Abstract
More than 30 years ago, a calcium-dependent enzyme activity was described that generates N-acyl phosphatidylethanolamines (NAPEs), which are precursors for N-acyl ethanolamine (NAE) lipid transmitters, including the endocannabinoid anandamide. The identity of this calcium-dependent N-acyltransferase (Ca-NAT) has remained mysterious. Here, we use activity-based protein profiling to identify the poorly characterized serine hydrolase PLA2G4E as a mouse brain Ca-NAT and show that this enzyme generates NAPEs and NAEs in mammalian cells.
N-Acyl ethanolamines (NAEs) are a class of bioactive lipids that include the endogenous cannabinoid (endocannabinoid) anandamide (C20:4 NAE, or AEA) 1. Much is known about NAE degradation, which is predominantly mediated in the brain by the integral membrane enzyme fatty acid amide hydrolase (FAAH), with additional contributions from the lysosomal enzyme N-acylethanolamine-hydrolyzing acid amidase (NAAA) in peripheral systems and FAAH-2 in primates2,3. The biosynthesis of NAEs is less well understood, but these lipids appear to originate from an unusual class of triacylated lipids termed N-acyl phosphatidylethanolamines (NAPEs) (Fig. 1a)2-4. NAPEs can be converted to NAEs by multiple pathways that include: i) direct release by phospholipase-D enzymes, such as NAPE-PLD5; ii) the combined action of phospholipase A1/A2 enzymes, such as ABHD4, and lyso-PLDs or glycerophosphodiesterases (e.g., GDE1)6,7; and iii) phospholipase C-mediated formation of phospho-NAEs and subsequent phosphatase-dependent conversion to NAEs8 (Fig. 1a).
Figure 1.
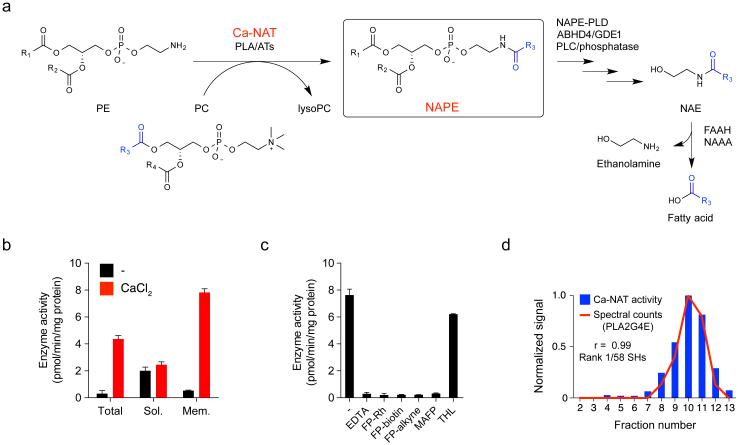
Characterization of calcium-dependent N-acyltransferase (Ca-NAT) activity in mouse brain. (a) Metabolic routes for N-acyl phosphatidylethanolamines (NAPEs) and N-acyl ethanolamines (NAEs). (b) Ca-NAT activity in mouse brain lysates (40 µg protein). Total homogenate (Total) and soluble (Sol.) and membrane (Mem.) brain lysates were incubated with sn-1, sn-2-dipalmitoyl-phosphatidylcholine (DPPC; 250 μM) and sn-1, sn-2-dioleoyl-phosphatidylethanolamine (DOPE, 250 µM) for 1 h at 37 °C with or without CaCl2 (3 mM) and N-C16:0 DOPE was measured. (c) Inhibition of Ca-NAT activity in mouse brain by EDTA and fluorophosphonate (FP) probes. Mouse brain membrane Ca-NAT activity was measured as in part b (+ 3 mM CaCl2) with or without the addition of EDTA (10 µM) or after pre-treatment with MAFP (3 µM), FP probes (1 µM each), or THL (3 µM) for 30 min at 25 °C. Data in parts b and c represent mean values ± s. d. for 3 biological replicates. (d) Correlation of spectral counts for PLA2G4E (determined for each fraction obtained by sucrose gradient (5-40%) centrifugation of detergent-solubilized mouse brain membrane lysates by labeling with FP-biotin (5 µM) followed by avidin enrichment and LC-MS analysis) with Ca-NAT activity of each fraction. A Pearson correlation coefficient of 0.99 was calculated for PLA2G4E. Data represent activity measurements from one experiment representative of two biological replicates.
The origin of NAPEs themselves, however, which beyond serving as precursors of NAEs may also have independent biological functions4, has remained mysterious. In the 1980s, Schmid and colleagues described an enzymatic activity capable of generating NAPEs from dog heart and brain tissue9,10. Key features of this enzymatic activity included: 1) dependence on calcium; 2) strong preference for the transfer of the sn-1 O-acyl chain of phosphatidylcholine (PC) to the amine of phosphatidylethanolamine (PE); and 3) predominant distribution to particulate fractions of tissues. Thereafter, this calcium-dependent N-acyltransferase (Ca-NAT) activity was shown to be highly expressed in rodent brain11, and treatment of cultured rat neurons with calcium ionophores or depolarizing agents was found to promote formation of NAPEs and NAEs12, presumably through stimulating the Ca-NAT activity. Most recently, a family of calcium-independent NATs (HRAS-like phospholipase A/acyltransferase (PLA/AT) family) has been discovered and shown to contribute to NAPE and NAE formation in mammalian cells13. But, molecular characterization of the brain Ca-NAT that generates NAPEs still represents an important and elusive goal.
In pursuit of the brain Ca-NAT, we first established a liquid chromatography-mass spectrometry (LC-MS) assay for monitoring enzymatic formation of N-C16:0 NAPE from 1,2-dipalmitoyl-sn-3-glycerophosphocholine (C16:0/C16:0-PC) and 1,2-dioleoyl-sn-3-glycerophoshoethanolamine (C18:1/C18:1-PE) as O-acyl donor and N-acyl-acceptor substrates, respectively. We found, as expected based on past research9,10,12, that NAPE formation in mouse brain lysates was greatly enhanced by CaCl2 (Fig. 1b) and blocked by EDTA (Fig. 1c). This Ca-NAT activity was also much higher in the particulate (membrane) versus soluble fraction of mouse brain (Fig. 1b) and showed the expected selectivity for sn-1 O-acyl transfer as measured with PC substrates harboring distinct acyl-chains in the sn-1 and sn-2 positions (Supplementary Results, Supplementary Fig. 1).
Considering that other characterized enzymes that transfer O-acyl chains from PC to acceptor lipids (e.g., lecithin-cholesterol acyltransferase14) are serine hydrolases, we next asked whether the brain membrane Ca-NAT activity was sensitive to broad-spectrum serine hydrolase inhibitors and probes. Treatment of brain membrane lysates with methoxy arachidonyl fluorophosphonate (MAFP) or the activity-based probes FP-biotin, FP-rhodamine, or FP-alkyne15, completely blocked the Ca-NAT activity (Fig. 1c). In contrast, the β-lactone lipase inhibitor tetrahydrolipstatin (THL) did not affect brain membrane Ca-NAT activity (Fig. 1c). These data indicated that the Ca-NAT activity could originate from a membrane-associated serine hydrolase with the potential to be labeled and enriched from brain tissue using FP activity-based probes.
A detergent screen revealed that the brain membrane Ca-NAT activity could be solubilized with IGEPAL CA-630 (0.5%) (Supplementary Fig. 2), but attempts to further purify detergent-solubilized Ca-NAT using traditional column chromatography methods, such as ion exchange, were thwarted by near-complete loss of enzyme activity. We therefore instead separated detergent-solubilized mouse brain membrane lysates by sucrose gradient (5-40%), where 13 fractions were collected and assessed for: 1) Ca-NAT activity by LC-MS, and 2) serine hydrolase content using FP probes. The Ca-NAT activity was enriched in fractions 8-12 with peak activity in fraction 10 (Fig. 1d and Supplementary Fig. 3a), while individual serine hydrolase activities were differentially distributed throughout the gradient as revealed by activity-based protein profiling (ABPP) using the FP-rhodamine probe (Supplementary Fig. 3b). The ABPP gel did not reveal any serine hydrolases with distributions that matched that of the brain Ca-NAT activity, possibly indicating the responsible enzyme was too low in abundance to visualize by SDS-PAGE. We therefore proceeded to enrich and identify serine hydrolase activities in each fraction using the FP-biotin probe and avidin enrichment followed by LC-MS/MS analysis on an LTQ-Orbitrap mass spectrometer. The quantity of each serine hydrolase in each fraction was estimated by spectral counting and related to the Ca-NAT activity in these fractions, furnishing a global correlation analysis that designated the enzyme PLA2G4E as having the strongest correlation coefficient (0.99) with Ca-NAT activity out of 58 serine hydrolases quantified by the LC-MS/MS-ABPP analysis (Fig. 1d and Supplementary Table 1).
PLA2G4E is a poorly characterized member of the PLA2G4 clan of cytosolic phospholipases16,17. Previous research has revealed that recombinant PLA2G4E exhibits very low phospholipase activity16,18. We expressed mouse PLA2G4E by transient transfection in HEK293T cells and found that the enzyme was robustly labeled by FP-rhodamine, and this labeling was enhanced by CaCl2 (Fig. 2a and Supplementary Fig. 4a). PLA2G4E-transfected cell lysates also showed much higher Ca-NAT activity compared to mock-transfected cell lysates (Fig. 2b), and this activity was blocked by EDTA (Supplementary Fig. 4b). We further evaluated two other serine lipases – PNPLA6 and PNPLA7 – that showed lower, but still good correlations with Ca-NAT activity in the sucrose gradient experiment (Supplementary Table 1) and found that neither of these recombinant enzymes showed Ca-NAT activity (Supplementary Fig. 5). PLA2G4D, which shares 43% sequence identity with PLA2G4E, also did not exhibit Ca-NAT activity (Supplementary Fig. 6).
Figure 2.
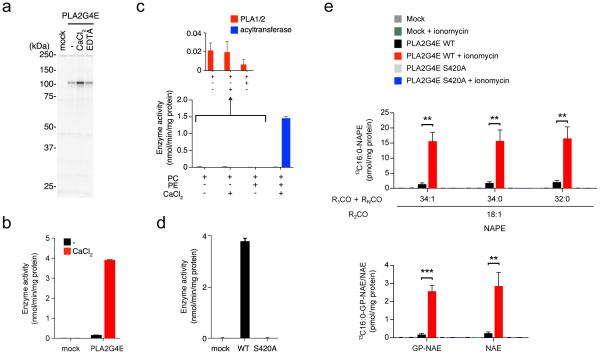
Recombinant PLA2G4E exhibits Ca-NAT activity and generates NAPEs and NAEs in mammalian cells. (a) Gel-based ABPP analysis demonstrating calcium-enhanced FP-rhodamine labeling of recombinant mouse PLA2G4E in transfected, detergent-solubilized HEK293T cell lysates. See Supplementary Fig. 4 for quantification of FP-labeled PLA2G4E band intensities. (b) Ca-NAT activity of transfected HEK293T cell lysates (2 µg) as measured by N-C16:0 DOPE production in reactions with DPPC (40 μM) and DOPE (75 µM) for 30 min at 37 °C with or without CaCl2 (3 mM) added to the reaction mixture. (c) Comparison of phospholipase vs. NAT activity for recombinant PLA2G4E. PLA2G4E-transfected HEK293T lysates were incubated with the indicated combination of sn-1, sn-2-diheptadecanoyl-phosphatidyl choline (PC; 40 µM), DOPE (PE; 75 µM), and CaCl2 (3 mM) for 30 min at 37 °C. The production of C17:0 fatty acid and N-C17:0 DOPE was measured to determine the relative phospholipase and NAT activities, respectively. (d) Ca-NAT activity of WT- and S420A-PLA2G4E-transfected HEK293T cell lysates (10 µg) as measured in b. (e) Production of 13C16:0-containing NAPEs, GP-NAEs, and NAEs in mock, WT-PLA2G4E, and S420A-PLA2G4E-transfected HEK293T cells. Cells were simultaneously fed 250 μM 13C16-palmitic acid and incubated in the presence or absence of 2 μM ionomycin for 4 h. Lipids were then extracted and analyzed by LC-MS/MS. For a-e, data represent mean values ± s. d. for 3 biological replicates. For e, **, p < 0.01, *** p < 0.001 by two-sided Student’s t-test for ionomycin-treated vs control (DMSO)-treated WT-PLA2G4E-transfected cells.
Consistent with previous studies showing that the brain Ca-NAT is activated by high-µM concentrations of calcium10,19, we found that mouse brain Ca-NAT and recombinant PLA2G4E were stimulated by CaCl2 with similar half-maximal values of 0.49 and 0.16 mM, respectively (Supplementary Fig. 7a). Like the brain Ca-NAT activity (Fig. 1c and Supplementary Fig. 1), recombinant PLA2G4E was inhibited by FP agents but not THL (Supplementary Fig. 4b), showed a strong preference for transferring the sn-1 O-acyl chain from mixed PC substrates (Supplementary Fig. 7b), was activated by calcium over other divalent metal cations11 (Supplementary Fig. 7c), and required DTT for maximal activity11 (Supplementary Fig. 7d). PLA2G4E also generated the anandamide precursor lipid N-C20:4 NAPE from a di-C20:4-PC substrate (Supplementary Fig. 8a). Notably, and consistent with previous research16,18, reactions performed without PE revealed that PLA2G4E exhibits very little PC-lipase activity either in the presence or absence of CaCl2 (Fig. 2c). These reactions did reveal, however, that PLA2G4E can catalyze the calcium-dependent formation of NAPEs using PE as the acyl chain donor (forming, in this instance, N-C18:1 NAPE; Supplementary Fig. 8b). Finally, a PLA2G4E variant in which the predicted catalytic serine nucleophile was mutated to alanine (S420A-PLA2G4E) lacked FP-rhodamine reactivity (Supplementary Fig. 9) and Ca-NAT activity (Fig. 2d).
Consistent with previous reports16, we found by RT-PCR analysis that PLA2G4E was most strongly expressed in mouse brain, heart, skeletal muscle, and testis (Supplementary Fig. 10a), and more highly expressed in neonatal versus adult brain (Supplementary Fig. 10b). This distribution generally matched that measured for Ca-NAT activity across the mouse tissues (Supplementary Fig. 10c,d), as well as that reported previously for rat Ca-NAT activity11,20. Among the tissues evaluated, only testis showed strong calcium-independent acyltransferase activity (Supplementary Fig. 10c), possibly reflecting the high expression of PLA-AT121 and PLA-AT522 in this organ. Similarly, previous ABPP23 and RNA-seq24 data indicated that PLA2G4E is expressed in mouse neurons, but not astrocytes or microglia (Supplementary Fig. 11a), and, consistent with these findings and other research12, we detected Ca-NAT activity only in cultured neurons, but not glia (Supplementary Fig. 11b).
Targeted lipidomic experiments revealed that PLA2G4E-transfected cells generated large quantities of NAPEs and the downstream metabolites glycerophospho (GP)-NAEs and NAEs, including anandamide, compared to either mock-transfected cells (Supplementary Fig. 12a) or cells transfected with the inactive S420A-PLA2G4E mutant (Supplementary Fig. 12b). Treating cells with the calcium ionophore ionomycin caused a further modest, but significant increase in PLA2G4E-dependent NAPE and NAE production (Supplementary Fig. 13a). We hypothesized that NAPEs and NAEs may have accumulated over multi-day transfection period in PLA2G4E-expressing cells and therefore obscured the measurement of ionomycin-induced production of these lipids. We addressed this question by performing metabolic labeling experiments, where the Ca2+-dependent formation of 13C-isotopically labeled NAPEs, GP-NAEs, and NAEs was measured in WT-versus S420A-PLA2G4E-transfected cells 4 h after simultaneous treatment with ionomycin (or DMSO control) and 13C16-palmitic acid. These experiments revealed a striking ionomycin-induced production of 13C-NAPEs, GP-NAEs, and NAEs in WT-, but not S420A-PLA2G4E- or mock-transfected cells (Fig. 2e). Control experiments confirmed that 13C-NAPEs, GP-NAEs, and NAEs were not observed in PLA2G4E-transfected cells without added 13C16-palmitic acid (Supplementary Fig. 13b).
Our findings, taken together, provide strong evidence that PLA2G4E represents the elusive Ca-NAT first described several decades ago from mammalian tissues. One of the most striking features of PLA2G4E is its strong preference for performing N-acyltransferase reactions over phospholipid hydrolysis. How PLA2G4E functions as a transacylase using a serine hydrolase mechanism and avoids water-mediated hydrolysis of the presumed acyl-enzyme intermediate in the absence of co-substrate is unclear. Future structural studies of PLA2G4E may shed light on this unusual property, as well as how calcium activates the enzyme. Considering we found that calcium stimulated both the acyltransferase activity and FP reactivity of PLA2G4E, we speculate that calcium binding, presumably to the C2 domain of the enzyme, may optimize the conformation of the catalytic domain to promote binding to substrates/probes and/or enhance the nucleophilicity of the active site catalytic serine Ser420.
Cells expressing PLA2G4E, but not the inactive S420A mutant, showed massive accumulation of NAPEs and NAEs, and these lipids were further elevated by ionomycin treatment. Considering the near-complete dependence of recombinant PLA2G4E on calcium for catalytic activity in vitro, it is not yet clear whether the large basal increases in NAPEs observed in PLA2G4E-transfected cells reflect the activation of PLA2G4E by physiological calcium or an ability of the enzyme to generate NAPEs in situ through other mechanisms.
Projecting forward, the discovery of PLA2G4E as a brain Ca-NAT provides a potentially key molecular linchpin for deciphering the functions of NAPEs and NAEs, including the endocannabinoid anandamide, in vivo. Mouse models in which PLA2G4E is inactivated by targeted gene disruption, as well as the development of PLA2G4E-selective inhibitors, should provide genetic and pharmacological systems to assess the contribution of PLA2G4E to endocannabinoid/NAE signaling in the brain, as well as to potential endocannabinoid-independent functions of NAPEs4. That mammals also possess a constitutive pathway (PLA/ATs) for producing NAPEs and NAEs13, some of which share tissue distribution with PLA2G4E21, suggests PLA2G4E may regulate a specific and dynamic pool of these lipids coupled to signaling events that mobilize intracellular calcium in vivo. The disruption of this PLA2G4E-regulated pathway may in turn provide a means to suppress aberrant forms of endocannabinoid signaling implicated in human disorders like addiction, obesity, and neurodegeneration1,2.
Online Methods
Materials
Unless otherwise noted, all chemicals and solvents were purchased from Sigma or Fisher Scientific. All lipids were purchased from Avanti Polar Lipids, except 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and anandamide-d4, which were obtained from Cayman Chemicals. 13C16-palmitic acid was purchased from Cambridge Isotope Libraries. 1,2-Dioleoyl-sn-glycero-3-phospho (N-nonadecenoyl) ethanolamine and 1,2-dihydroxy-sn-glycero-3-phospho (N-pentadecenoyl) ethanolamine were synthesized as previously described25, 26 Horseradish peroxidase-linked anti-mouse IgG was obtained from BioRad. SuperSignal West Pico chemiluminescent substrate was purchased from Thermo Fisher.
Enzyme preparation from mouse tissues
Adult male (10-week-old, n = 4) and neonatal (1-day-old, n = 4) C57BL/6J mice were anesthetized with isoflurane and killed by cervical dislocation. Frozen brains and other tissues were homogenized in 5 volumes (v/w) of 50 mM Tris-HCl (pH 8.0) containing 320 mM sucrose using a Dounce homogenizer. The extract was centrifuged at 100,000xg for 15 min. The supernatant obtained was used as a cytosolic fraction. The pellet was washed with 50 mM Tris-HCl (pH 8.0) containing 1 M NaCl, and centrifuged again. The resultant pellet was resuspended in 50 mM Tris-HCl (pH 8.0) and used as the membrane fraction. For the purpose of partial purification, the pellet was suspended in 50 mM Tris-HCl (pH 8.0) containing 0.5% IGEPAL CA-630 and centrifuged again at 100,000xg for 15 min. The clear supernatant was used for the sucrose density gradient centrifugation.
All mouse studies were performed following protocols that received approval from The Scripps Research Institute-Institutional Animal Care and Use Committee Office.
Sucrose density gradient centrifugation
1 mL of the membrane solubilized fraction (~5 mg) was loaded onto a 11 mL preformed 5-40% sucrose gradient in 50 mM Tris-HCl (pH 8.0), 2 mM DTT and 3 mM CaCl2 and centrifuged at 38,000xg for 16 h at 4ºC in an SW 40Ti rotor. After centrifugation, fractions (1 mL each) were collected from the top of the tube. The bottom of the tube was washed with 1 mL of 50 mM Tris-HCl (pH 8.0). Aliquots of each fraction were reacted with 1 µM FP-rhodamine and separated by SDS-PAGE, and visualized by gel fluorescence scanner (Bio-Rad).
ABPP sample preparation and protein identification and quantification
For the ABPP samples, 0.5 mL of each sucrose density gradient fraction (fractions 2-13) were labeled with FP-biotin (5 µM) in 50 mM TrisHCl (pH 8.0) containing 2 mM DTT and 3 mM CaCl2 for 30 min at room temperature. After labeling, the fractions were precipitated with methanol/chloroform precipitation, then resuspended in 50 mM Tris-HCl (pH 8.0) containing 6 M urea, 2% SDS and 10 mM DTT, followed by alkylation with 20 mM iodoacetamide. The biotinylated proteins were enriched with avidin agarose beads (20 µL; Sigma-Aldrich) by rotating at room temperature for 1.5 h in PBS with 0.2% SDS. The beads were washed sequentially with PBS containing 0.2% SDS. On-bead digestion was performed using trypsin (500 ng; Promega) in 2 M urea in 50 mM Tris-HCl (pH 8.0). Resulting peptides were desalted with StageTips and dissolved with 10 µL of 5% formic acid. An aliquot of the peptide mixtures (4 µL) was injected into the LC-MS/MS system consisting of LTQ-Orbitrap Velos coupled to Agilent 1200 HPLC (Agilent). The peptides were eluted from the in-house packed tip column (150 mm × 100 µm I.D., 5 µM Gemini C18; Phenomenex) with a linear gradient of acetonitrile in 0.1% formic acid at a flow rate of 300 nL/min. Each full-scan mass spectrum (350-2000 m/z) was followed by top 20 data-dependent MS/MS scans. Dynamic exclusion was used with an exclusion list of 500, repeat time of 20 s.
MS data analysis
Raw files were extracted with RawConverter and MS/MS spectra searched with ProLuCID against the mouse UniProt database downloaded November 9, 2012 using target-decoy approach in which each protein sequence was reversed and concatenated to the normal database. Search parameters were set to 50 ppm precursor mass tolerance, carbamidomethylation of cysteine residue (+57.0215 Da) as a static modification and oxidation of methionine (+15.9949 Da) as a variable modification. Search results were filtered with DTASelect version 2.0.47, allowing for tryptic peptides only and a peptide false discovery rate of less than 1%.
Proteomic correlation profiling
All identified proteins were quantified by spectral counting. Pearson correlation coefficient was calculated between total spectral counts for each protein and Ca-NAT activity.
Enzyme assays
For the Ca-NAT assay, an aliquot of sample was incubated at 37 ºC for 30 min or 1 hour with 250 µM DPPC and 250 µM DOPE for mouse tissue samples and the sucrose density gradient fractions or 40 µM DPPC and 75 µM DOPE for recombinant proteins in 40 µL of 50 mM TrisHCl (pH 8.0), 2 mM DTT and 0.1% IGEPAL CA-630 with or without 3 mM CaCl2. Ca-independent activities were subtracted in the calculations of Ca-dependent activity in the sucrose density gradient fractions. For assays directly comparing the calcium and divalent cation sensitivity of brain and recombinant protein, all samples were incubated at 37 ºC for 1 hour with 250 µM DPPC and 250 µM DOPE in 40 µL of 50 mM TrisHCl (pH 8.0), 2 mM DTT and 0.1% IGEPAL CA-630. For the assay comparing Ca-NAT and PLA activity, an aliquot of lysate was incubated at 37 ºC for 30 min with 40 µM 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine (di17:0 PC) with or without 75 µM DOPE in 40 µL of 50 mM TrisHCl (pH 8.0), 2 mM DTT, and 0.1% IGEPAL CA-630 with or without 3 mM CaCl2. Incubations were stopped by adding 150 µL of methanol/chloroform (2:1) containing 10 pmol N-C19:1 DOPE as an internal standard (and 10 pmol C19:1 FA for PLA activity measurements), followed by the addition of 50 µL of chloroform and 50 µL of 0.9% KCl (2:2:1.8). After centrifugation, 100 µL of lower organic phase was mixed with 50 µL of methanol. The resulting sample was injected in the LC-MS system consisting of a 1200 LC/MSD (Agilent Technologies). The analyte was separated on an analytical column, a 50 × 4.6 mm 5 µm Gemini C18 column (Phenomenex) coupled to a guard column (Gemini: C18: 4 × 3 mm). The LC method consisted of 0.1 mL/min of 90% buffer A [95:5 (v/v) H2O:MeOH plus 0.1% (v/v) of 28% ammonium hydroxide] for 0.5 min, 0.4 mL/min isocratic mode of 100% buffer B [60:35:5 (v/v/v) iPrOH:MeOH:H2O plus 0.1% (v/v) of 28% ammonium hydroxide] for 6.5 min, and equilibration with 0.5 mL/min 90% buffer A for 2 min (9 min total run time). The analyte was detected with the electrospray ionization (ESI) source operated in negative ion mode. MS data were acquired in selected ion monitoring mode at m/z 980.8 and m/z 1020.8 for N-C16:0 DOPE and N-C19:1 DOPE, respectively, for Ca-NAT activity measurements. For assays comparing Ca-NAT and PLA activity, MS data were acquired in selected ion monitoring mode at m/z 269.3 and m/z 994.8 for 17:0 free fatty acid and N-C17:0 DOPE, respectively.
RT-PCR
Total RNA was isolated from various organs of C57BL/6J mice using RiboZol (Amresco). cDNAs were then prepared from total RNA (1 µg) using ThermoScript RT (Invitrogen) and oligo dT primer and subjected to PCR amplification by Phusion DNA polymerase (New England Biolab). The primers used for PLA2G4E mRNA were 5’-ATTCTGCCTGCTTCCACTAC-3’ and 5’-GCCATACAAGGAGACCATAGAC-3’ (nucleotides 794-814 and 1197-1219, respectively, GenBank accession number NM_177845), and those for mouse glyceraldehyde-3-phosphate dehydrogenase mRNA were 5’- AGGTCGGTGTGAACGGATTTG-3’ and 5’- TGTAGACCATGTAGTTGAGGTCA-3’ (nucleotides 100-120 and 200-222, respectively, in NM_001289726). The PCR conditions used were as follows: for PLA2G4E, denaturation at 98 °C for 10 s, annealing at 61 °C for 20 s, and extension at 72 °C for 15 s (35 cycles); and for glyceraldehyde-3-phosphate dehydrogenase, denaturation at 98 °C for 10 s, annealing at 58 °C for 20 s, and extension at 72 °C for 10 s (25 cycles).
Construction of expression vectors
Full-length mouse PLA2G4E was amplified by PCR from mouse heart cDNA with primers PLA2G4E forward 5’-AAAACTCGAGCCATGCAGTCTATTCCACACTCCG-3’ and reverse 5’-AAAAGCGGCCGCGGAGGGACACTGGCTCCTCATG-3’ and was cloned into the pRK5 vector with C-terminal FLAG tag using Sal I and Not I sites. The mouse PLA2G4D expression vector was purchased from Origene (Cat. No. MR220078).
Expression of recombinant proteins in HEK293T cells
HEK293T cells (ATCC CRL-3216) were grown at 37 ºC to 70-80% confluency in Dulbecco’s Modified Eagle’s medium containing 10% fetal bovine serum and transfected with plasmid DNA using polyethylenimine (Polysciences). After 48 h, the cells were washed with PBS, scraped, resuspended in lysis buffer (50 mM TrisHCl pH 8.0, 0.5% IGEPAL CA-630), and sonicated. The lysates were spun at 100,000xg for 15 min to remove insoluble proteins.
Western blotting
Cell proteomes were separated by SDS-PAGE, transferred to nitrocellulose membrane (60 V for 90 min), and blocked by 5% milk in TBS-Tween. An anti-FLAG M2 antibody (Mouse, Sigma-Aldrich, F1804, 1:5,000) was used as the primary antibody. An anti-mouse IgG (H+L) horseradish peroxide conjugate (Goat, Bio-Rad, 170-6516, 1:20,000) was used as the secondary antibody, and signal was visualized using the Pierce ECL Western Blotting substrate (Thermo Scientific).
Targeted metabolite profiling
For studies with ionomycin, HEK293T cells were transfected with plasmid DNA as described above. After 48 h, fresh serum-containing media with or without 2 µM ionomycin was added to the cells. The cells were then incubated at 37 ºC for either 30 min or 4 h prior to lipid extraction. For feeding studies with isotopically labeled fatty acid, 250 µM of 13C16-palmitic acid (Cambridge Isotope Libraries) was additionally added to the media so that both the acid and ionomycin were simultaneously presented to the cells.
Targeted analyses of NAPEs, NAEs, and GP-NAEs were performed generally as described.27 Lipids were extracted from cells by slight modifications to the Bligh and Dyer method. In brief, cells were suspended in 760 µL of a mixture of methanol/chloroform/0.9% KCl (2:1:0.8 v/v/v), containing 100 pmol each 1,2-Dioleoyl-sn-glycero-3-phospho (N-nonadecenoyl) ethanolamine, 1,2-dihydroxy-sn-glycero-3-phospho (N-pentadecenoyl) ethanolamine and anandamide-d4 as internal standards, and sonicated. After standing for 10 min on ice, the mixture was centrifuged at 15,000xg for 1 min. The supernatant was withdrawn and the resultant pellet was mixed with 380 µL of methanol/chloroform/0.9% KCl (2:1:0.8), sonicated and centrifuged. Supernatants were combined, and 300 µL each of chloroform and 0.9% KCl was added to the sample to produce phase separation. After centrifugation of the mixture, the organic lower phase was withdrawn. 12 uL of formic acid was added to the upper layer to acidify the aqueous phase and mixed with 600 µL chloroform, and the mixture was centrifuged. Both the organic extracts were pooled and dried under a stream of N2. The metabolites were resolubilized in 100 µL of chloroform/methanol (2:1 v/v), and 10 µL were used for the targeted LC-MS/MS analysis.
Metabolite analysis in this study was performed by multiple reaction monitoring (MRM) methods on an Agilent Technologies 6460 Triple Quadrupole mass spectrometer. The solvents and LC separation conditions were the same as those described above for the enzyme assay. To assist in ion formation, 0.1% (v/v) of 28% ammonium hydroxide or 10 mM ammonium formate was added to the buffers for negative or positive mode, respectively. The LC method consisted of 0.1 mL/min 0% buffer B for 1 min, 0.4 mL/min linear gradient over 5 min to 100% buffer B, 0.4 mL/min 100% buffer B for 4 min, and 0.5 mL/min equilibration with 0% buffer B for 3 min. For NAPEs and pNAPEs, MRM transition from [M-H]− to the fragment ion [R2COO]− with collision energy 45 V was used. For GP-NAEs, MRM transition from [M-H]− to the fragment ion at m/z 171 with collision energy 24 V was used. For NAEs, MRM transition from [M+H]+ to m/z 62 with collision energy 11 V was used. Lipid species were quantified by measuring areas under the curve in comparison to the appropriate unnatural standard and then normalizing to the protein amount.
Statistical analyses
Statistical analyses were performed using the GraphPad Prism 6 software. All data are shown as mean values ± standard deviation. Student’s t-test (two-tailed) was used to measure statistically significant differences between study groups. A p value of <0.05 was considered statistically significant for this study.
Supplementary Material
Acknowledgments
We are grateful to H. Y. Lee for assistance with enzyme assays and G. Simon for numerous helpful discussions and critical reading of the manuscript. This work was supported by the NIH (DA033760), a Hewitt Foundation for Medical Research Fellowship (to W.H.P.) and the 9th Irving S. Sigal Postdoctoral Fellowship from the American Chemical Society (to S.S.K.).
Footnotes
Author Contributions
Y.O. and B.F.C. conceived the project. Y.O, W.H.P., S.S.K., and B.F.C. designed experiments. Y.O. characterized Ca-NAT activity in brain and performed the correlation profiling. Y.O., W.H.P., and S.S.K. characterized the activity of heterologously expressed PLA2G4E and performed the feeding studies. Y.O. and B.F.C. wrote the paper. W.H.P. edited the paper.
Competing Financial Interests
The authors report no competing financial interests.
References
- 1.Petrosino S, Ligresti A, Di Marzo V. Endocannabinoid chemical biology: a tool for the development of novel therapies. Curr. Opin. Chem. Biol. 2009;13:309–320. doi: 10.1016/j.cbpa.2009.04.616. [DOI] [PubMed] [Google Scholar]
- 2.Rahman IA, Tsuboi K, Uyama T, Ueda N. New players in the fatty acyl ethanolamide metabolism. Pharmacol. Res. 2014;86:1–10. doi: 10.1016/j.phrs.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Blankman JL, Cravatt BF. Chemical probes of endocannabinoid metabolism. Pharmacol. Rev. 2013;65:849–871. doi: 10.1124/pr.112.006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellner N, Diep TA, Janfelt C, Hansen HS. N-acylation of phosphatidylethanolamine and its biological functions in mammals. Biochim. Biophys. Acta. 2013;1831:652–662. doi: 10.1016/j.bbalip.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto Y, Morishita J, Tsuboi K, Tonai T, Ueda N. Molecular characterization of a phospholipase D generating anandamide and its congeners. J. Biol. Chem. 2004;279:5298–5305. doi: 10.1074/jbc.M306642200. [DOI] [PubMed] [Google Scholar]
- 6.Simon GM, Cravatt BF. Characterization of mice lacking candidate N-acyl ethanolamine biosynthetic enzymes provides evidence for multiple pathways that contribute to endocannabinoid production in vivo. Mol. Biosyst. 2010;6:1411–1418. doi: 10.1039/c000237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun YX, et al. Biosynthesis of anandamide and N-palmitoylethanolamine by sequential actions of phospholipase A2 and lysophospholipase D. Biochem. J. 2004;380:749–756. doi: 10.1042/BJ20040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, et al. A biosynthetic pathway for anandamide. Proc. Natl. Acad. Sci. USA. 2006;103:13345–13350. doi: 10.1073/pnas.0601832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy PV, Natarajan V, Schmid PC, Schmid HH. N-Acylation of dog heart ethanolamine phospholipids by transacylase activity. Biochim. Biophys. Acta. 1983;750:472–480. doi: 10.1016/0005-2760(83)90187-x. [DOI] [PubMed] [Google Scholar]
- 10.Natarajan V, Schmid PC, Reddy PV, Zuzarte-Augustin ML, Schmid HHO. Biosynthesis of N-acylethanolamine phospholipids in dog brain preparations. J. Neurochem. 1983;41:1303–1312. doi: 10.1111/j.1471-4159.1983.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 11.Cadas H, di Tomaso E, Piomelli D. Occurence and biosynthesis of endogenous cannabinoid precursor, N-arachidonyl phosphatidylethanolamine, in rat brain. J. Neurosci. 1997;17:1226–1242. doi: 10.1523/JNEUROSCI.17-04-01226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadas H, Gaillet S, Beltramo M, Venance L, Piomelli D. Biosynthesis of an endogenous cannabinoid precursor in neurons and its control by calcium and cAMP. J. Neurosci. 1996;16:3934–3942. doi: 10.1523/JNEUROSCI.16-12-03934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uyama T, et al. Generation of N-acylphosphatidylethanolamine by members of the phospholipase A/acyltransferase (PLA/AT) family. J. Biol. Chem. 2012;287:31905–31919. doi: 10.1074/jbc.M112.368712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piper DE, et al. The high-resolution crystal structure of human LCAT. J. Lipid Res. 2015;56:1711–1719. doi: 10.1194/jlr.M059873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niphakis MJ, Cravatt BF. Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem. 2014;83:341–377. doi: 10.1146/annurev-biochem-060713-035708. [DOI] [PubMed] [Google Scholar]
- 16.Ohto T, Uozumi N, Hirabayashi T, Shimizu T. Identification of novel cytosolic phospholipase A(2)s, murine cPLA(2)δ, ε, and ζ, which form a gene cluster with cPLA(2)β. J. Biol. Chem. 2005;280:24576–24583. doi: 10.1074/jbc.M413711200. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh M, Tucker DE, Burchett SA, Leslie CC. Properties of the Group IV phospholipase A2 family. Prog. Lipid Res. 2006;45:487–510. doi: 10.1016/j.plipres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Ghomashchi F, et al. Interfacial kinetic and binding properties of mammalian group IVB phospholipase A2 (cPLA2β) and comparison with the other cPLA2 isoforms. J. Biol. Chem. 2010;285:36100–36111. doi: 10.1074/jbc.M110.165647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natarajan V, Schmid PC, Schmid HHO. N-Acylethanolamine phospholipid metabolism in normal and ischemic rat brain. Biochem. Biophys. Acta. 1986;878:32–41. doi: 10.1016/0005-2760(86)90341-3. [DOI] [PubMed] [Google Scholar]
- 20.Moesgaard B, Petersen G, Jaroszewski JW, Hansen HS. Age dependent accumulation of N-acyl-ethanolamine phospholipids in ischemic rat brain: a 31P NMR and enzyme activity study. J. Lipid Res. 2000;41:985–990. [PubMed] [Google Scholar]
- 21.Shinohara N, et al. Enzymological analysis of the tumor suppressor A-C1 reveals a novel group of phospholipid-metabolizing enzymes. J. Lipid Res. 2011;52:1927–1935. doi: 10.1194/jlr.M015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin XH, et al. Discovery and characterization of a Ca2+-independent phosphatidylethanolamine N-acyltransferase generating the anandamide precursor and its congeners. J. Biol. Chem. 2007;282:3614–3623. doi: 10.1074/jbc.M606369200. [DOI] [PubMed] [Google Scholar]
- 23.Viader A, et al. A chemical proteomic atlas of brain serine hydrolases identifies cell type-specific pathways regulating neuroinflammation. eLife. 2016;5 doi: 10.7554/eLife.12345. doi: 10.7554/eLife.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon GM, Cravatt BF. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway. J. Biol. Chem. 2006;281:26465–26472. doi: 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- 26.Simon GM, Cravatt BF. Anandamide biosynthesis catalyzed by the phosphodiesterase GDE1 and detection of glycerophospho-N-acyl ethanolamine precursors in mouse brain. J. Biol. Chem. 2008;283:9341–9349. doi: 10.1074/jbc.M707807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HC, Simon GM, Cravatt BF. ABHD4 regulates multiple classes of N-acyl phospholipids in the mammalian central nervous system. Biochemistry. 2015;54:2539–2549. doi: 10.1021/acs.biochem.5b00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.