Abstract
Histone deacetylase 6 (HDAC6) is a critical target for drug design due to its role in oncogenic transformation and cancer metastasis, and is unique among all histone deacetylases in that it contains tandem catalytic domains designated CD1 and CD2. We now report the crystal structures of CD2 from Homo sapiens and CD1 and CD2 from Danio rerio HDAC6, and we correlate these structures with activity measurements using a panel of 13 different substrates. The catalytic activity of CD2 from both species exhibits broad substrate specificity, whereas that of CD1 is highly specific for substrates bearing C-terminal acetyllysine residues. Crystal structures of substrate complexes yield unprecedented snapshots of the catalytic mechanism. Additionally, crystal structures of complexes with 8 different inhibitors, including Belinostat and Panobinostat (currently used in cancer chemotherapy), the macrocyclic tetrapeptide HC toxin, and the HDAC6-specific inhibitor N-hydroxy-4-(2-[(2-hydroxyethyl)(phenyl)amino]-2-oxoethyl)benzamide, reveal surprising new insight regarding changes in Zn2+ coordination and isozyme-specific inhibition.
Reversible protein acetylation in living systems is a ubiquitous posttranslational modification that rivals phosphorylation.1–3 The cellular functions of nuclear and cytoplasmic proteins can be regulated by lysine acetylation-deacetylation cycles in epigenetics,4,5 cell signaling,6 and metabolism.7,8 Three classes of proteins are involved in the chemical biology of the acetylome: “writers”, or lysine acetyltransferases (KATs); “readers”, i.e., bromodomain-containing proteins that specifically recognize the acetyllysine moiety; and “erasers”, or histone deacetylases (HDACs, also known as lysine deacetylases, KDACs).9,10 The HDACs are particularly noteworthy in that upregulated activity is associated with tumorigenesis, and the HDAC inhibitors Romidepsin, Vorinostat, Belinostat, and Panobinostat are currently approved for clinical use.11–13 HDACs may also serve as therapeutic targets for other diseases, including neurodegenerative diseases and immune disorders.14,15
Four classes of HDAC isozymes are found in humans: class I HDACs 1, 2, 3, and 8; class IIa HDACs 4, 5, and 7, and class IIb HDACs 6 and 10; class III HDACs, designated sirtuins 1-7; and the class IV enzyme HDAC11.16 Class I, II, and IV HDACs are Zn2+-dependent enzymes that adopt the arginase-deacetylase fold,17 whereas class III HDACs (sirtuins 1-7) are NAD+-dependent enzymes that adopt an unrelated fold.18 Class IIb isozymes are predominantly cytoplasmic, so HDAC6 and HDAC10 are quite distinct in terms of their cellular localization and biological function. Furthermore, HDAC6 is unique in that it contains tandem catalytic domains CD1 and CD2,19 as well as a ubiquitin binding domain20 (Fig. 1a) through which polyubiquitinated misfolded protein cargo is recruited to dynein motors for transport to aggresomes.21 Additionally, HDAC6 CD1 exhibits E3 ubiquitin ligase activity.22
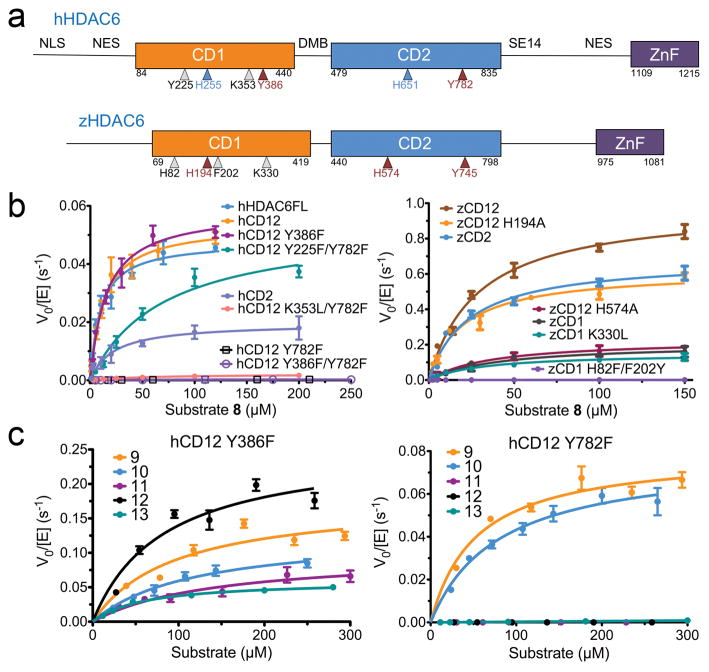
Fig. 1. Human and zebrafish HDAC6 domains and activity.
(a) Domain organization: NLS, nuclear localization signal; NES, nuclear export signal; CD1, catalytic domain 1; DMB, dynein motor binding; CD2, catalytic domain 2; SE14, serine-glutamate tetradecapeptide repeat; ZnF, zinc-finger ubiquitin binding domain. Amino acid sequence identities/similarities are 62%/77% for hCD1 and zCD1, and 59%/75% for hCD2 and zCD2. Approximate positions of residues subject to mutagenesis in HDAC6 constructs are indicated: catalytic residues, maroon arrows; Zn2+ ligands, blue arrows; hCD1 and zCD1 active site residues, gray arrows. (b) Steady-state kinetics of human and zebrafish HDAC6 constructs assayed with fluorogenic substrate 8 (substrate 8 is illustrated in Supplementary Fig. 1a; data represent mean values ± s.d. (n = 3)). (c) Steady-state kinetics of human HDAC6 constructs assayed with nonfluorogenic substrates 9–13 (substrates are illustrated in Supplementary Fig. 1a; data represent mean values ± s.d. (n = 3)).
Anchored in the cytoplasm by serine/glutamate-rich repeat motifs,23 HDAC6 catalyzes the deacetylation of K40 in the α-tubulin subunit of the microtubule,24 thereby regulating microtubule dynamics: HDAC6 inhibition leads to hyperacetylation of α-tubulin and suppression of microtubule dynamics, while overexpression of HDAC6 reduces tubulin acetylation levels and increases cell motility.25 Other HDAC6 substrates include Hsp9026 and the microtubule-associated protein Tau, implicating HDAC6 in the pathology of Alzheimer’s disease.27,28
Curiously, it is not clear that both CD1 and CD2 domains of HDAC6 are fully functional. Initial studies indicated that both domains are catalytically active toward histone substrates, with only CD2 exhibiting tubulin deacetylase activity,19,29 whereas subsequent studies suggested that only CD2 is catalytically active.30 While both domains may be required for activity,31 it is possible that CD1 could serve as a microtubule binding domain.32 Despite these unresolved structure-function relationships, no crystal structures of HDAC6 catalytic domains have been available until now.
Here, we report X-ray crystal structures of HDAC6 CD2 from Homo sapiens (human) and HDAC6 CD1 and CD2 from Danio rerio (zebrafish), demonstrating that the zebrafish enzyme is a valid and robust surrogate for the human enzyme. Additionally, we report activity measurements showing that CD2 from both species exhibits broad substrate specificity, whereas CD1 is highly specific for the hydrolysis of C-terminal acetyllysine substrates. Crystal structures of zebrafish CD2 mutants complexed with acetyllysine-containing peptide substrates provide snapshots of catalysis, including the metastable tetrahedral intermediate. Finally, crystal structures of zebrafish CD2 complexed with numerous inhibitors provide key insight regarding the molecular basis of affinity, including an alternative hydroxamate-Zn2+ coordination mode that characterizes isozyme-specific inhibition.
RESULTS
Substrate specificity and activity of HDAC6 CD1 and CD2
We prepared recombinant human and zebrafish protein constructs hCD1 and zCD1, hCD2 and zCD2, and tandem domain constructs hCD12 and zCD12. We assayed these proteins using fluorogenic peptide substrates 1–8; additionally, nonfluorogenic peptide substrates 9–13 were studied to examine potential bias due to the bulky aminomethylcoumarin chromophore (all substrates are illustrated in Supplementary Results, Supplementary Fig. 1). To isolate the catalytic activities of individual domains in the tandem domain constructs hCD12 and zCD12, we inactivated each domain by mutating the catalytic tyrosine or histidine (Y386 in hCD1, Y782 in hCD2, H194 in zCD1, H574 in zCD2), or the histidine Zn2+ ligand (H255 in hCD1, H651 in hCD2). We opted to mutate the catalytic tyrosines and histidine Zn2+ ligands to inactivate the human enzyme because mutants of the catalytic histidines had already been described previously.19,30
Fluorogenic peptide substrates 1–8 exhibited comparable trends in specific activity (Supplementary Fig. 2), showing that mutation of hCD1 does not significantly affect catalysis, whereas mutation of hCD2 essentially abolished catalytic activity. Detailed steady-state kinetics measured with representative substrates 1 and 8, derived from α-tubulin and histone H4, respectively (Fig. 1b, Supplementary Fig. 3, Supplementary Table 1), indicated that mutation of hCD2, but not hCD1, leads to more than 400-fold decreased catalytic efficiency (kcat/KM). The individual hCD2 domain was slightly less active (5-fold) than hCD12, whereas no activity was detected with hCD1 at up to 5 μM protein. These results were consistent with the binding of a fluorescent inhibitor to only a single high-affinity site in hCD12 (Supplementary Fig. 4). However, significant hCD1 activity in Y782F hCD12 was observed with C-terminal acetyllysine peptide substrates 9 and 10, indicating that hCD1 was indeed catalytically active, but exhibited strict substrate specificity for substrates bearing C-terminal acetyllysine residues (Fig. 1c, Supplementary Fig. 2). Steady-state kinetics showed that hCD1 (Y782F hCD12) exhibited robust catalytic efficiency (~1,000 M−1s−1) compared to hCD2 (Y386F hCD12) with substrates 9 and 10 (Supplementary Table 1). The hydrolysis of C-terminal acetyllysine substrates by hCD1 required an intact tandem domain assembly, since hCD1 was not catalytically active as an isolated domain. This behavior was similar to that observed for murine HDAC6, for which both domains were required for catalytic activity.31 Presumably, the hCD2 domain, even though inactivated by the Y782F substitution, structurally stabilized the hCD1 domain in hCD12 for optimal catalytic activity.
We also examined the catalytic activity of the zebrafish orthologue against substrates 1–13 (Supplementary Fig. 2), and characterized steady-state kinetics using substrates 1 and 8–13 (Fig. 1b, Supplementary Fig. 3, Supplementary Table 1). In contrast to hHDAC6, both catalytic domains of zHDAC6 were active when assayed with fluorogenic substrates, even when assayed as individual domains. However, zCD1 was less active than zCD2 (kcat/KM is reduced 21-fold and 7-fold for substrates 1 and 8, respectively). Even so, zCD1 similarly preferred substrates 9 and 10, which bear C-terminal acetyllysine residues. Steady-state kinetics showed that zCD1 was as active as zCD2 with substrates 9 and 10, but less active than zCD2 with substrates 11–13 (20–380-fold reduced kcat/KM).
In addition, both hCD2 and zCD2 exhibited broad substrate specificity. Steady-state kinetics showed that the catalytic efficiency was reduced by only 3–4-fold when the least favored substrates 11 and 13 were compared to the optimal substrate 12 (Supplementary Table 1). Taken together, these results demonstrated that in the full-length human and zebrafish enzymes, CD2 exhibited broad substrate specificity for endo- and exo-acetyllysine peptide substrates X-K(Ac)-X, X-K(Ac)-Pro-X, and X-K(Ac)-CO2−, whereas hCD1 exhibited strict substrate specificity for exo-acetyllysine peptide substrates X-K(Ac)-CO2−; substrate specificity for zCD1 was somewhat more relaxed. The structural basis for the strict substrate specificity of hCD1 is described below.
Snapshots of the catalytic mechanism
The crystal structure of hCD2 as a fusion construct with maltose binding protein was similar to that of zCD2 (59%/75% sequence identity/similarity) (Supplementary Fig. 5), with an r.m.s. deviation of 0.43 Å for 314 Cα atoms; both were cocrystallized with the inhibitor trichostatin A (TSA; all inhibitor structures are shown in Supplementary Fig. 1, and all crystallographic statistics are recorded in Supplementary Tables 2–4). Although zCD2 exhibited ~10-fold enhanced catalytic activity compared with hCD2 (Fig 1b), their active site contours were highly similar. In the zCD2 active site, only N530 and N645 were not conserved, appearing as D567 and M682 in hCD2; moreover, these residues were at the outer rim of the active site and were quite distant from bound inhibitors. Therefore, the zebrafish enzyme was judged to be a valid surrogate for the human enzyme. Furthermore, zCD2 consistently yielded higher quality crystals, which facilitated X-ray structural analysis.
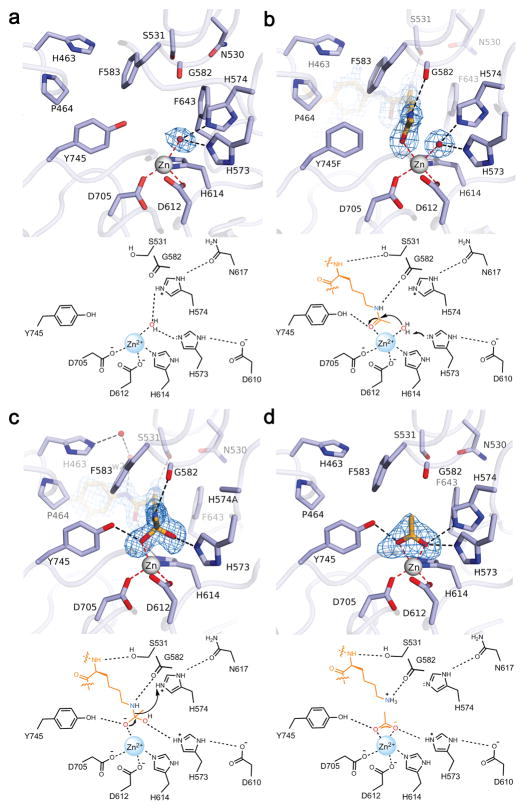
Crystal structures of zCD2 provided snapshots of all key steps along the reaction coordinate of catalysis, comprising the most thorough view of structure-mechanism relationships ever achieved for a histone deacetylase. First, the crystal structure of the resting enzyme showed that the catalytic Zn2+ ion resides at the base of a ~10 Å deep tunnel, liganded by D612, H614, D705, and a water molecule with distorted tetrahedral coordination geometry. The Zn2+-bound water molecule also formed hydrogen bonds with H573 and H574. A seemingly vacant coordination site on Zn2+ would allow for the binding of a ligand that could also interact with Y745 (Fig. 2a).
Fig. 2. Snapshots of catalysis by HDAC6 zCD2.
Simulated annealing maps are contoured at 3.0–4.0σ; metal coordination and hydrogen bond interactions are indicated by red and black dashed lines, respectively. Underneath each structure, a mechanistic scheme illustrates the corresponding complex in catalysis by the wild-type enzyme. (a) Active site of unliganded zCD2, showing the nucleophilic Zn2+-bound water molecule hydrogen bonding with H573 and H574; a vacant coordination site is observed on the “Y745 side” of Zn2+. (b) The scissile acetyllysine carbonyl of substrate 1 (derived from α-tubulin) coordinates to the formerly vacant site on Zn2+ without displacing the Zn2+-bound water molecule in Y745F zCD2. (c) In the active site of H574A zCD2, the acetyllysine carbonyl of substrate 8 (derived from histone H4) is activated by Zn2+ and Y745 to undergo nucleophilic attack, yielding a tetrahedral intermediate. The oxyanion is stabilized by Zn2+ and Y745; the hydroxyl group, which corresponds to the former Zn2+-bound water molecule, is more distant from Zn2+ and hydrogen bonds with H573. (d) In the wild-type enzyme, collapse of the tetrahedral intermediate yields products lysine and acetate. Acetate coordinates to Zn2+ nearly symmetrically; this structure represents a product complex in which lysine has already dissociated from the active site.
To study the precatalytic enzyme-substrate complex, we prepared Y745F zCD2, which exhibited compromised activity due to the loss of substrate activation by Y745. This enabled cocrystallization with the intact form of substrate 1 derived from the α-tubulin K40 acetylation site. The 1.82 Å-resolution structure of the enzyme-substrate complex revealed that the scissile carbonyl of acetyllysine coordinates to the formerly vacant site on Zn2+ without displacing the Zn2+-bound water molecule, resulting in a pentacoordinate metal ion poised for catalysis (Fig. 2b).
Surprisingly, the next snapshot of catalysis – the tetrahedral intermediate – was provided by the 1.80 Å-resolution structure of the complex between substrate 8 (derived from histone H4) and H574A zCD2. This mutant exhibited compromised activity due to the loss of the catalytically obligatory general acid. The electron density map indicated that the scissile carbonyl of acetyllysine underwent nucleophilic attack by a water molecule activated by H573 and Zn2+ to yield a tetrahedral intermediate, trapped in the active site due to the loss of the general acid H574 (Fig. 2c). The structure revealed that oxyanion atom O1 is stabilized by coordination to Zn2+ (2.1 Å) and a hydrogen bond with Y745; hydroxyl group O2 is somewhat distant from Zn2+ for inner sphere coordination (2.6 Å), but hydrogen bonds with H573. Since the Y745F mutation yielded an unreacted complex with substrate 1 and the H574A mutation yielded a tetrahedral intermediate with substrate 8, both Zn2+ and Y745 must be necessary to activate the substrate carbonyl for nucleophilic attack. In this complex, H573 could serve as a general base to assist Zn2+ in activating the nucleophilic water molecule, so the tandem histidines in the HDAC6 zCD2 active site may serve separate general base-general acid functions. If so, this contrasts with HDAC8, in which the second histidine of the tandem pair serves as a single general base-general acid.33
In H574A zCD2, the substrate carbonyl was sufficiently activated to undergo nucleophilic attack, but the resulting tetrahedral intermediate was locked in place, deep in the active site pocket – it could not collapse without general acid H574 to protonate the leaving amino group. Substrate 8 likely remained bound as a metastable tetrahedral intermediate because an enzyme generally favors the binding of a transition state-like structure over the binding of a substrate-like structure; the same principle applies for the stabilization of ketones as hydrated gem-diols in the zCD2 active site (see below).34 Parenthetically, we note that substrate binding to zCD2 does not trigger any major conformational changes in the L1 and L2 loops surrounding the active site, so the active site is essentially pre-formed for substrate binding (Supplementary Fig. 6a,b).
The tetrahedral intermediate was also mimicked by the binding of a trifluoroketone inhibitor35 as a tetrahedral gem-diol with Ki = 0.8 μM (Supplementary Fig. 6c). The binding of the gem-diol moiety was comparable to that observed for the binding of the tetrahedral intermediate with substrate 8, making asymmetric metal coordination interactions with Zn2+-O1 and Zn2+-O2 distances of 2.0 Å and 2.5 Å, respectively. The stronger interaction of O1 with Zn2+ may indicate ionization to an oxyanion, which would also be inductively stabilized by the adjacent trifluoromethyl group.36 Additionally, O1 formed a hydrogen bond with Y745, and O2 formed a hydrogen bond with H573 and H574. Finally, similar interactions were observed for the binding of the product acetate anion with Zn2+-O1 and Zn2+-O2 distances of 2.2 Å and 2.1 Å, respectively (Fig. 2d). Nearly symmetric bidentate carboxylate-Zn2+ coordination was facilitated by the negative charge delocalized over both carboxylate oxygen atoms.
Structural basis of peptide recognition by CD2
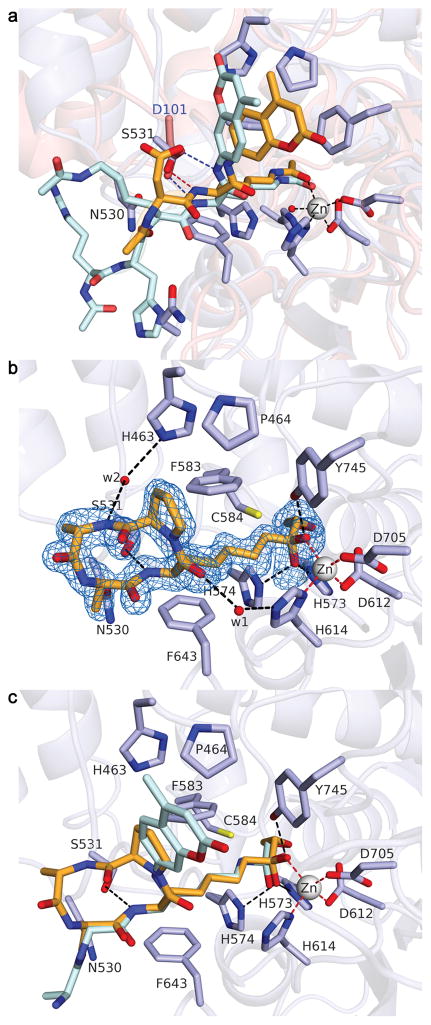
Binding interactions of peptide substrates to HDAC6 zCD2 and the well-studied class I enzyme HDAC8 differ significantly. In HDAC8, highly conserved D101 accepts hydrogen bonds from the two backbone amide groups flanking the scissile acetyllysine residue.37,38 However, the corresponding residue in zCD2, N530, was offset such that S531 of zCD2 structurally aligned with D101 of HDAC8 and accepted a hydrogen bond from the backbone NH group of acetyllysine in substrates 1 and 8 (Fig. 3a). The backbone amide group following acetyllysine hydrogen bonded only with a water molecule, suggesting that any amino acid – including proline – could be accommodated at this position. Accordingly, we observed significant catalytic activity for HDAC6 zCD2, but not HDAC8, using substrates with Lys(Ac)-Pro linkages (Supplementary Fig 2).
Fig. 3. Peptide binding to zCD2 exemplified by substrates and HC toxin.
(a) Comparison of substrate binding to HDAC6 (light blue; orange substrate 1) and HDAC8 (salmon; cyan substrate; PDB entry 2V5W). (b) Simulated annealing omit map (blue mesh, 2.5σ) showing HC-toxin (orange stick figure). The ketone carbonyl is hydrated to form a gem-diol(ate) coordinated to Zn2+. (c) Superposition of HC toxin with substrate 8.
To confirm the importance of S531 for HDAC6-substrate recognition, and to test the proposal that the conserved, adjacent aspartate/asparagine is not important for substrate binding, we prepared the N530A and S531A mutants of zCD2, and the D567A and S568A mutants of hCD2 (Supplementary Fig. 3). Consistent with our structural conclusions, mutations of zCD2 N530 and hCD2 D567 did not significantly affect activity, whereas mutations of hCD2 S568 and zCD2 S531 decreased catalytic efficiency by 10-fold and 258-fold, respectively. Therefore, this serine residue, which is strictly conserved in both catalytic domains of HDAC6 orthologues across all species (Supplementary Fig. 7), is crucial for positioning the scissile acetyllysine residue in the enzyme-substrate complex.
Additional inferences on peptide molecular recognition were provided by the structure of the zCD2 complex with HC toxin (cyclo-(L-Ala-D-Ala-L-Aoe-D-Pro); Aoe = 2-amino-8-oxo-9,10-epoxydecanoic acid), a cyclic tetrapeptide isolated from the maize pathogen Helminthosporium carbonum (Fig. 3b and Supplementary Fig. 8a).39,40 The carbonyl group of the Aoe side chain is isosteric with the carbonyl group of acetyllysine, and the adjacent epoxide moiety is a potent electrophile capable of covalently modifying a suitably positioned nucleophile in HDAC active sites. Accordingly, HC toxin exhibits irreversible inhibition against maize HD1-B (a class I HDAC), but reversible inhibition against HD1-A (a class II HDAC).39 Similarly, trapoxin contains the Aoe moiety and is a potent irreversible inhibitor of HDAC1, but it is a reversible inhibitor of HDAC6.40 Thus, there must be slight differences in the positioning of an active site nucleophile in the HDAC6 active site compared with class I HDACs.
No major conformational changes of HDAC6 accompanied the binding of HC toxin to zCD2 (Ki = 0.35 μM) (Supplementary Fig. 8b). The crystal structure revealed that the Aoe side chain extends into the active site tunnel, partially mimicking the substrate acetyllysine; the epoxide ring is clearly intact, but oriented toward the thiol side chain of C584 (Supplementary Fig. 8c). Unexpectedly, the ketone carbonyl of Aoe was hydrated so that the inhibitor binds as a gem-diol(ate), much like that observed for the trifluoroketone (Supplementary Fig. 6c) or for the tetrahedral intermediate with substrate 8 (Fig. 2c). The crystal structure revealed that the Aoe O1 atom coordinates to Zn2+ (2.0 Å) and hydrogen bonds with Y745; the O2 atom makes a weak coordination interaction with Zn2+ (2.5 Å), and hydrogen bonds with H573 and H574. It was surprising that the unactivated ketone of the α,β-epoxyketone moiety underwent hydration in the zCD2 active site. Notably, however, such hydration behavior was first observed for the binding of unactivated aldehyde- and ketone-based substrate analogues to carboxypeptidase A.41,42 The active site of a Zn2+ hydrolase has evolved to enable nucleophilic attack of water at virtually any carbonyl group, preferentially stabilizing the resulting tetrahedral species.
Significantly, interactions of the macrocyclic peptide backbone of HC toxin were similar to interactions with the substrate peptide backbone (Fig. 3c). The backbone NH group of Aoe donated a hydrogen bond to S531, and water-mediated hydrogen bond networks were observed between the backbone carbonyl of Aoe and H614, and the backbone NH group of HC toxin L-Ala and H463 (Fig. 3b). Interestingly, the L-Ala–D-Ala and the L-Aoe–D-Pro peptide linkages of HC toxin adopted cis configurations, giving the cyclic tetrapeptide unusual alternating cis-trans-cis-trans peptide bond geometry. This distinguishes HC toxin from other cyclotetrapeptides of known structure, such as trapoxin A (Supplementary Fig. 8a), which contain just one cis peptide linkage.
Based on substrate specificity studies (Fig. 1, Supplementary Figs. 2 and 3), the crystal structures of zCD2 complexes with HC toxin and substrates, and the structural homology between zCD2 and hCD2, we suggest that the amide bond following the scissile acetyllysine residue is dispensable for substrate binding to zCD2 and hCD2, since it makes no direct interactions with active site residues. Accordingly, we hypothesize that the L-Aoe-D-Pro-L-Ala fragment of HC toxin partially mimics a potential substrate site located in a reverse turn, such as X-Lys(Ac)-Pro-X, as well as other proline-containing loop structures or segments containing cis-Lys(Ac)-X linkages.
Basis of CD1 specificity for X-K(Ac)-CO2− substrates
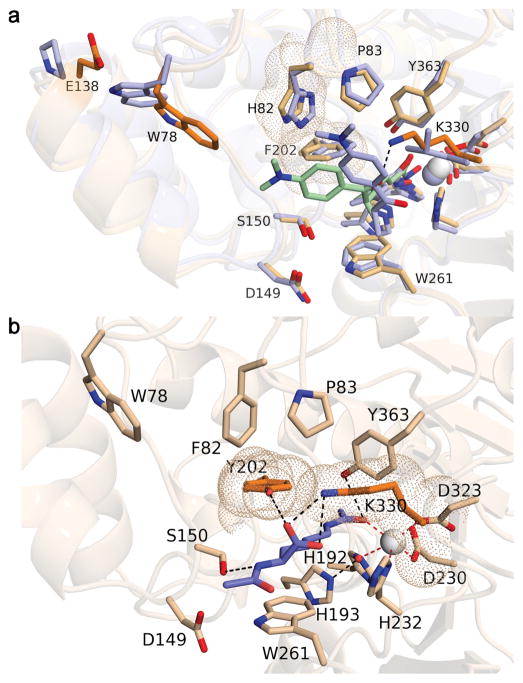
Curiously, despite significant overall sequence identity/similarity of 46%/64% and conservation of all residues important for substrate binding and catalysis, zCD1 and hCD1 exhibited low or no activity when assayed with traditional fluorogenic substrates 1–8, but instead were very specific for the hydrolysis of exo-acetyllysine substrates 9 and 10, X-K(Ac)-CO2− (Fig. 1c, Supplementary Figs. 2, 3). The crystal structure of the zCD1-TSA complex (Supplementary Fig. 9) suggested a likely explanation. The structure of this complex was similar to that of the zCD2-TSA complex, with an r.m.s. deviation of 0.58 Å for 311 Cα atoms. The Zn2+ ions, catalytic residues, and L1 and L2 loops flanking the active sites aligned well. However, the inhibitor TSA adopted a notably different binding orientation in each active site (Fig. 4a). In zCD1, the active site was slightly more constricted, due in part to the protruding side chain of K330, which donates a hydrogen bond to the carbonyl group of the “cap” moiety of TSA and sterically displaces it toward the L1 loop. The corresponding residue in zCD2 was L712, which does not protrude as far into the active site. Another difference evident in the active site of zCD1 was the conformational change of W78 toward the bound inhibitor, resulting from a steric interaction with E138 on a nearby helix. This residue appeared as P519 in zCD2 and did not trigger a conformational change of the corresponding tryptophan residue, W459. Based on the interaction of K330 with the carbonyl group of TSA, we hypothesized that K330 is a “gatekeeper” that confers specificity toward C-terminal acetyllysine substrates by hydrogen bonding with the α-carboxylate group of acetyllysine. Consistent with this hypothesis, K330L zCD1 exhibited more relaxed substrate specificity in comparison with wild-type zCD1 (Supplementary Fig. 2).
Fig. 4. Structural basis of CD1 substrate specificity.
(a) Superposition of the zCD1-TSA complex (wheat) and the zCD2-TSA complex (light blue). Zn2+ ions are white and light blue spheres, respectively. TSA binds with different orientations to zCD1 (light green) and zCD2 (light blue); K330 in zCD1, which corresponds to the smaller residue L712 in zCD2, is responsible for the alternative orientation of TSA. Also highlighted with van der Waals surfaces are zCD1 residues F202 and H82, which are found as Y225 and F105 in hCD1. (b) Model of acetyllysine docked in the active site of H82F/F202Y zCD1 (these mutations “humanize” zCD1 to serve as a surrogate for hCD1). The α-carboxylate of the C-terminal acetyllysine substrate is proposed to hydrogen bond with Y202 and K330 (black dashed lines).
Two amino acid substitutions further constrict the active site of hCD1 in addition to K353 (which corresponds to K330 of zCD1): F202 and H82 in zCD1 (Fig. 4a) appear as Y225 and F105 in hCD1. To “humanize” zCD1, we prepared and assayed H82F/F202Y zCD1. These amino acid substitutions abolished activity with endo-acetyllysine peptide substrate 8 (Fig. 1b) and conferred significant specificity for substrates 9 and 10, which bear an exo-acetyllysine group (Supplementary Fig. 2). We hypothesized that the hydroxyl group of Y202 hydrogen bonds with the α-carboxylate group of the acetyllysine substrates to abolish the binding of endo-acetylysine substrates, such that both K330 and Y202 (K353 and Y225 in hCD1) enforce specificity for substrates bearing an exo-acetyllysine group, X-K(Ac)-CO2−, as modeled in Fig. 4b. Indeed, Y225F/Y782F hCD12 exhibited nearly 100-fold enhanced activity with endo-acetyllysine fluorogenic substrates when compared to hCD12 Y782F, whereas K353L/Y782F hCD12 exhibited a more modest 10-fold activity enhancement (Fig. 1b, Supplementary Table 1). Therefore, Y225 in hCD1 is another “gatekeeper” that confers specificity toward C-terminal acetyllysine substrates.
HDAC6 inhibitor selectivity
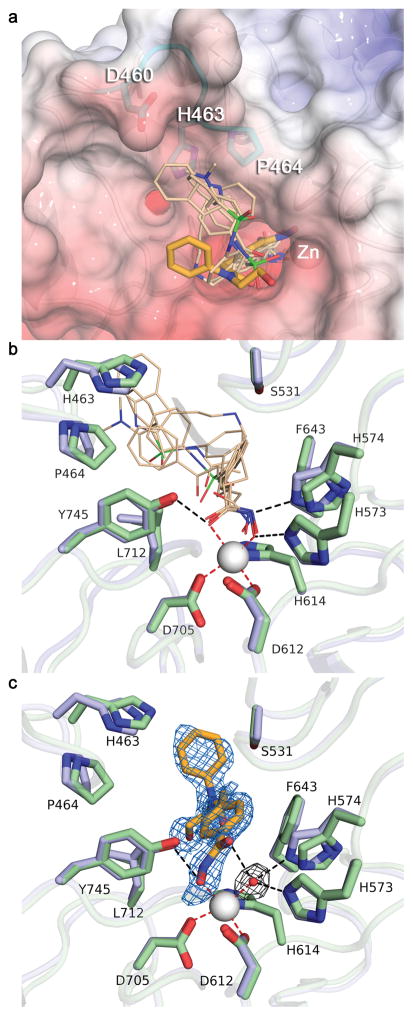
In addition to the previously described structures of hCD2, zCD1, and zCD2 complexed with TSA, we also determined crystal structures of zCD2 complexed with clinically-approved broad-specificity hydroxamate inhibitors SAHA, Belinostat, and Panobinostat; Oxamflatin; and the HDAC6-selective inhibitor N-hydroxy-4-(2-[(2-hydroxyethyl)(phenyl)amino]-2-oxoethyl)benzamide (HPOB) (Supplementary Fig. 10).43 Structural comparisons of these complexes revealed critical determinants of affinity and selectivity. First, the “cap” moieties of broad-specificity inhibitors clustered around a “hot spot” on the L1 loop (D460-P484), making contacts with H463 and P464 (Fig. 5a). Additionally, the hydroxamate groups of all broad-specificity inhibitors tended toward bidentate Zn2+ coordination (Fig. 5b and Supplementary Table 5), displacing the Zn2+-bound water molecule of the resting enzyme, with the hydroxamate OH and C=O groups additionally interacting with H573 and Y745, respectively.
Fig. 5. Inhibitor binding to zCD2.
(a) Superposition of broad-specificity HDAC hydroxamate inhibitors (thin wheat stick figures) and the HDAC6-specific inhibitor HPOB (thick orange stick figure). The “hot spot” L1 loop segment that accommodates the capping group of broad-specificity inhibitors is colored cyan (this segment is strictly conserved in hCD2). The backbone of zCD2 is shown as white cartoon and the molecular surface of hCD2 is shown for comparison (color coded by electrostatic potential, red to blue, −10 kT/e to 10 kT/e). (b) Close-up view of the Zn2+ binding site in zCD2 complexes with broad-specificity inhibitors. Both zCD2 (light blue) and hCD2 (pale green) are shown for comparison. (c) Close-up view of the Zn2+ binding site in the zCD2-HPOB complex. The simulated annealing omit map (blue mesh, 2.5σ) clearly defines the binding mode of the inhibitor, which coordinates to Zn2+ only through its hydroxyl group. Inhibitor binding does not displace the Zn2+-bound water molecule, as confirmed in a simulated annealing omit map (grey mesh, 3.0σ). Both zCD2 (light blue) and hCD2 (pale green) are shown for comparison. Metal coordination and hydrogen bond interactions are shown as red and black dashed lines, respectively.
In contrast, the hydroxamate group of the HDAC6-selective inhibitor HPOB (Ki = 1.0 nM) coordinated to Zn2+ only through its OH group, and it did not displace the Zn2+-bound water molecule; the Zn2+ ion is pentacoordinate with square pyramidal geometry (Fig. 5c). The hydroxamate C=O group hydrogen bonded with the Zn2+-bound water molecule. The short, bulky linker of HPOB sterically precluded a closer approach of the hydroxamate group to Zn2+, which may also have prevented the “cap” moiety of HPOB from interacting with the L1 loop, an interaction that appears to be more important for binding to HDACs 1-3 than to HDAC6. Accordingly, the potency of HPOB against HDAC6 is comparable to SAHA-like inhibitors, but HPOB potency against other isozymes is significantly reduced.43
DISCUSSION
Our studies have illuminated the structural basis of substrate specificity and catalysis for the CD1 and CD2 domains of the class IIb deacetylase HDAC6. In particular, we have demonstrated new modes of peptide substrate recognition by HDAC6 CD1 and CD2 that contrast with substrate binding modes previously observed for the well-studied class I isozyme HDAC8. The CD2 domains of human and zebrafish HDAC6 can accommodate proline at the +1 position relative to the scissile acetyllysine residue, whereas HDAC8 cannot based on the hydrogen bond between D101 and the backbone NH of the +1 residue. In contrast, the CD1 domains of human and zebrafish HDAC6 exhibit stringent specificity for acetyllysine substrates bearing a free α-carboxylate group, likely due to hydrogen bond interactions with gatekeeper residues K353 and Y225 (hCD1 numbering) as well as a more constricted active site contour. However, it remains unclear as to what biological substrates would undergo reversible acetylation at C-terminal lysine residues in vivo. Regardless, in view of the highly specialized binding activity and catalytic function of hCD1, we suggest that drug discovery programs employing in vitro assays of HDAC6 using traditional fluorogenic substrates will yield “hits” exclusively targeting hCD2 that miss the highly specific activity of hCD1.
Significantly, HDAC6-peptide recognition is similar for substrates as well as the macrocyclic peptide inhibitor HC toxin. Moreover, in binding as the gem-diol, the α,β-epoxyketone moiety of HC toxin binds as a tetrahedral transition state analogue rather than a substrate analogue with an intact ketone carbonyl. This likely represents the binding mode of other Aoe-containing cyclic tetrapeptides such as trapoxin A to all other HDAC isozymes, whether or not these inhibitors bind reversibly or irreversibly. In general, zinc hydrolases activate a substrate carbonyl group for direct nucleophilic attack by a water molecule using Zn2+ coordination and a hydrogen bond interaction to polarize the carbonyl group; the nucleophilic water molecule is activated by Zn2+ coordination and a general base.44,45 Thus, any carbonyl, whether that of a peptide, ester, ketone, or aldehyde, is subject to facile hydration in the active site of a zinc hydrolase as long as the carbonyl is isosteric with that of the natural substrate. Since the carbonyl group of the Aoe side chain is isosteric with the scissile carbonyl of acetyllysine, the binding of Aoe-containing peptides to any HDAC should enable facile hydration of the Aoe ketone carbonyl, as observed in the zCD2-HC toxin complex. Moreover, the structure of the zCD2-HC toxin complex reveals an important clue regarding the irreversible inhibition of class I HDAC isozymes by Aoe-containing cyclic tetrapeptides. Based on the geometry observed in the zCD2-HC toxin complex, the likely active site nucleophile is the strictly conserved residue C584. The thiol side chain of this residue is oriented for nucleophilic attack at the less hindered epoxide carbon. Presumably, inhibitor binding to class I HDAC active sites allows for a closer contact between the nucleophilic thiol and the electrophilic epoxide to enable covalent bond formation.
Finally, comparison of different zCD2-inhibitor structures highlights the structural basis for HDAC6-specific inhibitor design, which exploits a previously unobserved Zn2+ coordination mode by the hydroxamate group of HPOB: inhibitor binding does not require displacement of Zn2+-bound water. Additionally, the L1 and L2 loops flanking the active site are relatively rigid, as revealed in the 12 zCD2 structures reported herein, making HDAC6 an ideal receptor for in silico docking experiments. In closing, we expect that these structural data will enable and accelerate advances in structure-based drug design in the continuing search for new therapies for human disease.
ONLINE METHODS
No statistical methods were used to predetermine sample size.
General
PCR reactions were performed using the PfuUltra High-Fidelity DNA polymerase (Agilent Technologies). Restriction enzymes were purchased from New England Biolabs and used according to the manufacturer’s instructions. Custom oligonucleotides were synthesized by Integrated DNA Technologies. Escherichia coli strain NEB5α (New England Biolabs) was used for cloning procedures. Assay substrates 1–2 and 7–13 were purchased from GenScript®, and assay substrates 3–6 were purchased from Enzo® Life Sciences. HC toxin was purchased from Cayman Chemicals. A sample of 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptan-2-one was synthesized according to published procedures35 and determined to be >95% pure by mass spectrometry, NMR spectroscopy, and X-ray crystallographic structure validation. All other HDAC inhibitors were purchased from ApexBio®. All substrates and inhibitors were purchased as >95% pure preparations and used without further purification.
Protein expression and purification
For human HDAC6 protein expression, different expression constructs were made by subcloning the corresponding gene from the HDAC6Flag plasmid (a gift from Prof. Eric Verdin, University of California, San Francisco; Addgene plasmid #13823)46 into a modified pET28a(+) vector (a gift from Dr. Scott Gradia, University of California, Berkeley; Addgene plasmid #29656) in-frame with a TEV-cleavable N-terminal His-MBP-tag (MBP, maltose binding protein) using ligation independent cloning. Human HDAC6 proteins were all expressed in E. coli BL21 (DE3) Codon Plus RIL (Stratagene) in 2x YT medium in the presence of 50 mg/L kanamycin and 30 mg/L chloramphenicol. Expression was induced by 75 μM isopropyl β-D-1-thiogalactopyranoside (Carbosynth) when OD600 reached 1.0, and cell cultures were grown for an additional 18 h at 16 °C. The growth medium was supplemented with 200 μM ZnSO4 30 min before induction. Cells were harvested by centrifugation and resuspended in lysis buffer [50 mM K2HPO4 (pH 8.0), 300 mM NaCl, 10 mM MgCl2, 10% glycerol, 0.1 mg/mL lysozyme (Sigma), 50 μg/ml DNase I (Sigma), and protease inhibitor tablets (Roche Applied Science)]. Cells were lysed by sonication and the cell lysate was cleared by centrifugation at 26,000 g for 1 h at 4 °C. The supernatant was loaded onto a Ni-NTA affinity column (Qiagen) and the target protein was eluted with buffer A [50 mM K2HPO4 (pH 8.0), 1 mM TCEP, 300 mM NaCl, 300 mM imidazole, 5% glycerol]. Peak fractions were directly loaded onto an Amylose resin column (New England Biolabs) and digested on-column using TEV protease to remove the His-MBP tag in buffer B [20 mM Tris (pH 8.0), 100 mM NaCl, 1 mM TCEP, 5% glycerol]. Untagged proteins were further purified by anion-exchange chromatography (HiTrap Q, GE Healthcare) and then size-exclusion chromatography in buffer C [50 mM HEPES (pH 7.5), 100 mM KCl, 1 mM TCEP, 5% glycerol] with a HiLoad superdex 200 column. The human CD1-CD2 construct (hCD12) was concentrated to ~10 mg/mL and flash-frozen in liquid nitrogen, whereas the individual human CD1 and CD2 domains (hCD1 and hCD2, respectively) were concentrated to 2 mg/mL and used immediately after purification without further storage. Proteins precipitated after freezing or being concentrated over 2 mg/mL.
Due to the poor solubility of hCD1 and hCD2, the intact MBP-tagged proteins were used for crystallization. However, extensive trials did not yield any crystals. We reasoned that this was probably due to the linker peptide (containing a TEV cleavage site) between MBP and each HDAC6 domain, which could introduce unwanted flexibility and thus inhibit crystal formation. Therefore, we fused the MBP tag to the N-terminus of each human HDAC6 domain with a shorter linker (3 alanine residues) by overlap extension PCR. Three charged residues in the C-terminal alpha helix of MBP (E350, K363, D364) were also replaced by alanines, as first described in the crystallization of human T cell leukemia virus type 1 (HTLV-1) envelope protein gp21.47,48 The chimeric constructs MBP-hCD1 and MBP-hCD2 were cloned into a modified pET28a(+) vector (a gift from Dr. Scott Gradia, University of California, Berkeley; Addgene plasmid #29653) in-frame with a TEV cleavable N-terminal His-tag using ligation independent cloning, and expressed and purified in the same way but without TEV cleavage.
The zebrafish HDAC6 gene (residues 60-798, Uniprot F8W4B7) was synthesized with codon optimization by Genscript. The tandem catalytic domains (residues 60-798) and each individual catalytic domain (zCD1, residues 60-419; zCD2, residues 440-798) were subcloned into the same expression vector in fusion with a TEV-cleavable N-terminal His-MBP tag, in similar manner as outlined above for human HDAC6. The zebrafish HDAC6 proteins were expressed and purified in the same way except that E. coli BL21 (DE3) cells (Stratagene) were used as expression host.
Human HDAC8 was expressed, purified, and assayed as previously reported.49
Crystallization
The MBP-hCD2–trichostatin A (TSA) complex was crystallized using the sitting drop vapor diffusion method at 4 °C. Purified MBP-hCD2 (13 mg/mL) was first incubated with 2 mM TSA, 5% DMSO, 10 mM maltose, and 30 mM Gly-Gly-Gly in buffer C on ice for 30 min, and then 0.4 μL of protein solution was mixed with precipitant solution [0.2 M potassium sodium tartrate, 20% PEG 3350]. Plate-like crystals of the MBP-hCD2–TSA complex appeared after 6 months.
The zCD1–TSA complex was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of purified zCD1 [10 mg/mL protein preincubated with 2 mM TSA in 5% DMSO] on ice with 0.35 μL precipitant solution [0.2 M ammonium sulfate, 0.1 M MES (pH 6.5), 25% PEG monomethyl ether 5,000]. Plate-like crystals appeared in 2 weeks.
The zCD2 complex with suberoylanilide hydroxamic acid (SAHA) was crystallized using the sitting drop vapor diffusion method at 21 °C by mixing 0.35 μL of purified zCD2 [10 mg/mL protein preincubated with 4 mM SAHA in 5% DMSO] with 0.35 μL precipitant solution [0.1 M Bis-Tris (pH 6.5), 0.2 M MgCl2, 25% PEG monomethyl ether 5,000]. Plate-like crystals appeared after 1 week.
The zCD2–TSA complex was crystallized using the sitting drop vapor diffusion method at 21 °C by mixing 0.35 μL of purified zCD2 [16 mg/mL protein preincubated with 2 mM TSA in 5% DMSO] with precipitant solution [0.1 M ammonium iodide, 20% PEG 3350]. Thick plate-like crystals appeared in 4 days.
The zCD2–Belinostat complex was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of purified zCD2 [10 mg/mL protein preincubated with 4 mM Belinostat in 5% DMSO] with precipitant solution [0.1 M Bis-Tris (pH 5.5), 25% PEG 3350]. Plate-like crystals appeared after 6 days.
The zCD2 complex with the HDAC6-specific inhibitor N-hydroxy-4-(2-[(2-hydroxyethyl)(phenyl)amino]-2-oxoethyl)benzamide (HPOB)41 was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of purified zCD2 [10 mg/mL protein preincubated with 4 mM HPOB in 5% DMSO] with precipitant solution [0.1 M Tris (pH 8.5), 25% PEG 3350]. Large, thin, plate-like crystals appeared after 4 days.
The zCD2-Panobinostat complex was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.3 μL of purified zCD2 [8 mg/mL protein preincubated with 4 mM Panobinostat in 5% DMSO] with precipitant solution [0.1 M MES (pH 6.5), 12% PEG 20,000]. Thin plate-like crystals appeared after a month.
The zCD2-HC toxin complex was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.3 μL of purified zCD2 [8 mg/mL protein preincubated with 4 mM HC toxin in 5% DMSO] with precipitant solution [0.2 M sodium nitrate, 20% PEG 3350]. Thick plate-like crystals appeared after three weeks.
Unliganded zCD2 was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of purified zCD2 [10 mg/mL protein preincubated with 4 mM Rocilinostat in 5% DMSO] with precipitant solution [0.1 M sodium acetate, 0.1 M sodium formate, 20% PEG 3350]. Crystals appeared after 2 weeks (subsequent structure determination revealed that Rocilinostat was not bound to the enzyme).
The zCD2 complex with the trifluoroketone transition state analogue inhibitor was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of purified zCD2 (10 mg/mL protein preincubated with 5 mM inhibitor in buffer C) with precipitant solution [0.2 M sodium bromide, 20% PEG 3350]. Crystals appeared after 4 days.
The zCD2-acetate complex was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of purified zCD2 (10 mg/mL) with precipitant solution [0.2 M sodium acetate, 30% PEG 8000]. Crystals appeared after 3 days.
The H574A zCD2–substrate 8 complex was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of protein solution [10 mg/mL protein preincubated with the histone H4-based tripeptide assay substrate Ac-Arg-Gly-Lys(Ac)-aminomethylcoumarin in 30 mM Tricine (pH 7.3), 100 mM KCl, 5% glycerol, 1 mM TCEP, 5% DMSO] with 0.35 μL precipitant solution [0.1 M Bis-Tris (pH 6.5), 25% PEG 3350]. Clusters of crystal plates appeared after two weeks; single crystals for X-ray diffraction analysis were obtained by breaking up crystal clusters.
The Y745F zCD2–substrate 1 complex was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of protein solution [5 mg/mL protein preincubated with the α-tubulin K40 based tripeptide assay substrate Ac-Ser-Asp-Lys(Ac)-aminomethylcoumarin in buffer C supplemented with 5% DMSO] with 0.35 μL precipitant solution [0.1 M Bicine (pH 9.0), 2% v/v 1,4-dioxane, 10% PEG 20000]. Thick crystal plates appeared overnight.
The zCD2–oxamflatin complex was crystallized using the sitting drop vapor diffusion method at 4 °C by mixing 0.35 μL of protein solution [9 mg/mL protein preincubated with oxamflatin in buffer C supplemented with 5% DMSO] with 0.35 μL precipitant solution [0.1 M HEPES (pH 7.5), 10% v/v 2-propanol, 20% PEG 4000]. Small thin crystal plates appeared after 3 days.
All crystals were soaked in a cryoprotectant solution comprised of mother liquor supplemented with 25–30% glycerol prior to flash-freezing in liquid nitrogen.
Crystal structure determination
For the zCD1-TSA complex, the zCD2-TSA complex, and the zCD2-SAHA complex, X-ray diffraction data were recorded at the Stanford Synchrotron Radiation Lightsource (SSRL), beamline 14-1 (λ = 1.28184 Å). For the MBP-hCD2-TSA complex, unliganded zCD2, the H574A zCD2-substrate 8 complex, and the zCD2-Belinostat complex, X-ray diffraction data were recorded at the Advanced Photon Source (APS), beamline NE-CAT 24-ID-E (λ = 0.97918 Å). For all other structures, X-ray diffraction data were recorded at the Advanced Light Source (ALS), beamline 4.2.2 (λ = 1.00003 Å). Data reduction and integration for all datasets was achieved with HKL2000;50 data collection and reduction statistics are recorded in Supplementary Tables 2–4. Although Rmerge values were relatively high for some datasets, analysis of CC1/2 values indicated that these datasets were of sufficient quality for satisfactory structure determination and refinement.
All structures were solved by molecular replacement using the program Phaser.51 For the structure of the zCD2–SAHA complex, a model of the HDAC4 catalytic domain in a closed-loop conformation (PDB entry 4CBT)52 was used as the search probe for rotation and translation function calculations. For all other zCD1 and zCD2 structures, the structure of the zCD2-SAHA complex less inhibitor and water molecules was used as a search probe. For the structure determination of the fusion protein MBP-hCD2–TSA complex, maltose binding protein (PDB entry 4EDQ) and the zCD2–TSA complex less ligands and solvent molecules were used as search probes. The graphics program Coot was used for model building53 and Phenix was used for crystallographic refinement.54 Refinement statistics for each final model are recorded in Supplementary Tables 2–4. The quality of each model was verified with PROCHECK55 and MolProbity.56 Figures were prepared with Pymol and UCSF Chimera.57 The Ramachandran statistics for each model are as follows: zCD1-TSA complex: 90.3% allowed, 9.4% additionally allowed; zCD2-TSA complex: 91.6% allowed, 7.8% additionally allowed; MBP-hCD2-TSA complex: 88.5% allowed, 10.8% additionally allowed; unliganded zCD2: 91.1% allowed, 8.6% additionally allowed; H574A zCD2-substrate 8 complex: 92.2% allowed, 7.3% additionally allowed; Y785F zCD2-substrate 1 complex: 90.6% allowed, 8.7% additionally allowed; Y785F zCD2-substrate 1 complex: 90.6% allowed, 8.7% additionally allowed; zCD2-HC toxin complex: 90.6% allowed, 8.8% additionally allowed; zCD2-trifluoroketone inhibitor complex: 90.5% allowed, 9.0% additionally allowed; zCD2-acetate complex: 90.4% allowed, 9.1% additionally allowed; zCD2-SAHA complex: 91.4% allowed, 8.0% additionally allowed; zCD2-Belinostat complex: 91.1% allowed, 8.5% additionally allowed; zCD2-HPOB complex: 91.0% allowed, 8.7% additionally allowed; zCD2-Panobinostat complex: 89.9% allowed, 9.6% additionally allowed; zCD2-Oxamflatin complex: 89.3% allowed, 10.0% additionally allowed. No backbone torsion angles adopt disallowed conformations in any structure.
HDAC6 activity assay
Assays were performed in triplicate at room temperature. Briefly, substrates and enzymes were diluted into HDAC assay buffer [50 mM Tris-HCl (pH 8.0), 137 mM NaCl, 2.7 mM KCl, 1.0 mM MgCl2] at varying concentrations. To initiate the reaction, 25 μL of substrate solution was added to 25 μL of enzyme solution (3 – 250 nM). The reaction was stopped by adding the developer solution [1 μM trypsin and 10 μM TSA in HDAC assay buffer]. After 10–30 min, the reaction mixture was transferred to a NUNC 384 well optical bottom black plate (Thermo Fisher Scientific) and the fluorescence was measured using a Tecan Infinite M1000Pro plate reader (λex = 360 nm, λem = 455 nm). Enzyme activity was calculated based on the standard curve using Fluor de lys® deacetylated standard (Enzo® Life Sciences). Inhibition of zCD2 activity was measured by using 3 nM enzyme with 16 μM substrate 8, inhibition of zCD1 activity was measured by using 12 nM enzyme with 30 μM substrate 8, inhibition of MBP-hCD2 was measured by using 6 nM enzyme with 16 μM substrate 8 (Supplementary Fig. 11). Data were analyzed by logistic regression for IC50 determination and the inhibition constant Ki was calculated based on the Cheng-Prusoff equation, Ki = IC50/(1+[S]/KM).58
Evaluation of specific activity of HDAC6 with nonfluorogenic peptide substrates 9–13 (Supplementary Fig. 1a) was based on the reaction of fluorescamine with liberated primary amino groups after deacetylation.59 Briefly, 25 μL of enzyme (0.05 – 2 μM) in phosphate buffer [20 mM K2HPO4 (pH 7.8), 100 mM NaCl, 5 mM KCl] was added to 25 μL of substrate solution (2 mM) to initiate reaction. After incubation for 15–60 min at room temperature, the reaction was quenched by the addition of 50 μL fluorescamine (2 mM) in DMSO; fluorescence was recorded with a Tecan Infinite M1000Pro plate reader (λex = 390 nm, λem = 470 nm). Assays were performed in triplicate at room temperature.
For the study of steady-state kinetics of HDAC6 with nonfluorogenic peptide substrates, a discontinuous liquid chromatography-mass spectrometry (LC-MS) was developed. Briefly, 4 μL of enzyme (0.03 – 10 μM) in HEPES buffer [20 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM KCl, 1 mM MgCl2] was added to 36 μL of substrate solution to initiate reaction. After incubation for 15–60 min at room temperature, the reaction was quenched by addition of 50 μL of acetonitrile followed by 10 μL of NaHCO3 (1.6 M, pH 10.0). To efficiently separate the deacetylation products of substrate 9–12, the deacetylation products were derivatized with dansyl chloride (10 mM in acetonitrile) at 50 °C for 1 hr. The deacetylation products were detected by liquid chromatography-mass spectrometry (LC-MS) using a Waters SQD equipped with an Acquity UPLC (Waters, Milford, MA, USA) (Supplementary Fig. 12), and quantified by using the standard curves generated from the mass signals of the corresponding deacetylated synthetic peptide. Note that the product of the reaction with substrate 13 can be efficiently separated from the reaction mixture for MS characterization; thus, product dansylation was not required for this particular assay (Supplementary Fig. 13). Assays were performed in triplicate at room temperature.
HDAC6 Fluorescence Anisotropy Assay
The fluorescence anisotropy assay was performed with fluorescein-labeled SAHA (fl-SAHA, a gift kindly provided by Prof. Carol Fierke, University of Michigan) and measured using a TECAN infinite F200Pro plate reader (λex = 485 nm, λem = 535 nm).60 Briefly, 30 nM fl-SAHA was titrated with enzyme (0–15 μM) and incubated at room temperature for 20 minutes. The Kd values were determined by fitting the binding isotherm using GraphPad Prism software. Equation (1) was used for one-site binding. In the case of two nonequivalent ligand binding sites, as observed for hCD12, equation (2) was used to account for the second low-affinity binding site, which corresponded to a nonspecific binding site. In equations (1) and (2), Af is the fluorescence anisotropy of unbound fl-SAHA, AB is the fluorescence anisotropy of protein bound fl-SAHA. Assays were performed in triplicate at room temperature.
| (1) |
| (2) |
Supplementary Material
Acknowledgments
We thank R. Marmorstein, X. Li, and M. Chen for helpful discussions. We thank the NIH for grant GM49758 to D.W.C. in support of this research. We thank V. Stojanoff at beamline 14-1 at the Stanford Synchrotron Radiation Lightsource, D. Neau at NE-CAT beamline 24 ID-E at Advanced Photon Source, and J. Nix at beamline 4.2.2 of the Advanced Light Source for assistance with data collection. Finally, D.W.C. thanks the Radcliffe Institute for Advanced Study for the Elizabeth S. and Richard M. Cashin Fellowship.
Footnotes
Author contributions Y.H. and D.W.C. designed the project; Y.H. performed experiments; Y.H. and D.W.C. interpreted experimental results and prepared the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper.
Accession codes. Protein Data Bank (PDB): zCD1-TSA complex, 5EEF; zCD2-TSA complex, 5EEK; MBP-hCD2-TSA complex, 5EDU; unliganded zCD2, 5EEM; H574A zCD2-substrate 8 complex, 5EFN; Y745F zCD2-substrate 1 complex, 5EFK; zCD2-HC toxin complex, 5EFJ; zCD2-trifluoroketone inhibitor complex, 5EFH; zCD2-acetate complex, 5EFG; zCD2-SAHA complex, 5EEI; zCD2-Belinostat complex, 5EEN; zCD2-HPOB complex, 5EF7; zCD2-Panobinostat complex, 5EF8; zCD2-Oxamflatin complex, 5EFB.
References
- 1.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norvell A, McMahon Rise of the rival. Science. 2000;327:964–965. doi: 10.1126/science.1187159. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 4.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenet. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 7.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 10.Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma N, et al. Selective histone deacetylase inhibitors with anticancer activity. Curr Top Med Chem. 2015;16:415–426. doi: 10.2174/1568026615666150813145629. [DOI] [PubMed] [Google Scholar]
- 13.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 14.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 15.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 16.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deaectylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Lombardi PM, Cole KE, Dowling DP, Christianson DW. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr Opin Struct Biol. 2011;21:735–743. doi: 10.1016/j.sbi.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan H, Marmorstein R. Structural basis for sirtuin activity and inhibition. J Biol Chem. 2012;287:42428–42435. doi: 10.1074/jbc.R112.372300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouyang H, et al. Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. J Biol Chem. 2012;287:2317–2327. doi: 10.1074/jbc.M111.273730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi Y, et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSα. Mol Cell. 2014;55:31–46. doi: 10.1016/j.molcel.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertos NR, et al. Role of the tetradecapeptide repeat domain of human histone deacetylase 6 in cytoplasmic retention. J Biol Chem. 2004;279:48246–48254. doi: 10.1074/jbc.M408583200. [DOI] [PubMed] [Google Scholar]
- 24.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 25.Szyk A, et al. Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell. 2014;157:1405–1415. doi: 10.1016/j.cell.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovacs JJ, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Cohen TJ, et al. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Comm. 2011;2:252. doi: 10.1038/ncomms1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min SW, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou H, Wu Y, Navre M, Sang BC. Characterization of the two catalytic domains in histone deacetylase 6. Biochem Biophys Res Commun. 2006;341:45–50. doi: 10.1016/j.bbrc.2005.12.144. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Gilquin B, Khochbin S, Matthias P. Two catalytic domains are required for protein deacetylation. J Biol Chem. 2006;281:2401–2404. doi: 10.1074/jbc.C500241200. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 33.Gantt ML, et al. General base-general acid catalysis in human histone deacetylase 8. Biochemistry. 2016;55:820–832. doi: 10.1021/acs.biochem.5b01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pauling L. Molecular architecture and biological reactions. Chem Eng News. 1946;24:1375–1377. [Google Scholar]
- 35.Decroos C, Bowman CM, Christianson DW. Synthesis and evaluation of N8-acetylspermidine analogues as inhibitors of bacterial acetylpolyamine amidohydrolase. Bioorg Med Chem. 2013;21:4530–4540. doi: 10.1016/j.bmc.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang TC, Abeles RH. Complex of α-chymotrypsin and N-acetyl-L-leucyl-L-phenylalanyl trifluoromethyl ketone: structural studies with NMR spectroscopy. Biochemistry. 1987;26:7603–7608. doi: 10.1021/bi00398a011. [DOI] [PubMed] [Google Scholar]
- 37.Vannini A, et al. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 2007;8:879–84. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brosch G, Ransom R, Lechner T, Walton JD, Loidl P. Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell. 1995;7:1941–1950. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furumai R, et al. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc Natl Acad Sci USA. 2001;98:87–92. doi: 10.1073/pnas.011405598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christianson DW, Lipscomb WN. Binding of a possible transition state analogue to the active site of carboxypeptidase A. Proc Natl Acad Sci USA. 1985;82:6840–6844. doi: 10.1073/pnas.82.20.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christianson DW, David PR, Lipscomb WN. Mechanism of carboxypeptidase A: hydration of a ketonic substrate analogue. Proc Natl Acad Sci USA. 1987;84:1512–1515. doi: 10.1073/pnas.84.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JH, et al. Development of a histone deacetylase 6 inhibitor and its biological effects. Proc Natl Acad Sci USA. 2013;110:15704–15709. doi: 10.1073/pnas.1313893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews BW. Structural basis of the action of thermolysin and related zinc peptidases. Acc Chem Res. 21:333–340. [Google Scholar]
- 45.Christianson DW, Lipscomb WN, Carboxypeptidase A. Acc Chem Res. 22:62–69. [Google Scholar]
- 46.Fischle W, et al. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 47.Center RJ, et al. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 1998;7:1612–1619. doi: 10.1002/pro.5560070715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smyth DR, Mrozkiewicz MK, McGrath WJ, Listwan P, Kobe B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003;12:1313–22. doi: 10.1110/ps.0243403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Decroos C, et al. Compromised structure and function of HDAC8 mutants identified in Cornelia de Lange Syndrome spectrum disorders. ACS Chem Biol. 2014;9:2157–2164. doi: 10.1021/cb5003762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burli RW, et al. Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington’s disease. J Med Chem. 2013;56:9934–54. doi: 10.1021/jm4011884. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 56.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (IC50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 59.Hildmann C, et al. A new amidohydrolase from Bordetella or Alcaligenes strain FB188 with similarities to histone deacetylases. J Bacteriol. 2004;186:2328–2339. doi: 10.1128/JB.186.8.2328-2339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim B, Pithadia AS, Fierke CA. Kinetics and thermodynamics of metal-binding to histone deacetylase 8. Protein Sci. 2015;24:354–365. doi: 10.1002/pro.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.