Abstract
The field of small self-cleaving nucleolytic ribozymes has been invigorated by the recent discovery of the twister, twister-sister, pistol and hatchet ribozymes. We report on the crystal structure of the env25 pistol ribozyme, which adopts a compact tertiary architecture stabilized by an embedded pseudoknot fold. The G-U cleavage site adopts a splayed-apart conformation with in-line alignment of the modeled 2′-O of G for attack on the adjacent to-be-cleaved P-O5′ bond. Highly conserved residues G40 (N1 position) and A32 (N3 and 2′-OH positions) are aligned to act as general base and general acid respectively to accelerate cleavage chemistry, with their roles confirmed from cleavage assays on mutants, and an increased pKa of 4.7 for A32. Our structure of the pistol ribozyme defines how the overall and local topologies dictate the in-line alignment at the G-U cleavage site, with cleavage assays on mutants identifying key residues participating in acid-base catalyzed cleavage chemistry.
Introduction
Small self-cleaving ribozymes are widely distributed in nature1,2 and are essential for rolling-circle-based replication of satellite and pathogenic RNAs3,4 and processing of repetitive RNA species5. Members of this nucleolytic ribozyme class include hammerhead6,7, hairpin8,9, glmS10, hepatitis delta11,12 and Varkud satellite5,13 ribozymes. that mediate cleavage chemistry through nucleobase-specific acid-base catalysis. X-ray structural studies on the hammerhead14, hairpin15,16, glmS17,18, HDV19,20 and Varkud satellite21 ribozymes have defined the role of hydrogen bond networks, ionic and hydrophobic interactions for generation of catalytic pockets, which capitalize on steric constraints to generate in-line cleavage alignments involving attack of the 2′-OH on the adjacent 3′-phosphate and general acid-base chemistry to catalyze site-specific cleavage of the phosphodiester backbone1,22-25. Key challenges relate to which RNA residues contribute to ribozyme-mediated rate enhancement through involvement in nucleophilic activation of the 2′-OH and protonation of the P-O5′ oxygen at the cleavage step, as well as what factors contribute to stabilization of the transition state. Additional insights have emerged into how the transition state (vanadate mimics) can be preferentially accommodated within the catalytic pocket, though there remains uncertainty regarding the role of divalent cations in mediating cleavage chemistry.
The recent discovery of twister, twister sister, pistol and hatchet self-cleaving ribozymes by comparative genomic analysis2,26 has opened a unique opportunity to undertake structure-function studies to comparatively assess the architectural diversity, catalytic cores and mechanism of action of these four small self-cleaving ribozymes. To this end, recent chemical26, structural27-29, biochemical26,30 and computational31 studies have provided insights into the topological constraints contributing to catalysis by the twister ribozyme.
A recent study has reported on a small self-cleaving ribozyme composed of three stems, a hairpin and an internal loop, named the pistol ribozyme, shown schematically in Fig. 1a2,32. Highly conserved nucleotides (shown in red in Fig. 1a) are primarily located in the internal loop and the linker segment (AAA trinucleotide), while segments of the hairpin and internal loops are predicted to be involved in 5-base pair pseudoknot formation2. Cleavage occurs at a modestly conserved G-U dinucleotide junctional site (see arrow in Fig. 1a), whose length is strictly conserved. RNA cleavage was shown to be dependent on Mg2+ and the cleavage rate increased with increasing pH. Studies using phosphorothioate substitution at the cleavage site suggest that the pistol ribozyme exploits a contact involving one of two non-bridging phosphate oxygens to facilitate catalysis. RNA strand scission occurs with a rate constant of 10 min-1 under physiological pH and Mg2+ conditions32.
Figure 1. Secondary and tertiary structure of the env25 pistol ribozyme.
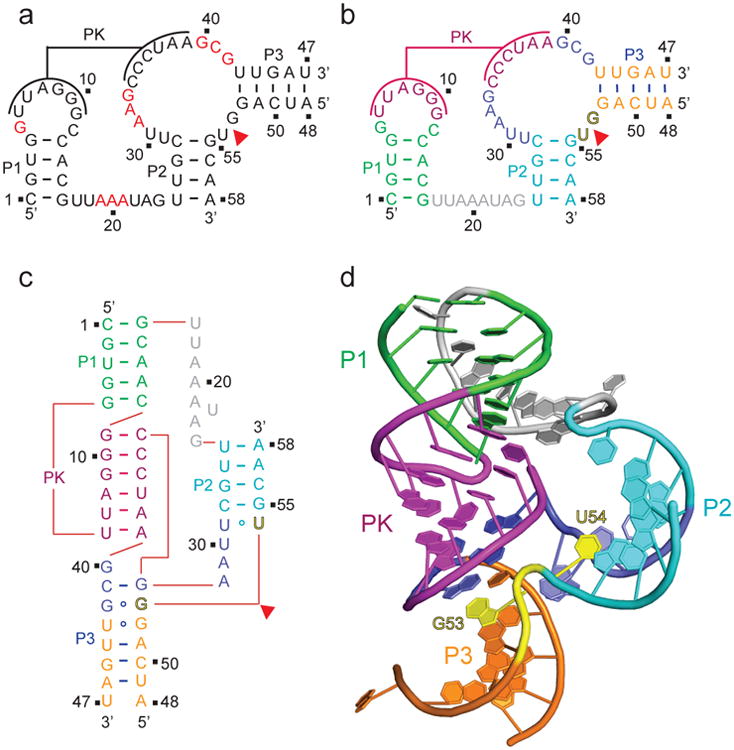
a, Secondary structure of the env25 pistol ribozyme showing highly conserved residues in red as adapted from ref. 2. PK stands for proposed pseudoknot. In this two-strand construct, the longer ribozyme strand is labeled from 1 to 47 and the shorter substrate strand that contains the G53-U54 cleavage site is labeled from 48 to 58. b, A color-coded representation of the secondary structure of the env25 pistol ribozyme. c, A schematic of the folding topology based on the crystal structure of the env25 pistol ribozyme (see panel d). d, The 2.7 Å structure of the pistol ribozyme. The color code is the same as in panel b, with the G53-U54 cleavage site colored in yellow.
There are currently no indications of either the tertiary fold of the pistol ribozyme, or which residues line the catalytic pocket, to facilitate cleavage at the G-U site within the short arm of the internal loop (Fig. 1a). Of particular interest is whether the conserved AAA trinucleotide has the potential to form stacked A-minor triples with the minor groove of a duplex segment. Metal ion dependent studies have been interpreted to conclude that the pistol ribozyme does not require inner-sphere coordination of divalent cation for catalysis32.
This paper reports on a structure-function study of the env25 pistol ribozyme. The structure defines a splayed-out orientation and in-line alignment of the internucleotide linkage at the G-U cleavage site, while also defining the relative alignment of nearby nucleotides and divalent cations with the potential for contributing to the catalytic mechanism. We have monitored the cleavage propensity of structure-guided mutants of the pistol ribozyme to deduce insights into the principles underlying site-specific cleavage chemistry. In addition, we have capitalized on NMR site-specific labeling approaches to measure the pKa of an adenine lining the catalytic pocket, while fluorescence approaches have been applied to measure precise cleavage rates as a function of temperature. Finally, we have compared our structure of the env25 pistol ribozyme with a recent structure of the RzB hammerhead ribozyme.
Results
The ten highly conserved (>97% invariant) residues in the pistol ribozyme secondary structure are highlighted in red in Fig. 1a2. Our efforts have focused on the pistol ribozyme termed env25, whose sequence and color-coded secondary structure are shown in Fig.1b. We have generated the env25 pistol ribozyme by annealing two chemically synthesized strands, the shorter substrate strand is a 11-mer (residues 48-58) containing the G53-U54 cleavage site, while the longer ribozyme strand is a 47-mer (residues 1-47) as shown schematically in Fig. 1b. We synthetically incorporated dG53 in the substrate strand at the G-U site so as to prevent cleavage.
Structure of the pistol ribozyme
We have solved the crystal structure of the bimolecular construct of the dG53-containing env25 pistol ribozyme at 2.7 Å resolution. The tertiary structural fold is shown schematically in Fig. 1c and in a ribbon representation in Fig. 1d (x-ray statistics listed in Supplementary Results, Supplementary Table 1). The crystals belong to space group P3221, with one molecule in each asymmetric unit.
We observe formation of a six-base pair pseudoknot involving complementary loop segments between the hairpin and internal loops, as predicted previously2, with the pseudoknot duplex positioned between stems P1 and P3 (Fig.1d). There is continuous stacking of stem P3 (in orange), a small segment involving non-canonical pairing interactions (in blue), pseudoknot (PK)stem (in magenta) and stem P1 (in green) (Fig. 1d and Supplementary Fig. 1a). Stem P2 (in cyan) folds back and is positioned opposite the pseudoknot stem, resulting in an overall compact fold, containing the splayed apart G53-U54 (in yellow) site (Fig. 1d)
Pairing alignment of conserved residues
The highly conserved A31-A32-G33 and G40-C41-G42 segments (Fig. 1a) together with G53 at the cleavage site of the pistol ribozyme are brought into proximity (Fig. 2a) as a result of pairing alignments involving G42 (Fig. 2b) and G40•(G33-C41) triple formation (Fig. 2c). Notably, the G40-C41 step forms an in-plane platform (Fig. 2c), while G40, which adopts a C2′-endo sugar pucker, is involved in a major groove-aligned triple with the Watson-Crick G33-C41 pair (Fig. 2c), thereby anchoring it in place. The Watson-Crick edge of G42 is directed towards the sugar of A32 (Fig. 2b).
Figure 2. Structural details of the env25 pistol ribozyme in the vicinity of the G53-U54 cleavage site.
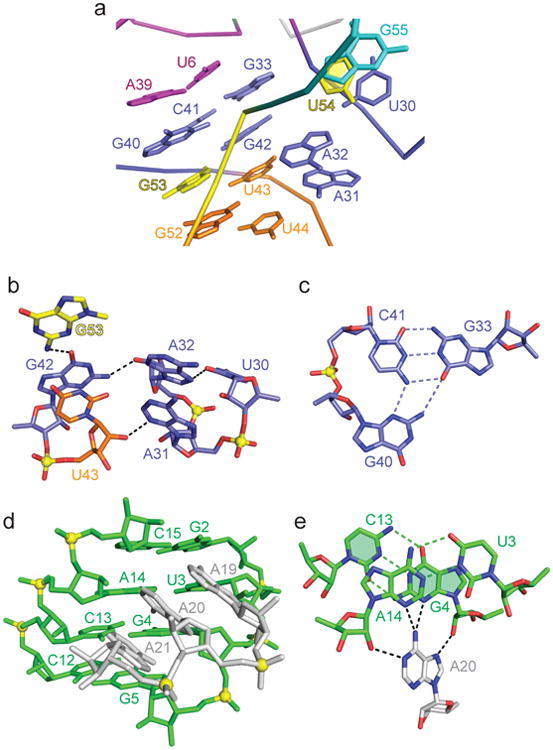
a, Representation of bases surrounding the G53-U54 cleavage site. The color code is the same as in Fig. 1b and 1c. b, Hydrogen bonding alignments involving the G42-U43, U20-A31-A32 and G53 segments. c, The major groove-aligned G40•(G33-C41) triple involving highly conserved residues G40 and C41. Note the platform at the G40-C41 step. d, The alignment of the highly conserved A19-A20-A21 segment within the minor groove of stem P1. The A-A-A segment is tilted relative to the base pairs of stem P1. e, Bifurcated hydrogen binds between the Hoogsteen edge of A20 and the minor groove of adjacent base pairs of stem P1.
The A19-A20-A21 segment is also highly conserved within pistol ribozyme sequences2 (Fig. 1a). The A19-A20-A21 segment is positioned in the minor groove of stem P1, where it forms consecutive A-minor triples (Supplementary Fig. 1b), with this stacked A-rich segment tilted with respect to the base pairs of stem P1 (Fig. 2d). Each adenine forms A-minor triples with adjacent base pairs of stem P1, as shown for A20 in Fig. 2e.
The remaining highly conserved residue is G5 positioned adjacent to stem P1 (Fig. 1a, b) in the pistol ribozyme. Stem P1 is extended through formation of a Watson-Crick G5-C12 base pair, with G5 anchored in place through formation of a minor groove triple involving the sugar edge of G24 and the G5-C12 pair (Supplementary Fig. 1c).
In-line alignment at G53-U54 cleavage site
The G53-U54 internucleotide linkage targeted for hydrolysis adopts an extended splayed out conformation (two alternate views in Fig. 3a). G53 is maximally stacked between G40 and G52 (Fig. 3a, top panel), while U54 is partially stacked between U30 and G55 (Fig. 3a, bottom panel). Both G53 and U54 adopt anti alignments at their glycosidic bonds with sugar puckers adopting C2′-endo at G53 and C3′-endo at U54.
Figure 3. In-line alignment at the G53-U54 cleavage site in the env25 pistol ribozyme and comparison with the structure of the RzB hammerhead ribozyme.
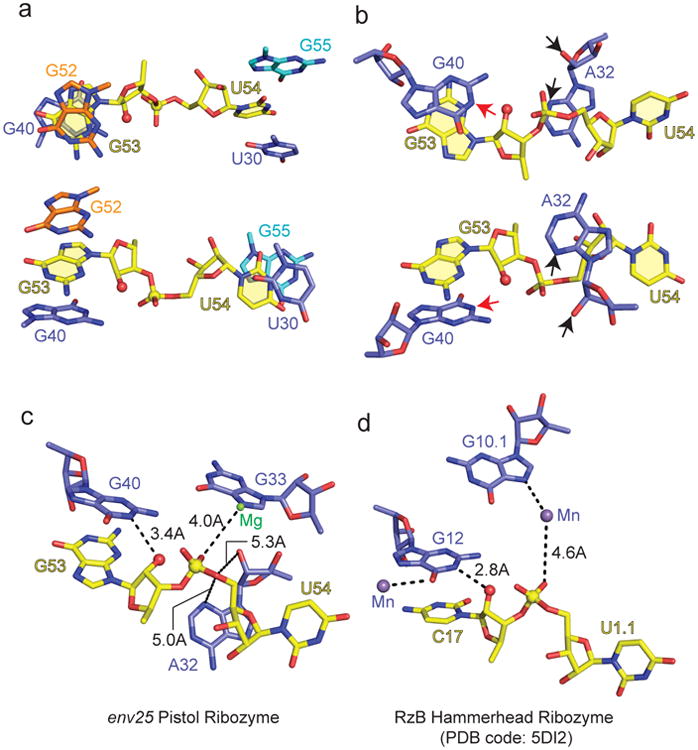
a, Base stacking with G53 (top panel) and U54 (bottom panel) at the G53-U54 cleavage site. The 2′-OH oxygen of G53 was modeled given that the pistol ribozyme construct contained dG53 to prevent cleavage. b, The positioning of G40 and A32 in the proximity of the G53-U54 cleavage site. The N1 of G53 is labeled by a red arrow, while the N3 and 2′-OH oxygen of A32 are labeled by black arrows. c, d, Cleavage sites of the env25 pistol ribozyme (panel c) and RzB hammerhead ribozyme (PDB code: 5DI2) (panel d) emphasizing the conformation of the dinucleotide step at the cleavage site, alignment of adjacent nucleotides and divalent cations, as well as critical distances.
Our structure of the pistol ribozyme contained a deoxy sugar (2′-H) at G53 (dG53), so as to prevent cleavage at the G53-U54 site by an active ribozyme. Incorporation of dG52 could possibly contribute to its C2′-endo sugar pucker, a characteristic of deoxy sugars. Following structure refinement, the 2′-H was replaced by 2′-OH at G53, to check for in-line alignment required for cleavage of the P-O5′ bond of the intervening phosphate at the G53-U54 site. The modeled 2′-OH structure exhibited a 2′-O (of G53) to P (at the G53-U54 site) distance of approximately 2.8 Å and a 2′-O (of G53) to P-O5′ angle (at G53-U54 site) of approximately 167° (Fig. 3b, two alternate views in top and bottom panels).
Bases and cations in vicinity of the cleavage site
Bases G40 and A32 are positioned close to the splayed apart G53-U54 cleavage site (Fig. 3b). Importantly, the separation between the N1 of G40 (red arrow) and the modeled nucleophilic 2′-OH oxygen of G53 (red ball) is approx. 3.4 Å. Notably, G40 is anchored in place through stacking (Fig. 3b) and also through formation of a base triple with the major groove of the G33-C41 pair (Fig. 2c). The bridging P-O5′ oxygen at the G53-U54 cleavage site is approximately 5.3 Å from the 2′-OH of A32 (black arrow) and approximately 5.0 Å from the N3 of A32 (black arrow) as shown in Fig. 3c. These distances can be compared with their counterparts in the pH 8.0 structures of the RzB hammerhead ribozyme (PDB code: 5DI2) (Fig. 3d).
We observe a bound hydrated Mg2+ ion in the vicinity of the G53-U54 cleavage site (Supplementary Fig. 1d), which was verified by soaking the crystal in Mn2+-containing solution and collecting an anomalous data set (Supplementary Fig. 1e). The hydrated Mg2+ cation is coordinated directly to the N3 of G33, but not directly coordinated to either non-bridging phosphate oxygens (Mg2+-O distances of 4.0 and 4.9 Å) of the G53-U54 site.
Mutations and cleavage activities
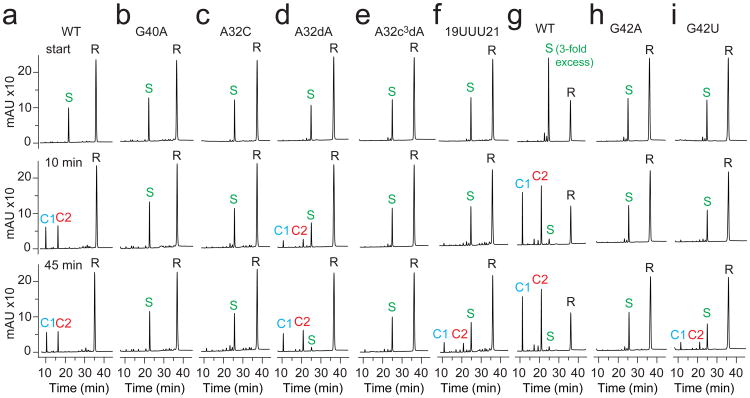
We performed cleavage assays of several single-mutation pistol variants using ion-exchange high-performance liquid chromatography (HPLC; Fig. 4). Under single-turnover conditions, the wild-type env25 pistol ribozyme cleaves its substrate to completeness in less than 10 min (Fig. 4a). As anticipated, mutations of G40 and A32 that are closest to the cleavage site had significant impact on the cleavage activities. Mutation of G40 to adenine completely abolished cleavage (Fig. 4b). It should be noted that replacement of G40 by A would also disrupt the G40•(G33-C41) base triple (Fig. 2c). When we mutated A32 to cytosine, cleavage was completely abolished (Fig. 4c). Presumably, replacement of the purine A by the smaller pyrimidine C alters the base heteroatom to O5′ separation,thereby impacting on cleavage chemistry. By contrast, the A32U mutated ribozyme had some activity with about 50% yield after 45 min (Supplementary Fig. 2c), while the A32G mutated ribozyme cleaved its substrate to almost 100% in less than 10 min (Supplementary Fig. 2d).
Figure 4. Self-cleavage of the env25 pistol ribozyme.
a, wild type (WT) and mutants b, G40A, c, A32C, d, A32dA, e, A32c3dA, and f, A19U-A20U-A21U. g, WT (3-fold excess substrate over ribozyme and mutants), i, G42A and j, G42U. Cleavage analyzed at 55 μM RNA each strand; 2 mM MgCl2, 100 mM KCl, 30 mM HEPES, pH 7.5, 23 °C. R 47-nt ribozyme, S 11-nt substrate; C1 and C2, 6-nt and 5-nt cleavage products. HPLC conditions: Dionex DNAPac colum (4 × 250 mm), 80 °C, 1 mL min-1, 0–60% buffer B in 45 min. Buffer A: Tris–HCl (25 mM), urea (6 M), pH 8.0. Buffer B: Tris–HCl (25 mM), urea (6 M), NaClO4 (0.5 M), pH 8.0.
To further test the role of A32 we investigated a A32dA mutant, with this mutant showing only about 25% cleavage yield after 10 min and about 90% after 45 min (Fig. 4d). Furthermore, to learn more about a potential role of the N3 in dA32, we investigated the corresponding 3-deaza nucleoside. No cleavage at all was observed for the A32c3dA mutant after 45 min (Fig. 4e).
We also replaced the conserved AAA trinucleotide within the linker segment by its UUU counterpart. No cleavage was observed after 10 min, with about 50% cleavage after 45 min (Fig. 4f). G42, which is positioned adjacent to the G53-U54 cleavage site plays a key role, since compared to wild-type control (Fig. 4g), mutation of G42 to A was deleterious (Fig. 4h). Mutation of G42 to U also severely decreased activity (Figure 4i).
NMR experiments – pKa of A32
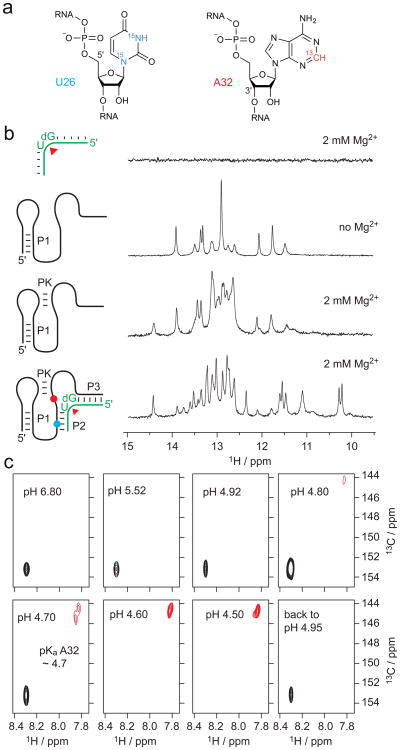
To better understand the participation of A32 in the mechanism of phosphodiester cleavage of pistol ribozymes, we set out for NMR spectroscopic determination of its pKa value, as we have recently shown for a twister ribozyme30. By solid-phase RNA synthesis, we prepared the same construct as used for X-ray structure determination, but with a site-specifically 15N-labeled uridine at position 26 and a 13C2-labeled adenine at position 32, respectively (Fig. 5a). While the 11-nt substrate strand displayed no 1H imino proton signals, the 47-nt ribozyme strand was already partly pre-folded under the no added Mg2+ buffer conditions used (Fig. 5b). Only in the presence of Mg2+, the pseudoknot fold was completed, according to the significantly increased number and distribution of resonances (Fig. 5b). When the substrate was added in the presence of Mg2, complex formation was confirmed by additional signals (Fig. 5b) and the appearance of a correlation in the 1H–15N HSQC spectrum of the 15N-labeled U26 probe (Supplementary Fig. 3a)that senses P2 duplex formation with the substrate because of reduced imino proton exchange with the solvent. The crosspeak was observed in the typical chemical shift range of A-U base pairs consistent with the formation of Watson Crick base pairs in the ribozyme's substrate binding arms (Supplementary Fig. 3b).
Figure 5. NMR spectroscopic analysis of the env25 pistol ribozyme in solution.
a, Chemical structures of 15N1/3-labeled U26 and 13C2-labeled A32 used. b, Complex formation verified by 1H NMR imino proton spectroscopy; spectra assigned to cartoons of RNA strands used. c, 1J(1H,13C) HSQC spectra of the 13C-A32 pistol complex at varying pH values. Conditions: c(RNA) = 80 to 120 μM; 25 mM NaCl, 2 mM MgCl2, 15 mM sodium phosphate, H2O/D2O 9:1, 298 K.
Next, we conducted pH-dependent NMR spectroscopic experiments with the 13C2-A32 labeled (Supplementary Fig. 4a), non-cleavable ribozyme complex (containing dG53) to determine the pKa of this adenine. In the 1J(1H,13C) HSQC spectra, we observed a very sharp, highly-defined pH dependence of the A32 C2-13C chemical shift with the transition between protonated and deprotonated forms (in slow exchange) in the pH 4.6 to 4.8 range, indicative of an estimated pKa value of 4.7 (Fig. 5c). By contrast, we have measured a pKa in the 3.6 to 3.85 range for A in the duplex (Supplementary Fig. 4b) and single strand (Supplementary Fig. 4c) context33. Conclusively, the pKa shifts by one pH unit towards neutrality for A32 in the pistol ribozyme.
Fluorescence assays: kinetics and conformational changes
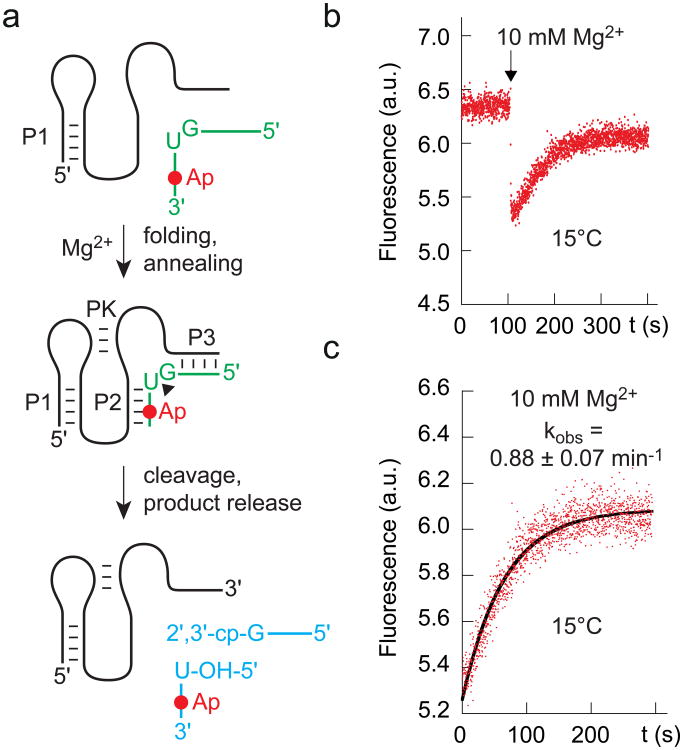
We utilized a fluorescence-based, real-time cleavage assay34,35 to monitor cleavage kinetics (Fig. 6). 2-Aminopurine (Ap) (Supplementary Fig. 5a) was integrated into a base pair of stem P2 (A57Ap). Ap becomes stacked (and hence fluorescence-quenched) when this stem forms through substrate binding upon addition of Mg2+. After cleavage has occurred, the 5-nt fragment (containing Ap) dissociates because of the rather weak remaining base pairing interactions (Fig. 6a). This experimental setup resulted in the expected immediate fluorescence decrease upon Mg2+ addition (folding and annealing), followed by an increase that was attributed to the release of the cleaved pentamer (Fig. 6b). The response followed single-exponential behavior, with the observed rate kobs of 0.88 ± 0.07 min-1 when measured at 15 °C (Fig. 6c). The value increased to 2.72 ± 0.38 min-1 when the temperature was raised to 20 °C (Supplementary Fig.5b).
Figure 6. Cleavage kinetics of the env25 pistol ribozyme analyzed by 2-aminopurine (Ap) fluorescence.
a, Schematics of the fluorescence assay that reports on folding and annealing, and cleavage product release. b, Qualitative assay: Fluorescence time course of the A57Ap variant upon MgCl2 addition (black arrow); conditions: cRNA = 0.5 μM, 50 mM KMOPS, 100 mM KCl, 15 °C, pH 7.5; mixing was performed manually in less than 2 s resulting in 10 mM Mg2+ concentration. c, Quantitative assay for rate determination at 15 °C based on data in panel b.
Importantly, cleavage in our assays was initiated by the addition of Mg2+ at saturating concentrations of 10 mM. This concentration guarantees that folding of the ribozyme into the catalytically active conformation occurs almost instantaneously [in less than 1–2 s corresponding to the immediate fluorescence decrease that is consistent with U26-Ap57 base pair formation (Fig.6b)].
Discussion
The pistol ribozyme is unusual both in terms of its small size and limited secondary structure (Fig. 1a) relative to other nucleolytic ribozymes. Thus, it remained to be discovered which features of its topology would facilitate site-specific self-cleavage at the G53-U54 site located within the short arm of the internal loop linking stems P2 and P3. We do observe formation of the predicted pseudoknot2, where it is embedded within the fold of the pistol ribozyme (Fig. 1d). The pseudoknot, together with its adjacent short non-canonical pairing segment, is positioned between flanking P1 and P3 stems (Fig. 1c and Supplementary Fig. 1a). Such coaxial stacking of helical segments (Supplementary Fig. 1a) is a common feature of higher-order RNA folds. Cleavage site residue G53 is part of this continuous helical stacking alignment (Fig. 1c and Supplementary Fig. 1a), while cleavage site residue U54 is displaced and stacks with G55 of outwardly directed stem P2 (Fig. 1d), thereby explaining the splayed-out alignment observed for the G53-U54 cleavage site.
A Mg2+ cation is bound proximal to the cleavage site within the fold of the pistol ribozyme (Supplementary Fig. 1d), but given that it is not directly coordinated to either non-bridging phosphate oxygens at the G53-U54 cleavage site, it most likely contributes to electrostatic stabilization rather than catalysis. This result contrasts with the observation of a Mg2+ bound directly to the non-bridging phosphate oxygen at the cleavage site in the twister ribozyme29,30.
The conserved residues in the pistol ribozyme2,32 are dispersed between G5 in the stem-loop, A19-A20-A21 in the linker segment and A31-A32-G33 and G40-C41-G42 on opposite sides of the internal loop. They play key stabilizing and catalytic roles in the structure of the pistol ribozyme. Both G5 (Supplementary Fig. 1c) and G40, as well as C41 (Fig. 2c) participate in base triple formation, with G5 involved in stabilizing the junction between the P1 and pseudoknot stems, while the G40 and C41 are involved in stabilizing the junction between the pseudoknot and P3 stems bridged by the short non-canonical paired segment of the internal loop. Notably, both A31-A32-G33 and G40-C41-G42 segments are positioned in the proximity of the G53-U54 cleavage site (Fig. 2a), thereby sculpting the arrangement of nucleotides that surround the cleavage pocket. In addition, the A19-A20-A21 segment is positioned within the minor groove of stem P1, where it forms stabilizing twisted A-minor triple interactions (Fig. 2d, e). Importantly, conserved G40 and A32 are positioned to play key roles in catalyzing the cleavage chemistry (see below).
In addition, G42 appears to be significant for proper positioning of G40 and A32 bases as a consequence of formation of a hydrogen bond between its 2-NH2 and A32 2′-O (2.7 Å), and additionally, between its O6 and G40 2-NH2 (2.8 Å) and G53 2-NH2 (2.7 Å) (Fig. 2b). This conclusion is supported by G42 mutation data (Fig. 4g, i).
We observe a splayed-apart arrangement at the G53-U54 cleavage site in the pistol ribozyme, with both G53 and U54 anchored in place by being sandwiched between flanking bases (Fig. 3a). Such extended splayed-apart alignments at the cleavage site has been reported previously for other nucleolytic ribozymes1. We observe a near in-line alignment (angle of 167°) at the G53-U54 cleavage site (Fig. 3b), similar to what we reported previously for the cleavage site in the twister ribozyme29. Such in-line alignments are a general feature critical for cleavage chemistry and have been reported earlier for the hammerhead14 and hairpin15,16 ribozymes.
The conserved G40 nucleotide (Fig. 1a) is positioned proximal to the G53-U54 cleavage site in the structure of the pistol ribozyme (Fig. 2a) and anchored and aligned in place though base triple formation (Fig. 2c). G40 is the closest nucleotide to the modeled 2′-OH of G53 at the G53-U54 cleavage site, with the separation between the N1 of G40 and the 2′-OH oxygen of G53 being 3.4 Å. Strikingly, we observed no cleavage for the G40A mutant (Fig. 4b), indicative of G40 being a strong candidate for the general base positioned to abstract the 2′-OH proton of G53, thereby impacting on acid-base catalysis at the G53-U54 site in the pistol ribozyme.
The conserved A32 nucleotide (Fig. 1a) is also positioned proximal to the G53-U54 cleavage site in the structure of the pistol ribozyme (Fig. 2a). A32 is anchored in place through hydrogen bond formation between its N7 atom and the base edge of U30 and between its 2′-OH and the 2-NH2 group of G42 (Fig. 2b). A32 is the closest nucleotide to the bridging P-O5′ oxygen at the G53-U54 cleavage site, with the separation between the P-O5′ oxygen and the N3 and 2′-OH of A32 in the 5.0 to 5.3 Å range. Our results indicate that A32 is a strong candidate for general acid during cleavage of the G53-U54 site, with contributions from both the base N3 atom and the ribose 2′-OH group.
Several lines of evidence support a role for N3 of A32 in the catalytic mechanism. Replacement of A32 by G had no impact on cleavage rate (Supplementary Fig. 2d), a result consistent with both purines having an N3 atom. By contrast, replacement by the smaller pyrimidines resulted either in no cleavage for the A32C mutation (Fig. 4c) or reduced cleavage for the A32U mutant (Supplementary Fig. 2c). Most importantly, replacement of N3 by C3 resulted in no cleavage for the A32c3dA mutant (Fig. 4e). In addition, the measured pKa of 4.7 for A32 in the pistol ribozyme (Fig. 5c) has shifted by one pKa unit compared to the pKa value of 3.6 for free adenosine36 and 3.6 to 3.8 at the single-strand and duplex level (Supplementary Fig.4). Notably, this shift in pKa is comparable to the pKa shift of 1.4 units reported previously for the adenine at the cleavage site of the twister ribozyme30, consistent with participation of partially protonated A32 in phosphodiester cleavage chemistry.
Similarly, support for participation of the 2′-OH of A32 in cleavage chemistry is cemented by the reduced cleavage activity for the A32dA mutant (Fig. 4d) relative to the wild-type (Fig. 4a) pistol ribozyme. Since no cleavage is observed for A32c3dA mutant (Fig. 4e) relative to partial cleavage observed for the A32dA mutant (Fig. 4d), it appears that the N3 base position of A32 makes a larger contribution to catalysis than does the 2′-OH in the pistol ribozyme.
Our real-time fluorescence cleavage assay revealed kinetics for the pistol ribozyme (Fig. 6) that are consistent with previously reported rates in the range of 1–10 min-1.2,32 Importantly, we show that temperature has a significant impact (Fig. 6c and Supplementary Fig. 5b), which likely correlates to multiple factors, including proper pseudoknot folding and substrate annealing, formation of the splayed out conformation of GpU, but also to release of the cleavage product. The obtained rates also compare well with the rates of other fast-cleaving ribozymes (e.g. twister ribozyme cleaves with 1.41 ± 0.16 min-1 at 15°C and 2.44 ± 0.31 min-1 at 20 °C under the same conditions)29. We further note that cleavage under single-turnover conditions proceeds in quantitative yield for env25 pistol ribozyme; even a 3-fold excess of substrate resulted in entire cleavage consistent with multiple turnover (Fig. 4g). This was not observed for the previously investigated env22 twister ribozyme29; single-turnover conditions gave at most 80% cleavage, likely because of a more stably paired cleavage fragment that caused product inhibition (14-nt fragment with 10 base pairs formed in twister as opposed to 6-nt fragment with 5 base pairs formed in pistol)29.
Recently, three crystal structures have been reported for the RzB hammerhead ribozyme, namely on crystals grown at pH 5.0 and 8.037 and on a transition state vanadate mimic38. These structural snapshots of the hammerhead ribozyme establish that the conformation centered about the cleavage site undergoes a substantial structural transition for crystals from pH 5.0 to 8.0, and a lesser transition for crystals from the pH 8.0 to the transition state vanadate mimic. These changes include repositioning of the putative general base (guanine) and a pair of divalent cations. We compare our structure of the env25 pistol ribozyme (Fig. 3c) with the pH 8.0 structure of the RzB hammerhead ribozyme (Fig. 3d) for the segments surrounding the cleavage site. For both ribozymes, the Watson-Crick edge of a guanine base is directed towards the to-be-abstracted 2′-OH proton, with the 2′-O positioned for in-line alignment relative to the to-be-cleaved P-O5′ bond (Fig. 3c, d). A divalent cation is coordinated to the O6 of the guanine (G12) in the hammerhead ribozyme (Fig. 3d), presumably with the potential to impact on its pKa. The situation is different between the two ribozymes for residues aligning with the 5′-O at the cleavage site. The pistol ribozyme aligns an adenine (A32) whose N3 and 2′-OH are positioned towards the 5′-O at the cleavage site (Fig. 3c), thereby contributing to putative general acid catalysis. A similar alignment of an adenine N3 relative to the O5′ oxygen was observed in the twister ribozyme28,29. In addition, both ribozymes contain a hydrated divalent cation that makes outer-sphere coordination to the pro-R non-bridging phosphate oxygen, while being directly coordinated to the N7 of a guanine base (Fig. 3c, d). It has been proposed that the divalent cation-coordinated water ligand could serve as a general acid mediating catalysis in the hammerhead ribozyme38. It should be noted that a divalent cation was inner-sphere coordinated to the non-bridging phosphate oxygen at the cleavage site in the twister ribozyme29. Further insights into the catalytic cleavage mechanism of nucleolytic ribozymes should emerge following comparison of transition state vanadate mimics of the pistol (not yet available) and hammerhead38 ribozymes.
It is very likely that the structure of the pistol ribozyme reported in this study reflects a pre-catalytic ground state conformation. It is anticipated that formation of a catalytically cleavage-competent state will require small conformational transitions given that the distances between the N3 and 2′-OH of A32 and the O5′ oxygen at the cleavage site are in the 5 Å range (Fig. 3c), requiring bringing them closer together for the adenine (A32) to be effective as a putative general acid in the pistol ribozyme.
Methods
Solid-phase synthesis of oligonucleotides
Standard phosphoramidite chemistry was applied for RNA solid-phase synthesis using 2′-O-TOM standard RNA nucleoside phosphoramidite building blocks (ChemGenes) and polystyrene support (GE Healthcare, Custom Primer Support™, 80 μmol/g; PS 200). All oligonucleotides were synthesized on a ABI 392 Nucleic Acid Synthesizer following standard methods: detritylation (80 sec) with dichloroacetic acid/1,2-dichloroethane (4/96); coupling (2.0 min) with phosphoramidites/acetonitrile (0.1 M × 130 μL) and benzylthiotetrazole/acetonitrile (0.3 M × 360 μL); capping (3 × 0.4 min, Cap A/Cap B =1/1) with Cap A: 4-(dimethylamino)pyridine in acetonitrile (0.5 M) and Cap B: Ac2O/sym-collidine/acetonitrile (2/3/5); oxidation (1.0 min) with I2 (20 mM) in THF/pyridine/H2O (35/10/5). The solutions of amidites and tetrazole, and acetonitrile were dried over activated molecular sieves (4 Å) overnight.
5′-O-(4,4′-Dimethoxytrityl)-2′-deoxyguanosine phosphoramidite was purchased from ChemGenes.5′-O-(4,4′-Dimethoxytrityl)-3-deaza-2′-deoxyadenosine (c3dA) phosphoramidite was synthesized according to reference39 or purchased form GlenResearch. 13C2-adenosine phosphoramidite was synthesized according to reference40
Deprotection of oligonucleotides
The solid support was treated each with MeNH2 in EtOH (33%, 0.5 mL) and MeNH2 in water (40%, 0.5 mL) for 7 h at room temperature. The supernatant was removed from and the solid support was washed 3 × with ethanol/water (1/1, v/v). The supernatant and the washings were combined with the deprotection solution of the residue and the whole mixture was evaporated to dryness. To remove the 2′-silyl protecting groups the resulting residue was treated with tetrabutylammonium fluoride trihydrate (TBAF·3H2O) in THF (1 M, 1 mL) at 37°C overnight. The reaction was quenched by the addition of triethylammonium acetate (TEAA) (1 M, pH 7.4, 1 mL). The volume of the solution was reduced and the solution was desalted with a size exclusion column (GE Healthcare, HiPrep™ 26/10 Desalting; 2.6 × 10 cm; Sephadex G25) eluting with H2O, the collected fraction was evaporated to dryness and dissolved in 1 ml H2O. Analysis of the crude RNA after deprotection was performed by anion-exchange chromatography on a Dionex DNAPac® PA-100 column (4 mm × 250 mm) at 80°C. Flow rate: 1 mL/min, eluant A: 25mM Tris·HCl (pH 8.0), 6 M urea; eluant B: 25 mM Tris·HCl (pH 8.0), 0.5 M NaClO4, 6 M urea; gradient: 0-60 % B in A within 45 min or 0-40 % B in 30 min for short sequences up to 15 nucleotides, UV-detection at 260 nm.
Purification of RNA
Crude RNA products were purified on a semipreparative Dionex DNAPac® PA-100 column (9 mm × 250 mm) at 80°C with flow rate 2 ml/min. Fractions containing RNA were loaded on a C18 SepPak Plus® cartridge (Waters/Millipore), washed with 0.1-0.15 M (Et3NH)+HCO3−, H2O and eluted with H2O/CH3CN (1/1). RNA containing fractions were lyophilized. Analysis of the quality of purified RNA was performed by anion-exchange chromatography with same conditions as for crude RNA; the molecular weight was confirmed by LC-ESI mass spectrometry. Yield determination was performed by UV photometrical analysis of oligonucleotide solutions.
Mass spectroscopy of RNA
All experiments were performed on a Finnigan LCQ Advantage MAX ion trap instrumentation connected to an Amersham Ettan micro LC system. RNA sequences were analyzed in the negative-ion mode with a potential of -4 kV applied to the spray needle. LC: Sample (200 pmol RNA dissolved in 30 μL of 20 mM EDTA solution; average injection volume: 30 μL); column (Waters XTerra®MS, C18 2.5 μm; 1.0 × 50 mm) at 21°C; flow rate: 30 μL/min; eluant A: 8.6 mM TEA, 100 mM 1,1,1,3,3,3-hexafluoroisopropanol in H2O (pH 8.0); eluant B: methanol; gradient: 0-100 % B in A within 30 min; UV-detection at 254 nm.
Crystallization
The sample for crystallization was generated by annealing the two purified strands of env25 pistol ribozyme at 70 °C for 3 min in a buffer containing 50 mM K-HEPES, pH 6.8, 100 mM KCl and 2 mM MgCl2 followed by incubation at room temperature for 5 min and then cooling on ice for 30 min before setting up crystallization trials. To prevent the cleavage of the ribozyme during crystallization, G53 was replaced by deoxy-G in the substrate strand.
The crystals of the env25 pistol ribozyme were grown at 20 °C over a period of 2 week using the sitting-drop vapor diffusion approach after mixing the RNA at an equimolar ratio with the reservoir solution containing 0.1 M Na-cacodylate, pH 6.5, Mg(OAc)2 0.2 M and 32% MPD. For data collection, crystals were quickly flash-frozen in liquid nitrogen.
For Ir(NH3)63+ soaking experiments, crystals were transferred into the crystallization solution containing 0.1 M Na-cacodylate, pH 6.5, Mg(OAc)2 0.2 M and 32% MPD and supplemented with 5 mM Ir(NH3)63+Cl3 at 4 °C for 24 h.
For Mn2+ soaking experiments, crystals were transferred into the crystallization solution containing 0.1 M Na-cacodylate, pH 6.5, 0.05 M Mg(OAc)2 and 32% MPD and supplemented with 150 mM MnCl2 at 4 °C for 24 h.
X-ray data collection and refinement
The Ir(NH3)63+ soaked crystals diffraction data were collected at 100K at beamline 14-1 located at the Stanford Synchrotron Radiation Light Source, using Rayonix Mx325 CCD detector. The native crytsal diffraction data were collected at 100K at NE-CAT beamline 24-ID-C located at the Advanced Photon Source, using Pilatus 6M detector. Data were processed using the HKL2000 (HKL Research) and XDS programs. The crystals belong to space group P3221 (Supplementary Table 1). Based on the molecular weight of 20,400 Da, one molecule is expected per asymmetric unit with 55% solvent, which is typical for RNA molecules. Using the iridium SAD data, two iridium sites were located using SHELXC and SHELXD as implemented in HKL2MAP program41. These heavy atom sites were refined using the program PHASER. Iterations of model building, phase combination and density modification enabled us to build the complete model using PHENIX42 and COOT43. The model was refined employing PHENIX using 2.7 Å native data set (Supplementary Table 1). Metal ions and their coordinated waters were identified based on 2Fo-Fc and Fo-Fc maps guided by the coordination geometries. Mg2+ sites were verified by soaking the crystal in Mn2+-containing solution and collect the anomalous data set (Supplementary Fig. 1e). The X-ray data statistics of the native and iridium hexamine-containing and Mn2+-containing crystals are listed in Supplementary Table 1.
Cleavage assays
Aliquots from aqueous millimolar stock solutions of the two RNA strands (R: ribozyme; S: substrate) were mixed and lyophilized. After addition of reaction buffer (30 mM HEPES, pH 7.5, 100 mM KCl, 2 mM MgCl2) to yield a final concentration of cRNA = 55 μM (each strand) in a total volume of 20 μL, the reaction was stopped by the addition of EDTA solution (20 μL; 3 mM) after 10, 45, and 120 min, stored at 4°C, and subsequently and analyzed by anion exchange HPLC (analytical Dionex DNAPac column) using the conditions as described above.
NMR experiments
All NMR spectra were recorded on a Bruker 600 MHz Avance II+ at 298 K equipped with a Prodigy TCI probe. One-dimensional 1H-NMR spectra were acquired with excitation sculpting for solvent suppression (Bruker pulse program: zgesgp), recording 512 scans.Two-dimensional 1H-13C-HSQC NMR spectra were recorded using a phase-sensitive ge-2D HSQC with water flip-back pulses and PEP with gradients in back-INEPT (Bruker pulse program hsqcetfpgpsi2): number of scans 72, spectral width in proton dimension 10 ppm, spectral width in carbon dimension 20 ppm, 2048×32 complex data points, 1J(1H,13C) = 200 Hz, interscan delay 1.0 s yielding an acquisition time of approx. 30 minutes per HSQC spectrum.
NMR sample preparation
The purified RNA (see above) was diluted with an equal volume of 100 mM sodium chloride solution and applied onto a C18 SepPak cartridge (Waters/Millipore). The cartridge was rinsed with water, followed by elution of the RNA with H2O-acetonitrile 1:1. After lyophilization, the RNA sodium salt was dissolved in 420 μL NMR buffer (15 mM sodium phosphate, 25 mM sodium chloride in 9/1 H2O/D2O. Final pistol ribozyme RNA concentrations were between 80 and 120 μM. Magnesium chloride was added from a 1M stock solution. The complex was formed by adding 1.2 equivalents of unlabeled substrate strand (with dG53 modification). Samples containing the bimolecular pistol complex were heated to 90 °C for two minutes and slowly cooled to room temperature to facilitate annealing of the two RNA strands.
For pH dependent experiments, the pH of the sample was determined directly in the 5 mm NMR tube using a Sigma-Aldrich® micro pH combination glass electrode and the pH was gradually adjusted by the addition of μL-amounts of 100 and 500 mM hydrochloric acid and 100/500 mM sodium hydroxide solutions, respectively. The pH of the solution was checked before and after the measurement. While the NMR spectrum was collected, the electrode was submerged in the pH 7 (for data between pH 6.8 and 5.5) or pH 4 (for data below pH 5.5) standard buffer.
Aminopurine fluorescence assays: cleavage kinetics
Rate constants kobs for the A57Ap variant were measured under pseudo-first-order conditions with Mg2+ in excess over RNA. Stock solutions were prepared for the A57Ap variant [concentration of each strand cRNA = 0.5 μM in 50 mM potassium 3-(N-morpholino)propanesulfonate (KMOPS), pH 7.5, 100 mM KCl] and for MgCl2 (concentration cMgCl2 = 1.0 M in 50 mM KMOPS; 100 mM KCl pH 7.5). Mixing of 120 μl RNA stock solution and of 1.2 μl MgCl2 stock solution manually, resulted in a final concentration of 0.5 μM RNA and 10 mM MgCl2. Spectra were recorded on a Varian Cary Eclipse spectrometer at 15 and 20 °C (as indicated) using the following instrumental parameters: excitation wavelength, 308 nm; emission wavelength, 372 nm; increment of data point collection, 0.2 s; slit widths, 10 nm. The fluorescence data were fit to a single-exponential equation: F = A1 + A2 e–k't (A1: final fluorescence; A2 e–k't: change in fluorescence over time (t) at the observed rate k′). The observed rate value provided is an arithmetic mean, determined from at least three independent measurements. All data processing was performed using Kaleidagraph software (Synergy Software).
Supplementary Material
Acknowledgments
We acknowledge assistance by staff at NE-CAT beamlines at the Advanced Photon Source and the experimental station 14-1 at the Stanford Synchrotron Radiation Light Source. X-ray diffraction studies were conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by a grant from the National Institute of General Medical Sciences (P41 GM103403) from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. A.R. was supported in part by new faculty start-up funds from Zhejiang University and the Thousand Young Talents Plan of China. The research was supported by US National Institutes of Health grant 1 U19 CA179564 to D.J.P., by the Memorial Sloan-Kettering Cancer Center Core Grant (P30 CA008748), and the Austrian Science Fund FWF (P27947, I1040 to R.M, P26550, P28725 to C.K.), and the Swiss National Foundation SNF (Early Postdoc.Mobility to J.G.).
Footnotes
Author Contributions: A.R. undertook all of the crystallographic experiments and the structure analysis with the assistance of P.G. under the supervision of D.J.P. while N.V, J.G, and M.J, were involved in nucleoside phosphoramidite synthesis, RNA preparation, and cleavage assays under the supervision of R.M. C.K. designed and performed NMR measurements. The paper was written jointly by A.R., N.V. C.K., R.M. and D.J.P. with input from the remaining authors.
Competing financial interests: The authors declare no competing financial interests.
Additional Information and Accession Codes: Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
Accession codes: Protein Data Bank (PDB): The atomic coordinates and structure factors have been deposited under the following accession codes: 5K7C for dG53-containing env25 twister ribozyme, 5K7D for same ribozyme crystals bound to Ir(NH3)63+Cl3 and 5K7E for same ribozyme crystals in Mn2+ soaks.
References
- 1.Jimenez RM, Polanco JA, Luptak A. Chemistry and Biology of Self-Cleaving Ribozymes. Trends Biochem Sci. 2015;40:648–61. doi: 10.1016/j.tibs.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinberg Z, et al. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol. 2015;11:606–10. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–80. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 4.Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–40. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–96. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 6.Perreault J, et al. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol. 2011;7:e1002031. doi: 10.1371/journal.pcbi.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb CH, Riccitelli NJ, Ruminski DJ, Luptak A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. [Google Scholar]
- 9.Fedor MJ. Structure and function of the hairpin ribozyme. J Mol Biol. 2000;297:269–91. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- 10.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–6. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 11.Watson PY, Fedor MJ. The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat Chem Biol. 2012;8:963–5. doi: 10.1038/nchembio.1095. [DOI] [PubMed] [Google Scholar]
- 12.Webb CH, Luptak A. HDV-like self-cleaving ribozymes. RNA Biol. 2011;8:719–27. doi: 10.4161/rna.8.5.16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lilley DM. The Varkud satellite ribozyme. Rna. 2004;10:151–8. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–20. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rupert PB, Ferre-D'Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–6. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 16.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–4. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 17.Klein DJ, Ferre-D'Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–6. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 18.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferre-D'Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–74. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 20.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–5. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 21.Suslov NB, et al. Crystal structure of the Varkud satellite ribozyme. Nat Chem Biol. 2015;11:840–6. doi: 10.1038/nchembio.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doherty EA, Doudna JA. Ribozyme structures and mechanisms. Annu Rev Biochem. 2000;69:597–615. doi: 10.1146/annurev.biochem.69.1.597. [DOI] [PubMed] [Google Scholar]
- 23.Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc Chem Res. 2008;41:1027–35. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 24.Ferre-D'Amare AR, Scott WG. Small self-cleaving ribozymes. Cold Spring Harb Perspect Biol. 2010;2:a003574. doi: 10.1101/cshperspect.a003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilley DM. Mechanisms of RNA catalysis. Philos Trans R Soc Lond B Biol Sci. 2011;366:2910–7. doi: 10.1098/rstb.2011.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth A, et al. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol. 2014;10:56–60. doi: 10.1038/nchembio.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eiler D, Wang J, Steitz TA. Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. Proc Natl Acad Sci U S A. 2014;111:13028–33. doi: 10.1073/pnas.1414571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Wilson TJ, McPhee SA, Lilley DM. Crystal structure and mechanistic investigation of the twister ribozyme. Nat Chem Biol. 2014;10:739–44. doi: 10.1038/nchembio.1587. [DOI] [PubMed] [Google Scholar]
- 29.Ren A, et al. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat Commun. 2014;5:5534. doi: 10.1038/ncomms6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosutic M, et al. A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class. Angew Chem Int Ed Engl. 2015;54:15128–33. doi: 10.1002/anie.201506601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaines CS, York DM. Ribozyme Catalysis with a Twist: Active State of the Twister Ribozyme in Solution Predicted from Molecular Simulation. Journal of the American Chemical Society. 2016;138:3058–3065. doi: 10.1021/jacs.5b12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris KA, Lunse CE, Li S, Brewer KI, Breaker RR. Biochemical analysis of pistol self-cleaving ribozymes. Rna. 2015;21:1852–8. doi: 10.1261/rna.052514.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Z, Tinoco I., Jr Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–36. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 34.Kirk SR, Luedtke NW, Tor Y. 2-Aminopurine as a real-time probe of enzymatic cleavage and inhibition of hammerhead ribozymes. Bioorg Med Chem. 2001;9:2295–301. doi: 10.1016/s0968-0896(01)00123-7. [DOI] [PubMed] [Google Scholar]
- 35.Jeong S, Sefcikova J, Tinsley RA, Rueda D, Walter NG. Trans-acting hepatitis delta virus ribozyme: catalytic core and global structure are dependent on the 5′ substrate sequence. Biochemistry. 2003;42:7727–40. doi: 10.1021/bi034627g. [DOI] [PubMed] [Google Scholar]
- 36.Kapinos LE, Operschall BP, Larsen E, Sigel H. Understanding the acid-base properties of adenosine: the intrinsic basicities of N1, N3 and N7. Chemistry. 2011;17:8156–64. doi: 10.1002/chem.201003544. [DOI] [PubMed] [Google Scholar]
- 37.Mir A, et al. Two Divalent Metal Ions and Conformational Changes Play Roles in the Hammerhead Ribozyme Cleavage Reaction. Biochemistry. 2015;54:6369–81. doi: 10.1021/acs.biochem.5b00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mir A, Golden BL. Two Active Site Divalent Ions in the Crystal Structure of the Hammerhead Ribozyme Bound to a Transition State Analogue. Biochemistry. 2016;55:633–6. doi: 10.1021/acs.biochem.5b01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erlacher MD, et al. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2′-OH at A2451 of 23S rRNA. J Am Chem Soc. 2006;128:4453–9. doi: 10.1021/ja0588454. [DOI] [PubMed] [Google Scholar]
- 40.Wunderlich C. PhD thesis. Leopold-Franzens University; Innsbruck, Austria: 2015. Advanced stable isotope labeling for NMR of RNA. [Google Scholar]
- 41.Schneider TPTR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 42.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Wilcox JL, Bevilacqua PC. pKa shifting in double-stranded RNA is highly dependent upon nearest neighbors and bulge positioning. Biochemistry. 2013;52:7470–6. doi: 10.1021/bi400768q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.