Abstract
Myelination occurs selectively around neuronal axons to increase the efficiency and velocity of action potentials. While oligodendrocytes are capable of myelinating permissive structures in the absence of molecular cues, structurally permissive neuronal somata and dendrites remain unmyelinated. Utilizing a purified spinal cord neuron-oligodendrocyte myelinating coculture system, we demonstrate that disruption of dynamic neuron-oligodendrocyte signaling by chemical crosslinking results in aberrant myelination of the somatodendritic compartment of neurons. We hypothesize that an inhibitory somatodendritic cue is necessary to prevent non-axonal myelination. Using next-generation sequencing and candidate profiling, we identify neuronal Junction Adhesion Molecule 2 (JAM2) as an inhibitory myelin-guidance molecule. Taken together, our results demonstrate that the somatodendritic compartment directly inhibits myelination, and suggest a model in which broadly indiscriminate myelination is tailored by inhibitory signaling to meet local myelination requirements.
eTOC
Redmond, et al. demonstrate that JAM2 is a somatodendritic inhibitor of oligodendrocyte myelination. JAM2 is necessary and sufficient to inhibit myelin wrapping in vitro, and loss of JAM2 leads to aberrant myelination of dorsal horn PAX2+ interneuron cell bodies in the spinal cord dorsal horn.
The central nervous system (CNS) greatly benefits from oligodendrocyte myelination. Myelin supports the clustering of ion channels to nodes of Ranvier and increases the resistance across the axonal membrane. These properties generate the saltatory conduction of action potentials, ensuring rapid and efficient neural communication over longer distances in a more compact space (Zalc et al., 2008). It has long been appreciated that myelin, with very few exceptions, forms selectively around axons (Braak et al., 1977; Lubetzki et al., 1993; Blinzinger et al., 1972; Remahl and Hildebrand, 1985; Cooper and Beal, 1977). This begs the fundamental question: how does an oligodendrocyte, the myelin-forming cells of the CNS, target each of its many myelin segments exclusively to axons?
Very little is currently known about axon selection by oligodendrocytes. More well understood are the consequences of when myelin fails to form correctly around axons, such as in leukodystrophies, or when myelin is attacked by the immune system, as in multiple sclerosis. In such cases, neuronal function is impaired and neurons eventually degenerate if myelin is not reformed (Fancy et al., 2011; Franklin and ffrench-Constant, 2008; Trapp and Nave, 2008; Yuen et al., 2014). In development, the coordinated differentiation and myelination of thousands of spatially distributed oligodendroglia is thought to be extrinsically determined, perhaps by neuronal activity. Indeed, recent findings suggest that neuronal activity is involved in modulating axon selection and the extent of myelin segment formation (Gibson, 2014; Hines et al., 2015; Mensch et al., 2015).
An unexplored aspect of myelination is the avoidance of selecting non-axonal targets. In CNS gray matter, oligodendrocytes must select axons while avoiding neuronal cell bodies, dendrites and processes of other glial cells. How is this specificity accomplished? Little evidence of non-axonal oligodendrocyte myelination exists (Blinzinger et al., 1972; Braak et al., 1977). Curiously, in vitro, oligodendrocyte myelination is not limited to axons: oligodendrocytes produce myelin membranes on planar glass coverslips, cylindrical polystyrene nanofibers (Bechler et al., 2015; Lee et al., 2012), and conical glass micropillars (Mei et al., 2014). These observations suggest that myelin substrate selection is not cell-intrinsically limited to physiologically relevant axon-like geometries, and that an inductive cue is not strictly required for differentiation and myelination. Oligodendrocyte cell processes are thus likely sensitive to cell-extrinsic ‘myelin guidance’ cues that ensure only correct selection of pre-myelinated axons.
We propose three potential mechanisms that would ensure correct axonal selection (Fig 1a). An attractive signal may be expressed by the neuronal axon (Fig 1a, ‘Attraction’) to specifically promote myelin formation. Alternatively, proper myelination of axons may be achieved by expression of an inhibitory cue by the neuron’s somatodendritic compartment (Fig 1a, ‘Inhibition’). Here, oligodendrocyte processes form myelin segments around permissive axons but not on inhibitor-expressing structures. However, the Attraction and Inhibition models are not mutually exclusive, and both modes of signaling may be at play to ensure proper targeting of myelin segments (Fig 1a, ‘Mixed’). Which of these signaling mechanisms are active during developmental myelination? By utilizing a purified spinal cord neuron-oligodendrocyte myelinating coculture system, we demonstrate that disruption of dynamic neuron-oligodendrocyte signaling results in aberrant myelination of the somatodendritic compartment of neurons indicative of an active inhibitory mechanism. Using next-generation sequencing and candidate profiling, we identify Junction Adhesion Molecule 2 (JAM2) as a somatodendritic protein necessary and sufficient to inhibit oligodendrocyte myelination. Taken together, our results demonstrate that the somatodendritic compartment directly inhibits myelination, and provide a model in which broadly indiscriminate myelination is tailored by inhibitory signaling to meet local myelination requirements.
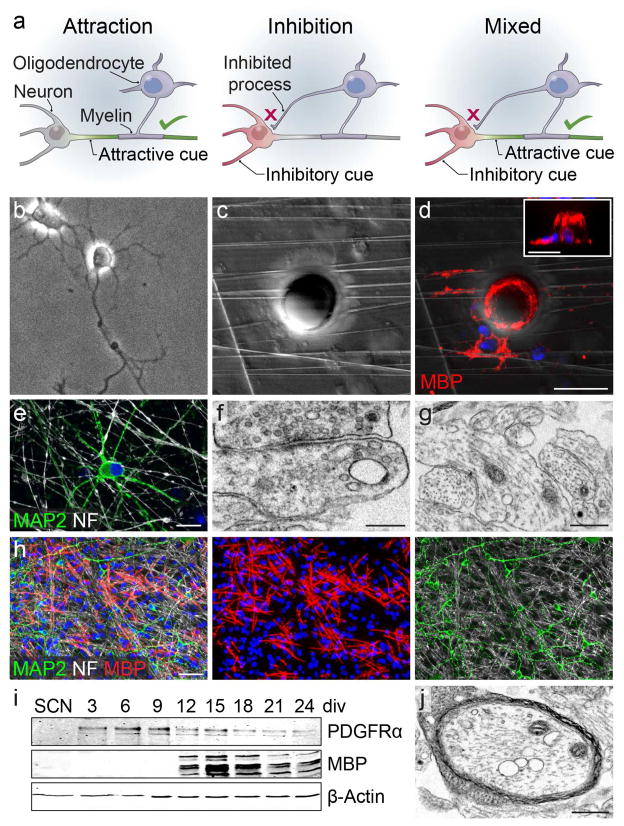
Figure 1. Myelin target selection by oligodendroglia.
(a) Models of oligodendrocyte axon selection and myelination. Attraction: an attractive cue is expressed on the axon (green), resulting in its myelination by the oligodendrocyte, while no myelination cues are present on non-myelinated structures (gray). Inhibition: non-myelinated structures express an inhibitory cue (red), resulting in oligodendrocyte process inhibition and myelination of inhibition-free axons (gray). Mixed: Inhibitory (red) and attractive (green) cues result in the correct myelination of axons. (b) a phase image of a spinal cord neuron (SCN) in culture. (c) a phase image of nanofibers and a 20-micrometer diameter polystyrene bead. (d) an oligodendrocyte ensheathing myelin basic protein-positive (MBP+, red) membranes on nanofibers and the bead shown in (c) with a side-view shown in the inset. (e) a neuron at eight days in vitro immunostained for microtubule associated protein 2 (MAP2) and neurofilament (NF). (f) electron micrograph of a synapse, and (g) neurofilament-positive axons in SCN cultures. (h) low-magnification image of neurons and oligodendroglia in a myelinating coculture. (i) timecourse of oligodendrocyte progenitor cell (OPC) and oligodendrocyte protein expression in SCN myelinating cocultures. (j) electron micrograph of a myelinated axon in a myelinating spinal cord neuron coculture. Scale bars = 20 micrometers (d), 10 micrometers (e), 250 nanometers (f), 500 nanometers (g, i), 50 micrometers (h).
Results
The somatodendritic compartment is biophysically permissive to myelin ensheathment
To understand the nature of selectivity and fidelity of the myelination process, we reasoned that the presence of a somatodendritic inhibitor would only be required if oligodendrocytes are capable of ensheathing the spherical shape of a neuron cell body (Fig 1b). To test this possibility, we cultured primary rat oligodendrocyte precursor cells (OPCs) on nanofibers with 20-micrometer diameter polystyrene beads that mimic neuron cell body geometry (Fig 1c). We find that differentiated myelin basic protein-positive (MBP+) oligodendrocytes readily ensheathe myelin membranes around both nanofibers and beads (Fig 1d).
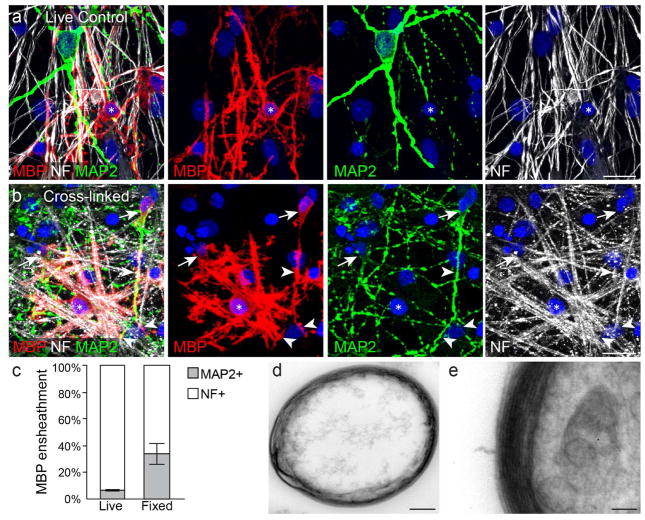
We then asked whether the somatodendritic compartment remains unmyelinated due to lack of attraction, or expression of inhibition? To investigate this question, we developed a primary purified spinal cord neuron (SCN) and oligodendroglial myelinating coculture system (Fig 1b,e–i). The SCN cultures robustly exhibited extensive axons, dendrites, and formed neuronal synapses over the course of three weeks in vitro (Fig 1e–g). Under normal conditions, purified OPCs seeded onto mature SCN cultures proliferate, differentiate after approximately one week in vitro (Fig 1h–i), and form compact layers of myelin membrane around axons (Fig 1j). Oligodendrocytes wrap myelin membrane around axons while avoiding neuron somata and dendrites (Fig 2a) (Lubetzki et al., 1993), recapitulating gray matter myelination patterns in vitro. We then asked whether dynamic neuron-oligodendrocyte signaling is required to prevent myelination of the somatodendritic compartment. Prior to seeding neuron cultures with OPCs, we treated the neurons with the chemical cross-linker paraformaldehyde (PFA) (Rosenberg et al., 2008). Surprisingly, oligodendrocytes wrapped myelin membranes on fixed neuron somata and dendrites in addition to axons (Fig 2b–c), and form multiple compacted layers of myelin membrane around cellular processes (Fig 2d–e). It is important to note, that paraformaldehyde cross-linking and the extended period of coculturing, subcellular structures are not well maintained (Rosenberg et al., 2008), making it impossible to determine whether myelinated structures are axons or dendrites. Based on our observations that (1) cultured oligodendroglia ensheathe bare nanofibers and microbeads, (2) avoid ensheathing neural somatodendritic compartments in vitro and in vivo, yet (3) aberrantly ensheathe cross-linked somata and dendrites, we hypothesize that neurons must express inhibitory molecules to prevent somatodendritic myelination.
Figure 2. Oligodendrocytes wrap somatodendritic compartments of chemically cross-linked neurons.
(a) an oligodendrocyte (asterisk) forms MBP+ myelin segments on SCN axons (NF, white) but not dendrites (MAP2, green). (b) an oligodendrocyte (asterisk) forms myelin segments on paraformaldehyde cross-linked SCN axons, dendrites (arrowheads), and somata (arrows). (c) quantification of MBP+ ensheathments on either MAP2+ dendrites or NF+ axons in live and cross-linked (‘Fixed’) cocultures. (d–e) multiple layers of myelin membrane formed in a cross-linked myelinating coculture. DAPI, blue. Bar graph in (c) represents means with s.e.m. error bars. n = 4 experiments quantified from 26–30 20× fields per condition per experiment. Significance was calculated using one-tailed paired Students t-test. P = 0.04. Scale bars = 20 micrometers (a,b), 500 nanometers (d), 100 nanometers (e).
Differential RNA-seq and candidate profiling for myelination-inhibitory molecules
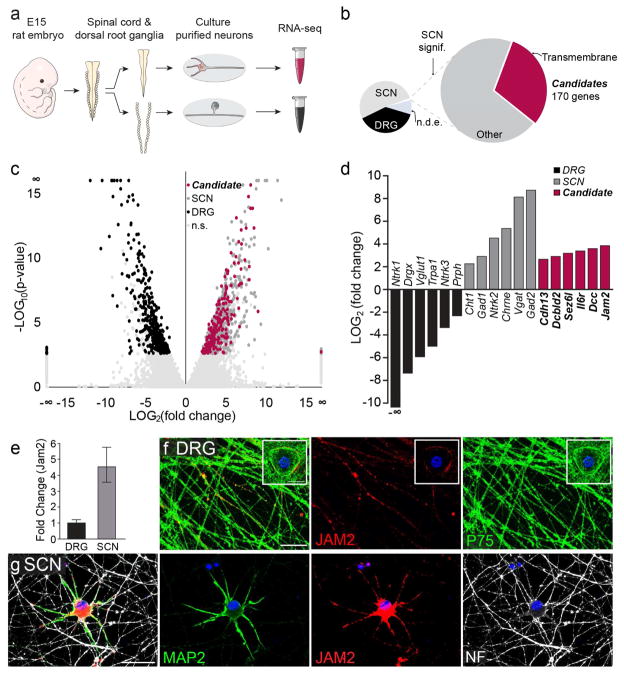
To identify candidate inhibitory molecules, we conducted next-generation RNA-sequencing of SCN as well as dorsal root ganglion neuron (DRG) cultures (Fig 3a). We reasoned that differential expression analysis would increase the likelihood of identifying a somatodendritic inhibitor of myelination because SCN cultures develop extensive dendritic arbors around their somata, while pseudo-unipolar DRG neurons lack true dendrites. Candidate inhibitory proteins were chosen based on the following criteria: proteins that have (1) significantly higher expression in SCNs as compared to DRGs, and (2) transmembrane domains (Fig 3b). The differential RNA-seq analysis yielded two distinct populations of transcripts, representing higher expression in either DRGs (Fig 3b–d, black) or SCNs (Fig 3b–d, dark gray/pink). Several genes that have been described in the literature as expressed in either DRGs or SCNs were also differentially expressed in our analysis (Fig 3d, black and gray bars). Overall, 170 unique genes were identified as candidate inhibitory proteins (Fig 3b–d, pink). By profiling and prioritizing candidates from the RNAseq analysis, we identified the protein Junction Adhesion Molecule 2 (JAM2) as a differentially expressed (Fig 3d–e) potential inhibitor of myelination because of its low expression in DRG somata (Fig 3f, insert), little to no expression in the majority of DRG (Fig 3f) and SCN axons (Fig 3g), and its high expression and specific localization on the somatodendritic compartment of SCN (Fig 3g).
Figure 3. Differential RNA-Seq and candidate profiling.
(a) Experimental procedure for RNA extraction. Embryonic day 15 (E15) rat embryos were dissected and their spinal cords and dorsal root ganglia were isolated. SCN and dorsal root ganglion neurons (DRG) were separately cultured and their RNA was collected for sequencing (RNA-seq). (b) Results of differential RNA-seq. Candidate inhibitory proteins are SCN transcripts that are significantly differentially expressed (large pie chart, right), are localized to the plasma membrane and have transmembrane domains (pink). Transcripts detected also included other SCN transcripts (big pie chart, light gray), SCN enriched, but not significantly differentially expressed (small pie chart, gray), DRG-enriched transcripts (black) and not differentially expressed transcripts (n.d.e.). (c) Volcano plot of DRG (black) and SCN (dark gray), SCN candidate (“Candidate”, pink), and not significantly differentially expressed (“n.s.”, light gray) transcripts. (d) Examples of differentially expressed transcripts from (c). (e) qPCR amplification of jam2 from cultured DRG and SCN, normalized to DRG expression. (f) DRG somata and axons (P75, green) immunostained for JAM2 (red), and (g) Cultured SCN somatodendritic compartment (MAP2, green) and axons (NF, white), and DAPI, blue. Bar graph in (e) represents mean and s.e.m. from n = 4 biological replicates. Gapdh was used as an internal control. Scale bars = 20 micrometers.
JAM2 is sufficient to inhibit oligodendroglial wrapping
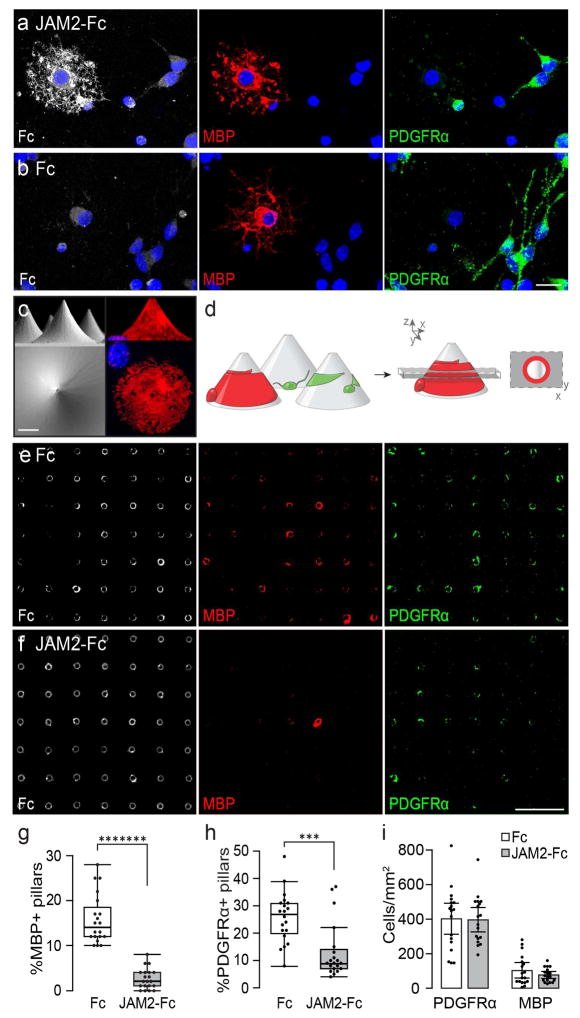
JAM2 is a single-pass transmembrane protein with two extracellular immunoglobulin-like domains (Arcangeli et al., 2013). We found that the extracellular portion of JAM2 fused to the Fc region of immunoglobulins (‘JAM2-Fc’) strongly binds to MBP-positive oligodendrocytes and weakly to PDGF receptor alpha-positive (PDGFRα+) OPCs (Fig 4a) compared to an Fc only negative control (Fig 4b). This suggests that oligodendroglia express at least one JAM2 receptor, and the expression of JAM2 receptor(s) is upregulated as oligodendroglia differentiate. We hypothesized that if JAM2 acts to inhibit myelination, oligodendrocytes will avoid wrapping JAM2-coated permissive structures in vitro. To test this possibility, we adapted the micropillar array platform (Mei et al., 2014) (Fig 4c) to assay the activity of Fc-fusion proteins on oligodendroglial wrapping. Wildtype OPCs were seeded onto micropillars coated with either JAM2-Fc or Fc alone. Under control conditions, OPCs and oligodendrocytes associate with micropillars and wrap them with PDGFRα+ or MBP+ membranes. When a single optical section is taken through the micropillar array, immunostained OPC and oligodendrocyte wrapping is visualized as a ring (Figure 4d). Compared to Fc alone, oligodendrocytes wrap significantly fewer JAM2-Fc coated micropillars (Fig 4e–h, 85.7% reduction). Interestingly, even OPC wrapping of micropillars is significantly reduced in the JAM2-Fc condition compared with Fc alone (Fig 4h; 66.6% reduction). We reasoned that we might observe a decrease in OPC and oligodendrocyte wrapping if oligodendroglia failed to adhere or differentiate on JAM2-Fc coated surfaces. Due to technical limitations of imaging cells within the micropillar fields, we quantified the densities of OPCs and oligodendrocytes on JAM2-Fc or Fc coated areas directly adjacent to the micropillar arrays. We found no significant difference between the densities of OPCs or oligodendrocytes between JAM2-Fc and Fc conditions (Fig 4i). Taken together, these results show that the extracellular portion of JAM2 is sufficient to specifically inhibit oligodendroglial wrapping without affecting cell density or oligodendrocyte differentiation.
Figure 4. JAM2 is sufficient to inhibit oligodendrocyte wrapping.
(a) JAM2-Fc or (b) Fc protein (white) was cultured with MBP+ oligodendrocytes (red) and PDGFRα+ OPCs (green) to demonstrate JAM2-Fc binding. (c) scanning electron micrograph micropillar array platform (left column) and immunostaining of a cultured oligodendrocyte wrapping a micropillar (MBP, right column) from the x–z plane/side view (top row) and x–y plane/top-down view (bottom row). (d) experimental setup: oligodendroglia are cultured on coated micropillars (left). Myelin membrane (red) wraps of differentiated oligodendrocytes are visualized as rings by taking an optical section (dashed gray box) through immunostained cultures (red ring in x–y plane, far right). Optical sections of (e) Fc- and (f) JAM2-Fc-coated micropillars immunostained for oligodendrocyte (MBP, red) and OPC (PDGFRα, green) wrapping, and micropillars (Fc protein, white). (g) The percentage of micropillars with a MBP+ or (h) PDGFRα+ ring, and the (i) densities of oligodendoglia on flat areas adjacent to micropillars. Box plots (g,h) are defined by the center median line, 25th and 75th percentiles determined by R software, whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, and dots outside of whiskers are defined as outliers. n = 20 fields of 100 micropillars (dots) each condition pooled from four independent experiments. Significance was calculated using Wilcoxon Rank-Sum test. (g) *******, P = 6.3×10−8; (h) ***, P = 3.75×10−4. Bar graphs in (i) represent means with s.e.m. error bars. n = 18 10× fields (dots) each condition pooled from three independent experiments. Significance was calculated using two-tailed Students t-test. Scale bars: (a–c) 10 micrometers, (e–f) 400 micrometers.
JAM2 is necessary to prevent somatodendritic myelin wrapping
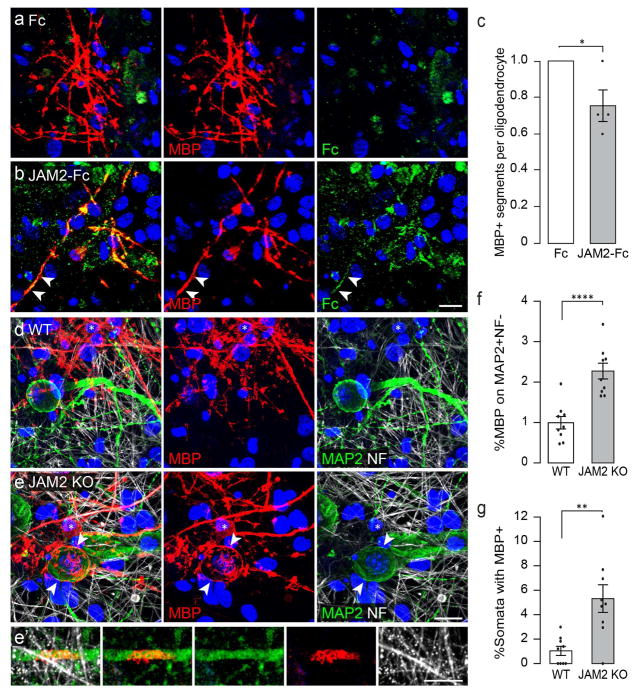
Given that oligodendrocytes wrap nanofibers, beads, and Fc coated micropillars, but significantly fewer JAM2-Fc coated micropillars, we next asked whether JAM2 signaling is sufficient to inhibit myelin segment formation in vitro. We established myelinating SCN cocultures in the presence of either soluble Fc or JAM2-Fc. After seven days, the cocultures were immunostained for MBP and Fc, and the number of MBP+ segments per oligodendrocyte was analyzed. Consistent with Fc binding analysis, myelinating oligodendrocytes in the control condition do not show specific Fc labeling (Fig 5a), while oligodendrocytes in the JAM2-Fc condition show co-labeling of MBP and JAM2-Fc (Fig 5b), indicating that myelinating oligodendrocytes express a JAM2 receptor. Interestingly, we find that the number MBP+ segments formed per oligodendrocyte is significantly reduced in the presence of JAM2-Fc as compared to Fc alone (Fig 5c, 24.6% reduction).
Figure 5. JAM2 is sufficient to reduce myelin segment formation and is necessary to prevent somatodendritic wrapping in vitro.
Oligodendrocytes in SCN cocultures form myelin segments in the presence of soluble (a) Fc or (b) JAM2-Fc protein. (c) the relative number of MBP+ segments formed in the presence of Fc or JAM2-Fc. (d) wildtype or (e) JAM2 KO neurons cocultured with wildtype OPCs for seven days in vitro. Arrowheads indicate myelin membranes on neuron soma and asterisks mark oligodendrocyte cell bodies. (f) the percentage of myelin membrane structures colocalized with MAP2+ and NF- somatodendritic compartments. (g) the percentage of neuron somata wrapped with MBP+ myelin membranes. (h) a wildtype MBP+ oligodendrocyte (asterisk) wraps membranes on a JAM2 KO MAP2+ neuron in vitro, clustering neuronal CASPR proteins (green). Bar graphs represent means with s.e.m. error bars. (c) n = 4 independent experiments (dots) representing median number of MBP+ segments formed per oligodendrocyte from 59–79 oligodendrocytes quantified per experiment per condition. (f–g) n = 9 coverslips (dots) pooled from three independent experiments. Significance was calculated using one-tailed paired (c) or unpaired (f–g) Students t-test. (c) *, P = 0.032; (f) ****, P = 7.1×10−5; (g) **, P = 0.0012. DAPI, blue. Scale bars: 20 micrometers.
Next, we investigated whether JAM2 is necessary to prevent somatodendritic myelin wrapping of SCNs. First, we established myelinating cocultures with wildtype SCN and OPCs, and allowed them to interact normally for five days in vitro. During the final two days of coculture, at the onset of OPC differentiation and initiation of myelination, we applied a function-blocking antibody against JAM2 (Fig S1a–c). In control conditions, oligodendrocyte segments are overwhelmingly wrapped on axons (Fig S1a). However, in the presence of JAM2 function-blocking antibody, we found that oligodendrocytes wrap significantly more MBP+ segments on neuronal dendrites (Fig S1b–c, 321.8% increase).
To understand whether neuronal JAM2 is necessary to ensure that the somatodendritic compartment remains unmyelinated, we established myelinating cocultures with wildtype OPCs and either JAM2-knockout (JAM2 KO) or wildtype (WT) SCNs. We observed that compared to wildtype SCN controls (Fig 5d), wildtype oligodendrocytes wrapped significantly more MBP+ segments on JAM2-deficient somatodendritic compartments (Fig 5e–f, 226.4% increase). Strikingly, we frequently observed large spheroidal ‘bubbles’ of MBP+ membranes in JAM2 KO neuron cocultures (Fig 5e, MBP panel arrowheads). These structures stood out against the typically cylindrical myelin segments on axons and colocalized with neuronal somata (Fig 5e, MAP2 panel arrowheads). We quantified the relative frequency in which neuron cell bodies are wrapped with myelin membranes, and find that JAM2-knockout somata are five-fold more likely to be wrapped by oligodendrocytes compared with wildtype controls (Fig 5g).
During normal axonal myelination, oligodendrocyte adhesion proteins cause the clustering of neuronal proteins and ion channels to nodes of Ranvier, enabling saltatory conduction of action potentials. We next asked whether aberrant somatic wrapping could affect the molecular organization of JAM2-knockout neurons? Unexpectedly, we observe that MBP+ membrane wrapping of neuronal somata induced the clustering of neuronal contactin-associated protein (CASPR) (Fig S2a, inset 1). Wrapped neuronal dendrites also exhibited clustered CASPR (Fig S2a, inset 2), but unwrapped somata and dendrites showed no CASPR clusters (Fig S2a, inset 3). This pattern of molecular organization is indicative of a direct neuroglial molecular interaction normally restricted to paranodes at the node of Ranvier (Einheber et al., 1997; Eisenbach et al., 2009; Pedraza et al., 2009).
Loss of JAM2 results in myelination of the somatodendritic compartment in vivo
In the more complex environment of the CNS, is inhibitory JAM2 signaling required to prevent myelination of the somatodendritic compartment? At postnatal day 30–31 (P30–31) in the mouse, most neurons in the cervical spinal cord express JAM2. We examined a JAM2 reporter and JAM2 protein expression in spinal cord cross-sections of both jam2:beta-galactosidase knock-in mice (Fig S3a–b), as well as heterozygous and JAM2 KO mice, respectively (Fig S3c–d, Fig S4a–b;). The CNS is heavily myelinated and individual myelin segments are generally difficult to resolve by immunostaining. One exception to this technical limitation is the dorsal horn, where very few axons are myelinated. In this area, we observed many beta-galactosidase/NeuN-positive neurons (Fig S3a), and robust gray matter JAM2 immunostaining in heterozygous but not JAM2 KO mice (Fig S4a–b), confirming the specificity of the antibody staining. In P31 littermate wildtype and JAM2 KO mice, we found no significant differences in the average densities of NeuN-positive neurons or CC1-positive oligodendrocytes within the dorsal horn region (Fig S3c–f), suggesting that the development of these two cell types is normal in the absence of JAM2.
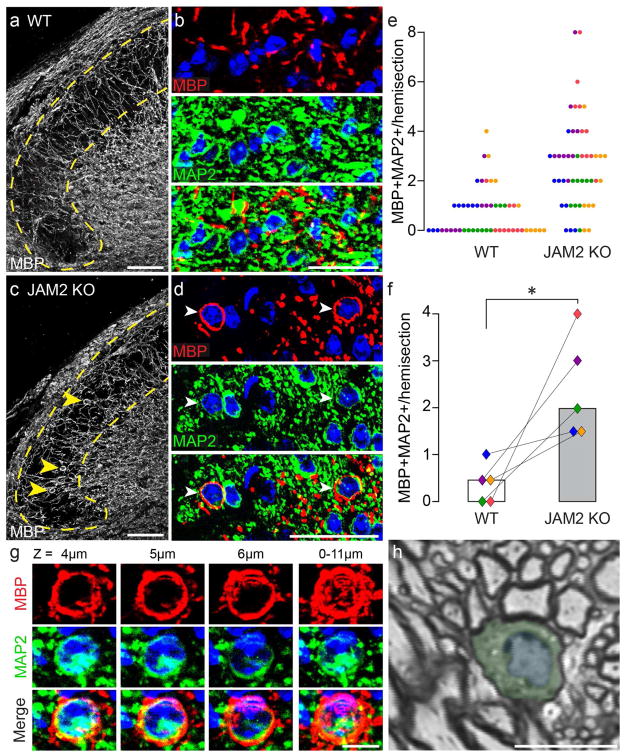
When we examined myelination patterns in dorsal horns of littermate wildtype and JAM2 KO mice (Fig 6a–d), we observed conspicuous MBP+ membrane ‘bubbles’ around neuronal somata in JAM2 KO dorsal horns (Fig 6c–d, arrowheads). Seventy-two percent of JAM2 KO hemisections contained more than one myelin-wrapped neuron, compared to just 16% of wildtype hemisections (Fig 6e). Overall, the median number of wrapped neurons per hemisection was four-fold higher in JAM2 KO mice compared to wildtype littermates (Figure 6f). Optical sectioning of immunostained tissue revealed that wrapped neurons could be wrapped with MBP-positive membrane around their entire circumference through several micrometers in the z-axis (Fig 6g), reminiscent of myelin-wrapped beads (Fig 1d). One of the hallmarks of myelination is the clustering of CASPR to paranodes. Like myelin-wrapped JAM2 KO neurons in vitro (Fig S2a), we find that the molecular organization of CASPR on JAM2 KO neurons is dramatically affected by somatic myelination in vivo. CASPR protein clustering is evident on MBP+ membrane-wrapped JAM2-knockout neurons (Fig S2b). Consistent with our immunostaining observations, semi-thin sections through a JAM2 KO dorsal horn neuron show myelination around the total circumference of a soma (Fig 6h). This particular cell is wrapped across three different semi-thin sections (Fig S2c–e).
Figure 6. JAM2 is necessary to inhibit somatic wrapping in vivo.
(a) dorsal horn (dotted yellow line) in a coronal section of a wildtype spinal cord immunostained for MBP. (b) higher magnification of dorsal horn myelin (top, red), neurons (middle, green), and merged (bottom). (c–d) same as (a–b) but in JAM2 KO mice. Arrowheads indicate MBP+ wrapped MAP2+ neurons. (e) number of myelin wrapped dorsal horn neurons per hemisection (one dot = one hemisection, colors indicate littermate pairs). (f) bar graph of median number of wrapped neurons per hemisection per mouse. Connected diamonds indicate littermate pairs, and fill color corresponds to dots in (e). n = 5 littermate pairs, median taken from ten hemisections per mouse. *, P = 0.034; Wilcoxon Paired Signed-Ranks test. (g) a myelin wrapped neuron from a JAM2 KO dorsal horn imaged through an 11 micrometer thick z-stack. Individual columns labeled by z-plane depth, and a maximum intensity projection is shown in the far-right column. (h) a toluidine blue stained semi-thin section from a JAM2 KO mouse dorsal horn shows a myelin-wrapped neuron (pseudo-colored green cytoplasm and blue nucleus). Scale bars: (a–d) 50 micrometers, (g–h) 10 micrometers. DAPI, blue.
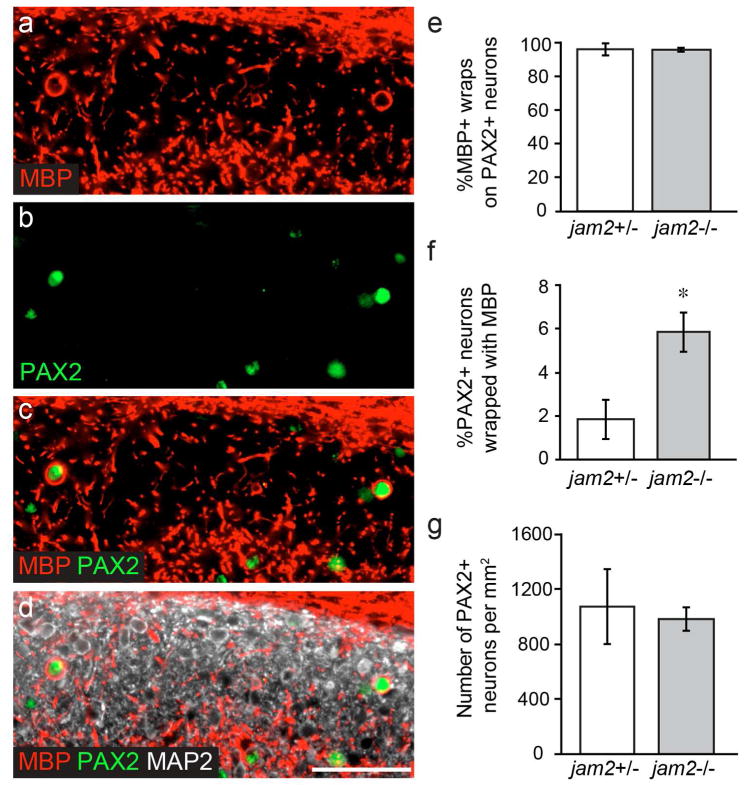
We wanted to further understand the identity of the wrapped Jam2-knockout neurons. Based on our observations that Jam2-knockout neurons are generally wrapped around their entire circumference in vivo, are small (10–20 micrometer diameter) and have little cytoplasm (Fig 6c,d,g–h; Fig S2b–c), we screened dorsal horn interneuron markers, and found that MBP+ wrapped neurons are exclusively positive for the paired box gene 2 (PAX2) transcription factor (Figure 7a–e). PAX2+ dorsal horn neurons are inhibitory interneurons and their dendrites and axons exhibit islet morphology oriented along the rostral-caudal axis of the spinal cord (Smith et al., 2015). We find that approximately six-percent of PAX2+ neurons are wrapped in JAM2-knockout mice, significantly more than the under two-percent observed in JAM2-heterozygous mice (Fig 7f). We find no difference in the density of PAX2+ neurons in the dorsal horn between genotypes (Fig 7g). Finally, we find that PAX2+ neurons receive very little presynaptic innervation of their somata (Fig S5a–b) with many cells of both genotypes exhibiting no presynaptic puncta (VGLUT1: 64.4%; VGLUT2: 23.3%; VGAT: 30.6%). We also never observed synaptic puncta between the wrapping MBP+ sheath and the PAX2+ neuron cell body (Fig S5c, 0/480 PAX2+ neurons). Taken together, our observations support the conclusion that oligodendrocyte myelin membranes are in direct contact with neural somata as they wrap.
Figure 7. Pax2+ neurons are wrapped in JAM2 knockout mice.
(a) dorsal horn immunostained for MBP, (b) Pax2, (c) label a subset of wrapped (d) MAP2+ neurons. (e) quantification of the percent of MBP+ ‘bubbles’ on Pax2+ neurons. (f) quantification of the percent of Pax2+ neurons wrapped by MBP+ ‘bubbles’. (g) number of Pax2+ neurons per square millimeter in Jam2 +/− and Jam2−/− littermate pairs. n = 4 littermate pairs. Bar graphs represent: (e–f) median calculated as (e) total wrapped Pax2+ out of total wrapped cells per mouse, or (f) percent per field from 10 20× fields per mouse. Error bars represent median absolute deviation; (g) mean density per mouse calculated from 10 20× fields per mouse. Error bars represent s.e.m. *, P = 0.034; Wilcoxon Paired Signed-Ranks test. Scale bar: 50 micrometers.
Discussion
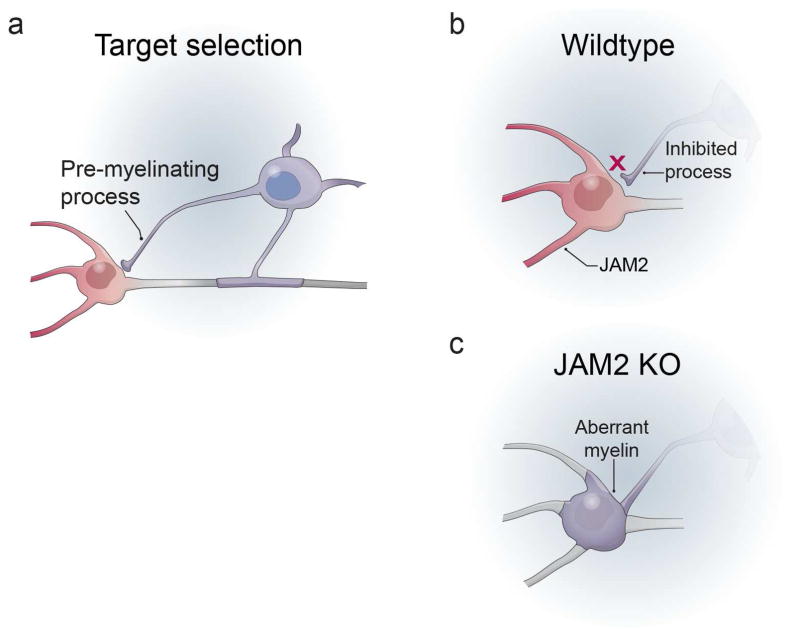
Overall, we show that somatodendritic JAM2 is necessary and sufficient to inhibit oligodendrocyte wrapping. Our results support a model of myelin target selection and wrapping in which pre-myelinating oligodendrocyte processes (Fig 8a) are normally inhibited by somatodendritic cues, including JAM2 (Fig 8b). When such inhibitors are missing, as in JAM2 KO mice, oligodendrocytes form aberrant somatodendritic myelin wraps (Fig 8c). As growing neurons utilize both attractive and repulsive axon guidance cues to precisely target correct brain areas and postsynaptic neurons, our findings indicate that oligodendrocytes utilize somatodendritic inhibition, potentially in addition to axonal attraction, to target myelin segments specifically to axons (Fig 1a, ‘Inhibition’, ‘Mixed’).
Figure 8. Model of JAM2 function on oligodendrocyte wrapping.
(a) a pre-myelinating process of an oligodendrocyte comes into contact with the somatodendritic compartment of a neuron. (b) in wildtype mice, somatodendritic JAM2 inhibits the oligodendrocyte process from wrapping. (c) when JAM2 is absent or blocked, the oligodendrocyte process is not inhibited, and continues to wrap myelin membranes on the neuron cell body.
Inhibitory signals guide oligodendrocyte myelination
The need for inhibitory signaling in tailoring myelination patterns becomes more apparent in neuron-free myelination assays. When oligodendroglia are cultured in the presence of permissive nanofibers, micropillars and spherical beads, they wrap MBP+ myelin membranes around each structure type, including the 50-micrometer diameter circular base of a micropillar (Figs 1c–d, 4c). These results suggest that oligodendroglia have few geometric requirements to form myelin. Indeed, the study of myelination by culturing oligodendrocytes on planar glass coverslips has historically been a successful approach because the cells readily differentiate and form membrane sheets with myelin-like composition in the absence of inductive substrate signaling. Here we use the oligodendrocyte’s flexible wrapping requirements to our advantage by adapting the high-throughput micropillar array platform to screen integral membrane protein activity on oligodendrocyte wrapping (Fig 4c–i). Under identical geometric conditions, and in the absence of any other molecular cues, oligodendrocytes and OPCs are dramatically less likely to wrap micropillars coated with JAM2-Fc (Fig 4e–h), and oligodendrocytes form significantly fewer myelin segments in the presence of soluble JAM2-Fc (Fig 5a–c). Interestingly, the magnitude of JAM2 inhibition is greater on micropillars (66.6%) than in SCN cocultures (24.6%), opening the possibility that axons may express an attractive signal that is able to partially overcome JAM2 inhibition. These results demonstrate the sufficiency of JAM2 to inhibit wrapping, and support the model that oligodendroglial target selection can be regulated by negative and positive microenvironment cues (Fig 1a ‘Mixed’).
Furthermore, JAM2’s inhibition of wrapping is not the result of changes in oligodendroglial cell density or differentiation (Figs 4i; S4c–f). In contrast, previously described inhibitors of axonal myelination negatively affect oligodendroglial cell adhesion in vitro and/or oligodendrocyte differentiation in vitro and in vivo (e.g. LINGO-1, LSAMP) (Lee et al., 2007; Mi et al., 2005; Sharma et al., 2015). It would not necessarily be advantageous for an inhibitor of somatodendritic myelination to prohibit oligodendrocyte differentiation. Oligodendrocytes would still need to differentiate and myelinate gray matter axons that are tightly intermingled with neuron cell bodies and dendrites.
Additional inhibitory myelin guidance molecules
What additional signal(s) prevent aberrant wrapping of other spinal cord neurons and dendrites? JAM2 is widely expressed in the young adult mouse spinal cord (Figs S3, S4a–b). Here we describe the aberrant wrapping of dorsal horn neuron cell bodies. Additional EM analysis of heavily myelinated gray matter areas may reveal that other neuron somatodendritic compartments are wrapped in JAM2 KO mice that are not detectable by immuno- or toluidine blue staining. Nevertheless, it is entirely possible that other neuron types may express other inhibitory molecules in addition to, or instead of JAM2. But even among JAM2-expressing spinal cord motor neurons, it is notable that their dendrites and cell bodies are densely covered with presynaptic terminals (Molofsky et al., 2014), another normally unmyelinated neuronal structure. Axon terminals, then, may present either a structural barrier preventing oligodendroglial contact with somatodendritic surfaces, or may themselves express a myelination-inhibitory cue that is sufficient to prevent oligodendrocyte wrapping with or without somatodendritic expression of JAM2. Unlike ventral motor neurons, dorsal horn PAX2+ neurons are largely devoid of presynaptic innervation of their somata (Fig S5). Importantly, our data does not distinguish between the possibilities that somatic myelin wrapping removes synapses, or whether only neurons devoid of somatic synaptic terminals are vulnerable to myelin wrapping in the absence of JAM2. In addition to neural sources of myelin inhibition, astrocytes, microglia, oligodendroglia and endothelial cells also remain unmyelinated, and may each express a molecular cue to prevent wrapping.
Oligodendroglial JAM2 receptors
Many receptors in oligodendroglia may be responsible for transducing myelin guidance signals, especially if JAM2 is one of many inhibitory molecules in the CNS. Known JAM2 receptors include other JAM family members JAM1, 2 and 3, as well as integrin alpha-4 beta-1, which may act alone or in combination to bind JAM2 (Arcangeli et al., 2013). In this study, the identity of the JAM2 receptor(s) in oligodendrocytes remains an open question. But if oligodendrocyte differentiation and wrapping are indeed dissociable events as our results suggest (Fig 4e–i), the understanding of downstream mechanisms that separately govern these two processes will be of high interest for future study.
Inhibitory myelin guidance molecules and failure of myelin repair
Promoting efficient myelin repair in diseases like multiple sclerosis (MS) has long been a clinical goal. While current therapies aim to control immune infiltration into the CNS, more attention is now being given to directly promoting oligodendrocyte differentiation and remyelination (Mei et al., 2014; Ruckh et al., 2012). However, it has also been observed in human postmortem MS lesions that differentiated oligodendrocytes are present, but fail to remyelinate axons (Chang et al., 2002). Could inhibitory myelin guidance molecules be aberrantly upregulated or mislocalized in MS lesions and prevent efficient repair? If so, it is essential to identify such signals, as they may be feasible targets to block and promote myelin repair. As additional myelin guidance molecules are identified, it will be important to understand their roles in both myelin formation and regeneration.
Experimental Procedures
Animals
All animals used in this study were housed and handled with the approval of the University of California San Francisco Institutional Animal Care and Use Committee (IACUC). Timed-pregnant Sprague-Dawley rats were procured from Charles River Laboratory (Strain No: 400, RRID:RGD_734476). Dorsal root ganglion neuron and spinal cord neuron dissections were done on embryonic day 15. Junction Adhesion Molecule 2 (JAM2) knockout mice are as previously described (Arcangeli et al., 2011, MGI:5779544). The jam2:beta-galactosidase knock-in mice were purchased from Jackson Laboratory (B6N(Cg)-Jam2tm1.1(KOMP)Mbp/J, Stock No: 024055, MGI:5522546). Jam2 knockout mice were used to generate data in Figures 5–6, S3–S4, and B6N(Cg)-Jam2tm1.1(KOMP)Mbp/J mice were used to generate data in Figures 7, S2 and S5.
Immunohistochemistry
Cell cultures were immunostained with standard techniques, discussed elsewhere in detail (Lee et al., 2013). Briefly, cultures were fixed with 4% paraformaldehyde (Electron Microscopy Tools) in DPBS (Life Technologies), washed with PBS, air dried, then blocked and permeabilized in 20% normal goat serum (NGS) (Sigma-Aldrich) plus 0.1% Triton X-100 (Sigma-Aldrich) in DPBS. The cultures were incubated in 20% NGS in dPBS plus primary antibodies. Cultures were washed in PBS, and incubated in 20% NGS in dPBS plus secondary antibodies. The cultures were washed in PBS then distilled water, air dried and then mounted on microscope slides with Fluorescence Mounting Medium (Dako).
Spinal cord tissue was collected from P30–33 mice using standard techniques. Briefly, after transcardial perfusion with PBS and then 4% paraformaldehyde (Electron Microscopy Tools), the spinal cord was dissected out and post-fixed in 4% paraformaldehyde overnight at 4° Celsius and cryoprotected by incubation in 30% sucrose in PBS until tissue sank. Coronal sections were cut in 30-micrometer thick slices from the cervical spinal cord (C5–C8) using a freezing microtome (Microm HM 450 and KS 34, Thermo Scientific). Sections were immunostained with the same methods as cell cultures above.
Images were collected using a Zeiss Axio Imager Z1 Apotome or M2 Apotome.2 fluorescence microscope with either Axiovision or Zen software (Zeiss). Images are shown as a maximum intensity projection of optical section z-stacks. Micropillar cultures (Mei et al., 2014) and semi-thin sections were imaged using a Zeiss LSM-700 confocal microscope and Zen software.
Antibodies
Antibodies used for immunostaining are: microtubule associated protein 2 (chicken anti-MAP2, 1:1,000, Millipore Cat# AB5543, RRID:AB_571049); neurofilament (mouse anti-NF, 1:200, Covance Cat# SMI-312R, RRID:AB_10119994); junction adhesion molecule 2 (rabbit anti-JAM2, 1:100–200, ThermoFisher Scientific Cat# PA5-21576, RRID:AB_11156435); myelin basic protein (rat anti-MBP, 1:100, Millipore Cat# MAB386, RRID:AB_94975); p75-neurotrophin receptor (mouse anti-p75NTR, 1:25 hybridoma cell line gift of E.M. Shooter lab, clone MC192, commercially available as RRID:AB_528539); contactin associated protein (mouse anti-CASPR, 1:500, E. Peles laboratory, RRID:AB_2314218), neuronal nuclei (rabbit anti-NeuN, 1:2000 Abcam Cat# ab177487, RRID:AB_2532109), adenomatous polyposis coli (mouse anti-APC/CC1, 1:200 Millipore Cat# OP80, RRID:AB_2057371), beta-galactosidase (chicken anti-βGal, 1:1000 Aves Labs Cat# BGL-1040, RRID:AB_2313507), vesicular glutamate transporter 2 (guinea pig anti-VGLUT2, 1:4,000 Millipore Cat# AB2251, RRID:AB_1587626), vesicular glutamate transporter 1 (guinea pig anti-VGLUT1, 1:4,000 Synaptic Systems Cat#135 304, RRID: AB_887878), vesicular GABA transporter (guinea pig anti-VGAT, 1:500 Synaptic Systems Cat# 131 004, RRID:AB_887873), paired box gene 2 (rabbit anti-PAX2, 1:1,000–4,000 Abcam Cat# ab79389, RRID:AB_1603338), and platelet derived growth factor receptor-alpha (rabbit anti-PDGFRα, 1:8,000, generous gift of W.B. Stallcup, Sanford-Burnham Medical Research Institute, Cancer Center, La Jolla, CA USA, RRID:AB_2315173). Secondary antibodies were Alexa Fluor raised in goat against rat, mouse, rabbit, human or chicken in the following wavelengths: 488, 594, 647 (1:1,000 Life Technologies) and nuclei were stained with DAPI. The antibodies used in function-blocking experiments (Figure S1): rat anti-JAM2 (5μg/mL, R&D Systems MAB9881, RRID:AB_2128924) or rat IgG (5μg/mL, R&D Systems 6-001-F, RRID:AB_2616570 ).
Primary neuron and glia isolation and culture
Spinal cord neuron (SCN) isolation and culture has been described previously (Camu and Henderson, 1992; Molofsky et al., 2014). Briefly, embryonic day 15 rat or mouse embryo spinal cords were dissected and meninges removed as much as possible. Spinal cords were chopped with a sterile scalpel blade and the tissue was incubated in 0.05% trypsin-EDTA (Life Technologies) for 15 minutes at 37° Celsius. The tissue was dissociated into a single-cell suspension and incubated in a series of immunopanning dishes; two negative selection dishes coated with monoclonal hybridoma antibodes rat neural antigen-2 (Ran-2, ATCC) and Galactocerebrocide (Gal-C (Ranscht et al., 1982)); and one positive selection plate against p75 neurotrophin receptor (p75NTR, rat cells: MC192 hybridoma; mouse cells: rabbit anti-p75 function blocking antibody ‘REX’, a generous gift of L.F. Reichardt (Weskamp and Reichardt, 1991)). Adherent cells were released from the final immunopanning plate with a brief application of 0.05% trypsin-EDTA and 200,000–300,000 cells were seeded on 25mm round coverslips prepared with 200μL of 1:25 Matrigel (Corning, 356230) in DMEM (Life Technologies). SCN were left to adhere overnight, and flooded with growth medium plus 5-fluoro-2′-deoxyuridine (FDU) to kill mitotic cells, and 80% of growth medium was changed every three days for 3–4 weeks: DMEM (Life Technologies), B27 (Life Technologies), N2 (Life Technologies), penicillin-streptomycin (Life Technologies), N-acetylcisteine (NAC, Sigma-Aldrich), forskolin (EMD), insulin (Sigma-Aldrich), brain-derived neurotrophic factor (BDNF, Peprotech), ciliary neurotrophic factor (CNTF, Peprotech), glial-derived neurotrophic factor (GDNF, Peprotech).
Dorsal root ganglion (DRG) neuron cultures were established as previously described (Mei et al., 2014). Oligodendrocyte precursor cells (OPCs) were isolated (Lee et al., 2013) and cultured on micropillars as previously described (Mei et al., 2014).
OPCs were cocultured with 3–4 week old SCN as follows: OPCs were isolated with hybridoma anti-A2B5 or anti-O4-coated panning dishes, and 400,000 OPCs were seeded on each SCN culture in chemically-defined medium (Lee et al., 2013), which was changed every three days. For Jam2-Fc or Fc cocultures (Figure 5), 10 micrograms/mL protein was added to culture medium.
Oligodendrocyte nanofiber and microbead culture
Aligned polycaprolactone nanofibers were purchased in an 8-chamber slide format (Nanofiber Solutions cat 0802). 20 micrometer-diameter NH2 polystyrene beads (Kisker Biotech Gmbh & Co cat PPS-20.0NH2) were sparsely seeded into the nanofiber chambers, coated with poly-L-lysine and dried to promote adhesion of beads to the nanofibers. OPC culture on beads and nanofibers was performed essentially as described (Chong et al., 2011; Lee et al., 2013). OPCs were seeded into the chambers and cultured for 3–4 days, then fixed and immunostained as above.
Oligodendrocyte coculture on live and cross-linked SCNs
SCN cultures were established as above. Prior to OPC seeding, SCN cultures were chemically cross-linked as described (Rosenberg et al., 2008) with 4% paraformaldehyde for 10 minutes at room temperature. Fixed neuron cultures were extensively washed in L15 medium + 10% FBS (Gibco), and finally washed with coculture medium. One million OPCs were seeded onto either live or cross-linked neurons and cocultured for 5 days (Figure 2) or 7–12 days (Figure 1) in a chemically defined medium, then immunostained as above.
Micropillar protein coating
Micropillar arrays were sterilized with 100% ethanol and dried. Micropillars were coated with poly-L-lysine for one hour at room temperature, washed three times with water and dried. Each micropillar array of 1000 pillars was then coated with 2μg of either Fc protein (R&D Systems 110-HG, RRID:AB_276244) or JAM2-Fc protein (R&D Systems 988-VJ-050) in 27μL Tris HCl pH 9.5 overnight at room temperature. The micropillars were washed three times with water and twice with oligodendrocyte culture medium and kept wet with medium until 40,000 OPCs were seeded.
JAM2-Fc and Fc protein binding assay
OPCs were seeded onto poly-L-lysine coated coverslips in PDGF-containing culture medium overnight. On div 3, coverslips were washed with DMEM and incubated in 2μg/mL JAM2-Fc or Fc in DMEM for 30 minutes at room temperature. The coverslips were washed three times with DPBS, fixed with paraformaldehyde and immunostained as above.
Western Blot Analysis
Western blotting was performed essentially as previously described (Rosenberg et al., 2008). 25mm coverslips containing either SCN alone or myelinating SCN cocultures were collected in 100μL/coverslip of ice cold RIPA buffer, kept on ice and frozen immediately at −80°C. Samples were thawed on ice and homogenized. Protein concentration was determined by BCA Protein Assay (Thermo Scientific 23225). Proteins were transferred to pure nitrocellulose membranes and probed with primary antibodies (see Antibodies) and Alexa Fluor 680 secondary antibodies raised in goat (Life Technologies). Imaging was conducted with an Odyssey infrared system (LI-COR).
RNA isolation, RNA sequencing and bioinformatics
Spinal cords and dorsal root ganglia were dissected from a single litter of embryonic-day 15 rat embryos, SCN and DRG neurons were isolated and cultured for three weeks as described above. RNA was isolated using Trizol Reagent and manufacturer’s protocol (Life Technologies). Sequencing was performed with Illumina Genome Analyzer Sequencing System. Over 142M single end reads of length 100 bases were sequenced per sample. Sequences were produced with CASAVA (version 1.8.1.) and aligned to Rattus norvegicus rn4 genome build using TopHat (version 1.3.0) (Trapnell et al., 2009). TopHat was run separately for each RNA-Seq library with the options “--segment-mismatches 1 --min-intron-length 30 --max-multihits 10 --solexa-quals -o”. Transcriptome was assembled using Cufflinks (Roberts et al., 2011; Trapnell et al., 2010) (version 1.1.0) and RefSeq reference annotation file (using the options –g). The reference was downloaded from the UCSC tables. The two assembled transcriptomes were merged with the tool Cuffmerge (version 1.1.0.). Cuffdiff was used to detect differentially expressed genes and transcripts between the two samples. Identification of the rat transcripts coding for membrane and transmembrane proteins was done with Ingenuity® Pathway Analysis (IPA®). The criteria for selection of differentially expressed transcripts was a higher expression in SCNs as compared to DRGs, and q values <=0.05.
Quantitative PCR
Total RNA was extracted from three-week-old spinal cord neuron or dorsal root ganglion neuron cultures using Trizol Reagent (Life Technologies), and precipitated using either chloroform and isopropanol or PureLink RNA Mini Kit (Life Technologies, cat. 12183020). RNA was reverse transcribed using the RETROscript kit oligo-dT primers (Thermo Fisher, cat. AM1710). cDNA was amplified in triplicate using Power SYBR Green PCR master mix (Thermo Fisher, cat. 4367659) and primers for either jam2 (Forward 5′ TACTGTGAAGCCCGCAACTC 3′, Reverse 5′ GCAGAAATGACGAAGGCCAC 3′, product length 122 base pairs), or gapdh (Forward 5′ GTGCCAGCCTCGTCTCATAG 3′, Reverse 5′ AGAGAAGGCAGCCCTGGTAA 3′, product length 91 base pairs). qPCR was run on a 7500 Real Time PCR system and analyzed with 7500 software version 2.0.6 (Applied Biosystems).
Electron microscopy
Transmission electron microscopy of cell cultures is as described previously (Lee et al., 2013; Rosenberg et al., 2008). Briefly, cultures were fixed with 4% PFA (Electron Microscopy Tools), incubated in 1% osmium tetroxide (Electron Microscopy Tools), then counterstained with 1% uranyl acetate. Samples were dehydrated in a series of ethanol dilutions and embedded in 1:1 EMBed-812 resin (Electron Microscopy Sciences) to propylene oxide (PPO, Electron Microscopy Sciences), then incubated in a 2:1 resin to PPO mixture. Cultures were detached from the coverslip with a razor blade and rolled tightly before embedding into the final 100% resin block. Ultrathin sections were cut at the W.M. Keck Foundation Advanced Microscopy Laboratory and imaged with a JEM1400 Electron Microscope (JEOL) in the Zilkha Neurogenetic Institute. Scanning electron microscopy of micropillars is as described previously (Mei et al., 2014).
Image analysis and statistics
The UCSF department of Biostatistics was consulted to identify suitable statistical tests. Statistical significance was determined at P-values < 0.05. Statistical tests were applied as described in figure legends. Data normality was determined using the Kolmogorov–Smirnov test. N values are indicated in the figure legends.
Supplementary Material
Highlights.
Neuronal JAM2 is necessary and sufficient to inhibit myelination of the somatodendritic compartment
JAM2 knockout mice exhibit aberrantly myelinated neuronal cell bodies
Myelinated neuronal cell bodies are distinctly PAX2+ inhibitory interneurons
Oligodendrocytes utilize non-axonal inhibitory signaling in myelination
Acknowledgments
We thank Dr. W.B. Stallcup and Dr. L.F. Reichardt for antibodies, M.L. Wong for ultrathin sectioning of electron microscopy samples, J. Wong for semi-thin sectioning and toluidine blue staining (Gladstone Institutes Electron Microscopy Core, San Francisco, CA). We also thank Dr. K.J. Chang for technical assistance, Dr. S.Y.C. Chong for efforts on development of SCN culture conditions, members of the Chan Laboratory for critical reading of the manuscript, and the UCSF Clinical and Translational Science Institute, for biostatistics consultation (NIH UL1 TR000004). This work was supported by NMSS Research Grants (RG4541A3 and RG5203A4), NIH/NINDS (R01NS062796, R01NS097428) and the Rachleff Family Professorship to J.R.C.; NIH/NINDS (R01NS50220), and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Incumbent of the Hanna Hertz Professorial Chair for Multiple Sclerosis and Neuroscience to E.P.; NSERC PGS D to L.A.O.; and NIH/NINDS Ruth L. Kirschstein NRSA (F31NS081905) to S.A.R.
Footnotes
Author Contributions
S.A.R., F.M., Y.E.E., L.A.O., D.L., Y.A.S., J.N.K. and J.R.C. performed experiments. Y.E.E., M.A.L., E.P. and J.R.C. provided reagents. S.A.R., F.M., Y.E.E., L.A.O., Y.A.S., J.N.K., D.A.L., E.P. and J.R.C. provided intellectual contributions. S.A.R., D.L. and J.R.C. analyzed the data. S.A.R. and J.R.C. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arcangeli ML, Frontera V, Bardin F, Obrados E, Adams S, Chabannon C, Schiff C, Mancini SJC, Adams RH, Aurrand-Lions M. JAM-B regulates maintenance of hematopoietic stem cells in the bone marrow. Blood. 2011;118:4609–4619. doi: 10.1182/blood-2010-12-323972. [DOI] [PubMed] [Google Scholar]
- Arcangeli ML, Frontera V, Aurrand-Lions M. Function of junctional adhesion molecules (JAMs) in leukocyte migration and homeostasis. Arch Immunol Ther Exp (Warsz) 2013;61:15–23. doi: 10.1007/s00005-012-0199-5. [DOI] [PubMed] [Google Scholar]
- Bechler ME, Byrne L, Ffrench-Constant C. CNS Myelin Sheath Lengths Are an Intrinsic Property of Oligodendrocytes. Curr Biol CB. 2015;25:2411–2416. doi: 10.1016/j.cub.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinzinger K, Anzil AP, Müller W. Myelinated nerve cell perikaryon in mouse spinal cord. Z Für Zellforsch Mikrosk Anat Vienna Austria 1948. 1972;128:135–138. doi: 10.1007/BF00306893. [DOI] [PubMed] [Google Scholar]
- Braak E, Braak H, Strenge H. The fine structure of myelinated nerve cell bodies in the bulbus olfactorius of man. Cell Tissue Res. 1977;182:221–233. doi: 10.1007/BF00220591. [DOI] [PubMed] [Google Scholar]
- Camu W, Henderson CE. Purification of embryonic rat motoneurons by panning on a monoclonal antibody to the low-affinity NGF receptor. J Neurosci Methods. 1992;44:59–70. doi: 10.1016/0165-0270(92)90114-s. [DOI] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med. 2002;346:165–173. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- Chong SYC, Rosenberg SS, Fancy SPJ, Zhao C, Shen YAA, Hahn AT, McGee AW, Xu X, Zheng B, Zhang LI, et al. Neurite outgrowth inhibitor Nogo-A establishes spatial segregation and extent of oligodendrocyte myelination. Proc Natl Acad Sci. 2011;109:1299–1304. doi: 10.1073/pnas.1113540109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MH, Beal JA. Myelinated granule cell bodies in the cerebellum of the monkey (Saimiri sciureus) Anat Rec. 1977;187:249–255. doi: 10.1002/ar.1091870210. [DOI] [PubMed] [Google Scholar]
- Einheber S, Zanazzi G, Ching W, Scherer S, Milner TA, Peles E, Salzer JL. The axonal membrane protein Caspr, a homologue of neurexin IV, is a component of the septate-like paranodal junctions that assemble during myelination. J Cell Biol. 1997;139:1495–1506. doi: 10.1083/jcb.139.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbach M, Kartvelishvily E, Eshed-Eisenbach Y, Watkins T, Sorensen A, Thomson C, Ranscht B, Barnett SC, Brophy P, Peles E. Differential clustering of Caspr by oligodendrocytes and Schwann cells. J Neurosci Res. 2009;87:3492–3501. doi: 10.1002/jnr.22157. [DOI] [PubMed] [Google Scholar]
- Fancy SPJ, Harrington EP, Yuen TJ, Silbereis JC, Zhao C, Baranzini SE, Bruce CC, Otero JJ, Huang EJ, Nusse R, et al. Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. Nat Neurosci. 2011;14:1009–1016. doi: 10.1038/nn.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RJM, ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- Gibson EM. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 2014;344:1252304. doi: 10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JH, Ravanelli AM, Schwindt R, Scott EK, Appel B. Neuronal activity biases axon selection for myelination in vivo. Nat Neurosci. 2015;18:683–689. doi: 10.1038/nn.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Leach MK, Redmond SA, Chong SYC, Mellon SH, Tuck SJ, Feng ZQ, Corey JM, Chan JR. A culture system to study oligodendrocyte myelination processes using engineered nanofibers. Nat Meth. 2012;9:917–922. doi: 10.1038/nmeth.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chong SYC, Tuck SJ, Corey JM, Chan JR. A rapid and reproducible assay for modeling myelination by oligodendrocytes using engineered nanofibers. Nat Protoc. 2013;8:771–782. doi: 10.1038/nprot.2013.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee X, Yang Z, Shao Z, Rosenberg SS, Levesque M, Pepinsky RB, Qiu M, Miller RH, Chan JR, Mi S. NGF regulates the expression of axonal LINGO-1 to inhibit oligodendrocyte differentiation and myelination. J Neurosci Off J Soc Neurosci. 2007;27:220–225. doi: 10.1523/JNEUROSCI.4175-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubetzki C, Demerens C, Anglade P, Villarroya H, Frankfurter A, Lee VM, Zalc B. Even in culture, oligodendrocytes myelinate solely axons. Proc Natl Acad Sci U S A. 1993;90:6820–6824. doi: 10.1073/pnas.90.14.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Fancy SPJ, Shen YAA, Niu J, Zhao C, Presley B, Miao E, Lee S, Mayoral SR, Redmond SA, et al. Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat Med. 2014;20:954–960. doi: 10.1038/nm.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensch S, Baraban M, Almeida R, Czopka T, Ausborn J, El Manira A, Lyons DA. Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nat Neurosci. 2015;18:628–630. doi: 10.1038/nn.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Kelley KW, Tsai HH, Redmond SA, Chang SM, Madireddy L, Chan JR, Baranzini SE, Ullian EM, Rowitch DH. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature. 2014;509:189–194. doi: 10.1038/nature13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza L, Huang JK, Colman D. Disposition of axonal caspr with respect to glial cell membranes: Implications for the process of myelination. J Neurosci Res. 2009;87:3480–3491. doi: 10.1002/jnr.22004. [DOI] [PubMed] [Google Scholar]
- Ranscht B, Clapshaw PA, Price J, Noble M, Seifert W. Development of oligodendrocytes and Schwann cells studied with a monoclonal antibody against galactocerebroside. Proc Natl Acad Sci U S A. 1982;79:2709–2713. doi: 10.1073/pnas.79.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remahl S, Hildebrand C. Myelinated non-axonal neuronal elements in the feline olfactory bulb lack sites with a nodal structural differentiation. Brain Res. 1985;325:1–11. doi: 10.1016/0006-8993(85)90296-3. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinforma Oxf Engl. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Rosenberg SS, Kelland EE, Tokar E, De la Torre AR, Chan JR. The geometric and spatial constraints of the microenvironment induce oligodendrocyte differentiation. Proc Natl Acad Sci U S A. 2008;105:14662–14667. doi: 10.1073/pnas.0805640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckh JM, Zhao JW, Shadrach JL, van Wijngaarden P, Rao TN, Wagers AJ, Franklin RJM. Rejuvenation of Regeneration in the Aging Central Nervous System. Cell Stem Cell. 2012;10:96–103. doi: 10.1016/j.stem.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch UK, Philips MA, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Boyle KA, Madden JF, Dickinson SA, Jobling P, Callister RJ, Hughes DI, Graham BA. Functional heterogeneity of calretinin-expressing neurons in the mouse superficial dorsal horn: implications for spinal pain processing. J Physiol. 2015;593:4319–4339. doi: 10.1113/JP270855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma Oxf Engl. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Nave KA. Multiple sclerosis: an immune or neurodegenerative disorder? Annu Rev Neurosci. 2008;31:247–269. doi: 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- Weskamp G, Reichardt LF. Evidence that biological activity of NGF is mediated through a novel subclass of high affinity receptors. Neuron. 1991;6:649–663. doi: 10.1016/0896-6273(91)90067-a. [DOI] [PubMed] [Google Scholar]
- Yuen TJ, Silbereis JC, Griveau A, Chang SM, Daneman R, Fancy SPJ, Zahed H, Maltepe E, Rowitch DH. Oligodendrocyte-encoded HIF function couples postnatal myelination and white matter angiogenesis. Cell. 2014;158:383–396. doi: 10.1016/j.cell.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalc B, Goujet D, Colman D. The origin of the myelination program in vertebrates. Curr Biol CB. 2008;18:R511–R512. doi: 10.1016/j.cub.2008.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.