Abstract
During protein synthesis, tRNA and mRNA are translocated from the A to P to E sites of the ribosome thus enabling the ribosome to translate one codon of mRNA after the other. Ribosome translocation along mRNA is induced by the universally conserved ribosome GTPase, elongation factor G (EF‐G) in bacteria and elongation factor 2 (EF‐2) in eukaryotes. Recent structural and single‐molecule studies revealed that tRNA and mRNA translocation within the ribosome is accompanied by cyclic forward and reverse rotations between the large and small ribosomal subunits parallel to the plane of the intersubunit interface. In addition, during ribosome translocation, the ‘head’ domain of small ribosomal subunit undergoes forward‐ and back‐swiveling motions relative to the rest of the small ribosomal subunit around the axis that is orthogonal to the axis of intersubunit rotation. tRNA/mRNA translocation is also coupled to the docking of domain IV of EF‐G into the A site of the small ribosomal subunit that converts the thermally driven motions of the ribosome and tRNA into the forward translocation of tRNA/mRNA inside the ribosome. Despite recent and enormous progress made in the understanding of the molecular mechanism of ribosome translocation, the sequence of structural rearrangements of the ribosome, EF‐G and tRNA during translocation is still not fully established and awaits further investigation. WIREs RNA 2016, 7:620–636. doi: 10.1002/wrna.1354
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
The ribosome translates the sequence of codons in mRNA to synthesize proteins in all living organisms. mRNA codons are decoded by the binding of tRNA molecules charged with amino acids. Both the small and large ribosomal subunits contain three tRNA binding sites: the A (aminoacyl) site, the P (peptidyl) site and the E (exit) site (Figure 1(a)). To extend the polypeptide chain by one amino acid, the ribosome undergoes an elongation cycle that begins with binding of an aminoacyl‐tRNA to the A site followed by the catalysis of peptide transfer from the P‐ to the A‐site tRNA. The elongation cycle is completed when the resulting peptidyl A‐site and deacylated P‐site tRNAs are translocated to the P and E sites, respectively. tRNA translocation is coupled to the movement of the associated codons of the mRNA through the ribosome and is catalyzed by a universally conserved elongation factor (EF‐G in prokaryotes and EF‐2 in eukaryotes). Ribosomal translocation is an essential facet of protein synthesis in all organisms. Additionally, studies of the molecular mechanism of ribosomal translocation contribute to the understanding of the general physical and structural principles underlying the mechanics of macromolecules and macromolecular complexes that undergo unidirectional movement in the cell. Because of the fundamental importance of translocation for protein synthesis and the complexity of the translocation mechanism, this problem remains one of the most fascinating and popular topics in the field of protein synthesis. The emergence of high‐resolution cryo‐EM and X‐ray crystal structures of the ribosome as well as single‐molecule Förster resonance energy transfer (smFRET) and optical tweezers approaches has led to tremendous progress in the understanding of the translocation mechanism in recent years. Nevertheless, a number of important details remain obscure and require further investigation.
Figure 1.

Structural organization of the ribosome and elongation factor G. (a) Crystal structure of the 70S ribosome (Protein Data Bank ID [PDBID] 4V6F1). Large, 50S subunit and small, 30S subunit are colored in light blue and light green, respectively. A‐site, P‐site, and E‐site tRNAs are shown in yellow, orange, and red, respectively. mRNA is colored purple. A box diagram of the ribosome showing tRNAs bound in the A, P, and E sites of the 50S and 30S subunits is shown below the crystal structure of the 70S ribosome. (b) Crystal structure of ribosome‐free EF‐G (PDBID 1DAR2) with domains color‐coded: G′ domain (dark blue), G domain (green), domain II (dark red), domain III (orange), domain IV (magenta), and domain V (light blue).
Below we summarize recent structural and biochemical studies of the translocation of bacterial 70S ribosomes. The functional core of the ribosome, which includes sites of tRNA and EF‐G binding, is conserved throughout all branches of life. Hence, the main features of the translocation mechanism discovered in bacteria are likely similar in archaea and eukaryotes.
FUNDAMENTALS OF RIBOSOMAL TRANSLOCATION
Translocation Is Augmented by the Binding of EF‐G · GTP to the Ribosome
EF‐G is a five‐domain protein2, 3 that accelerates translocation by ~50,000‐fold.4, 5 Domain I of EF‐G (Figure 1(b)) comprises the G′ and G subdomains; the latter hydrolyses GTP and is structurally similar to the G‐domains in other G‐proteins.2, 3 Most published reports suggest that EF‐G binds to the ribosome with high affinity and induces translocation only in GTP‐bound form.6, 7 EF‐G · GDP and nucleotide‐free EF‐G do not show significant translocation activity.6, 8, 9, 10 EF‐G has low intrinsic GTPase activity, which is dramatically enhanced via interaction of the G domain of EF‐G with the universally conserved sarcin‐ricin loop (SRL) of the 23S rRNA of the large ribosomal subunit.11, 12, 13 GTP hydrolysis and the subsequent release of inorganic phosphate trigger EF‐G dissociation from the ribosome.7, 14
Although GTP hydrolysis precedes mRNA/tRNA translocation, the release of inorganic phosphate after GTP hydrolysis and mRNA translocation occur at similar rates. Hence, these two events may be coupled through conformational changes in EF‐G that are induced by Pi release.14 However, inhibiting the ability of EF‐G to hydrolyze GTP by amino acid substitutions in the G domain of EF‐G,15, 16, 17, 18 or replacing GTP with non‐hydrolyzable analogues of GTP4, 9, 19 severely hamper EF‐G turnover but only moderately slow down the rate of a single round of translocation (by 2‐ to 50‐fold, depending on experimental conditions4, 9, 19). Furthermore, a number of antibiotics, such as viomycin or hygromycin B, strongly inhibit translocation by binding to the ribosome without impeding the binding of EF‐G, GTP hydrolysis or inorganic phosphate (Pi) release4, 20, 21 (Table 1). Hence, GTP hydrolysis and translocation do not appear to be directly coupled. Consistent with this model, smFRET experiments show that not every EF‐G binding event results in translocation17, 35, 36 suggesting that EF‐G dissociation triggered by Pi release sometimes occurs before tRNA/mRNA are translocated. Thus, binding of EF‐G · GTP to the ribosome makes a major contribution to the catalysis of translocation while GTP hydrolysis mainly serves to stimulate EF‐G release.
Table 1.
Inhibitors of Ribosomal Translocation
| Inhibitor | Domain of Life | Mechanism |
|---|---|---|
| Aminoglycosides (gentamycin, hygromycin B, kanamycin, neomycin, paromomycin) | B, A, E | Aminoglycosides inhibit both EF‐G‐induced and spontaneous translocation by increasing the affinity of peptidyl‐tRNA to the A site.22 |
| Spectinomycin | B, A | Spectinomycin is thought to inhibit translocation by interfering with swiveling of the head of the small ribosomal subunit.22 |
| Tuberactinomycins (viomycin and capreomycin) | B | Viomycin and capreomycin inhibit both EF‐G‐induced and spontaneous translocation by increasing the affinity of peptidyl‐tRNA to the A site and locking the ribosome in the rotated hybrid state.20, 25, 26 |
| Pactamycin | B, A, E | Pactamycin inhibits translocation by occluding the mRNA‐binding channel in the E site of the small ribosomal subunit.27, 28 |
| Thiostrepton | B, A | Thiostrepton inhibits binding of EF‐G to the ribosome.26, 29, 30 |
| Fusidic acid | B, A, E | Fusidic acid inhibits EF‐G/EF‐2 release after GTP hydrolysis.31 |
| Sordarin | E(fungi) | Inhibits EF‐2 release from the ribosome after GTP hydrolysis.32 |
| EF‐2 kinase | E | EF‐2 kinase phosphorylates a conserved threonine residue (Thr 56 in human EF‐2) in domain I of EF‐2, which hampers EF‐2 binding to the ribosome.33 |
| Diphtheria toxin | E(human) | Diphtheria toxin abrogates EF‐2 activity by catalyzing ADP‐ribosylation of the diphtamide residue (i.e. posttranslationallly‐modified His714 in human EF‐2).34 |
| α‐sarcin and ricin‐like ribotoxins | B, A, E | These ribotoxins can cleave (α‐sarcin) or depurinate (ricin) the SRL of the rRNA of the large subunit, which activates GTPase activity of EF‐G (EF‐2) and EF‐Tu (EF‐1A).22 |
The column ‘Domain of life’ indicates whether the translocation inhibitors are active in bacteria (B), archaea (A) or eukaryotes (E).
Translocation of mRNA Is Driven by tRNA Movement
Toeprinting37 and single‐molecule optical tweezers experiments38 show that mRNA is translocated inside the ribosome three nucleotides at a time in a codon‐by‐codon manner without detectable intermediates, suggesting that the translocation of mRNA is coupled to the movement of the associated anticodon stem‐loops (ASLs) of tRNAs. Indeed, mRNA translocation requires the presence of an ASL bound in the A site of the small subunit and a full‐length deacylated tRNA bound in the P site of the ribosome.37 Binding of EF‐G · GTP to ribosomes containing a single tRNA bound in the P site does not induce translocation.37 By contrast, translocation of tRNAs through the ribosome can be induced by EF‐G in the absence of an mRNA template.39 Mutations in tRNAs that allow the tRNA anticodon loop to form four instead of three basepairs with the nucleotides of mRNA result in the translocation of mRNA by four nucleotides.40, 41 These observations provide further support for the idea that mRNA translocation is passive and is driven by tRNA movement. Nevertheless, structural rearrangements of the small ribosomal subunit may also play a role in directing and facilitating mRNA movement.42
Translocation Is an Intrinsic Feature of the Ribosome
Under certain experimental conditions, translocation of mRNA and tRNA inside the ribosome can occur in absence of EF‐G, albeit very slowly.43, 44 Spontaneous translocation is stimulated by a decrease in the concentration of magnesium ions,44 modification of ribosomal proteins with thiol‐specific reagents45, 46 or removal of ribosomal proteins S12 and S13.47 Furthermore, single‐round translocation can be induced by the peptidyl‐transferase inhibitors, sparsomycin, linkomycin, and chloramphenicol.48, 49
A number of antibiotics bind to the ribosome and impede both EF‐G‐induced and spontaneous translocation47, 48, 49 (Table 1) providing evidence for underlying similarities between the mechanisms of these phenomena and suggesting that tRNA/mRNA translocation is, to a degree, an inherent property of the ribosome. Indeed, structural studies showed that the movement of tRNA/mRNA is accompanied by conformational changes of the ribosome, which will be discussed in detail below.
STRUCTURAL INTERMEDIATES OF RIBOSOMAL TRANSLOCATION
Hybrid‐State Intermediate of Translocation
The translocation of tRNAs appears to occur in a stepwise fashion. Early mapping of tRNA binding sites on the ribosomes using chemical probing revealed that the transpeptidation reaction triggers the spontaneous movement of the acceptor stems of the resulting peptidyl‐tRNA and deacylated tRNA from A and P to P and E sites of the large subunit, respectively, while tRNA ASLs remain in the A and P sites of the small subunit.50 Thus, the tRNAs move into the hybrid A/P and P/E sites50 (Figure 2(a) and (b)). FRET, chemical probing and cryo‐EM experiments suggested that, in pretranslocation ribosomes, the spontaneous movement of tRNAs into the hybrid state is coupled to rotation of the platform and body domains of the small ribosomal subunit by 8–10°25, 51, 52, 53 (Figure 3(a)). The intersubunit rotation is often described as a ratchet‐like movement or simply ‘ratcheting’.57 smFRET experiments show that, in the absence of EF‐G, pretranslocation ribosomes spontaneously fluctuate between nonrotated, classical and rotated, hybrid state conformations.58, 59, 60, 61 smFRET experiments also suggest that the nonrotated, classical and the rotated, hybrid state conformations of pretranslocation ribosomes have similar thermodynamic stabilities.58, 60, 61, 62
Figure 2.
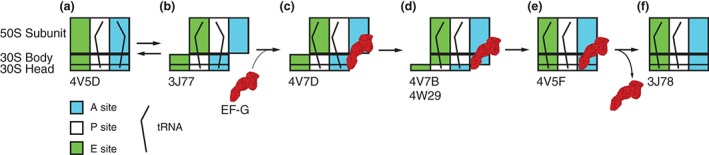
Scheme of tRNA rearrangements during EF‐G‐catalyzed ribosome translocation. Diagrams show tRNA positions relative to the A, P, and E sites on the 50S subunit and 30S head and body. (a) peptidyl‐ and deacylated tRNAs are bound in A/A and P/P classical states, (b) A/P and P/E hybrid states, (c) A/P* and P/E states in the presence of ribosome‐bound EF‐G, (d) ap/P and pe/E chimeric states in the presence of ribosome‐bound EF‐G, (e) classical P/P and E/E state in the presence of ribosome‐bound EF‐G, and (f) classical P/P and E/E state after EF‐G dissociation. Please see additional details in the text. PDB IDs corresponding to each structural state are indicated under the schematic.
Figure 3.
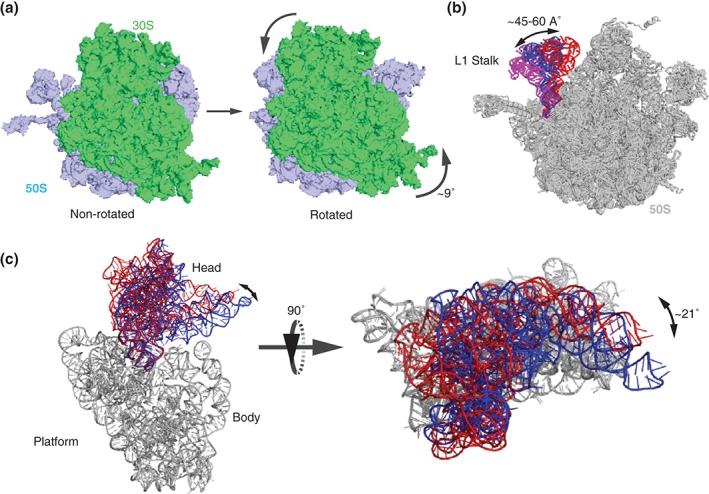
Structural rearrangements of the ribosome that accompany translocation. (a) Structures of the ribosome in the non‐rotated, classical state54 (PDBID 4V9D) and the rotated, hybrid state55 (PDBID 4V7C). The small subunit is shown in light green and the large subunit is shown light blue. The ribosome is viewed from the solvent side of the small subunit. Curved arrows indicate the counter‐clockwise rotation of the small subunit relative to the large subunit. (b) 50S L1 stalk (23S rRNA helices 76, 77, and 78) is shown in the closed54 (red, PDBID 4V9D), half‐closed1 (blue, PDBID 4V6F), and open54 (magenta, PDBID 4V9D) positions. The rest of the large subunit is shown in gray. Structures were superimposed by structural alignment of 23S rRNA. (c) The swiveling motion of the head domain of the small subunit is shown by structural alignment of the body and platform domains of 16S rRNA (shown in gray) of crystal structures of nonrotated ribosome containing classical A/A, P/P, and E/E site tRNAs (head domain in blue, PDBID 4V5156) and partially rotated ribosome containing a chimeric ap/ap and pe/E tRNAs (head domain in red, PDBID 4W2942). The 30S is viewed from its solvent side (left) and the ‘top’ of the small subunit head (right). Double‐headed arrows indicate the direction of head swiveling, which is perpendicular to the long axis of the small subunit.
Available structures of the ribosome suggest that the movement of deacylated tRNA from the classical P/P to the hybrid P/E state is strictly coupled to intersubunit rotation. By contrast, the peptidyl‐tRNA can spontaneously sample both classical A/A and hybrid A/P states in ribosomes that adopt the rotated conformation.59, 63 Furthermore, in rotated bacterial and mammalian ribosomes containing a deacylated tRNA bound in the hybrid P/E state, the peptidyl‐tRNA can adopt another conformation that is distinct from canonical A/A and A/P states. This distinct conformation was termed A/P*55 (A/P in Ref64 and ‘state 5’ in Ref65). Similar to the canonical A/P‐tRNA, the anticodon and CCA ends of the A/P* tRNA are positioned in the A and P sites of the small and large ribosomal subunits, respectively, whereas the tRNA elbow is moved around the A site finger (conserved helix 38 of 23S or 28S rRNA) by 20–25Å toward the P site relative to canonical A/P state55 (Figure 2(c)).
In the absence of EF‐G, spontaneous fluctuations of the pretranslocation ribosome between nonrotated, classical and rotated, hybrid states do not lead to efficient translocation of tRNA/mRNA on the small subunit, which evidently requires EF‐G. Furthermore, peptidyl‐tRNAs bound in the A/P state appear to react with the antibiotic puromycin, a 50S A‐site aminoacyl‐tRNA mimic, with lower efficiency than peptidyl‐tRNAs bound in the classical P/P state.50, 66, 67 Hence, the movement of the peptidyl‐tRNA on the large subunit into a fully puromycin‐reactive conformation also necessitates EF‐G.50, 66, 67 Mutations in rRNA that destabilize tRNA binding in the hybrid states only moderately affect the rate of mRNA and tRNA translocation.68, 69, 70 Therefore, formation of the hybrid state intermediate is not the rate‐limiting step of translocation. Additional EF‐G‐induced rearrangements of the ribosome and tRNAs are likely required to facilitate tRNA/mRNA translocation.
Coupling of Intersubunit Rotation and mRNA Translocation
Upon binding to the pretranslocation ribosome, EF‐G was shown to transiently stabilize the rotated, hybrid state10, 19, 57, 71 and subsequently induce the transition into the nonrotated, classical state conformation of the ribosome19, 35, 51 (Figure 2). In contrast to pretranslocation ribosomes containing a deacylated P‐site tRNA that spontaneously fluctuates between nonrotated, classical and rotated hybrid state conformations, posttranslocation ribosomes containing a peptidyl‐tRNA in the P/P state are predominately fixed in the nonrotated conformation.60 The intersubunit rotation and the movement of peptidyl‐tRNA from the classical P/P into the hybrid P/E state are likely blocked in posttranslocation ribosomes because the 50S E site binds only deacylated tRNA.72, 73 Therefore, the acylation state of the P‐site tRNA controls the structural dynamics of tRNAs and the ribosome.
Translocation of mRNA and tRNA on the small subunit was shown in kinetic experiments to occur at a rate similar to the rate of reverse rotation of the small subunit from the rotated back into the nonrotated conformation.19 Inhibition of translocation with the antibiotics spectinomycin or hygromycin B slows down mRNA translocation and the reverse movement of the small subunit to the same extent, further supporting the idea that the translocation of mRNA and ASLs of tRNA are coupled to the reverse rotation of the small subunit.19 Consistent with these results, the antibiotics viomycin and neomycin, inhibitors of translocation, were shown to perturb intersubunit rotation by trapping the ribosome in conformations in which subunits are rotated relative to each other by ~10°25, 55 and 6°,74 respectively. Furthermore, a cross‐link introduced between proteins S6 of the 30S and L2 of the 50S subunits to block intersubunit rotation was shown to completely abolish ribosomal translocation75 demonstrating that intersubunit rotation is essential for translocation.
Movement of the L1 Stalk of the Large Ribosomal Subunit
Intersubunit rotation and translocation of tRNAs are also coupled to the movement of a mobile domain of the large ribosomal subunit named the L1 stalk, which comprises ribosomal protein L1 and helices 76, 77 and 78 of 23S rRNA71 (Figure 3(b)). In the classical, nonrotated state of prestranslocation ribosomes containing tRNAs bound in A/A and P/P state the L1 stalk is predominantly positioned in the ‘open’ conformation, oriented away from the core of the 50S subunit.54, 61, 76 In the rotated, hybrid state of pretranslocation ribosomes, the L1 stalk is moved inward by 45–60Å (depending on the points chosen for distance measurements) into the ‘closed’ position and interacts with the elbow of the P/E tRNA54, 61, 71, 76 (Figure 3(b)). In the absence of EF‐G, the L1 stalk spontaneously fluctuates between the open and closed positions in a movement that is coupled to the fluctuations of tRNAs between classical and hybrid states, respectively.61, 76 As a result of EF‐G‐induced translocation, the L1 stalk adopts an intermediate position between the closed and open positions (i.e., ‘half‐closed’ state) moving outward by 15–20 Å relative to fully closed position76, 77 (Figure 3(b)). In this half‐closed conformation, the L1 stalk retains its interactions with the elbow of the deacylated tRNA, which is bound in the E/E classical state.1, 78 The L1 stalk moves into the open position when the deacylated E‐site tRNA disassociates from the ribosome.76, 79 It appears that the movement of the L1 stalk enables remodeling of the 50S E site between the E/E vacant, P/E hybrid and E/E classical states and facilitates the translocation and release of deacylated tRNA (Box 1).
BOX 1. INTERSUBUNIT ROTATION ACCOMPANIES TRANSLATION IN BOTH BACTERIA AND EUKARYOTES.
Nonrotated, classical and rotated, hybrid state conformations of the ribosome appear to be the two predominant conformations that the ribosome adopts during protein synthesis. In addition to sampling the classical and hybrid states during the elongation cycle, the ribosome adopts the nonrotated conformation in complex with release factors 1 and 2 (RF1 and RF2) during translation termination.23, 24, 80 The rotated conformation of the ribosome that contains a single deacylated tRNA bound in the hybrid P/E state is sampled during the termination and recycling phases of protein synthesis and is stabilized by bacterial release factor 3 (RF3)51, 80, 81 and ribosome recycling factor (RRF).54, 80 Furthermore, the rotated conformation of the ribosome and hybrid states of tRNA binding were also observed in yeast and mammalian ribosomes, suggesting that hybrid state formation is a universal feature of protein synthesis.64, 82, 83
Swiveling Motion of the 30S Head
The small ribosomal subunit comprises three structural domains: head, body and platform (Figure 3(c)). In a number of structures of both bacterial and eukaryotic ribosomes, a large rotation of the head domain of the small subunit by up to 21° relative to the rest of the small subunit was observed57, 84, 85 (Figure 3(c)). The axis of this rotation, termed head swivel, is perpendicular to the axis of intersubunit rotation. The head swivel moves the ASLs of tRNAs and the associated mRNA codons along the direction of translocation through the ribosome. Furthermore, the head swivel is thought to be essential for ribosomal translocation because it opens a wide (over 20 Å) path for tRNA translocation between the P and E sites on the small subunit that is otherwise constricted by the rRNA residues of the head and platform of the small subunit.85 Kinetic measurements of FRET between fluorophores introduced into the 30S head and body or 30S head and platform demonstrated that the 30S head swiveling indeed accompanies translocation.86 Moreover, the antibiotic spectinomycin is thought to inhibit translocation in bacteria by binding to one of the hinges connecting the head with the rest of the small subunit and, thus, trapping the head of the small subunit in a partially swiveled state.87, 88, 89 Analysis of available structures suggests that in contrast to intersubunit rotation and L1 stalk movement, the 30S head swivel does not correlate with spontaneous movement of tRNAs into A/P and P/E hybrids states.88 Consistent with this idea, ensemble kinetic and smFRET experiments suggest that spectinomycin stabilizes an intermediate of translocation that is distinct from the hybrid state intermediate.9, 36
Kinetic FRET experiments showed that the 30S head swivel is triggered by EF‐G binding and may follow the formation of the hybrid, rotated state but precede translocation of tRNAs on the small subunit.86 The back‐swiveling motion of the 30S head into the non‐swiveled conformation occurs at a rate that is similar to the rates of mRNA translocation and reverse intersubunit rotation.86 Although swiveling and back‐swiveling motions of the 30S head were resolved in these kinetic translocation experiments, an intermediate of translocation that shows a significant degree (≥8°) of both intersubunit rotation and 30S head swiveling in 2‐tRNA · ribosome complexes is yet to be visualized using cryo‐EM or X‐ray crystallography.
Several cryo‐EM and crystallographic studies revealed an EF‐G‐bound structural state of the ribosome, in which ribosomal subunits are rotated by less than 3.5°, while the 30S head is swiveled by 18–21°.42, 90, 91, 92 In this structural state of the ribosome, tRNAs are in unique binding sites called ‘chimeric’ ap/P (ap/ap) and pe/E states (Figure 2(e)). The acceptor arms of ap/P and pe/E tRNAs are bound in the posttranslocation P and E site on the large subunits. Their ASLs interact with posttranslocation P and E sites of the platform and the body of the 30S while retaining interactions with elements of the 30S A and P site located in the 30S head. Hence, ‘chimeric’ ap/P (ap/ap) and pe/E tRNAs are trapped midway between canonical hybrid A/P and P/E and posttranslocation classical P/P and E/E states. This structural state of the ribosome may represent a late intermediate of translocation, which is formed as a result of partial reverse rotation of the 30S platform and body from the rotated, hybrid state intermediate of translocation. Further kinetic studies are required to clarify the possible order and coupling of reverse intersubunit rotation, back‐swivel of the head domain of the small subunit and translocation of tRNA/mRNA on the small subunit.
Structural Rearrangements of EF‐G during Ribosomal Translocation
Structural and smFRET studies showed that in addition to conformational changes of the ribosome, translocation is also accompanied by rearrangements of ribosome‐bound EF‐G. Recent cryo‐EM and X‐ray crystallography studies allowed for visualization of EF‐G · ribosome · 2‐tRNA complexes in four different conformations: (1) EF‐G bound to the nonrotated pretranslocation ribosome containing A/A‐ and P/P‐ tRNAs93 (Figure 4(a)); (2) EF‐G bound to the fully rotated pretranslocation ribosome containing A/P*‐ and P/E‐ tRNAs55 (Figure 4(b)); (3) EF‐G bound to moderately rotated ribosomes containing tRNAs in chimeric ap/P‐(ap/ap) and pe/E‐ tRNAs42, 94 (Figure 4(c)) and (4) EF‐G bound to the nonrotated posttranslocation ribosome containing P/P‐ and E/E‐ tRNAs13 (Figure 4(d)).
Figure 4.
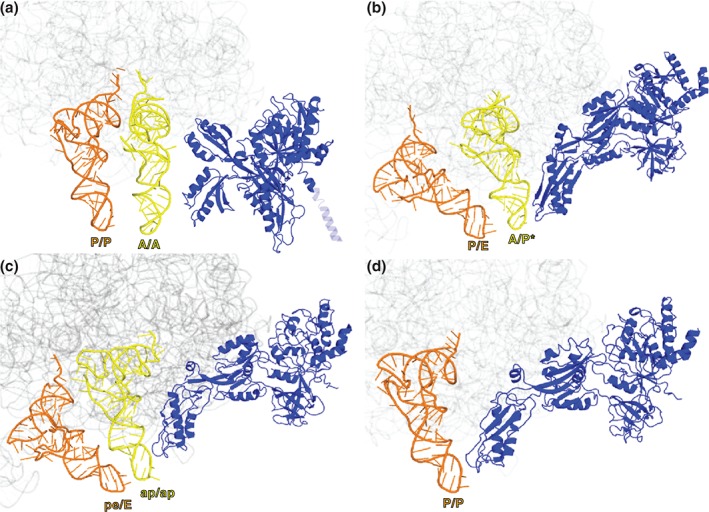
Structural rearrangements of EF‐G on the ribosome during translocation. (a) EF‐G (blue in all structures) bound to the nonrotated pretranslocation ribosome containing A/A‐ and P/P‐ tRNAs93 (PDBID 4WPO). The N‐terminal domain of large subunit protein L9 covalently linked to the N‐terminus of EF‐G is shown as a transparent blue. (b) EF‐G bound to the fully rotated pretranslocation ribosome containing A/P*‐ and P/E‐ tRNAs55 (PDBID 4V7D); (c) EF‐G bound to partially rotated ribosomes containing chimeric ap/ap and pe/E‐ tRNAs42 (PDBID 4 W29) and (d) EF‐G bound to the nonrotated posttranslocation ribosome containing P/P‐ and E/E‐ tRNAs13 (PDBID 4V5F). 23S rRNA in all structures is shown in gray.
The key difference between these structures is the conformation of domain IV of EF‐G. The elongated domain IV of EF‐G is critical for the catalysis of translocation. Deletion or mutation of domain IV abolishes the translocation activity of EF‐G.4, 95, 96 Furthermore, diphtheria toxin inhibits protein synthesis in humans by ADP‐ribosylation of a conserved post‐translationally modified histidine residue located at the tip of domain IV.34 In the structure of the posttranslocation ribosome, in which the A site of the ribosome is not occupied by tRNA, the elongated domain IV of EF‐G is docked in the 30S A site13, 71 (Figures 4(d) and 5). EF‐G bound to the ribosome containing tRNAs in the ap/P (ap/ap) and pe/E chimeric states adopts a conformation that is very similar to its posttranslocation conformation42, 94 (Figure 4(c)). However, domain IV of EF‐G is not fully docked in the 30S A site because of the large (~21°) swivel of the 30S head. In the pretranslocation EF‐G‐ribosome complexes captured in the presence of an inhibitor of translocation, the antibiotic viomycin, domain IV of EF‐G is positioned approximately 15–20 Å away from the A site relative to the postranslocation conformation of ribosome‐bound EF‐G55 (Figures 4(b) and 5). smFRET data suggest that a similar pretranslocation conformation of EF‐G, in which domain IV of EF‐G is positioned away from the A site, is sampled during translocation not perturbed by antibiotics.17 Evidently, tRNA translocation on the small subunit is accompanied by the movement of domain IV toward the 30S A site from the conformation observed in the viomycin‐trapped structure into the conformation observed in the structure of the postranslocation ribosome. This movement mostly results from the rotation of the entire EF‐G around the SRL of the 23S rRNA55 (Figure 5).
Figure 5.

Movement of EF‐G from the pre‐ to posttranslocation conformation on the ribosome. EF‐G (red, PDBID 4V7D55) bound to the viomycin‐trapped pretranslocation ribosome is superimposed with EF‐G bound to the posttranslocation ribosome (blue, PDBID 4V5F13). The superimposition is obtained by structural alignment of respective 23S rRNAs. A/P* tRNA is shown in yellow. 23S rRNA of pre and posttranslocation ribosomes are shown in transparent red and blue, respectively.
In addition to the movement relative to the ribosome, EF‐G undergoes interdomain rearrangement that involves the movement of two superdomains, one formed by domains III, IV and V and the other formed by domains I and II, relative to each other (Figure 6). Crystallographic, cryo‐EM and FRET studies showed that EF‐G adopts fairly similar extended, elongated conformations when bound to (1) hybrid‐state pretranslocation,55 (2) chimeric‐state pretranslocation42, 94 or (3) nonroated posttranslocation ribosomes13 (Figure 6). By contrast, ribosome‐free EF‐G assumes a more compact conformation where superdomain III‐IV‐V is positioned closer to superdomain I‐II2, 3 (Figure 6(a)). The rearrangements between ribosome‐bound, posttranslocation and ribosome‐free conformations result in a movement of the tip of domain IV by approximately 30 Å (Figure 6(a)). An even more dramatic interdomain rearrangement of EF‐G is observed in the structure of EF‐G bound to a pretranslocation ribosome captured in the nonrotated conformation in the presence of the antibiotic dityromycin that was recently solved using X‐ray crystallography.93 In this structure, the tip of domain IV of EF‐G is moved by approximately 100 Å relative to the conformation of EF‐G bound to the posttranslocation ribosome (Figure 6(b)).
Figure 6.
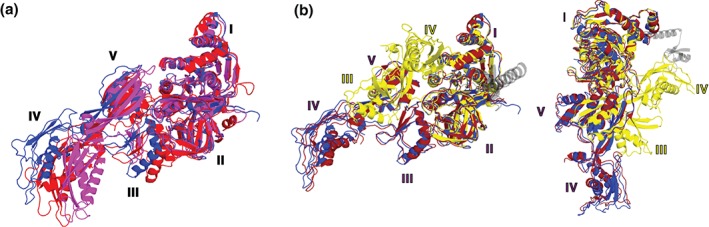
Interdomain rearrangements of EF‐G. (a) EF‐G in the posttranslocation conformation (blue, PDBID 4V5F13) is superimposed by structural alignment of domains I and II with ribosome‐free EF‐G•GDP (magenta, PDBID 1DAR2) and EF‐G bound to the viomycin‐trapped pretranslocation ribosome (red, PDBID 4V7D55), (b) EF‐G in the posttranslocation conformation (blue, PDBID 4V5F13) is superimposed by structural alignment of domains I and II with EF‐G bound to a ribosome containing a chimeric ap/ap tRNA (dark red, PDBID 4W2942) and EF‐G‐L9 fusion bound to the nonrotated, pretranslocation ribosome (yellow, PDBID 4WPO93). The N‐terminal domain of L9 covalently linked to EF‐G (PDBID 4WPO93) is shown in transparent gray. Two differently oriented views of EF‐G structures are shown. Domains of EF‐G are numbered as indicated.
It has been hypothesized that EF‐G promotes translocation by the movement of superdomain III‐IV‐V relative to superdomain I‐II from highly compact to the extended conformation observed in posttranslocation ribosomes.4, 93 Indeed, restricting the conformational dynamics of EF‐G by introducing an intramolecular disulfide crosslink between domains I and V was shown to abolish EF‐G translocation activity, suggesting the importance of interdomain rearrangements for translocation.97 However, recent smFRET experiments showed that neither abrogation of GTP hydrolysis nor inhibition of translocation by the antibiotic viomycin hamper the rearrangement of EF‐G from a compact to the extended conformation.98 Therefore, it is likely that the movement of EF‐G from the compact conformation, which EF‐G adopts in the absence of the ribosome, into a more extended conformation is not coupled to but likely precedes both GTP hydrolysis and mRNA/tRNA translocation.98
The minimal model for the structural rearrangements of EF‐G, the ribosome and tRNAs during translocation includes following steps (Figure 2): (1) Upon binding to the ribosome, EF‐G moves from a compact to an extended conformation and transiently stabilizes the rotated conformation of the ribosome with tRNAs bound in the hybrid state (A/P* and P/E states). (2) EF‐G binding also leads to the swiveling motion of the head domain of the small subunit. (3) Upon reverse rotation of the small subunit back into the nonrotated conformation and back‐swiveling motion of the head of the small subunit (the order or possible coupling of these two events is not fully established) tRNAs are translocated into classical P/P and E/E states. Translocation of tRNAs is accompanied by the movement of mRNA by one codon and docking of domain IV of EF‐G into the A site of the small subunit, which is vacated by the peptidyl‐tRNA. The movement of domain IV of EF‐G toward the A site of the small subunit and the reverse rotation of the small subunit are hampered by translocation inhibitors and, thus, directly coupled to mRNA/tRNA translocation. (4) By contrast, GTP hydrolysis is not directly coupled to any of these rearrangements. Nevertheless, the release of inorganic phosphate on average occurs concurrently with translocation and triggers EF‐G disassociation from the ribosome. Presented here, the model includes only those events that were detected in kinetic ensemble and single‐molecule biophysical experiments and visualized by cryo‐EM or crystallographic reconstruction of 2‐tRNA ribosome complexes. However, this minimal model of ribosome translocation is likely incomplete. Indeed, a number of distinct conformations of the ribosome were observed in structures of EF‐G bound to ribosomes that either were vacant or contained a single tRNA in the P site.90, 92, 99, 100 Hence, at least some of these structural states may represent additional intermediates of translocation.
THE MOLECULAR MECHANISM OF TRANSLOCATION
Mechanics of Translocation
Macromolecules and macromolecular complexes move in a unidirectional manner by converting the energy of a chemical reaction into mechanical movement. There are two fundamental mechanisms of such conversion. In the first mechanism, called the power stroke, chemical change occurs either concurrently with the movement or precedes it.101 In the second mechanism, called the Brownian ratchet, the movement occurs spontaneously and precedes the chemical change.101 In the Brownian ratchet mechanism, the chemical change traps the macromolecule in the post‐movement state thus acting like a pawl that rectifies the movement of the wheel of a mechanical ratchet. These two mechanisms can be distinguished by examination of the load dependence of the translocation rate.101 Recent optical tweezers measurements of ribosomal translocation against an applied force suggested that the ribosome likely translocates by the Brownian ratchet mechanism.102 It is noteworthy, however, that the Brownian ratchet and the power stroke mechanisms are idealized models; ribosomal translocation may combine features of both models.102
What is the chemical change that biases the thermally driven movements of tRNAs and thus, acts as the ‘pawl’ of the Brownian ratchet mechanism of translocation? Observation of spontaneous translocation implies that ribosomal translocation is a downhill process driven by the energy of peptide bond formation. Indeed, the maximum mechanical work generated by ribosomal translocation measured in optical tweezers experiments is comparable to the energy of peptide bond formation.102 Still, it is not clear how the energy of transpeptidation can be conveyed to promote tRNA and mRNA movements. On one hand, an early proposal suggested that peptide bond formation renders the translocation of tRNA thermodynamically favorable because it decreases the affinity of P‐site tRNA toward the P site due to deacylation.103 However, later measurements revealed that deacylation of tRNA does not significantly change its affinity to the P site, while the affinity of esterified tRNA to the A site is similar to the affinity of deacylated tRNA to the E site, thus, making net free energy change of tRNA translocation from A and P sites to P and E sites close to zero.72, 73, 104 Consistent with these data, it was demonstrated that spontaneous forward translocation is unfavorable in some tRNA/mRNA contexts since the reverse spontaneous translocation of tRNAs and mRNA from P and E to A and P sites is instead observed.105, 106 On the other hand, the deacylation of the P‐site tRNA during peptide bond formation dramatically alters ribosome and tRNA dynamics58, 60, 61 and, thus, may facilitate tRNA translocation. In any case, the reaction of transpeptidation is unlikely to be the only energy source of translocation because it would require an unusually high efficiency of conversion (~80%) of chemical energy into mechanical motion, which is untypical for molecular motors.102 Therefore, translocation is likely promoted by the energy stored in EF‐G · GTP, although, as discussed above, GTP hydrolysis itself may not be directly coupled to translocation. Hence, by binding to the ribosome, EF‐G · GTP probably acts as the ‘pawl’ of the Brownian ratchet of translocation.
The Role of Domain IV of EF‐G in Ribosome Translocation
Domain IV of EF‐G promotes tRNA translocation, at least in part, by biasing peptidyl‐tRNA movement and coupling it to ribosome dynamics. Comparison of the rotated pre‐ and nonrotated post‐translocation states of the EF‐G‐ribosome complex suggests that domain IV of EF‐G acts as a steric hindrance for the return of the peptidyl‐tRNA from the hybrid A/P* into the classical A/A state and, thus, promotes the movement of peptidyl‐tRNA into the P site of the small subunit upon reverse rotation of the small subunit (Figure 7). Likewise, domain IV of EF‐G creates a steric hindrance for the return of peptidyl‐tRNA from the chimeric ap/P‐(ap/ap) into the classical A/A state upon back‐rotation of the 30S head from the swiveled conformation. In addition, domain IV of EF‐G docks into the A site of the small ribosomal subunit concurrently or right after the movement of the peptidyl‐tRNA into the P site of the small subunit and, thus, prevents its reverse translocation (Figure 5).
Figure 7.
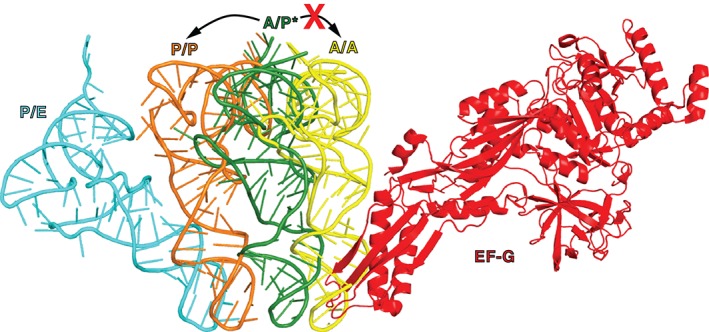
EF‐G may promote tRNA translocation by clashing with A‐site tRNA. A/A (yellow, PDBID 4V5D107), A/P* (dark green, PDBID 4V7D55), P/P (orange, PDBID 4V5D107), and P/E (cyan, PDBID 4V7C55) tRNAs are superimposed by structural alignment of 23S rRNA in the respective structures. EF‐G bound to the viomycin‐trapped pretranslocation ribosome is shown in red (PDBID 4V7D55). In the absence of EF‐G, spontaneous intersubunit rotation is coupled to the fluctuation of tRNAs between classical (A/A and P/P) and hybrid (A/P [A/P*] and P/E) states. In the presence of ribosome‐bound EF‐G, upon the reverse intersubunit rotation from the rotated to nonrotated conformations of the ribosome, the movement of peptidyl‐tRNA from A/P* to A/A state is disallowed because of the clash with domain IV of EF‐G and, thus, the peptidyl‐tRNA translocates into the P/P state.
The idea that EF‐G can promote translocation of tRNAs by biasing tRNA movements inside the ribosome is further supported by the phenomenon of antibiotic‐induced translocation. The antibiotics, sparsomycin, linkomycin and chloramphenicol, that bind to the A site of the large subunit were found to trigger mRNA and tRNA translocation on the small subunit.48, 49 It has been hypothesized that these antibiotics induce translocation by sterically blocking the reverse, nonproductive movement of the peptidyl‐tRNA from the hybrid A/P to classical A/A state during spontaneous intersubunit rotation.49 Therefore, the observation of antibiotic‐induced translocation is consistent with the idea that the intrinsic dynamics of the ribosome can be rectified into unidirectional translocation by ligand binding.
Diffusion of tRNAs inside the ribosome is hindered by their tight binding to the A and P sites of the small subunit as evident from the very slow rate of antibiotic‐induced translocation. Hence, in addition to biasing tRNA movement, EF‐G likely also induces conformational changes of the ribosome that facilitate the diffusion of tRNA/mRNA through the ribosome. This rate‐limiting step of translocation is often referred to as ‘unlocking.’ However, after almost five decades of intense studies of ribosomal translocation since the term ‘unlocking’ was first introduced,108 the identity of the conformational change induced by EF‐G that ‘unlocks’ the ribosome and, thus, facilitates tRNA diffusion remains unclear.
EF‐G may enable diffusion of tRNAs inside the ribosome by inducing conformational changes in the 30S decoding center and, thus, destabilizing the binding of peptidyl‐tRNA to the A site of the small ribosomal subunit.96, 109 In addition, EF‐G may also facilitate tRNA translation by inducing the head swivel of the small subunit, which opens the otherwise constricted path for the movement of tRNA from the P to the E site on the small subunit.85, 86 Additional studies are required to precisely identify the rearrangement of the ribosome that is rate‐limiting for translocation.
CONCLUSION
Concluding Remarks
Emerging over the last 15 years, X‐ray crystal and cryo‐EM structures of the ribosome revolutionized our understanding of translation and revealed dramatic conformational rearrangements of the ribosome, tRNA and EF‐G that accompany ribosome translocation. Ensemble kinetic measurements together with single‐molecule FRET and optical tweezers experiments provided important insights into the structural dynamics, kinetics and mechanics of translocation. However, structural and biophysical studies of translocation also raised new unanswered questions.
Instead of converging onto a few defined structural states that likely resemble intermediates of translocation, the dozens of cryo‐EM and crystal structures obtained to date reveal great structural heterogeneity of elongation‐like complexes of the ribosome. In particular, these structures show large variability in the degree of intersubunit rotation and the 30S head swivel.88 In equilibrium structural studies, transient intermediates of translocation are trapped by use of antibiotics inhibiting translocation (Table 1) and non‐hydrolysable GTP analogues impeding EF‐G release. Other factors such as crystal lattice in X‐ray crystallography and interactions of the ribosome with the water–air interface and the supporting film in cryo‐EM microscopy110 may also alter the relative stability of various structural states of the ribosome. Using the terminology of energy landscape theory, crystal and cryo‐EM structures likely represent energy minima (i.e., mesoscopic states) in an energy landscape111 that are perturbed by antibiotics and other experimental conditions. Here lies of one of the major challenges as at least some structures of elongation‐like ribosome complexes may represent off‐pathway states that are not sampled during translocation. In addition, translocation from the pre‐ to postranslocation state of the ribosome may occur through several alternative pathways.112, 113 Finally, the unperturbed energy landscape of translocation may be smooth and, thus, structural states of the ribosome captured by antibiotics and/or GTP analogues and visualized using X‐ray crystallography and cryo‐EM microscopy may represent artificially stabilized snapshots of the trajectory of structural changes associated with translocation rather than discrete meta‐stable intermediates.
Further kinetic ensemble and single‐molecule experiments are required to complement existing structural data in order to reconstruct the trajectory (or alternative trajectories) of the structural transitions of the ribosome, tRNA, mRNA and EF‐G during ribosomal translocation and identify authentic intermediates of translocation. However, kinetic biophysical studies do not reveal high‐resolution structural details. Hence, developing time‐resolved cryo‐EM114, 115 and molecular dynamic simulation approaches113 will likely be instrumental in the investigation of ribosome translocation in the future. Finally, the studies of the translocation mechanism in bacteria need to be further extended to the examination of ribosomal translocation in eukaryotes, which likely has a number of unique features.
ACKNOWLEDGMENTS
We apologize to all authors whose works has not been cited due to space limitation. We thank Andrei Korostelev for the comments on the manuscript. The work of the D.N.E lab was supported by grant from the US National Institute of Health no. GM‐099719 (to D.N.E.). C.L. was partially supported by NIH Training Grant in Cellular, Biochemical, and Molecular Sciences 5 T32 GM‐068411.
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
FURTHER READING
- Frank J, Gonzalez RL Jr. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem 2010, 79:381–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Molecular Machines in Biology: Workshop of the Cell. Cambridge and New York: Cambridge University Press; 2011. [Google Scholar]
- Moore PB. How should we think about the ribosome? Annu Rev Biophys 2012, 41:1–19. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem 2013, 82:203–236. [DOI] [PubMed] [Google Scholar]
REFERENCES
- 1. Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 2010, 17:555–560. [DOI] [PubMed] [Google Scholar]
- 2. AEvarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al‐Karadaghi S, Svensson LA, Liljas A. Three‐dimensional structure of the ribosomal translocase: elongation factor G from thermus thermophilus. EMBO J 1994, 13:3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J 1994, 13:3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 1997, 385:37–41. [DOI] [PubMed] [Google Scholar]
- 5. Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 2002, 41:12806–12812. [DOI] [PubMed] [Google Scholar]
- 6. Inoue‐Yokosawa N, Ishikawa C, Kaziro Y. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J Biol Chem 1974, 249:4321–4323. [PubMed] [Google Scholar]
- 7. Belitsina NV, Glukhova MA, Spirin AS. Translocation in ribosomes by attachment‐detachment of elongation factor G without GTP cleavage: evidence from a column‐bound ribosome system. FEBS Lett 1975, 54:35–38. [DOI] [PubMed] [Google Scholar]
- 8. Zavialov AV, Hauryliuk VV, Ehrenberg M. Guanine‐nucleotide exchange on ribosome‐bound elongation factor G initiates the translocation of tRNAs. J Biol 2005, 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell 2007, 25:519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid‐state conformation of the 70S ribosome. RNA 2007, 13:1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parmeggiani A, Sander G. Properties and regulation of the GTPase activities of elongation factors Tu and G, and of initiation factor 2. Mol Cell Biochem 1981, 35:129–158. [DOI] [PubMed] [Google Scholar]
- 12. Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF‐G and EF‐Tu with a conserved loop in 23S RNA. Nature 1988, 334:362–364. [DOI] [PubMed] [Google Scholar]
- 13. Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 2009, 326:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G‐induced ribosome rearrangement precedes tRNA‐mRNA translocation. Mol Cell 2003, 11:1517–1523. [DOI] [PubMed] [Google Scholar]
- 15. Mohr D, Wintermeyer W, Rodnina MV. Arginines 29 and 59 of elongation factor G are important for GTP hydrolysis or translocation on the ribosome. EMBO J 2000, 19:3458–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cunha CE, Belardinelli R, Peske F, Holtkamp W, Wintermeyer W, Rodnina MV. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation 2013, 1:e24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salsi E, Farah E, Dann J, Ermolenko DN. Following movement of domain IV of elongation factor G during ribosomal translocation. Proc Natl Acad Sci U S A 2014, 111:15060–15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koripella RK, Holm M, Dourado D, Mandava CS, Flores S, Sanyal S. A conserved histidine in switch‐II of EF‐G moderates release of inorganic phosphate. Sci Rep 2015, 5:12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol 2011, 18:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G‐dependent tRNA‐mRNA translocation. J Mol Biol 2004, 343:1183–1194. [DOI] [PubMed] [Google Scholar]
- 21. Wintermeyer W, Savelsbergh A, Semenkov YP, Katunin VI, Rodnina MV. Mechanism of elongation factor G function in tRNA translocation on the ribosome. Cold Spring Harb Symp Quant Biol 2001, 66:449–458. [DOI] [PubMed] [Google Scholar]
- 22. Wilson DN. The A‐Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol 2009, 44:393–433. [DOI] [PubMed] [Google Scholar]
- 23. Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature 2008, 454:852–857. [DOI] [PubMed] [Google Scholar]
- 24. Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 2008, 322:953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol 2007, 14:493–497. [DOI] [PubMed] [Google Scholar]
- 26. Modolell J, Vazquez. The inhibition of ribosomal translocation by viomycin. Eur J Biochem 1977, 81:491–497. [DOI] [PubMed] [Google Scholar]
- 27. Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P‐site RNA binding. Mol Cell 2004, 13:113–124. [DOI] [PubMed] [Google Scholar]
- 28. Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014, 513:517–522. [DOI] [PubMed] [Google Scholar]
- 29. Cameron DM, Thompson J, March PE, Dahlberg AE. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J Mol Biol 2002, 319:27–35. [DOI] [PubMed] [Google Scholar]
- 30. Walter JD, Hunter M, Cobb M, Traeger G, Spiegel PC. Thiostrepton inhibits stable 70S ribosome binding and ribosome‐dependent GTPase activation of elongation factor G and elongation factor 4. Nucleic Acids Res 2012, 40:360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bodley JW, Zieve FJ, Lin L. Studies on translocation. IV. The hydrolysis of a single round of guanosine triphosphate in the presence of fusidic acid. J Biol Chem 1970, 245:5662–5667. [PubMed] [Google Scholar]
- 32. Justice MC, Hsu MJ, Tse B, Ku T, Balkovec J, Schmatz D, Nielsen J. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J Biol Chem 1998, 273:3148–3151. [DOI] [PubMed] [Google Scholar]
- 33. Ryazanov AG, Shestakova EA, Natapov PG. Phosphorylation of elongation factor 2 by EF‐2 kinase affects rate of translation. Nature 1988, 334:170–173. [DOI] [PubMed] [Google Scholar]
- 34. Davydova EK, Ovchinnikov LP. ADP‐ribosylated elongation factor 2 (ADP‐ribosyl‐EF‐2) is unable to promote translocation within the ribosome. FEBS Lett 1990, 261:350–352. [DOI] [PubMed] [Google Scholar]
- 35. Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol 2013, 20:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF‐G and the ribosome regulate translocation. Nat Struct Mol Biol 2010, 17:1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joseph S, Noller HF. EF‐G‐catalyzed translocation of anticodon stem‐loop analogs of transfer RNA in the ribosome. EMBO J 1998, 17:3478–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 2011, 475:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Belitsina NV, Tnalina GZ, Spirin AS. Template‐free ribosomal synthesis of polylysine from lysyl‐tRNA. FEBS Lett 1981, 131:289–292. [DOI] [PubMed] [Google Scholar]
- 40. Phelps SS, Gaudin C, Yoshizawa S, Benitez C, Fourmy D, Joseph S. Translocation of a tRNA with an extended anticodon through the ribosome. J Mol Biol 2006, 360:610–622. [DOI] [PubMed] [Google Scholar]
- 41. Walker SE, Fredrick K. Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J Mol Biol 2006, 360:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A‐site tRNA to the P site during EF‐G‐catalyzed translocation. Science 2014, 345:1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pestka S. Studies on the formation of transfer ribonucleic acid‐ribosome complexes. 3. The formation of peptide bonds by ribosomes in the absence of supernatant enzymes. J Biol Chem 1968, 243:2810–2820. [PubMed] [Google Scholar]
- 44. Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS. Factor‐free (“non‐enzymic”) and factor‐dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J Mol Biol 1976, 101:537–552. [DOI] [PubMed] [Google Scholar]
- 45. Gavrilova LP, Spirin AS. Stimulation of “non‐enzymic” translocation in ribosomes by p‐chloromercuribenzoate. FEBS Lett 1971, 17:324–326. [DOI] [PubMed] [Google Scholar]
- 46. Southworth DR, Brunelle JL, Green R. EFG‐independent translocation of the mRNA:tRNA complex is promoted by modification of the ribosome with thiol‐specific reagents. J Mol Biol 2002, 324:611–623. [DOI] [PubMed] [Google Scholar]
- 47. Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell 2003, 12:321–328. [DOI] [PubMed] [Google Scholar]
- 48. Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science 2003, 300:1159–1162. [DOI] [PubMed] [Google Scholar]
- 49. Ermolenko DN, Cornish PV, Ha T, Noller HF. Antibiotics that bind to the A site of the large ribosomal subunit can induce mRNA translocation. RNA 2013, 19:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature 1989, 342:142–148. [DOI] [PubMed] [Google Scholar]
- 51. Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol 2007, 370:530–540. [DOI] [PubMed] [Google Scholar]
- 52. Agirrezabala X, Lei J, Brunelle JL, Ortiz‐Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell 2008, 32:190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Julian P, Konevega AL, Scheres SH, Lazaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A 2008, 105:16924–16927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 2011, 332:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci U S A 2013, 110:20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 57. Frank J, Agrawal RK. A ratchet‐like inter‐subunit reorganization of the ribosome during translocation. Nature 2000, 406:318–322. [DOI] [PubMed] [Google Scholar]
- 58. Blanchard SC, Kim HD, Gonzalez RL Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A 2004, 101:12893–12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell 2007, 25:505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell 2008, 30:578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fei J, Kosuri P, MacDougall DD, Gonzalez RL Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell 2008, 30:348–359. [DOI] [PubMed] [Google Scholar]
- 62. Ning W, Fei J, Gonzalez RL Jr. The ribosome uses cooperative conformational changes to maximize and regulate the efficiency of translation. Proc Natl Acad Sci U S A 2014, 111:12073–12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fu J, Munro JB, Blanchard SC, Frank J. Cryoelectron microscopy structures of the ribosome complex in intermediate states during tRNA translocation. Proc Natl Acad Sci U S A 2011, 108:4817–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell 2011, 44:214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time‐resolved electron cryomicroscopy. Nature 2010, 466:329–333. [DOI] [PubMed] [Google Scholar]
- 66. Sharma D, Southworth DR, Green R. EF‐G‐independent reactivity of a pre‐translocation‐state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA 2004, 10:102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Holtkamp W, Cunha CE, Peske F, Konevega AL, Wintermeyer W, Rodnina MV. GTP hydrolysis by EF‐G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J 2014, 33:1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol 2006, 13:234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA‐binding states in ribosomal translocation. Proc Natl Acad Sci U S A 2008, 105:9192–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fei J, Richard AC, Bronson JE, Gonzalez RL Jr. Transfer RNA‐mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol 2011, 18:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell 2003, 114:123–134. [DOI] [PubMed] [Google Scholar]
- 72. Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry 1986, 25:3245–3255. [DOI] [PubMed] [Google Scholar]
- 73. Schilling‐Bartetzko S, Franceschi F, Sternbach H, Nierhaus KH. Apparent association constants of tRNAs for the ribosomal A, P, and E sites. J Biol Chem 1992, 267:4693–4702. [PubMed] [Google Scholar]
- 74. Wang L, Pulk A, Wasserman MR, Feldman MB, Altman RB, Cate JH, Blanchard SC. Allosteric control of the ribosome by small‐molecule antibiotics. Nat Struct Mol Biol 2012, 19:957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci U S A 2007, 104:4881–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A 2009, 106:2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol 2008, 12:674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome‐tRNA complex reveals functional interactions and rearrangements. Cell 2006, 126:1065–1077. [DOI] [PubMed] [Google Scholar]
- 79. Fei J, Bronson JE, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL Jr. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A 2009, 106:15702–15707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL Jr. Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat Struct Mol Biol 2009, 16:861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 2007, 129:929–941. [DOI] [PubMed] [Google Scholar]
- 82. Ben‐Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 2011, 334:1524–1529. [DOI] [PubMed] [Google Scholar]
- 83. Svidritskiy E, Brilot AF, Koh CS, Grigorieff N, Korostelev AA. Structures of yeast 80S ribosome‐tRNA complexes in the rotated and nonrotated conformations. Structure 2014, 22:1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Spahn CM, Gomez‐Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 2004, 23:1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila‐Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science 2005, 310:827–834. [DOI] [PubMed] [Google Scholar]
- 86. Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A 2012, 109:20391–20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol 2007, 2:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mohan S, Donohue JP, Noller HF. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci U S A 2014, 111:13325–13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Carter AP, Clemons WM, Brodersen DE, Morgan‐Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407:340–348. [DOI] [PubMed] [Google Scholar]
- 90. Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra‐subunit tRNA hybrid sites. Nature 2010, 468:713–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ramrath DJ, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, et al. The complex of tmRNA‐SmpB and EF‐G on translocating ribosomes. Nature 2012, 485:526–529. [DOI] [PubMed] [Google Scholar]
- 92. Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF‐G‐ribosome complexes trapped in intermediate states of translocation. Science 2013, 340:1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell 2015, 160:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CM. Visualization of two transfer RNAs trapped in transit during elongation factor G‐mediated translocation. Proc Natl Acad Sci U S A 2013, 110:20964–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Martemyanov KA, Gudkov AT. Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett 1999, 452:155–159. [DOI] [PubMed] [Google Scholar]
- 96. Liu G, Song G, Zhang D, Zhang D, Li Z, Lyu Z, Dong J, Achenbach J, Gong W, Zhao XS, et al. EF‐G catalyzes tRNA translocation by disrupting interactions between decoding center and codon‐anticodon duplex. Nat Struct Mol Biol 2014, 21:817–824. [DOI] [PubMed] [Google Scholar]
- 97. Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell 2000, 6:501–505. [DOI] [PubMed] [Google Scholar]
- 98. Salsi E, Farah E, Netter Z, Dann J, Ermolenko DN. Movement of elongation factor G between compact and extended conformations. J Mol Biol 2015, 427:454–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science 2013, 340:1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 2013, 340:1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Howard J. Protein power strokes. Curr Biol 2006, 16:R517–R519. [DOI] [PubMed] [Google Scholar]
- 102. Liu T, Kaplan A, Alexander L, Yan S, Wen JD, Lancaster L, Wickersham CE, Fredrick K, Noller H, Tinoco I, et al. Direct measurement of the mechanical work during translocation by the ribosome. Elife 2014, 3:e03406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Holschuh K, Riesner D, Gassen HG. Steps of mRNA translocation in protein biosynthesis. Nature 1981, 293:675–677. [DOI] [PubMed] [Google Scholar]
- 104. Fahlman RP, Uhlenbeck OC. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P‐ and A‐sites. Biochemistry 2004, 43:7575–7583. [DOI] [PubMed] [Google Scholar]
- 105. Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell 2006, 24:931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Konevega AL, Fischer N, Semenkov YP, Stark H, Wintermeyer W, Rodnina MV. Spontaneous reverse movement of mRNA‐bound tRNA through the ribosome. Nat Struct Mol Biol 2007, 14:318–324. [DOI] [PubMed] [Google Scholar]
- 107. Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 2009, 16:528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Spirin AS. A model of the functioning ribosome: locking and unlocking of the ribosome subparticles. Cold Spring Harb Symp Quant Biol 1969, 34:197–207. [DOI] [PubMed] [Google Scholar]
- 109. Khade PK, Joseph S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol 2011, 18:1300–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chen B, Frank J. Two promising future developments of cryo‐EM: capturing short‐lived states and mapping a continuum of states of a macromolecule. Microscopy (Oxf) 2016, 65:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome's metastable energy landscape. Trends Biochem Sci 2009, 34:390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Whitford PC, Blanchard SC, Cate JH, Sanbonmatsu KY. Connecting the kinetics and energy landscape of tRNA translocation on the ribosome. PLoS Comput Biol 2013, 9:e1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Nguyen K, Whitford PC. Steric interactions lead to collective tilting motion in the ribosome during mRNA‐tRNA translocation. Nat Commun 2016, 7:10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lu Z, Shaikh TR, Barnard D, Meng X, Mohamed H, Yassin A, Mannella CA, Agrawal RK, Lu TM, Wagenknecht T. Monolithic microfluidic mixing‐spraying devices for time‐resolved cryo‐electron microscopy. J Struct Biol 2009, 168:388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chen B, Kaledhonkar S, Sun M, Shen B, Lu Z, Barnard D, Lu TM, Gonzalez RL Jr, Frank J. Structural dynamics of ribosome subunit association studied by mixing‐spraying time‐resolved cryogenic electron microscopy. Structure 2015, 23:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]


