival was examined via cutpoint analysis on the 92 cases that included complete survival data. Using a k-adaptive partitioning scheme (R package “kaps”), we found that a split value of 2 induced a statistically significant survival difference on the training cohort (44 cases, chosen randomly) (see Kaplan-Meier plot, Fig. 2 upper). The low survival group had a median survival of approximately 7 months and the high survival group had a median survival of approximately 19 months. This survival difference was confirmed in a validation set (the remaining 48 of 92 cases) (Fig. 2 lower). This split value partitioned the data into 2 groups (cases with volume-class:T1/FLAIR:hemorrhage combination ≤ 2, and those with volume-class:T1/FLAIR:hemorrhage combination > 2).
FIG. 2.

Survival differences induced by the cutpoint (i.e., split value of 2) in the training set and validation set. The difference in median survival and the corresponding p value are listed in adjoining panels. The combinatorial phenotype stratifies survival in the training set (p = 0.00441). Survival difference: 12 months (upper). Survival stratification in the validation set (p = 0.000181). Survival difference: 8 months (lower). Figure is available in color online only.
The group defined by a split value of “2” was also examined for differences in PFS. The p value of the log-rank test was 0.0577, suggesting a trend toward PFS stratification by this combinatorial phenotype.
The predictive performance (measured using area under the ROC curve, AUC) of the combinatorial phenotype is superior relative to the individual variables (volume-class, T1/FLAIR ratio, and hemorrhage). The AUCs for the combinatorial phenotypes and the individual variables, for 4 different survival times (12, 15, 24, and 36 months), are represented graphically in Fig. 3.
FIG. 3.
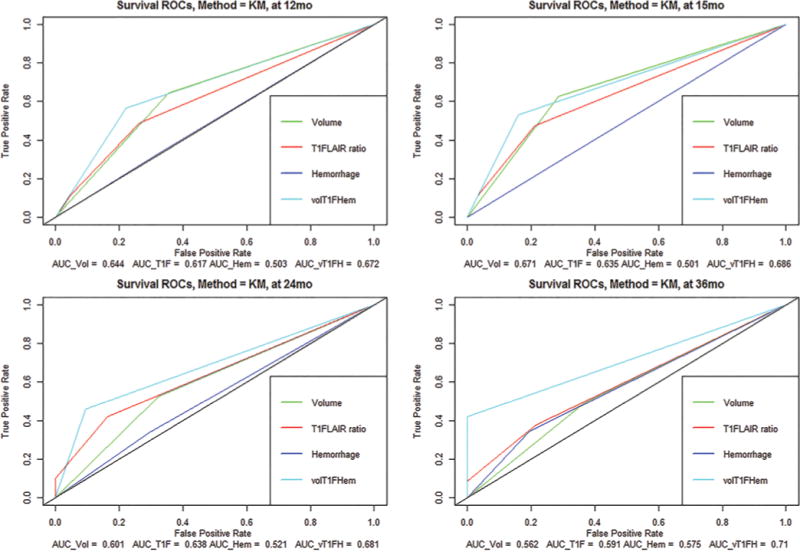
The time-dependent ROC curves for comparing the predictive value of each of the 4 features are shown. The black diagonal line in each of the 4 plots corresponds to random classification. The corresponding AUCs are listed under each plot. The AUC of the combination (in cyan) is superior to each of the individual variables. Hem = hemorrhage; KM = Kaplan-Meier; T1F = T1/FLAIR ratio; vol = volume; volT1FHem = combination of vol, T1F, and Hem. Figure is available in color online only.
Differential Expression Between Phenotypic Classes Suggests a Role for Metabolism and Immune-Associated Pathways
Application of the Comparative Marker Selection procedure to the mRNA and miRNA data yielded 384 genes and 23 miRNAs showing differential expression between the 2 phenotypic classes. Tables 1–3 show the ontological analysis of these 384 genes (using the IPA tool). The key pathways activated in this set are related to cell-to-cell signaling and interaction, cellular assembly and organization, mitochondrial dysfunction, oxidative phosphorylation (OXPHOS), and so on.
TABLE 1.
Gene ontology categories (top networks) for integrative miRNA:mRNA analysis in IPA
| Top Networks: ID No. | Associated Network Functions | Score |
|---|---|---|
| 1 | Cancer, organismal injury & abnormalities, reproductive system disease | 65 |
| 2 | Cell-to-cell signaling & interaction, cellular assembly & organization, nervous system development & function | 58 |
| 3 | Endocrine system disorders, gastrointestinal disease, metabolic disease | 53 |
| 4 | Cell-to-cell signaling & interaction, nervous system development & function, molecular transport | 50 |
| 5 | Connective tissue disorders, developmental disorder, skeletal & muscular disorders | 50 |
TABLE 3.
Gene ontology categories (top canonical pathways) for integrative miRNA:mRNA analysis in IPA
| Top Canonical Pathways | p Value | Ratio (%)* |
|---|---|---|
| Mitochondrial dysfunction | 1.34E–06 | 15:215 (0.07) |
| OXPHOS | 1.11E–05 | 11:120 (0.092) |
| Role of osteoblasts, osteoclasts, & chondrocytes in rheumatoid arthritis | 4.04E–05 | 15:250 (0.06) |
| GABA receptor signaling | 3.37E–04 | 6:56 (0.107) |
| Role of macrophages, fibroblasts, & endothelial cells in rheumatoid arthritis | 1.35E–03 | 15:342 (0.044) |
No. of molecules in list/no. of molecules in pathway.
To find pathways that were overrepresented between these 2 radiophenotype classes, we used GSEA. Several pathways (gene sets) associated with metabolism and immune function were found to be significant at an FDR cutoff of 0.1 (Table 4).
TABLE 4.
Top 10 ranked pathways enriched between phenotype 0 and phenotype 1*
| Pathways Enriched in Phenotype 0 vs Phenotype 1 | Pathways Enriched in Phenotype 1 vs Phenotype 0 |
|---|---|
| LI_INDUCED_T_TO_NATURAL_KILLER_DN | LI_AMPLIFIED_IN_LUNG_CANCER |
| ZWANG_DOWN_BY_2ND_EGF_PULSE | SANA_RESPONSE_TO_IFNG_DN |
| GAL_LEUKEMIC_STEM_CELL_UP | GOTZMANN_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_DN |
| GINESTIER_BREAST_CANCER_20Q13_AMPLIFICATION_DN | LU_AGING_BRAIN_DN |
| LEE_DIFFERENTIATING_T_LYMPHOCYTE | VERHAAK_GLIOBLASTOMA_MESENCHYMAL |
| BIOCARTA_PAR1_PATHWAY | YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_13 |
| HOEBEKE_LYMPHOID_STEM_CELL_UP | DAZARD_RESPONSE_TO_UV_NHEK_UP |
| REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | |
| MOOTHA_PGC | |
| MCCABE_HOXC6_TARGETS_CANCER_DN |
Based on normalized enrichment score. FDR < 0.1.
Integrated miRNA:mRNA Analysis Suggests Multiple Biological Processes Underlying Transition of This Combinatorial Phenotype
Using the IPA tool, we combined the miRNA expression results with the mRNA expression results and found that at least 8 differentially expressed miRNAs (miR-214–3p, miR-499–5p, miR-129–5p, miR-433–3p, miR-485–5p, miR-614, miR-617, and miR-637) target several differentially expressed genes while exhibiting concordant expression correlation (lowering of mRNA expression and elevation of miRNA expression; Table 5).
TABLE 5.
miRNA:mRNA relationships among the molecules differentially expressed between the 2 phenotypic classes*
| miRNA Symbol | Upregulated in Phenotype Group (0 or 1) | mRNA Symbol | Upregulated in Phenotype Group (0 or 1) |
|---|---|---|---|
| miR-214–3p (& other miRNAs w/seed CAGCAGG) | 0 | APH1A | 1 |
| miR-214–3p (& other miRNAs w/seed CAGCAGG) | 0 | ESRRG | 1 |
| miR-214–3p (& other miRNAs w/seed CAGCAGG) | 0 | LRRC59 | 1 |
| miR-214–3p (& other miRNAs w/seed CAGCAGG) | 0 | RIMBP2 | 1 |
| miR-214–3p (& other miRNAs w/seed CAGCAGG) | 0 | SV2B | 1 |
| miR-499–5p (& other miRNAs w/seed UAAGACU) | 0 | CNTNAP2 | 1 |
| miR-499–5p (& other miRNAs w/seed UAAGACU) | 0 | EIF5 | 1 |
| miR-499–5p (& other miRNAs w/seed UAAGACU) | 0 | GABRA1 | 1 |
| miR-499–5p (& other miRNAs w/seed UAAGACU) | 0 | PPP3CB | 1 |
| miR-499–5p (& other miRNAs w/seed UAAGACU) | 0 | SYT1 | 1 |
| miR-499–5p (& other miRNAs w/seed UAAGACU) | 0 | UTP11L | 1 |
| miR-499–5p (& other miRNAs w/seed UAAGACU) | 0 | YIPF6 | 1 |
| miR-129–5p (& other miRNAs w/seed UUUUUGC) | 1 | C1RL | 0 |
| miR-129–5p (& other miRNAs w/seed UUUUUGC) | 1 | IQCE | 0 |
| miR-129–5p (& other miRNAs w/seed UUUUUGC) | 1 | KLF13 | 0 |
| miR-129–5p (& other miRNAs w/seed UUUUUGC) | 1 | PHF21A | 0 |
| miR-129–5p (& other miRNAs w/seed UUUUUGC) | 1 | PLEKHA2 | 0 |
| miR-129–5p (& other miRNAs w/seed UUUUUGC) | 1 | RERE | 0 |
| miR-129–5p (& other miRNAs w/seed UUUUUGC) | 1 | SUN2 | 0 |
| miR-129–5p (& other miRNAs w/seed UUUUUGC) | 1 | ZNF395 | 0 |
| miR-433–3p (& other miRNAs w/seed UCAUGAU) | 1 | KLF13 | 0 |
| miR-433–3p (& other miRNAs w/seed UCAUGAU) | 1 | RAD52 | 0 |
| miR-433–3p (& other miRNAs w/seed UCAUGAU) | 1 | SLC6A4 | 0 |
| miR-433–3p (& other miRNAs w/seed UCAUGAU) | 1 | VASH2 | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | AFF1 | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | EFHC1 | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | EPB41L4A | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | FGD2 | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | KLF13 | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | PARP11 | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | PHF21A | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | SZT2 | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | TCF7 | 0 |
| miR-485–5p (& other miRNAs w/seed GAGGCUG) | 1 | TNKS | 0 |
| miR-614 (& other miRNAs w/seed AACGCCU) | 1 | EPB41L5 | 0 |
| miR-617 (& other miRNAs w/seed GACUUCC) | 1 | PLEKHA2 | 0 |
| miR-637 (& other miRNAs w/seed CUGGGGG) | 1 | GCK | 0 |
| miR-637 (& other miRNAs w/seed CUGGGGG) | 1 | GGA1 | 0 |
| miR-637 (& other miRNAs w/seed CUGGGGG) | 1 | ICOSLG | 0 |
| miR-637 (& other miRNAs w/seed CUGGGGG) | 1 | MYO9B | 0 |
| miR-637 (& other miRNAs w/seed CUGGGGG) | 1 | PCDHGB6 | 0 |
| miR-637 (& other miRNAs w/seed CUGGGGG) | 1 | RERE | 0 |
The third column represents the mRNA targeted by the miRNA in column 1. The group in which the molecule (miRNA or mRNA) is upregulated is listed in columns 2 and 4, respectively. Group 1 is the more aggressive phenotype (with vol-class:T1/FLAIR ratio:hemorrhage > 2). Group 0 is the less aggressive phenotype (with vol-class:T1/FLAIR ratio:hemorrhage ≤ 2). Only those combinations where an mRNA is downregulated and the corresponding miRNA is upregulated are listed here.
Several potentially interesting networks were indicated. The top-scoring canonical pathways pertained to immunological signaling (iCOS-iCOSL signaling in T helper cells, RXR activation, and PI3K signaling in B lymphocytes), mitochondrial dysfunction, and OXPHOS. The primary biological processes (based on Fisher exact tests for overrepresentation analysis) were “cell-to-cell signaling and interaction” and “cellular assembly and organization” (Table 1).
Discussion
In the present study, we show that the combinatorial radiographic phenotype (element-wise product), comprising volume-class, T1/FLAIR-envelope ratio, and hemorrhage, stratifies survival in GBM in a statistically significant manner. It also shows a trend toward stratifying PFS in a statistically significant manner.
To summarize, the volume is derived based on T2 abnormality. The T1/FLAIR ratio attribute compares the relative size of the abnormality in the T1-weighted images to the size in the T2/FLAIR images. Hemorrhage represents intrinsic hemorrhage in the tumor matrix (observed high signal in T1-weighted images or low signal in T2-weighted images). Prior work in this domain has focused on the identification of single image variables (i.e., contrast-enhancing tumor,9 nonenhancing tumor17) as prognostic factors and identified molecular processes that might underlie such disease-associated phenotypes (spanning processes such as invasion38 and hypoxia4). Independently, each of these imaging characteristics may be relevant to patient outcome, but together they become a much more powerful prognostic indicator. Patients with a particular imaging profile may be at risk for earlier recurrence of disease and poorer survival. Because of their clinical relevance, these imaging characteristics should be more objectively defined. For example, volumetric quantification is rarely, if ever, reported beyond a general description of size. Although the role of volume in stratifying survival is expected, we observed that combining this with T1/FLAIR ratio and hemorrhage has predictive value. The T1/FLAIR ratio is related to extent of tumor infiltration. Hemorrhage is potentially a histological manifestation (presumably related to extent of necrosis or microvascular proliferation). Examination of this combinatorial phenotype’s association with survival leads to observations that might have clinical consequences, e.g., low-volume, infiltrating disease portends better outcome than high-volume, moderately infiltrating disease; or low-volume, moderately infiltrating disease is worse than low-volume disease with hemorrhage. Based on the molecular and phenotypic correlates of this combined radiophenotype, we hypothesize that it might correlate with the aggressiveness of the tumor.
We sought to obtain a molecular characterization of the groups induced by the combinatorial phenotype, rather than based on survival characteristics alone. Examination of the miRNAs and mRNAs in Table 5 suggests the functional role of several of these molecules in glioma biology (spanning proliferation, invasion, and growth characteristics). Among the miRNAs, miR-499 has been shown to promote tumor metastasis and cellular invasion.20 miR-199a is known to regulate cellular proliferation and survival.31 miR-429 inhibits cellular growth, invasion, and epithelial-to-mesenchymal transition (EMT)34 and is seen to be upregulated in the less aggressive phenotype.
Furthermore, hsa-miR-146b and hsa-miR-193a are associated with survival in GBM.32 hsa-miR-146b was shown to be involved in glioma cell migration and invasion.36 Also, miR-499 regulates apoptosis, mitochondrial dynamics, and represses CNTNAP2, PPP3CB, and SYT1 (invasion-associated genes upregulated in the more aggressive phenotype). Upregulation of miR-146b is observed in the less aggressive phenotype (Group 0), concordant with the observation that it reduces migration and invasiveness in glioma.15,36 miR-125 and miR-129 were associated with tumor progression and glioma proliferation.19 Although other genes/miRNAs in Table 5 indicate phenotypic modulators of interest, their specific roles in GBM remain to be elucidated. Low expression of miR-200 family members has been linked to the epithelial-to-mesenchymal phenotype and subsequent metastatic disease.11,16,35
Several of the pathways obtained via GSEA point to interesting aspects of GBM biology (Table 4). An examination of pathways enriched at the top of the differentially expressed gene lists indicates the potential involvement of several immune-modulatory pathways underlying this phenotypic transition: natural killer cell activity, interferon gamma response, and T-cell lymphocyte differentiation. Additionally, EMT is associated with the more aggressive phenotype, an association further strengthened by the enrichment of the mesenchymal subtype signature in the same group.37 Molecules such as PAR1 and HOXC6 are implicated in tumor invasiveness.12 Furthermore, 20q13 region amplification is known to be relevant to glioma risk and GBM.2,17,23,27 However, an in-depth validation in a reliable in vivo model is crucial before drawing any conclusions pertinent to this specific combinatorial phenotype.
Similarly, several pathways, mostly related to cellular metabolism, are overrepresented in the differential enrichment list, e.g., PGC1alpha activity is associated with OXPHOS,5 which is involved in brain tumor progression.8 These sources all point to the potential association of metabolism-related activity with the transition of this combinatorial phenotype. Moreover, OXPHOS has been suggested to confer tumorigenic potential to GBM cancer stem cells.14 This finding is also concordant with that of altered metabolic activity underlying tumor-invasive characteristics in GBM.8
IPA indicated that the coordinated activity of several miRNAs and mRNAs might underlie this phenotype. Specifically, we observed the presence of several immune-modulatory genes and key signaling molecules. However, determining the roles of these molecules in modulating this phenotypic transition will require very careful study.
Molecular analysis of this combinatorial radiophenotype thus suggests the role of proliferation/invasion characteristics underlying its transition. Recent literature suggests that targeting invasion/proliferation mechanisms may hold therapeutic promise.3,6,21,22,28 Thus, this combinatorial radiophenotype might also be used as a screening tool for assessing response to drugs that target glioma cell proliferation and invasion characteristics. A potential avenue might also be the surveillance of this combinatorial phenotype to assess treatment response, thereby serving as a complement to existing response evaluation criteria (such as MacDonald, Response Evaluation Criteria In Solid Tumors [RECIST] criteria). In addition, because this phenotype has a survival correlate that is presumably related to the aggressiveness of the disease, modulating this phenotype might have consequences for treatment decision making. Indeed, there are several approaches targeting invasion,37 proliferation, EMT, and metabolic31 characteristics for GBM. The role of immune-modulatory processes underlying this combinatorial phenotype also suggests the possibility of targeting disease through immune modulation25 and for assessing response to immune-targeted therapeutics using this phenotype.
Our work has several limitations, providing abundant scope for further investigation. An unbiased search for other feature combinations (within the VASARI set) that can stratify survival might reveal other interesting characteristics. Beyond indicating interesting combinations of features, an integrative molecular analysis comprising mRNA, miRNA, methylation, and copy number from these phenotype-induced groups might identify other molecular processes relevant to glioma biology. This study focused exclusively on the association between combinatorial radiographic phenotypes and survival. In the future, assessment of the phenotype’s clinical value will require a multivariate model including clinical factors (e.g., age, Karnofsky Performance Scale score) and known molecular predictors such as MGMT methylation and molecular subtype.
Conclusions
Our results suggest that combining radiographic phenotypes provides additional variables that stratify survival. Specifically, we showed that combination of multiple MR image–derived features has more predictive value for survival than some individual ones. Because volume-class is important in this combinatorial predictor, exploring precise 3D volume measurement based on a 3D segmentation markup of the lesion data may be useful. Currently, only major and minor axis lengths are available. Further, it would be interesting to explore quantitative assessments of the hemorrhage component’s volume and T1/FLAIR ratio as additional predictors for stratifying survival, rather than the current categorical measurements.
TABLE 2.
Gene ontology categories (molecular and cellular functions) for integrative miRNA:mRNA analysis in IPA
| Type of Function | p Value | No. of Molecules |
|---|---|---|
| Cell-to-cell signaling & interaction | 1.88E-10–2.01E-02 | 57 |
| Molecular transport | 2.44E-05–2.01E-02 | 67 |
| Small-molecule biochemistry | 2.44E-05–2.01E-02 | 64 |
| Amino acid metabolism | 8.11E-05–2.01E-02 | 19 |
| Cell death & survival | 2.68E-04–2.01E-02 | 110 |
| Nervous system development & function | 1.88E-10–2.01E-02 | 55 |
| Tumor morphology | 4.02E-04–5.72E-03 | 2 |
| Behavior | 5.43E-04–2.01E-02 | 51 |
| Connective tissue development & function | 1.19E-03–2.01E-02 | 22 |
| Skeletal & muscular system development & function | 1.19E-03–2.01E-02 | 15 |
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We would like to thank Dr. Chad Holder from the TCGA Phenotype Research Group for guidance with data interpretation. We are grateful to Dr. Keith Baggerly and Dr. Rehan Akbani for discussions about the TCGA methylation and expression data. We also thank Dr. Rehan Akbani for helpful discussions about clustering categorical data. We are also grateful to Ms. Priyanka Raja and Mr. Suvindra Vijayan for assistance with coding the radiological attributes. In addition, Ms. Anisha Luthra assisted with the miRNA:mRNA analysis. Support of the TCGA Glioma Phenotype Research Group is gratefully acknowledged. We would also like to thank Ms. Audria Patrick and Mr. David Wildrick for editorial assistance. We gratefully acknowledge the support of institutional start-up funds, an MD Anderson Cancer Center institutional research grant (IRG), support from MD Anderson Cancer Center Support Grant P30 CA016672 (the Bioinformatics Shared Resource), and a Career Development Award from the Brain Tumor SPORE (granted to A.R.) for conducting this research.
ABBREVIATIONS
- AUC
area under the curve
- EMT
epithelial-to-mesenchymal transition
- FDR
false discovery rate
- GBM
glioblastoma
- GSEA
Gene Set Enrichment Analysis
- IPA
Ingenuity Pathway Analysis
- miRNA
microRNA
- OXPHOS
oxidative phosphorylation
- PFS
progression-free survival
- ROC
receiver operating characteristic
- TCGA
The Cancer Genome Atlas
- TCIA
The Cancer Imaging Archive
- VASARI
Visually AcceSAble Rembrandt Images
Footnotes
Disclosure
Mr. Kirby has direct stock ownership in Myriad Genetics.
Author Contributions
Conception and design: A Rao, G Rao. Acquisition of data: A Rao, G Rao, Gutman, Flanders, Hwang, Rubin, Colen, Zinn, Jain, Wintermark. Analysis and interpretation of data: A Rao, G Rao. Drafting the article: A Rao, G Rao. Critically revising the article: A Rao, G Rao, Flanders, Hwang, Rubin, Colen, Zinn, Jain, Wintermark, Kirby, Freymann, Jaffe. Reviewed submitted version of manuscript: all authors. Approved the final version of the manuscript on behalf of all authors: A Rao. Statistical analysis: A Rao. Administrative/technical/material support: A Rao, G Rao, Kirby, Freymann, Jaffe. Study supervision: A Rao, G Rao.
Current Affiliation
Dr. Wintermark: Department of Radiology, Neuroradiology Section, Stanford Medical Center, Stanford, CA.
References
- 1.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 2.Chen H, Chen Y, Zhao Y, Fan W, Zhou K, Liu Y, et al. Association of sequence variants on chromosomes 20, 11, and 5 (20q13.33, 11q23.3, and 5p15.33) with glioma susceptibility in a Chinese population. Am J Epidemiol. 2011;173:915–922. doi: 10.1093/aje/kwq457. [DOI] [PubMed] [Google Scholar]
- 3.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 4.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105:5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ermoian RP, Kaprealian T, Lamborn KR, Yang X, Jelluma N, Arvold ND, et al. Signal transduction molecules in gliomas of all grades. J Neurooncol. 2009;91:19–26. doi: 10.1007/s11060-008-9683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 7.Gower JC. General coefficient of similarity and some of its properties. Biometrics. 1971;27:857. [Google Scholar]
- 8.Griguer CE, Oliva CR. Bioenergetics pathways and therapeutic resistance in gliomas: emerging role of mitochondria. Curr Pharm Des. 2011;17:2421–2427. doi: 10.2174/138161211797249251. [DOI] [PubMed] [Google Scholar]
- 9.Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267:560–569. doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammoud MA, Sawaya R, Shi W, Thall PF, Leeds NE. Prognostic significance of preoperative MRI scans in glioblastoma multiforme. J Neurooncol. 1996;27:65–73. doi: 10.1007/BF00146086. [DOI] [PubMed] [Google Scholar]
- 11.Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–1326. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivkovic S, Beadle C, Noticewala S, Massey SC, Swanson KR, Toro LN, et al. Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol Biol Cell. 23:533–542. doi: 10.1091/mbc.E11-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain R, Poisson L, Narang J, Gutman D, Scarpace L, Hwang SN, et al. Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology. 2013;267:212–220. doi: 10.1148/radiol.12120846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janiszewska M, Suvà ML, Riggi N, Houtkooper RH, Auwerx J, Clément-Schatlo V, et al. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katakowski M, Zheng X, Jiang F, Rogers T, Szalad A, Chopp M. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Invest. 2010;28:1024–1030. doi: 10.3109/07357907.2010.512596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pope WB, Sayre J, Perlina A, Villablanca JP, Mischel PS, Cloughesy TF. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol. 2005;26:2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 18.Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95:190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 19.Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, et al. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One. 2013;8:e54652. doi: 10.1371/journal.pone.0054652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Zhang Z, Sun L, Chai N, Tang S, Jin J, et al. MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis. 2011;32:1798–1805. doi: 10.1093/carcin/bgr213. [DOI] [PubMed] [Google Scholar]
- 21.Mischel PS, Cloughesy TF. Targeted molecular therapy of GBM. Brain Pathol. 2003;13:52–61. doi: 10.1111/j.1750-3639.2003.tb00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikaki A, Piperi C, Papavassiliou AG. Role of microRNAs in gliomagenesis: targeting miRNAs in glioblastoma multiforme therapy. Expert Opin Investig Drugs. 2012;21:1475–1488. doi: 10.1517/13543784.2012.710199. [DOI] [PubMed] [Google Scholar]
- 23.Phillips JJ, Huillard E, Robinson AE, Ward A, Lum DH, Polley MY, et al. Heparan sulfate sulfatase SULF2 regulates PDGFRa signaling and growth in human and mouse malignant glioma. J Clin Invest. 2012;122:911–922. doi: 10.1172/JCI58215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pope WB, Mirsadraei L, Lai A, Eskin A, Qiao J, Kim HJ, et al. Differential gene expression in glioblastoma defined by ADC histogram analysis: relationship to extracellular matrix molecules and survival. AJNR Am J Neuroradiol. 2012;33:1059–1064. doi: 10.3174/ajnr.A2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prins RM, Soto H, Konkankit V, Odesa SK, Eskin A, Yong WH, et al. Gene expression profile correlates with T-cell infiltration and relative survival in glioblastoma patients vaccinated with dendritic cell immunotherapy. Clin Cancer Res. 2011;17:1603–1615. doi: 10.1158/1078-0432.CCR-10-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raza SM, Lang FF, Aggarwal BB, Fuller GN, Wildrick DM, Sawaya R. Necrosis and glioblastoma: a friend or a foe? A review and a hypothesis. Neurosurgery. 2002;51:2–13. doi: 10.1097/00006123-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Reichardt W, Jung V, Brunner C, Klein A, Wemmert S, Romeike BF, et al. The putative serine/threonine kinase gene STK15 on chromosome 20q13.2 is amplified in human gliomas. Oncol Rep. 2003;10:1275–1279. [PubMed] [Google Scholar]
- 28.Salhia B, Tran NL, Symons M, Winkles JA, Rutka JT, Berens ME. Molecular pathways triggering glioma cell invasion. Expert Rev Mol Diagn. 2006;6:613–626. doi: 10.1586/14737159.6.4.613. [DOI] [PubMed] [Google Scholar]
- 29.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 30.Shen H, Decollogne S, Dilda PJ, Hau E, Chung SA, Luk PP, et al. Dual-targeting of aberrant glucose metabolism in glioblastoma. J Exp Clin Cancer Res. 2015;34:14. doi: 10.1186/s13046-015-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. 2011;124:2826–2836. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- 32.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 34.Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu Z, et al. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol Cell Biochem. 2014;390:19–30. doi: 10.1007/s11010-013-1950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S, et al. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–165. doi: 10.1016/j.brainres.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 37.Zarkoob H, Taube JH, Singh SK, Mani SA, Kohandel M. Investigating the link between molecular subtypes of glioblastoma, epithelial-mesenchymal transition, and CD133 cell surface protein. PLoS One. 2013;8:e64169. doi: 10.1371/journal.pone.0064169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zinn PO, Mahajan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One. 2011;6:e25451. doi: 10.1371/journal.pone.0025451. (Erratum in PLoS One 7: 10.1371/annotation/b5267cb3-6aa7-47fc-a648-47f30a7cff3e, 2012) [DOI] [PMC free article] [PubMed] [Google Scholar]


