Abstract
The RNA Recognition Motif (RRM) is the largest family of eukaryotic RNA-binding proteins. Engineered RRMs with new specificity would provide valuable tools and an exacting test of our understanding of specificity. We have achieved the first successful re-design of the specificity of an RRM using rational methods and demonstrated re-targeting of activity in cells. We engineered the conserved RRM of human Rbfox proteins to specifically bind to the terminal loop of miR-21 precursor with high affinity and inhibit its processing by Drosha and Dicer. We further engineered Giardia Dicer by replacing its PAZ domain with the designed RRM. The reprogrammed enzyme degrades pre-miR-21 specifically in vitro and suppresses mature miR-21 levels in cells, which results in increased expression of PDCD4 and significantly decreased viability for cancer cells. The results demonstrate the feasibility of engineering the sequence-specificity of RRMs and of using this ubiquitous platform for diverse biological applications.
Introduction
RNA-binding proteins (RBPs) perform many essential functions, and being able to dial the specificity of RBPs would provide very valuable tools for research and therapeutics. Manipulating gene expression at the RNA level is often transient and reversible, suggesting artificial RBPs would complement permanent changes achievable by genome editing1,2,3. Despite significant efforts the only success has been with the Pumilio and FBF homology (PUF) motif, where a universal code for RNA-recognition was established and successfully applied to target single-stranded RNA (ssRNA)1,4,5. However, PUFs do not bind secondary structure motifs, and bind inefficiently (35 amino acids recognize each nucleotide), compared to RRMs (70-80 amino acids recognize 4-6 nucleotides) or ZFs (30 amino acids recognize three nucleotides). Therefore, it is necessary to explore other RNA-binding domains2,3.
RRM proteins are particularly attractive. They are nearly as common as zinc fingers and are modular, composed of compact RNA-binding domains to which functional domains are appended. However, engineering their specificity by rational or combinatorial means has met limited success6,7 because of poor understanding of recognition principles and the size of the interfaces. Many RRMs have weak binding activity3, which provide inefficient starting points to evolve proteins with higher binding potency. We chose the conserved RRM of human Rbfox proteins, a small family of tissue-specific alternative splicing regulators8. The structure of the Rbfox1-RRM in complex with a ssRNA (UGCAUGU) shows how high sequence-specificity and affinity (~ nM) are achieved9, one of only a few RRMs so far with these desirable properties.
We targeted microRNA (miRNA) precursor species. Sequence and structural features of primary (pri-miR) and precursor miRNAs (pre-miR) play critical roles in Drosha and Dicer processing10-11,12,10,11,12,13. Since RBPs recognize the terminal loops (TLs) of miRNA precursors and regulate their processing14-,15,16, we reasoned we could engineer the Rbfox1-RRM to affect miRNA processing by binding to its precursors sequence-specifically. MiR-21 was chosen as it is one of the best-studied oncogenic miRNAs, overexpressed in most tumors17. Its inactivation represses tumor growth, invasiveness and metastasis, making this RNA a promising target for anticancer therapy18.
We report the first successful rational engineering of the specificity of an RRM to generate a protein with high affinity for a new target. The re-designed RRM (referred to as Fox-RRM*) binds to the highly conserved TL of pri-/pre-miR-21 specifically and inhibits its processing by Drosha and Dicer. We further engineered Giardia intestinalis Dicer (G. Dicer) by replacing its PAZ domain, which recognizes the 2-nucleotide (nt) overhang at the 3′-end of double-stranded RNA (dsRNA)19, with Fox-RRM*. The resulting customized G. Dicer (called RRM*-Dicer) targets pre-miR-21 specifically, suppresses mature miR-21 levels in cells and reduces the viability of cancer cells by enhancing the expression of tumor suppressor protein PDCD4 (programmed cell death 4), a direct target of miR-2120 and a negative regulator of smooth muscle α-actin (SMA)21.
These results demonstrate the feasibility of engineering the sequence-specificity of RRMs and using them to inhibit the maturation of disease-related miRNAs, providing valuable tools in research and potentially therapy.
Results
Engineering the Rbfox1 RRM to target miR-21 precursors
A good starting point to engineer RRM specificity is a single domain protein with high binding affinity and specificity, for which a structure is available. Typically, single RRMs bind RNA only with μM affinity and limited specificity. Exceptions are the Rbfox-RRM (we refer to the conserved domain with a single name), which binds to UGCAUG with nM affinity9; and human U1A, which recognizes AUUGCAC very strongly within a secondary structure22. Many of the contacts formed by Rbfox RRM are directly to RNA bases9, rather than the backbone, making the Rbfox RRM a particularly good candidate to engineer sequence specificity. Furthermore, overlapping the structures of the Rbfox and U1A RRMs shows that the paths of the RNA on the β-sheet surfaces are very similar (Supplementary Results, Supplementary Fig. 1), suggesting that the Rbfox-RRM could bind to its cognate RNA within a stem loop.
The UGAAUC sequence in the TL of pre-miR-21 (nucleotides 27 and 32) is related to but not identical to the Rbfox target. Electrophoresis mobility shift assay (EMSA) studies with wild-type Rbfox-RRM show only a smear at 10 μM protein concentration (Supplementary Fig. 2a), indicating an extremely weak interaction. When the loop sequence is mutated to UGCAUG (pre-miR-21-LM1), the target sequence for Rbfox-RRM, binding is restored (Supplementary Fig. 2b and Table 1), consistent with the high specificity of this protein9. Two-nucleotide changes abolish binding of the Rbfox-RRM to the pre-miR-21 stem-loop.
In order to engineer the specificity of the Rbfox1-RRM to pre-miR-21, the binding pocket for C3 must be altered to accommodate an A and that for G6 must switch to C (Fig. 1a) and the domain must be adapted to bind to a stem-loop. We adopted a manual design strategy that utilizes a statistical analysis of protein-RNA interactions to examine individual amino acid substitutions23,24. When a new amino acid was introduced, following brief energy minimization, it was accepted if hydrogen bonds which satisfied the geometric criteria identified by us24 were formed with good Van der Waals interactions and protein stereochemistry. We then validated the stepwise changes by direct experimental measurement and retained a substitution when it improved binding.
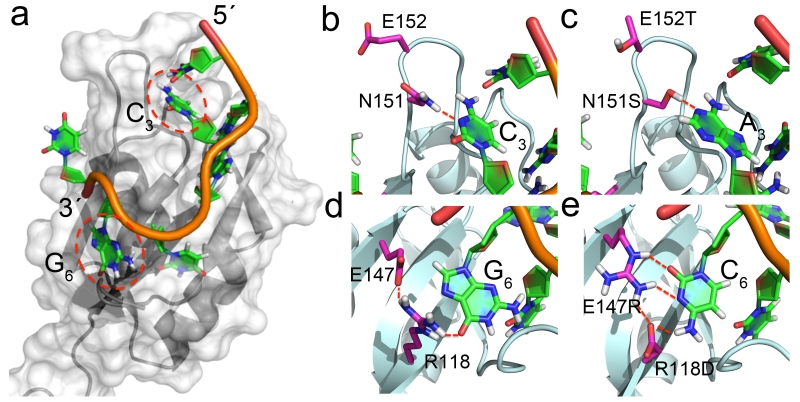
Figure 1.
Engineering the RNA-binding specificity of Rbfox-RRM by rational design. (a) Bird-eye view of the complex between Rbfox-RRM and the single-stranded UGCAUGU sequence (PDB: 2ERR); (b) Recognition of C3 in UGCAUG by N151 (PDB: 2ERR); (c) the N151S mutation is introduced to switch the specificity from C3 to A3 in UGAAUC, while E152T in the RRM β2β3 loop favors the loop insertion into the RNA stem-loop structure; (d) recognition of G6 in UGCAUG by R118 and E147; (e) R118D and E147R mutations switch the specificity from G6 to C6 in UGAAUC, with the second mutation buttressing the direct hydrogen bonds between R118 and the cytosine.
In the Rbfox complex9, the RNA-binding pocket for C3 is mostly constructed from the protein β2β3 loop (Fig. 1b). Recognition of C3 is mediated by intra-RNA hydrogen bond between U1 O2 and the H42 of C3, and intermolecular hydrogen bonds between N3 of C3 and N151. To change the binding specificity from C3 to A3, we mutated N151 to Ser to accommodate the larger base (Fig. 1c). Hydrogen-bonding interactions to adenine in protein-RNA complexes are formed mostly through the Ade-N1 and N6 positions, and Ser is often used to recognize it23. In the model generated by substituting Ser for Asn151, sequence-specific hydrogen-bonds are predicted to form, with good planar and linear geometry, between the Ser γ-OH and N1 of A3, and between H62 of A3 and O2 of U1. However, the N151S mutation only enhances binding affinity slightly (Kd improves from 1.1 to 0.86 μM) (Supplementary Table 1), perhaps due to residual flexibility of the β2β3 loop.
In order to switch the specificity from G6 to C6 (Fig. 1d,e), we mutated R118 to Asp and E147 to Arg. The Cyt-O2, N3 and N4 are often utilized for base-specific hydrogen bonds23, and the guanidinium group of Arg is most frequently observed for simultaneous interactions with the Cyt-O2 and N3. In the model, the cytosine is in the commom anti conformation. The model suggested that the guanidinium group would provide simultaneous interactions with the N3 and O2 of C6, while H42 and H41 would be recognized by carbonyl groups from both the side chain of D118 and the backbone of T192. The E147R mutation reduces unfavorable electrostatic interaction with the RNA phosphate backbone, and is predicted to form stabilizing hydrogen bonds with R118D (Fig. 1e). Buttressing of the direct protein-RNA interaction is required for high affinity binding. Remarkably, the combined mutation (R118D and E147R) increased binding affinity ~103 fold, while individual mutations (R118E and E147R) only increased binding affinity modestly (Supplementary Table 1).
The U1A complex is the only example of a single RRM bound to a stem-loop22. When the two structures (2ERR and 1URN) are superimposed (Supplementary Fig. 1), the binding paths of two RNAs are very similar, but the β2β3 loop of U1A inserts into the stem-loop and ‘locks’ the conformation of the complex by forming multiple intermolecular interactions with the loop-closing base pair. The Rbfox β2β3 loop is acidic. Therefore, we mutated E152 on the tip of β2β3 loop to Thr, one of the favorable amino acids observed in loops connecting β-sheets25 (Fig. 1b,c). The E152T mutation increased binding affinity to pre-miR-21 by an additional ~2 fold (Supplementary Table 1).
Altogether, compared to wild-type Rbfox-RRM (Supplementary Fig. 2e,f), the engineered Fox-RRM* containing all four mutations (R118D, N151S, E152T and E147R) shows dramatically increased binding affinity (Kd ~ 13 nM) towards pre-miR-21 (Supplementary Fig. 2g). The Fox-RRM* binds to pre-miR-21-LM1 ~12-fold weaker (Kd ~ 156 nM) (Supplementary Fig. 2h and Table 1). Thermodynamic parameters obtained through ITC are summarized in Supplementary Table 1. The quantitative results of the ITC analysis were confirmed by EMSA (Supplementary Fig. 2). Thus, through rational design, we obtained >10,000-fold improvement in binding affinity to pre-miR-21 and ~12-fold discrimination against the sequence targeted by the protein from which design was initiated. Further improvement in specificity could be achieved by optimization of the binding pocket at position 3, which currently can tolerate the wild-type C due to the intrinsic flexibility of the β2β3 loop.
In order to monitor RNA-binding on the protein, we titrated pre-miR-21 into 15N-labeled Fox-RRM* and recorded HSQC spectra (Supplementary Fig. 3). Formation of the complex caused large and specific chemical-shift changes, showing that RNA binding induces a tight intermolecular interface.
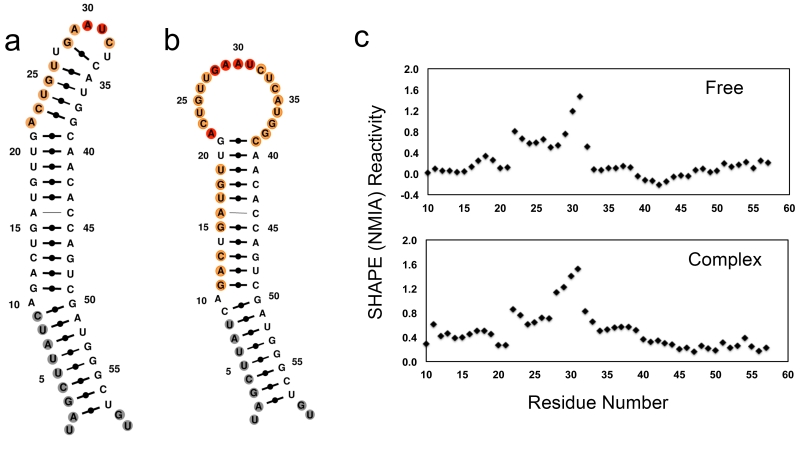
We used SHAPE26 chemistry to determine the secondary structures of pre-miR-21 in free and in complex with Fox-RRM*. The terminal stem-loop region (A22 to G38) of free pre-miR-21 is highly dynamic; the short double helix predicted by thermodynamic analysis is instead open (Fig. 2a). Significant changes in SHAPE reactivity were observed when pre-miR-21 was bound to Fox-RRM* (Fig. 2b,c). The nucleotides in the entire TL (A22 to G38) become more reactive, indicating increased dynamic and the complete opening of any transient base pair26. We have observed the same behavior in the complex of the Rbfox RRM bound to pre-miR-20b, closely related to the present system27. Altogether, these data demonstrate that Fox-RRM* binding to the TL of pre-miR-21 has profound effects on its local structure.
Figure 2.
Secondary structures of free pre-miR-21 and its complex with Fox-RRM* calculated with MC-Fold by incorporating information from SHAPE chemistry. (a) Free pre-miR-21. (b) Complex of pre-miR-21 with Fox-RRM*. Nucleotides are colored according to their normalized SHAPE reactivity with no data labeled in gray, low reactivity (< 0.4) in black, medium reactivity (0.4 ~ 0.85) in orange, and high reactivity (> 0.85) in red. The color coding reflects the single-stranded probability predicted by MC-Fold. (c) SHAPE reactivity traces for pre-miR-21 in free and complex forms; reactivates are normalized to untreated samples. All of the experiments were replicated at least three times, and representative results are shown.
Inhibition of miR-21 processing by engineered Fox-RRM*
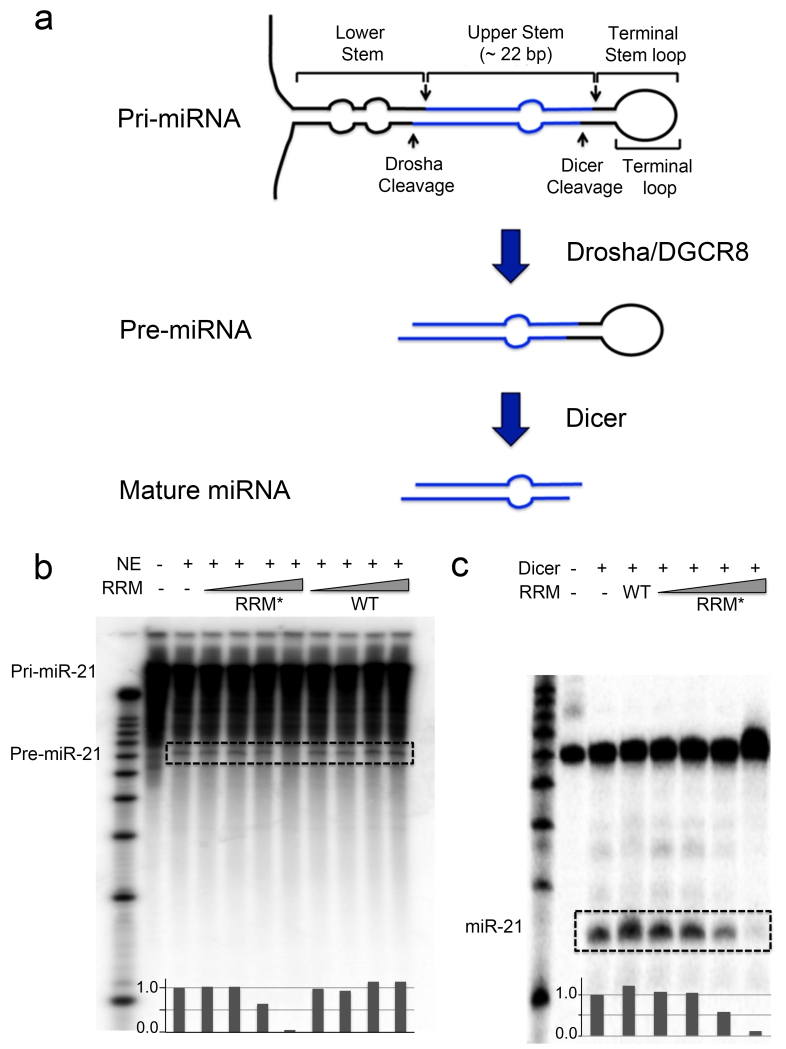
During miRNA maturation, the pri-miRNA is first processed by the Drosha/DGCR8 complex to a ~60-70-nt pre-miRNA, then transported to the cytoplasm and cleaved by Dicer to ~20-bp mature miRNA/miRNA*28 (Fig. 3a). We hypothesized that binding of Fox-RRM* to pri- and pre-miR-21 would affect Drosha and Dicer processing, and set up in vitro processing assays. The Drosha–DGCR8 microprocessor complex, present in the HeLa nuclear extracts, processed pri-miR-21 (> 200 nt) to generate pre-miR-21 (~ 60 nt) (Fig. 3b). With increasing concentration of Fox-RRM*, the band at ~ 60 nt (pre-miR-21) weakens and completely disappears at 10 μM concentration. This specific effect is not observed with wild-type Rbfox-RRM, which has much weaker binding affinity.
Figure 3.
Engineered Fox-RRM* inhibits primary and precursor miR-21 processing. (a) Schematic diagram illustrates the steps in canonical miRNA biogenesis. (b) Representative gel image from in vitro processing assays of pri-miR-21 in the presence of increasing concentrations (10 nM, 100 nM, 1 μM and 10 μM) of wild-type (WT) Rbfox-RRM or Fox-RRM*. Pri-miR-21 was internally labeled with [α-32P]-CTP. Quantification of the pre-miR-21 bands by ImageJ was normalized to nuclear extract (NE) only. (c) Representative gel image from in vitro processing assays of wild-type pre-miR-21 by recombinant human Dicer in the presence of Rbfox1-RRM (10 μM) or increasing concentrations of Fox-RRM* (10 nM, 100 nM, 1 μM and 10 μM). Pre-miR-21 was 5′-end-labeled with 32P. Quantification of the mature miR-21 bands by ImageJ was normalized to Dicer only. The processing products are highlighted with dashed boxes. All of the processing assays were replicated at least three times.
For in vitro pre-miRNA processing assays, pre-miR-21 was incubated with recombinant Dicer (Fig. 3c). Wild-type Rbfox-RRM has no effect on pre-miR-21 processing, while Fox-RRM* inhibits pre-miR-21 processing (~ 90% reduction at 10 μM protein).
Altogether, the results of these assays indicate that binding of Fox-RRM* to the TL of pri- and pre-miR-21 inhibits processing by both Drosha and Dicer. However, inhibition occurs at higher concentrations compared to binding. Drosha binds preferentially to the junction between the double- and single-stranded portions of the stem-loop, while DGCR8 interacts with the double helical region and an apical enhancer element (UGU)29. Although Fox-RRM* binds tightly to pre-miR-21, it does not directly block the interactions between pri-miR-21 and Drosha/DGCR8, which may explain the less efficient inhibition of processing by Fox-RRM*. Similarly, Lin-28, which binds to Let-7 precursor loop tightly (Kd ~ 1 nM), inhibits its processing at much high protein concentration (~ μM)30.
Engineered Giardia Dicer degrades pre-miR-21 specifically
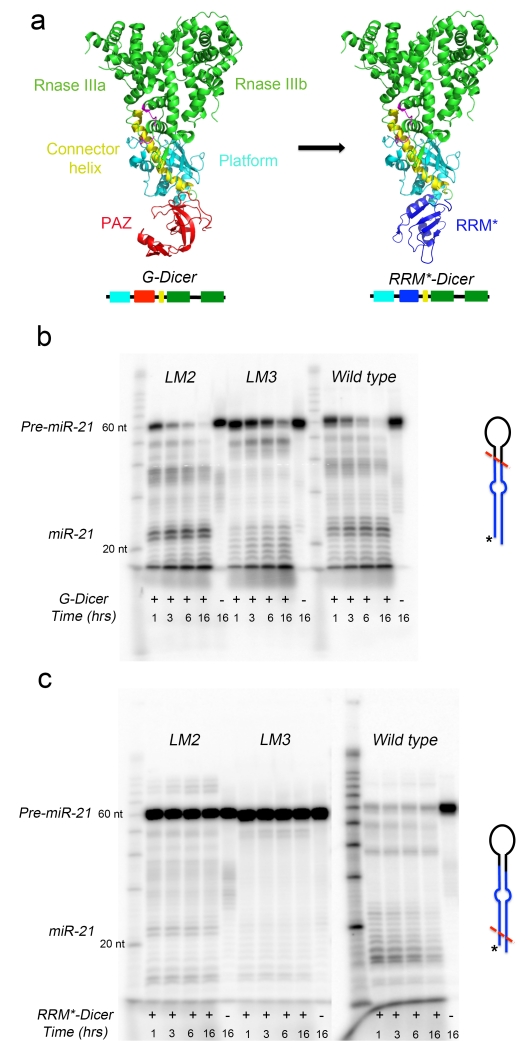
In order to engineer function beyond RNA-binding, we constructed a chimeric Dicer-like enzyme that would specifically cleave pre-miR-21. The minimal functional core of Dicer consists of the PAZ domain and two RNase III domains19. Double-stranded RNA (dsRNA) binding affinity is conferred by the positively charged platform domain (Fig. 4a). As a molecular ruler, Dicer cleaves ~25 base pairs from the helical end where PAZ binds. Since RRM and PAZ have similar size, we reasoned that G. Dicer could be reprogrammed to recognize specific pre-miRNAs by replacing the PAZ domain with the designer RRM (Fig. 4a), and the resulting engineered RRM*-Dicer would be able to cleave pre-miR-21 specifically by recognizing the TL as an anchor point. This strategy was demonstrated before in a study of G. Dicer function31, where the PAZ domain was replaced with U1A RRM to create an enzyme with altered end-recognition specificity.
Figure 4.
Engineering Giardia Dicer to target pre-miR-21 specifically. (a) Strategy used to engineer G. Dicer to target pre-miR-21. The PAZ domain (red) in native G. Dicer (PDB: 2QVW) was replaced with the engineered Fox-RRM* (blue) to generate RRM*-Dicer. (b) G. Dicer cleaves both pre-miR-21 and its loop mutants (LM2 and LM3) without specificity, although the UUCG-containing loop shows a different pattern of cleavage. The sequences of pre-miR-21, LM2 and LM3 are shown in Supplementary Figure 4. (c) In contrast, engineered RRM*-Dicer specifically degrades pre-miR-21 but does not affect its loop mutants significantly (-LM2 and -LM3). Shown here are representative gel images from time-dependent in vitro cleavage reactions, which were carried out at 37°C and stopped at the indicated time points. The proposed cutting sites (red dashed lines) for G. Dicer and RRM*-Dicer on pre-miRNAs are shown schematically, with 5′-end-labeled 32P indicated as *. All of the processing experiments were replicated at least three times.
We purified both the wild-type and engineered RRM*-Dicer proteins and, in order to probe specific activity and exclude the possibility of RNase contamination, we subjected wild-type and mutated pre-miR-21 loop sequences to processing (Supplementary Fig. 4). One mutant changes the UGAAUC sequence to UAAAUA (pre-miR21-LM2), the other replaces the entire loop with a UUCG tetraloop (pre-miR21-LM3). EMSA shows that Fox-RRM* binds specifically to the wild-type pre-miR-21 with nM affinity, but it does not bind appreciably to either pre-miR-21-LM2 or -LM3 (Supplementary Fig. 5). Time-dependent in vitro processing assays by G. Dicer and RRM*-Dicer were then carried out to compare enzymatic activities between pre-miR-21 and its two loop mutants (LM2 and LM3).
G. Dicer cleaves all three pre-miRNAs, with increased activity towards pre-miR-21 and -LM2 (Fig. 4b). Major processed bands of 21~22 nucleotides were observed after 15 minutes. Significantly reduced cleavage activity and altered cleaved products were observed for the variant containing the UUCG tetraloop. In contrast, RRM*-Dicer cleaves wild-type pre-miR-21 specifically and with high efficiency, with almost no cross reactivity towards -LM2 and -LM3 (Fig. 4c). A series of distinct bands between 10 and 25 nucleotides, similar to degradation products, are generated by RRM*-Dicer. Only very weak cleavage products at 21~22 nucleotides are observed for -LM2, suggesting that an alternative but inefficient processing mode for RRM*-Dicer may exist. In the cleavage of long RNA hairpin (33 and 37 base pairs), U1A Dicer produces major bands at 12 and 18 nucleotides, corresponding to 19-21 base pairs from the U1A recognition loop31. Using the TL of pre-miR-21 as anchor point for RRM*-Dicer to measure cleavage length is not as straightforward as for the perfectly base-paired dsRNA used for G. Dicer or U1A-Dicer cleavages31. The TL of pre-miR-21 is large and flexible when bound to RRM* and its imperfect stem of ~20 base pairs has multiple bulges and internal loops, both of which could disrupt the ruler mechanism of Dicer and contribute to the non-specific cleavage products. Because the RNA is 32P-labelled at the 5′-end, the leftover structures (19-21 base pairs) generated by RRM*-Dicer are not observed31, while cleavage bands observed at ~40 and ~50-nt length, in both G. Dicer and RRM*-Dicer processing, may be due to single cuts of 5′-32P-labeled pre-miR-21. Interestingly, processing of pre-miR-21 by RRM*-Dicer is much faster than for G. Dicer, as also observed for U1A Dicer, perhaps due to increased substrate affinity31. These results demonstrate that RRM*-Dicer cleaves pre-miR-21 by selectively targeting its UGAAUC loop sequence and degrades it.
Engineered RRM*-Dicer down-regulates miR-21 in cells
We surmised that both Fox-RRM* and RRM*-Dicer could suppress mature miR-21 levels in cells. Since Fox-RRM* would act as a competitive inhibitor for processing of pri-/pre-miRNA, good protein expression levels would be necessary to observe any effect on mature miRNA levels. However, protein expression was poor for both wild-type and designer RRMs in mammalian cells, despite using different vectors and protein tags. Satisfactory protein expressions were obtained instead for G. Dicer and RRM*-Dicer when cloned into vectors containing N-terminal SUMO protein tags32. Therefore, we evaluated the efficacy and specificity of designer RRM*-Dicer in cell-based assays.
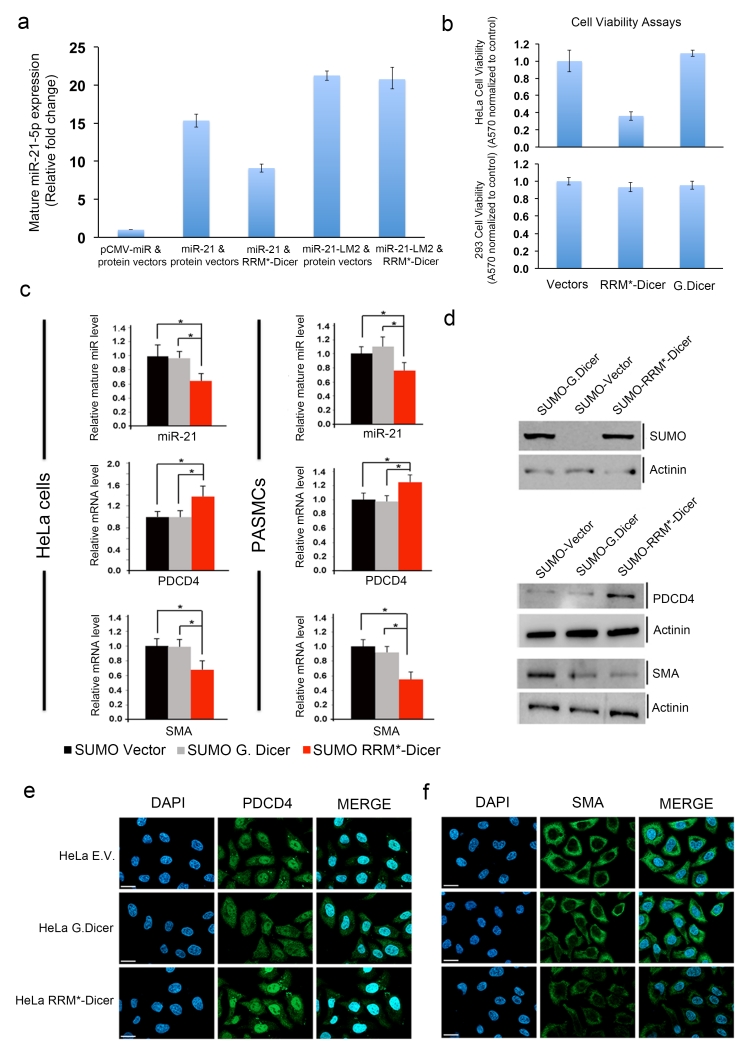
HEK293 cells were transiently transfected with plasmids containing RRM*-Dicer and pri-miR-21. A plasmid containing the loop mutant pri-miR-21-LM2 was used as specificity control. Levels of mature miR-21 increased 15-fold two days after co-transfection with the pri-miR-21 and empty protein vectors (Fig. 5a). When RRM*-Dicer and pri-miR-21 are co-transfected, the level of mature miR-21 increased only 9-fold, a 40% reduction. If pri-miR-21 is replaced with pri-miR-21-LM2, no difference in expression is observed in the presence or absence of RRM*-Dicer. No significant differences in mature miR-21 levels were observed when wild-type G. Dicer was co-transfected with vectors containing either pri-miR-21 or pri-miR-21-LM2 (Supplementary Fig. 6). These results demonstrate that engineered RRM*-Dicer reduces expression of mature miR-21 in cells. Considering that the transfected RRM*-Dicer has first to be expressed and then compete with endogenous Dicer in processing pre-miR-21, a 40% reduction of mature miR-21 level is significant33.
Figure 5.
Engineered RRM*-Dicer reduces cellular mature miR-21 levels and increases expression of PDCD4 and reduces cell viability. (a) RT-PCR analysis of mature miR-21-5p expression in HEK293 cells transiently transfected with expression vectors containing empty miRNA and protein; primary miR-21 and empty protein; primary miR-21 and RRM*-Dicer; primary miR-21-LM2 and empty protein; primary miR-21-LM2 and RRM*-Dicer. (b) Cell viability analysis for HeLa and HEK293 cells two days after transfection with G. Dicer or RRM*-Dicer expression vectors; expression of the engineered enzyme reduces the viability of HeLa cells but not 293 cells. Error bars in (a) and (b) represent the mean ± SD from at least 3 independent experiments. (c) RRM*-Dicer reduces endogenous mature miR-21-5p levels, which results in increased PDCD4 and decreased SMA mRNA levels (* p < 0.05) in both HeLa cells (left panel) and PASMCs (right panel). All experiments were done at three different times using independent sample preparation in triplicate. Mean values for individual experiments are expressed as mean ± SEM. (d) Immunoblot analysis of the expression of SUMO-tagged G. Dicer and RRM*-Dicer in HeLa cells, and their effects on endogenous PDCD4 and SMA protein levels. Expression of RRM*-Dicer increased endogenous PDCD4 and decreased SMA protein levels. Actinin (α-actinin) serves as a loading control. Full gels are shown as Supplementary Fig. 7. Immunofluorescence micrographs for endogenous (e) PDCD4 and (f) SMA proteins in HeLa cells. Cells were stained with anti-PDCD4 antibody (green) or with anti-SMA antibody (green) and 4,6-diamidino-2-phenylindole (DAPI; blue), 48 h after transfections with empty vector (E.V.), G. Dicer or RRM*-Dicer expression vectors. Scale bars indicate the size of 10 μm.
Since miR-21 is overexpressed in many cancers and knock down of miR-21 increases apoptosis and reduces invasiveness in cancer cell lines and small animal models33, we evaluated the effect of RRM*-Dicer on cell growth using the crystal violet cell viability assays. HEK293 cells, with low endogenous miR-21, were minimally affected by expression of RRM*-Dicer (Fig. 5b); while HeLa cells, with high levels of miR-2134, have significantly reduced cell viability. Neither HeLa nor 293 cells were affected by transient transfection of G. Dicer. The effect of RRM*-Dicer on endogenous mature miR-21 levels in HeLa cells was further quantified by qRT-PCR. Compared to transfections with empty vector alone or wild-type G. Dicer, RRM*-Dicer significantly decreases mature miR-21 levels (Fig. 5c).
The tumor suppressor gene PDCD4 is a direct target of miR-21 and acts as negative regulator of the smooth muscle cell (SMC) marker smooth muscle α-actin (SMA)21. Reduction of mature miR-21 by RRM*-Dicer in HeLa cells increased PDCD4 while decreasing SMA mRNA expression (Fig. 5c). Changes of PDCD4 and SMA protein expression levels were further confirmed by immunoblot analysis and immunofluorescence micrographs (Fig. 5d, e, f).
Previous studies showed that transforming growth factor β (TGF-β) and bone morphogenetic protein (BMP) signaling promote miR-21 processing and significantly increases its expression in human vascular SMCs, and reduction of miR-21 by anti-miR-21 in PASMCs (primary pulmonary artery smooth muscle cells) increased PDCD4 and decreased SMA protein expression21. In PASMCs induced with BMP-ligand (BMP4), engineered RRM*-Dicer significantly reduced endogenous mature miR-21, compared to empty SUMO vector and G. Dicer controls, and increased mRNA and protein expression for PDCD4, while decreasing SMA levels (Fig. 5c).
Altogether, we have demonstrated that engineered RRM*-Dicer significantly reduces mature miR-21 level in cells and increases protein expression of its downstream targets, consistent with the increased cancer cell death we observed.
Discussion
We demonstrate the first rational redesign of the specificity of an RRM protein, which could provide ideal counterparts to zinc finger DNA-binding domains for engineering RNA regulatory tools, because of their versatile modular structures35, ubiquitous presence in eukaryotes and efficient RNA binding capacity. However, it has so far not been possible to re-target with high affinity even the best-studied RRMs using either computational or combinatorial methods7, because of large interface area and complex physical properties of their interfaces36.
We chose the Rbfox-RRM because of its high intrinsic specificity and affinity, achieved with a single domain9. Unlike many other RRMs, where specific recognition is achieved through protein backbone interactions, it recognizes UGCAUG through multiple direct interactions between protein side-chains and the RNA bases9. Using the structure of the complex, we introduced by rational design several mutations in a step-wise manner to re-target its specificity to UGAAUC, found within the TL of pri-/pre-miR-21. Mutations were also introduced to improve the ability of the designed protein to bind to a stem-loop, rather than ssRNA. The resulting Fox-RRM* binds to pre-miR-21 >10,000 fold better than the wild-type protein from which it was derived and has >12-fold specificity against a pre-miR-21 loop mutant containing the sequence targeted by the wild-type protein. Because of its ubiquitous presence and diverse sequence specificity, our strategy to engineering Rbfox-RRM can be broadly applied to any single RRM. However, improvements in the quantification of the fitness of protein-RNA interfaces would greatly facilitate this task by allowing the fully automated computational sampling of sequence space instead of the systematic examination of individual amino acid substitutions, as we have done. The present design also provides an ideal system for the development and testing of atomic-level scoring functions aimed at these interfaces.
The TL of primary and precursor miRNAs controls the efficiency and specificity of miRNA processing10. By using the engineered Fox-RRM* to target pri-/pre-miR-21 specifically, we were able to down-regulate its processing, either by directly blocking the access of processing enzymes and their protein cofactors, and/or by altering the stem-loop structure of miR-21 precursors.
To demonstrate the functional versatility of the engineered RRM, we fused it to an effector domain with distinct function1,2. Namely, we replaced the PAZ domain used by G. Dicer for end-recognition of all siRNA precursors with Fox-RRM*, to generate a sequence- and structure-specific endonuclease engineered to cleave only pre-miR-21. The reprogrammed RRM*-Dicer degrades pre-miR-21 both in vitro and in cultured cells, and increases the expression of its downstream target PDCD4, consistent with the significantly reduced viability of transfected HeLa cells we observe. The current Fox-RRM* binds to sequences related to UGAAUC, such as UGCAUG targeted by wild-type protein, although with weaker affinity. This could lead to off-target effects for RRM*-Dicer which would be reduced by increased discrimination at the C3/A3 positions in future rounds of design and optimization.
Since miR-21 is a key regulator of oncogenic processes17, its knockdown increases apoptosis and reduces cell proliferation, invasiveness and metastasis20. Aptamers37 and peptoids38 block the maturation of miR-21 by targeting its terminal stem-loop. The results of our study suggest an alternative approach to pharmacologically inactivate the production of over-expressed oncogenic miRNAs, such as miR-21.
In summary, we have for the first time engineered by rational design the specificity of an RRM, by far the most abundant class of eukaryotic RNA-binding proteins. The engineered protein binds to the terminal loop of pri- and pre-miR-21 specifically and inhibits its processing by Drosha and Dicer. By replacing the PAZ domain with the designer RRM*, we engineered a customized Dicer enzyme to specifically degrade pre-miR-21 in vitro and suppresses mature miR-21 levels in cells, which significantly reduces the viability of cancer cells. Further improvements in the specificity of Fox-RRM* to target pre-miR-21 could be achieved through combinatorial approaches or by further cycles of structure-based design.
Online Methods
Protein preparation
Full-length Rbfox1 cDNA was obtained from Open Biosystems. DNA encoding the RRM of Rbfox1 protein (its sequence is identical to the RRM of Rbfox2; residues 109-208, Swissprot Q9NWB1) was PCR amplified and cloned into pET28a (with an N-terminal His-tag) vector. Site-directed mutagenesis for protein engineering was carried out using the QuikChange kit (Agilent). Both the wild-type and mutant Rbfox RRMs were expressed in transformed BL21 (DE3) E. Coli grown at 37°C in LB medium. Cells were grown to OD600 ≈ 0.6 and induced with 1 mM IPTG. Cells were harvested after 4 hours by centrifugation. After cell lysis, proteins were purified by nickel-affinity HisTrap HP column (GE Healthcare) equilibrated in 50 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl, 10 mM imidiazole and 5 mM β-Mercaptoethanol (β-ME). Bound proteins were eluted by an ascending imidazole gradient (up to 300 mM) in 50 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl and 5mM β-ME. The N-term 6×His tag was cleaved by overnight incubation at 4°C with thrombin, which was then removed by HiTrap Benzamidine FF column (GE Healthcare). Proteins were finally purified by HiTrap Heparin HP column (GE Healthcare) using an AKTA Purifier with an ascending NaCl gradient (up to 1.5 M) in 50 mM Tris-Cl (pH 8.0) and 5mM β-ME. Purified proteins were dialyzed and concentrated against assay buffer of 20 mM Tris-Cl (pH 7.5) and 50 mM NaCl. Protein concentrations were measured by UV absorbance at 280 nm and confirmed by Bradford assays.
The wild-type Giardia intestinalis Dicer (G. Dicer) gene was PCR amplified from Giardia genomic DNA, ordered from ATCC (50803D). The DNA template for engineered G. Dicer (referred to as RRM*-Dicer) was synthesized by Genescript. The PAZ domain (Ala137 to Pro251) of G. Dicer was replaced by the amino acid sequence Gly-Ser-Ser-Gly6, followed by amino acid residues 112-208 of Fox-RRM*, followed by Gly-Gly-Ser-Ser. For protein expression, the genes encoding the G. Dicer and RRM*-Dicer were inserted into a modified pFastBac1 vector (Invitrogen) which has an N-terminal glutathione-S-transferase (GST) tag. The GST fusion proteins were expressed in HiFive insect cells following standard procedures and isolated by glutathione affinity chromatography (GE healthcare: GSTrap FF) using buffer containing 1×PBS and 5 mM DTT. After on-column cleavage by Tobacco Etch Virus (TEV) protease overnight, the proteins were eluted by 1×PBS buffer with 5 mM DTT. The eluted proteins were diluted and further purified on a Resource Q column equilibrated in 20 mM Tris-Cl (pH 8.0), 50 mM NaCl. After running a linear gradient against 20 mM Tris-Cl (pH 8.0), 1 M NaCl, fractions containing the target proteins were concentrated and loaded onto Superdex 200 10/300 GL column equilibrated in the final buffer containing 20 mM Tris-Cl (pH 8.0), 150 mM NaCl.
The wild-type human dicer gene was ordered from Addgene and inserted into pFastBac 1vector (Invitrogen) with an N-terminal 6×His tag. The fusion proteins were expressed in HiFive insect cells and isolated by HisTrap HP column (GE Healthcare) equilibrated in 20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 20 mM imidiazole and 5 mM β-ME. Bound proteins were eluted with an ascending imidazole gradient (up to 300 mM) in buffer containing 20mM Tris-HCl (pH 8.0), 150 mM NaCl and 5 mM β-ME. TEV protease was added to remove the 6×His tag at 4°C overnight. The target proteins were further purified exactly as for the wild-type G. Dicer and RRM*-Dicer by anion exchange and gel filtration chromatography.
RNA preparation by in vitro transcription
The sequences of the primary and precursor miR-21 were obtained from miRbase (http://www.mirbase.org)39. Pre-miR-21 and its loop mutants were synthesized by in vitro transcription using T7 RNA polymerase with synthetic DNA templates obtained from Integrated DNA Technologies, Inc. (IDT). All RNA samples were purified as described40, lyophilized and dialyzed into buffers (typically containing 10mM sodium phosphate, pH6.5, 10 mM NaCl and 0.01 mM EDTA) before use. RNA annealing was carried out by heating the sample to 90°C for 3 minutes and snap-cooled on ice.
Analysis of protein-RNA interactions by EMSA and ITC
Rbfox1-RRM and its mutant protein were assayed for their RNA-binding affinities by using gel electrophoretic mobility shift assay (EMSA) and Isothermal Titration Calorimetry (ITC). The former technique provides visualization of aggregated or non-specific complexes (e.g. smeared bands) and the latter provides more thorough quantitation of thermodynamic parameters of binding. Truncated pre-miR-21 and its terminal loop (TL) mutant variants were used in these studies. The RNAs were 32P-labeled at 5′-end with [γ-32P]-ATP using T4 polynucleotide kinase (NEB) and purified by using NAP-10 columns (GE Healthcare). Serial dilutions of proteins were incubated with 20 pM of 32P-labeled RNAs in binding buffer (20 mM Tris-Cl, pH 7.4, 100 mM NaCl, 0.1 μg/μl yeast tRNA) at room temperature for 1 h. Samples were run on 10% native polyacrylamide gels in cold room and the radioactive signals were quantified by ImageJ analysis after overnight exposure41.
MicroCal iTC200 was used to carry out the ITC analysis. All RNAs and proteins were dialyzed into the same buffer containing 20 mM Tris-Cl (pH 7.4) and 120 mM NaCl. The titrations were carried out at 293 K, with 300 μl of RNA (10 μM) in the sample cell as titrate and 200 μl of protein (100 μM to 200 μM) in the syringe as titrant. Due to the weak binding affinity between pre-miR-21 and Rbfox1-RRM, 100 μM of RNA and 2 mM of protein were used. Data analysis was carried out using Origin 7.0 software provided by MicroCal. ITC data were fitted with a one-site binding model and the baselines were corrected automatically with minor manual adjustment.
Selective 2′OH acylation Analyzed by Primer Extension (SHAPE)
We performed SHAPE42 chemical probing on pre-miRNA-21 in both free and bound to engineered Fox-RRM*. The resulting chemical modifications were analyzed by primer extension with a radiolabeled DNA oligonucleotides and gel electrophoresis. Pre-miR-21 was transcribed with a 3′-end extension in order to accommodate primer binding. RNAs were denatured by heating at 95 °C for 3 mins, and snap-cooled on ice to refold prior to SHAPE analysis.
Free precursor miRNAs or protein-RNA complexes were diluted to 100 nM with final buffer content of 100 mM HEPES (pH 8.0) and 100 mM NaCl. Three 9 μL aliquots of the RNA solution were distributed to individual Eppendorf tubes and combined with 1 μL of either DMSO (control), or 65 mM NMIA (dissolved in DMSO), or 130 mM NMIA. Reactions were incubated at 37°C for 40 minutes. Following modification, the RNA solution was diluted to 100 μL, adjusted to 0.2 M NaCl, 20 μg glycogen, and 2 mM EDTA and finally precipitated with 3.5× ice-cold absolute ethanol. Precipitation was carried out on dry ice for 15 minutes and finally pelleted at 4°C by centrifugation. The RNA pellet was allowed to dry in air before re-suspension in 10 μL of water.
The DNA primer was 5′-end labeled with γ-[32P]-ATP using T4 polynucleotide kinase (PNK), then purified on a G50 resin spin column. NMIA-modified RNA (~ 0.5 pmol) was combined with 1.5 pmol 32P-labeled DNA primer and annealed by heating at 95°C for 5 minutes followed by 5 minutes on ice. Reverse transcription was performed using Superscript III (Invitrogen), followed by incubation at 52°C for 10 minutes. Each sequencing reaction was similarly prepared using 1 pmol unmodified RNA. The reaction was stopped by degrading the RNA with 200 mM NaOH at 95°C for 5 minutes. The cDNA mixture was then neutralized at 95°C for 5 minutes. Each sequencing and modification reaction was loaded onto a pre-warmed 8% denaturing polyacrylamide gel and separated by electrophoresis. The resulting gel was transferred to Whatman paper and fixed with a solution of 40% Methanol and 10% Acetic Acid. The fixed gel was dried and exposed overnight onto a storage phosphor screen.
The autoradiograph was imaged with a Typhoon laser scanner and manipulated with Semi-Automated Footprinting Analysis (SAFA) software43. Secondary structure analysis was conducted with MC-Fold44.
In vitro primary miRNA processing assays
The DNA segment containing the pri-miR-21 sequence (274 nucleotides, comprising the full pre-miR-21 sequence plus 109 nucleotides at the 5′-end and 107 at the 3′-end) was amplified from human genomic DNA by PCR, and cloned into a pUC19 vector along with a T7 promoter. The coding sequence was verified by sequencing. The DNA plasmid was linearized downstream of the pri-miRNA sequences using BamH1 and gel purified. The internally labeled pri-miR-21 was transcribed in vitro using T7 RNA polymerase (Riboprobe system, Promega) in the presence of [α-32P]-CTP, and purified with QIAGEN RNeasy Mini kit. Cell nuclear extracts (293 or HeLa) from ProteinOne were used for pri-miRNA processing assays, which were carried out (typically 104 ~ 105 cpm of labeled pri-miRNA, 25 μg cell nuclear extract per 20 μl reaction) at 30°C for 1 hour in buffer containing 20 mM Tris (pH7.5), 6.4 mM MgCl2, 100 mM KCl, 0.5 mM DTT and 0.05 μg/μl tRNA. Following incubation, samples were phenol/chloroform extracted, precipitated and re-suspended in RNA loading dye prior to denaturing polyacrylamide gel electrophoresis (PAGE) separation using 8% acrylamide-8M urea gel.
In vitro precursor miRNA processing assays
The precursor miR-21 sequence (Supplementary Fig. 4a) was derived from miRBase by removing the extra 5′- and 3′-end sequences flanking the mature miRNA sequences within the stem-loop, which are cleaved by Drosha from primary transcripts. Pre-miR-21 was prepared by standard in vitro transcription with T7 RNA polymerase using synthetic DNA templates (IDT). Transcribed pre-miR-21 was 5′-dephosphorylated, PAGE purified, and then 5′-end labeled by [γ-32P]-ATP using T4 Polynucleotide Kinase (NEB), and further purified by QIAGEN miRNeasy Mini kit. Recombinant human Dicer enzyme kit (Genlantis) was used for in vitro precursor miRNA processing assays according to the manufacturer’s instructions.
For in vitro pre-miRNA processing assays using wild-type and engineered G. Dicer proteins, the assays were carried out at 37°C with ~40 nM of 32P-labeled pre-miR-21 and 0.5 u of recombinant human Dicer in buffer containing 20mM Tris-Cl (pH 7.6), 50 mM NaCl, 3 mM MgCl2 and 1 mM DTT. At the indicted time points, 2× RNA loading dye (Fermentas) was added to stop the processing reactions. Processed pre-miRNA products were resolved by electrophoresis using a 12.5% acrylamide-8M urea gel. Before loading, samples were heated at 95°C for 5 min. A molecular weight marker of radiolabeled RNA oligonucleotides (Ambion® Decade™ Marker System) was used to determine the size of the processed miRNA products. After electrophoresis, the gel containing the separated RNAs were exposed to a phosphor-screen and visualized by autoradiography using a Storm PhosphorImager (GE Life Sciences). Band intensities were quantified using ImageJ software.
Cell culture and transfection
G. Dicer and RRM*-Dicer proteins were cloned into pM-SUMOstar vector (LifeSensors Inc.). HEK293 and HeLa cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) with L-glutamine, sodium pyruvate, and 10% fetal bovine serum with Pennicillin/Streptomycin in an atmosphere containing 5% CO2 at 37°C. Human primary PASMCs were purchased from Lonza (CC-2581; http://www.lonzabioscience.com/Lonza_Catnav.oid.734.prodoid.PASMC) and were maintained in Sm-GM2 media (Lonza) containing 5% fetal bovine serum. 293 cells were plated one day prior to transfection in 24 well plates at 2.0×105 cells/well. Transient transfections with DNA plasmid mixture (250 ng pM-SUMOstar protein vectors and 250 ng pCMV-miR vectors per well) were carried out at ~70% cell confluency, using FuGENE HD (Roche) transfection reagent at 6:1 ratio of reagent to total DNA plasmids. HeLa cells were plated one day before transfection in 12-well plates at 1.0×105 cells/well. Transient transfections with DNA plasmid mixture (1 μg pM-SUMOstar protein vectors) were carried out at ~50% cell confluence, using TransIT®-LT1 Transfection Reagent (Mirus). Cells were harvested 48 hours post transfection for either RNA extraction or western blotting. PASMCs were plated one day prior to transfection in 12-well plates at 2.0×105cells/well. Transient transfections with DNA plasmid mixture (1 μg pM-SUMOstar protein vectors) were carried out at ~70% cell confluence, using TransIT®-LT1 Transfection Reagent and after 6 hours cells were stimulated with BMP4 (3 nM) growth factor (kindly provided by Prof. Luciano Conti). PASMC cells were harvested 24 hours post transfection for either RNA extraction or Western blot analysis.
RNA preparation and qRT-PCR
Total RNA was extracted using either Qiagen miRNeasy or the RNeasy Plus mini kits and quantified by a nanodrop spectrophotometer (Thermo Sci.). For detection of mature miRNAs, the TaqMan® MicroRNA assay kit (Applied Biosystems) was used according to the manufacturer’s instructions. Mature miR-21 levels were assayed using RNU48 as internal control for normalization. Data are presented as fold changes in miR-21 levels (2-ΔΔCt). Relative expression of SMA and PDCD4 mRNA was normalized to GAPDH. The following primers were used: GAPDH 5′-ACCACAGTCCATGCCATCAC-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′ (reverse); SMA: 5′-CCAGCTATGTGTGAAGAAGAGG-3′ (forward) and 5′-GTGATCTCCTTCTGCATTCGGT-3′ (reverse). PDCD4: 5′-TATGATGTGGAGGAGGTGGATGTGA-3′ (reverse) and 5′-TATGATGTGGAGGAGGTGGATGTGA-3′ (reverse). Statistical analysis was performed by ANOVA followed by Bonferroni’s multiple comparisons test. An average of three experiments, each performed in triplicate with standard errors, is presented. The TaqMan® probes for mature miR-21-5p (Cat. # 4427975, ID 000397) and RNU48 (human) and TaqMan® Control miRNA Assay were purchased from Life Technologies.
Preparation of cell extracts, immunoblot analysis and antibodies
Cell extracts were prepared with radioimmunoprecipitation assay (RIPA) buffer (Sigma) supplemented with a protease-inhibitor cocktail (Roche). For immunoblot analysis, protein samples were resolved on a 10% polyacrylamide Bis-Tris gel and transferred onto a nitrocellulose membrane. The following primary antibodies were used: anti-SMA (clone 1A4, Sigma), anti-PDCD4 (SAB1407349 Sigma), anti-SUMO (GeneScript A00640) and anti-actinin (H-2) (sc-17829, 1:2000, Santa Cruz Biotechnology). Goat anti-rabbit IgG-HRP (1:3000, Santa Cruz Biotechnology) and goat anti-mouse IgG-HRP (sc-2005, 1:3000, Santa Cruz Biotechnology) were used as secondary antibodies. All Western blots were analyzed with the ChemiDoc XRS+ System (Bio-Rad).
Crystal violet proliferation assay
HeLa cells were plated in 24-well plates, including 3 control wells of medium only to provide the blanks for absorbance readings. Cells were transiently transfected in triplicate with 0.5 μg of control pM-SUMOstar vector, or pM-SUMO-RRM*-Dicer or pM-SUMO-G-Dicer vectors. Forty-eight hours after transfection, cells were fixed by Cytofix and stained with crystal violet solution. After two washes with water, crystal violet staining was solubilized by 1% SDS and measured by a plate reader spectrophotometer (Thermo Labsystems, Multiskan Spectrum) at a wavelength at 570 nm.
Immunofluorescence microscopy
Cell staining and immunofluorescence microscopy were carried out as previously described45. Briefly, HeLa and PASMC cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline. Cells were then incubated with the primary antibodies. Alexa-488–conjugated goat antibody against mouse IgG (A-11017, 1:500, Molecular Probes) was used as secondary antibody. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI, 5 μg/ml). Microscopy analysis was performed using the Zeiss Observer Z.1 microscope implemented with the Zeiss ApoTome device. Pictures were acquired using AxioVision imaging software package (Zeiss) and assembled with Adobe Photoshop CS3. Images were not modified other than for adjustments of levels, brightness and magnification.
Modeling the structures of protein-RNA complexes
The structural models of all engineered proteins, including Fox-RRM* and multiple other mutants, in complex with various RNA sequences were built based on the NMR structure of the Rbfox1-RRM in complex with the RNA heptamer UGCAUGU (PDB code: 2ERR). Insight II 2000 (Accelrys Software Inc, San Diego, CA, USA) was employed to mutate the nucleotides and amino acids in the structural complexes, and the modeled structures were further optimized by energy minimization using MOE2010 (Chemical Computing Group, Montreal, CA) with the MMFF94x force field (gradient 0.1).
The complete redesign of RNA-protein interfaces using fully automated tools, such as Rosetta, would be highly desirable but is also currently premature. Rosetta has not been tested in this application, and the relative paucity of structures in the PDB is insufficient to develop fully predictable potentials to evaluate the free energy of protein-RNA interfaces; when potentials are generated, these are ‘over-trained’ and fail at the task of generating new predictions46. Therefore, we adopted a manual design strategy that consisted of the following steps. 1: We identified a ‘designable’ interface, as is often done in protein design projects. Namely, we looked for a protein-RNA complex, Rbfox1, where intermolecular contacts were primarily from the protein side chain to the RNA bases, so that specificity could be switched by substitutions of amino acid side chains without requiring the complete re-design of the protein backbone. This is not a trivial task, since it is well established that a significant majority of protein-RNA contacts are mediated by the protein backbone. 2: Within the interface of Rbfox1 RRM in complex with the RNA heptamer UGCAUGU, the location of C3 and G6, residues that we wished to switch to alter the specificity of the protein towards miR-21, was also evaluated for ‘designability’. Namely, the two sites do not structurally interact (can be optimized independently) and intermolecular contacts are primarily mediated by protein side chains. An additional consideration was that not too many positions needed to be changed in order to switch the specificity to pre-miR21. Introducing a large number of changes would likely lead to a complete alteration of the entire interface. 3: Each of the two nucleotides was computationally replaced with the variant nucleotide (A and C, respectively). We notice that G6 is in the syn conformation in the structure, but C6 is anti (Pyr does not occupy the syn conformation). 4: Amino acid side chains that make direct contacts with the two bases were identified and systematically computationally changed to identify new residues that would form favorable hydrogen bonding interaction with the new bases. In practice, we focused on side chains known to form favorable base-specific interactions with the RNA from statistical surveys of protein-RNA interfaces. 5: For each substitution, the structure was energy minimized locally within AMBER and the quality of hydrogen bonding interactions was evaluated. The substitution was accepted if hydrogen bonds satisfied the geometric criteria (bond angle and length) identified by us in a statistical analysis of hydrogen bonds at protein-RNA interfaces, and good Van der Waals interactions were observed, together with satisfactory amino acid side chains. The highly directional hydrogen-bonding potential is a key component of the Rosetta program47, and, as we noticed in a related publication, hydrogen bonds at protein-RNA interfaces are even more restricted geometrically than at protein-protein interfaces or within protein hydrophobic cores48. 6: The N151S substitution was immediately identified at position 3; a systematic and unbiased analysis with Rosetta conducted retrospectively identifies the same switch (P. Bradley, personal communication). For the second position, when we examined the original structure, we observed that Glu147, which was buttressed by the side chain of Arg118, forms hydrogen bond to G6 O4 in the starting structure; when Arg118 was replaced to Asp or Glu in the mutant protein, their carbonyl groups would recognize H42 and H41 of C6. When R118D is combined with E147R, which forms favorable hydrogen bonds to O2 and N6 of C6, we observed similar stabilizing hydrogen-bonding interaction between R118D and E147R. The R118D substitution, together with E147R, increased binding affinity as desired. 7: Finally, we noticed that the NMR structure that was used as a template for design was for single-stranded RNA, but we wanted the new design to target the pre-miR-21 stem-loop. In the canonical U1A structure of an RRM bound to a stem-loop49, the β2-β3 loop makes critical interactions. In the Rbfox protein, this would be rendered difficult by the acidic E152 amino acid. Thus, we replaced it with Thr.
This protocol can be replicated for other protein-RNA interfaces with the following criteria: base recognition must be mediated by direct contacts with the side chains; the sites must be independent (at least to a first approximation); the global geometry of the interfaces should not be significantly altered by the substitutions.
Supplementary Material
Table 1.
Summary of binding affinities determined by ITC.
| Kd | Rbfox-RRM | Fox-RRM* |
|---|---|---|
| Pre-miR-21 | N/A | 13 ± 2 nM |
| Pre-miR-21-LM1 | 252 ± 43 nM | 156 ± 22 nM |
Acknowledgements
The work was supported by the National Institutes of Health Grant 1R01 GM103834 to G.V., and by the University of Trento (Progetto Biotecnologie) and by the Autonomous Province of Trento (Madelena Project) to P.M.. The authors are grateful for members in the lab for technical assistance, especially Jana Mandic. We thank the Analytical Biopharmacy Core for access to ITC in the School of Pharmacy at the University of Washington.
Footnotes
Accession codes
Sequence data for RRM*-Dicer was deposited under accession code KX060583 with GenBank.
Author Contributions
Y.C. conceived the project, performed protein design, biochemical and cell-based assays, analyzed the data and wrote the paper; F.Y. performed the analysis of protein-RNA interactions by ITC and NMR experiments; L.Z. and T.P. performed the cell biological assays; W.Y. cloned and expressed G. Dicer proteins; K.G. performed SHAPE analysis; M.W. performed Dicer processing assays. S.Z. performed protein structural modeling; P.M. analyzed the data and wrote the paper; G.V. conceived the project, analyzed the data and wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Wang Y, Wang ZF, Hall TMT. Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J. 2013;280:3755–3767. doi: 10.1111/febs.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackay JP, Font J, Segal DJ. The prospects for designer single-stranded RNA-binding proteins. Nat. Struct. Mol. Biol. 2011;18:256–261. doi: 10.1038/nsmb.2005. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Varani G. Engineering RNA-binding proteins for biology. FEBS J. 2013:3734–3754. doi: 10.1111/febs.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. USA. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filipovska A, Razif MFM, Nygård KKA, Rackham O. A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 2011;7:425–427. doi: 10.1038/nchembio.577. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal AA, McLaughlin KJ, Jenkins JL, Kielkopf CL. Structure-guided U1AF65 variant improves recognition and splicing of a defective pre-mRNA. Proc. Natl. Acad. Sci. USA. 2014;111:17420–17425. doi: 10.1073/pnas.1412743111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blakeley BD, McNaughton BR. Synthetic RNA recognition motifs that selectively recognize HIV-1 trans-activation response element hairpin RNA. ACS Chem. Biol. 2014;9:1320–1329. doi: 10.1021/cb500138h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell. Mol. Life. Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu S, et al. The loop position of shRNAs and pre-miRNA is critical for the accuracy of Dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhury NR, Michlewski G. Terminal loop-mediated control of microRNA biogenesis. Biochem. Soc. Trans. 2012;40:789–793. doi: 10.1042/BST20120053. [DOI] [PubMed] [Google Scholar]
- 13.Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1011–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castilla-Llorente V, Nicastro G, Ramos A. Terminal loop-mediated regulation of miRNA biogenesis: selectivity and mechanisms. Biochem. Soc. Trans. 2013;41:861–865. doi: 10.1042/BST20130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krichevsky AM, Gabriely G. miR-21, a small multi-faceted RNA. J. Cell. Mol. Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–91. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 19.MacRae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 20.Asangani IA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 21.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oubridge C, Ito N, Evans PR, Teo C-H, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:423–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 23.Allers J, Shamoo Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Kortemme T, Robertson T, Baker D, Varani G. A new hydrogen-bonding potential for the design of protein-RNA interactions predicts specific contacts and discriminates decoys. Nucleic Acids Res. 2004;32:5147–5162. doi: 10.1093/nar/gkh785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minuchehr Z, Gollaei B. Propensity of amino acids in loop regions connecting beta-strands. Protein Pept. Lett. 2005;12:379–382. doi: 10.2174/0929866053765617. [DOI] [PubMed] [Google Scholar]
- 26.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc. Chem. Res. 2011;44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, et al. Rbfox proteins regulate miRNA biogenesis by sequence specific binding to miRNA precursors and by regulating dicer expression. Nucleic Acids Res. 2016 Mar 21; doi: 10.1093/nar/gkw177. advance access published on. doi: 10.1093/nar/gkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen TA, et al. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Stru. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 32.Peroutka RJ, Elshourbagy N, Piech T, Butt TR. Enhanced protein expression in mammalian cells using engineered SUMO fusions: Secreted phospholipase A2. Protein Sci. 2008;17:1586–1595. doi: 10.1110/ps.035576.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Si M-L, et al. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 34.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughout method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auweter SD, Oberstrass FC, Allain FH-T. Sequence-specific binding of single-stranded RNA: is there a code for recognition. Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lünse CE, et al. An aptamer targeting the apical-loop domain modulated pri-miRNA processing. Angew. Chem. Int. Ed. 2010;49:4674–4677. doi: 10.1002/anie.200906919. [DOI] [PubMed] [Google Scholar]
- 38.Diaz JP, et al. Association of a peptoid ligand with the apical loop of pri-miR-21 inhibits cleavage by Drosha. RNA. 2014;20:528–539. doi: 10.1261/rna.042911.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 39.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price SR, Oubridge C, Varani G, Nagai K. Preparation of RNA–protein complexes for X-ray crystallography and NMR. In: Smith C, editor. RNA–Protein Interaction: Practical Approach. Oxford University Press; Oxford: 1998. pp. 37–74. [Google Scholar]
- 41.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 43.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 45.Tenzer S, Moro A, Kuharev J, Francis AC, Vidalino L, Provenzani A, Macchi P. Proteome-wide characterization of the RNA-binding protein RALY-interactome using the in vivo-biotinylation-pulldown-quant (iBioPQ) approach. J. Proteome Res. 2013;12:2869–2884. doi: 10.1021/pr400193j. [DOI] [PubMed] [Google Scholar]
- 46.Zheng SX, Robertson TA, Varani G. A knowledge-based potential function predicts the specificity and relative binding energy of RNA-binding proteins. FEBS J. 2007;274:6378–6391. doi: 10.1111/j.1742-4658.2007.06155.x. [DOI] [PubMed] [Google Scholar]
- 47.Kortemme T, Morozov AV, Baker D. An Orientation-dependent Hydrogen Bonding Potential Improves Prediction of Specificity and Structure for Proteins and Protein–Protein Complexes. J Mol Biol. 2003;326:1239–59. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Kortemme T, Robertson T, Baker D, Varani G. A new hydrogen-bonding potential for the design of protein–RNA interactions predicts specific contacts and discriminates decoys. Nucleic Acids Res. 2004;32:5147–62. doi: 10.1093/nar/gkh785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oubridge C, Ito N, Evans PR, Teo C-H, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.