Abstract
Embryonic dermal fibroblasts in the skin have the exceptional ability to initiate hair follicle morphogenesis and contribute to scarless wound healing. Activation of the Wnt signaling pathway is critical for dermal fibroblast fate selection and hair follicle induction. In humans, mutations in Wnt pathway components and target genes lead to congenital focal dermal hypoplasias with diminished hair. Despite its critical importance, gene expression signature of embryonic dermal fibroblasts during differentiation and its dependence on Wnt signaling is unknown. Here we applied Shannon entropy analysis to identify the gene expression signature of mouse embryonic dermal fibroblasts. We used available human DNase-seq and histone modification ChiP-seq data on various cell-types to demonstrate that genes in the fibroblast cell identity signature can be epigenetically repressed in other cell-types. We found a subset of the signature genes whose expression is dependent on Wnt/β-catenin activity in vivo. With our approach, we have defined and validated a statistically derived gene expression signature that may mediate dermal fibroblast identity and function in development and disease.
Keywords: fate specification, genomics, transcription, hair, dermis
INTRODUCTION
During a defined window in skin development, embryonic dermal fibroblast progenitors play an integral role in the determination and initiation of integumentary structures such as hair follicles and feathers (Dhouailly et al., 2004; Driskell et al., 2011; Driskell and Watt, 2014; Eames and Schneider, 2005; Millar, 2005; Sengel and Mauger, 1976). Similarly, only embryonic dermal fibroblasts have the capacity for scarless wound healing in skin (Mackool et al., 1998). Given the restricted temporal competence of these cells, defining their unique gene expression signature will be important in understanding the etiology of congenital skin defects and in advancing tissue engineering of skin equivalents with hair (Sriwiriyanont et al., 2012; Supp and Boyce, 2005).
Until recently, isolating the gene expression signature of mouse embryonic dermal fibroblast progenitors and genetically manipulating dermal fibroblasts in vivo was challenged by the lack of specific genetic tools (Cadau et al., 2013; Driskell and Watt, 2014; Rinkevich et al., 2015; Sennett et al., 2015a). Morphologically similar dermal fibroblasts vary in function, differentiation potential, and positional identity (Driskell et al., 2013; Driskell and Watt, 2014; Rinkevich et al., 2015; Rinn et al., 2008; Sorrell and Caplan, 2009). Mouse dermal fibroblasts are derived from diverse embryonic origins, further confounding identification of a gene expression signature (Ohtola et al., 2008; Atit et al., 2006a; Byrne et al., 2002; Tran et al., 2010a).
In the mouse embryo, mesenchymal Wnt/β-catenin signaling activity is visible between E11.5–14.5 and is required for specifying mouse upper dermal fibroblast fate (Andl et al., 2002; Atit et al., 2006a; Y. Zhang et al., 2009). Upon deletion of mesenchymal Wnt/β-catenin between E10.5–12.5, dermal progenitors ectopically express markers of cartilage fate and differentiate to cartilage (Ohtola et al., 2008; Tran et al., 2010a). Between E12.0–14.5, broad dermal Wnt/β-catenin activity precedes hair follicle formation and is required for initiation of hair follicles in the mouse embryo (Cadau et al., 2013; Chen et al., 2012; Fu and Hsu, 2012; Y. Zhang et al., 2009). Dysregulation of Wnt/β-catenin signaling has pronounced effects in human and murine dermal disease and development. Diminished Wnt signaling due to an inactivating mutation in PORCUPINE leads to focal dermal hypoplasia among other defects in humans and mice (Barrott et al., 2011; Grzeschik et al., 2007; X. Wang et al., 2007). Mutations in the Wnt target gene DERMO1/TWIST2 leads to bi-temporal dermal dysplasia in humans and mice (Franco et al., 2011; Tukel et al., 2010). Thus, identifying the Wnt/β-catenin gene expression signature of dermal fibroblasts is clinically relevant and will reveal new mediators for dermal fibroblast function.
The development of dermal fibroblast progenitor lineage-restricted genetic tools is needed to obtain cell-type specific expression data. In addition, multiple high-resolution genomics data sets are needed to confirm actively transcribed genes. As an alternative to the high cost of multiple genome-wide assays or for cases in which the amount of tissue is limiting, experimental strategies using statistical and comparative analyses of published datasets have emerged as feasible approaches for interrogating the cell-type specific gene expression signature. Statistical tools such as Shannon Entropy can be applied to available DNA microarray datasets to identify highly enriched and cell-type specific expression compared to the other cell-types assayed (Schug et al., 2005). Modified Shannon Entropy generates a Q-score value for each identified gene that is associated with enrichment and specificity of expression. DNaseI hyperpersensitivity sites that indicate accessible chromatin correlate with transcriptional start sites (TSS), regulatory elements, and histone modification markers of active transcription (Heintzman et al., 2009; 2007). Comparison of DNaseI hypersensitivity sites from DNase-seq data across various cell-types can be used to predict cell-type specific gene expression (Natarajan et al., 2012; Song et al., 2011). Finally, with the discovery of the “histone code”, comparison of genome-wide histone modification patterns across cell-types can be used to understand the regulatory basis of cell-type-specific gene expression that may govern cell identity (Heintzman et al., 2009; 2007).
In this study, we sought to define a actively regulated gene expression signature of dermal fibroblast progenitors and identify candidate effectors of Wnt/β-catenin signaling for dermal identity. We generated the gene expression profile of E12.5 embryonic dermal fibroblasts by microarray. We then subjected this microarray dataset to Shannon Entropy analysis and identified 63 genes that had statistically enriched expression in dermal fibroblasts compared to other cell-types. By comparing available DNase-seq and histone-modification tracks from various cell-types, we found a subset of the fibroblast gene expression signature that was epigenetically repressed in other related cell-types. Expression of a subset of these genes was positively dependent on Wnt/β-catenin signaling as determined by RNA-Sequencing of β-catenin mutant mouse embryonic dermal fibroblast progenitors. These genes were associated with human dermal and hair follicle defects. Thus, using published and original data sets, we have identified a statistically enriched gene-expression signature that could be relevant for embryonic fibroblast identity and hair follicle development.
RESULTS
Defining a putative embryonic dermal fibroblast gene expression signature
To obtain an accurate gene expression profile of embryonic dermal fibroblasts, we isolated E12.5 mouse dorsal dermal fibroblast progenitors by laser capture microdissection and obtained a gene expression profile by performing microarray analysis (GSE75913). Next, we used Shannon Entropy analysis to rank genes by the specificity of their expression levels in E12.5 dermal fibroblast progenitors compared to that in seven other cell-types. Of the 25,697 genes probed, 63 genes were identified to have a 1.4- to 2-fold elevated expression (Q-score below 9.246) compared to the other available cell-types (Figure 1, Table S1). The 63 genes were grouped into Mouse Genome Informatics (MGI) ontologies, with genes listed in order of ascending Q score, which corresponds to a decreasing fold difference (Blake et al., 2014; Smith et al., 2014) (Figure 1, Table S2). Notably, seven of these genes were the following transcription factor genes: Hoxc6, Twist1, Twist2, Nfatc4, Sp5, Tead2, and Pitx2 (Figure 1). Secreted factors such as Igf2, Bmp4, Wnt5a and extracellular matrix genes including Emid2/Collagen26a1, Collagen3a1, Collagen5a1, were identified in E12.5 dermal fibroblast progenitors (Figure 1). Other genes identified by Q-score as the highest-ranking genes of the remaining ontologies are shown in Figure 1. Thus, Shannon Entropy analysis revealed a statistically enriched expression signature for E12.5 dermal fibroblast progenitors genes that had a variety of cellular functions and were present in different sub-cellular locations. Subsequently we used several experimental and bioinformatics techniques to validate the gene expression signature, define the basis for cell type specific expression, and identify putative role in disease (Summarized in Table S8). Due to inherent limitations of each technique and few publicly available data sets in mouse tissues, we used all 63 genes in all our analyses.
Figure 1. Characterization of E12.5 mouse dermal fibroblast gene expression signature determined by Shannon Entropy analysis.
Genes with fold-change associated Q scores less than 9.25 were clustered into basic ontologies using Mouse Genome Informatic (MGI) batch query tool. Genes are listed in order of ascending Q score in each category.
Next we validated the results of our Shannon Entropy analysis in the transcriptome of E13.5 upper dermal fibroblast progenitors obtained by next generation whole genome RNA-sequencing (Table S4, GSE75944). We determined the expression levels and transcript abundance of all the 63 genes in E13.5 Engrailed1Cre (En1Cre); RRYFP lineage-marked dorsal dermal fibroblasts. We found that 42 of the 63 genes were expressed (FPKM≥1, Table S4). However, genes such as Mid1, Notum, Pitx2, and Sp5 that were expressed at FPKM <1 in our E13.5 RNA-seq results were present in the transcriptome of E14.5 dermal progenitors (Sennett et al., 2015b). We attributed the discrepancy between the RNA-seq data sets due to technical variation in genome wide coverage and library preparation.
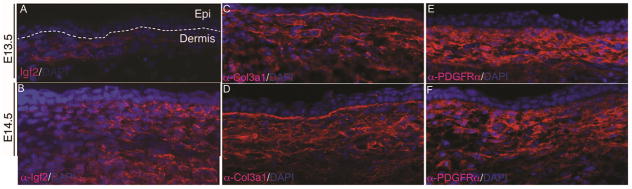
Since Insulin-like Growth factor 2 (IGF2), Collagen3a1, Crabp1, and Mest were most abundantly expressed by RNA-seq (FPKM≥106), we examined the protein expression of a known gene such as Col3a1 and a novel gene such as, IGF2. We found Collagen3a1 and IGF2 have restricted protein expression in the embryonic dermis similar to the pan-dermal fibroblast marker PDGFRa (Figure 2) (Driskell et al., 2013). Crabp1 was previously shown to have restricted expression in embryonic dermis (Collins and Watt, 2008).
Figure 2. Dermal restricted protein expression of Col3a1, IGF2, and PDGFRα.
Indirect immunofluorescence with DAPI stained nuclei was performed on transverse mouse E13.5 and E14.5 dorsal trunk skin. Protein expression of Igf2(A,B), Col3a1 (C,D), and PDGFRα (E,F) was visible in the upper dermis and excluded from the epidermis of the developing skin.
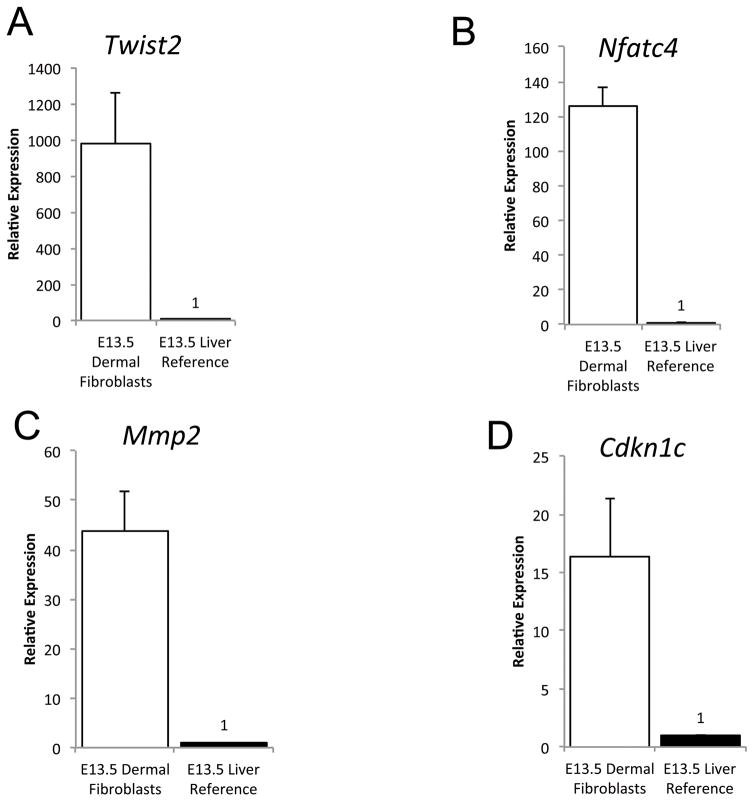
We also validated relative expression levels of certain new and known genes that were transcription factors or targets of the Wnt signaling pathway (Nfatc4, Twist2, Cdkn1c, Mmp2, Tead2, Pitx2, Fzd10). We used qRT-PCR to determine relative mRNA abundance in E13.5 primary dorsal dermal fibroblasts progenitors compared to E13.5 liver or brain (Table S3). Pitx2, Fzd10 were expressed <1FPKM by RNA-seq and remained undetectable by qRT-PCR. We were unable to validate expression of Tead2. However, relative expression of Twist2, Nfatc4, Mmp2, and Cdkn1c was consistently higher in the E13.5 dermis compared to liver and brain (Figure 3, data not shown). Twist2 is one of the earliest transcription factor expressed in mouse dermal fibroblast progenitors at E11.5 and a known target of the Wnt/β-catenin signaling pathway (Atit et al., 2006a; Tran et al., 2010b). Nfatc4, Mmp2, and Cdkn1c protein expression remain to be characterized in embryonic dermal fibroblasts. Thus, we were able to validate expression of known genes and identify new genes that are enriched in E12.5–13.5 dermal fibroblasts.
Figure 3. Tissue enrichment of putative embryonic dermal fibroblast signature genes.
Relative mRNA expression analysis of dermal fibroblast signature genes in E13.5 dermal fibroblasts and liver (n=3 for fibroblasts). The Y-axis represents relative expression in dermal fibroblasts compared to liver.
Regulation of fibroblast-specific gene expression
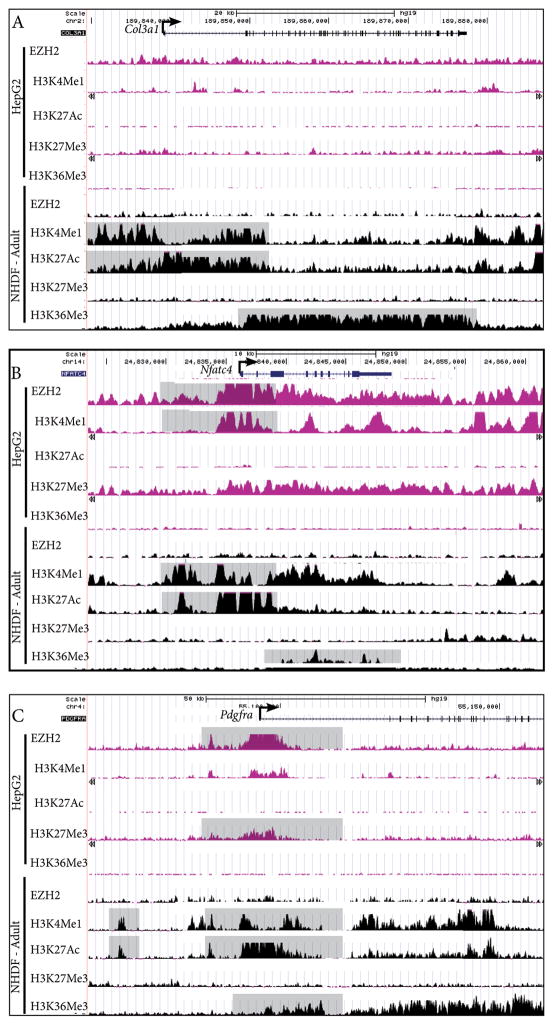
Next, we sought to define the basis of fibroblast cell-type specific expression by identifying differential histone modifications and DNaseI hypersensitivity at the promoter. Using publicly available histone modification datasets on UCSC genome browser (http://genome.ucsc.edu/), we located putative dermal fibroblast specific regulatory regions in silico. Actively transcribed enhancer elements correlate with mono-methylated lysine 4 of histone 3 (H3K4me1), and acetylated lysine 27 of histone 3 (H3K27ac). Tri-methylated lysine 36 on histone 3 (H3K36me3) is associated with the path of RNA Pol II elongation (Heintzman et al., 2009; 2007; Kent et al., 2002; Lupien et al., 2008; Raney et al., 2014; Zentner et al., 2011). Epigenetic repression is marked by trimethylated lysine 27 of histone 3 (H3K27me3), catalyzed by Enhancer of zeste homolog 2 (EZH2) in the Polycomb repressive complex 2 (PRC2) (Heintzman et al., 2009; 2007). Because of the relative abundance of published genome-wide histone modification data sets for human tissue-types and the limited number of similar datasets available for mouse tissues, we compared profiles of these predictive histone modifications ±50kb from the transcriptional start site (TSS) in normal adult human dermal fibroblasts (NHDF) cell line to liver carcinoma (HepG2). We selected three candidates that were validated by multiple techniques in our study: Col3a1, Nfatc4, and Pdgfrα. (Figure 4A–C). Expression of Col3a1 and Pdgfrα has been characterized in mouse embryonic dermis and Nfatc4 remains a novel gene that has previously not been implicated in embryonic dermal development (Figure 2) (Collins et al., 2011; X. Q. Zhang et al., 1998). At the human COL3a1 locus in NHDF, we found broad H3K36me3 binding downstream of the TSS and “blanket” type binding of H3K4me1 and H3K27Ac within 15kb of TSS (Figure 4A). These regions lacked histone modification for transcriptional activation and repression in the HepG2 cells, suggesting lack of tissue specific epigenetic regulation. At the human NFATC4 locus, 5 kilo bases (kb) upstream of the TSS for NFATC4, we found enrichment for H3K4me1 and H3K27ac and absence of EZH2 and H3K27me3 binding. We found broad binding of H3K36me3 within the gene, indicative of active transcription in NHDF cells. In contrast, the HepG2 cells had broad and strong EZH2 and H3K27Me3 binding profile and an absence of H3K27Ac binding in the equivalent region (Figure 4B), demonstrating active repression by an epigenetic mechanism. At the human PDGFRα locus, we identified three regions spanning 500-1000bp with at least 25-fold enrichment for both H3K4me1 and H3K27Ac located approximately 50 kb, 25 kb, and 10 kb upstream of the TSS, suggestive of regulatory elements (Figure 4C). In HepG2 cells, enrichment of EZH2 binding was in the 10kb upstream element and at the TSS, demonstrating transcriptional repression. There was depletion of H3K27Ac at the TSS and upstream putative regulatory elements, indicative of cell type specific regulatory elements (Figure 4C). Our proof of principle analysis using available human data sets reveals putative fibroblast-specific regulatory elements and suggests the presence of an actively regulated fibroblast identity program maintained in adult dermal fibroblasts.
Figure 4. Putative regulatory regions for dermal fibroblast gene expression.
ChIP-seq profiles of histone modifications and EZH2 binding in human dermal fibroblast (HDF) and liver cell (HepG2) lines at Collagen3a1, Nfatc4, and Pdgfrα loci (A–C). Y-axis values range from 0–50, and refer to a relative value of signal enrichment for each region of interest. Data were visualized using the UCSC Genome Browser. Putative transcriptional active regions are associated with H3K4me1, H3K27Ac, and H3K36Me3. Transcriptional repression is associated with EZH2 binding and H3K27me3 at the TSS.
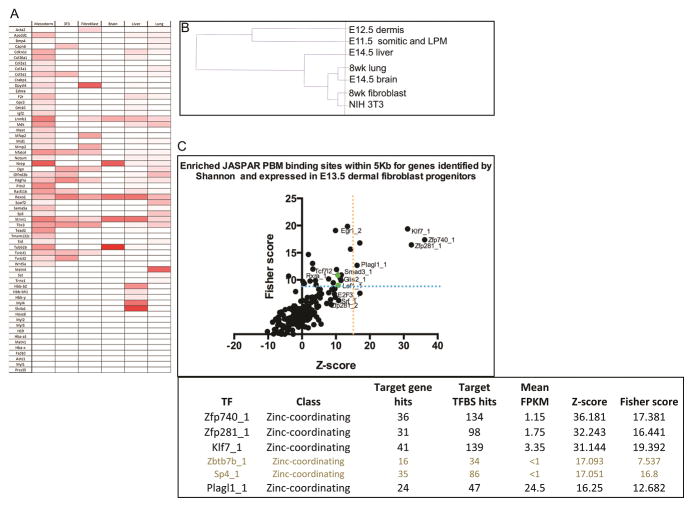
DNaseI hypersensitivity patterns at the promoter indicates active or poised for transcription. Comparison of DNaseI hypersensitivity patterns at the promoter across cell types can reveal cell-type specific gene expression (Heintzman et al., 2009; 2007; Song et al., 2011). Next, we analyzed several publicly available mouse DNase-seq data sets from various adult and embryonic cell-types. We identified DNaseI hypersensitivity peaks within +/− 2kb of the TSS of all the 63 genes using peak-finding and peak annotation tools (Heinz et al., 2010) (Figure 5A and Table S5). These analyses yielded several interesting results. The DNaseI hypersensitivity pattern near TSS of E11.5 of somitic and LPM cells was most closely related to E12.5 dermal fibroblasts gene expression signature, revealing lineage similarity. Using DNase hypersensitivity peak values, we performed hierarchical clustering of cell lines as previous described (Figure 5B) (Song et al., 2011). The clustering robustly reflected the closest lineage relationship with E11.5 somitic and LPM compared to the other cell types. We found E14.5 whole liver exhibited the next closest relationship with the expression signature and the other cell types were distantly related. Since we do not know the technical or biological reasons for the considerable overlap with E14.5 whole liver and brain we did not eliminate these genes from subsequent analyses. As expected, the gene expression signature of E12.5 dermal fibroblasts progenitors most closely resembles the precursor population in E11.5 somitic and lateral plate mesoderm.
Figure 5. Inferring cell-type-specific gene expression from regions of open chromatin.
Heatmap of peak scores assigned to DNase hypersensitive sites within 2kb of the TSS for the putative 63 dermal fibroblasts signature genes in related and unrelated cell-types in mouse. Default settings and threshold values were used for all HOMER analysis (A). Hierarchical clustering of 6 cell types based on DNAase HS value ≥10 and relationship to E12.5 dermal fibroblast progenitor (B). Transcription Factor overrepresentation analysis on 63 dermal fibroblast gene expression signature (C). Statistically significant transcription factors that are also expressed in E13.5 dermal fibroblasts are in black (C).
Next, we examined if the genes lacking DNaseI hypersensitivity sites at the TSS were also transcriptionally repressed by epigenetic mechanisms. Due to limited availability of genome-wide H3K27me3 repressive modification data sets for embryonic mouse tissues, we analyzed the H3K27me3 ChIP-seq data set for E14.5 mouse brain. Using HOMER, we calculated the peak scores of H3K27me3 modification within ± 2Kb for all 63 genes. We found 21 genes had PRC2-mediated H3K27me3 modifications near the TSS in brain tissue (Table S6) (Heinz et al., 2010). Notably, Nfatc4 promoter was epigenetically repressed in HepG2 cells and also in the mouse embryonic brain (Table S6, S8). We were unable to obtain and perform similar analysis on embryonic mouse mesenchyme and fibroblast. Analysis of other mouse embryonic cell types and epigenetic repression mechanisms will be required to obtain a more complete picture. Together, the absence of DNaseI hypersensitivity and additional presence of H3K27me3 at TSS demonstrates that a subset of genes in the fibroblast gene expression signature is epigenetically repressed in unrelated cell-types.
Identifying putative transcriptional regulators of dermal fibroblast gene expression signature
Next, we sought to identify candidate transcription factors that may regulate the dermal fibroblast gene expression signature. We performed over-representation analysis of predicted binding sites of transcription factors in promoter regions (+/− 5kb) of the 63 genes. Using single site analysis in OPOSSUM 3.0 with JASPAR PBM database, we identified relative enrichment of predicted binding sites of transcription factors (Ho Sui et al., 2007). We annotate over-represented transciption factors that were also expressed in E13.5 dermal fibroblasts (FPKM>1) by RNA-seq (Figure 5C). Because Wnt/β-catenin signaling is required for dermal fibroblasts progenitor idenitity, we hypothesized that TCF/Lef transcription factors would over represented. We found TCF7l2 (TCF4) and Lef1, as well as Smad3, the mediator of TGFβ signaling were over-represented, but were below the statistical threshold (Figure 5C). Klf7, Zfp281, Plagl1/Zac1, and Zfp740 transcription factors were represented above the statistical thresholds (Figure 5B) (Ho Sui et al., 2007). Klf7 has been linked directly to mesodermal differentiation using mouse embryonic fibroblasts (MEFs) (Caiazzo et al., 2010). Zfp281 is closely associated with the expression of β-catenin and Zfp281 is known to bind to the promoter region of β-catenin (Seo et al., 2013). Zfp281 can function as a master regulator of pluripotent stem cell development and regulates critical checkpoints in the commitment of cell development (Fidalgo et al., 2011; Wang et al., 2008). Plagl1/Zac1 is expressed in neuronal cell progenitors and functions in cardiac morphogenesis, chondrogenesis, and development of the embryonic mesoderm (Schmidt-Edelkraut et al., 2014; Tsuda et al., 2004; Valente et al., 2005; Yuasa et al., 2010). There is no known association between Zfp740 and mesoderm development or embryogenesis in general. Hence, our analysis reveals a set of new transcription factors that are poised to regulate the dermal fibroblast gene expression signature at the promoter.
Determining dermal identity genes in the embryonic fibroblast gene expression signature
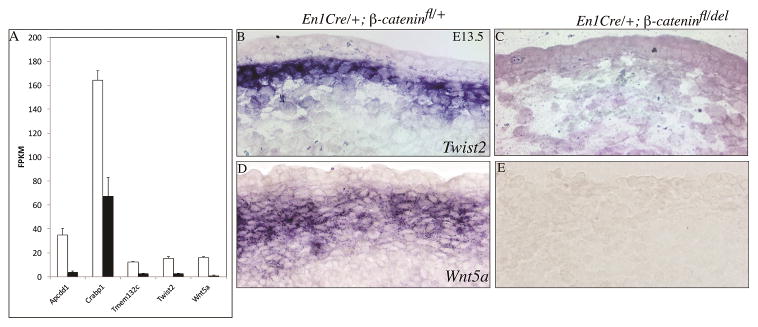
Without dermal mesenchymal Wnt/β-catenin signaling, dermal progenitors become cartilage (Ohtola et al., 2008; Tran et al., 2010b). Therefore, these mutants can serve as a model for dermal fibroblast identity. Next, we determined which of the 63 genes in the embryonic fibroblast gene expression signature were dependent on Wnt/β-catenin signaling. We profiled the transcriptome of E13.5 mouse embryonic dermal fibroblasts in En1Cre/+; R26YFP/+; β-catenindel/flox mutants and En1Cre/+; R26YFP/+ control by whole-genome RNA-seq (GSE75944). Given β-catenin signaling is a transcriptional regulator, we found a subset of the signature genes were either positively or negatively regulated in the β-catenin mutants. The following five genes were significantly down-regulated in β-catenin mutants by RNA-seq: Apccd1, Crabp1, Twist2, Tmem132c, and Wnt5a (Figure 6A). The following six genes were also up regulated in β-catenin mutants: Prss35, Grb10, Mest, Cdkn1c, Gpc3, Col2a1. In situ hybridization of Twist2 and Wnt5a mRNA confirmed expression in upper dermis at E13.5 controls and absence in conditional β-catenin mutants (Figure 6B–E). Thus, these Wnt/β-catenin signaling-responsive genes can be candidate mediators of embryonic dermal fibroblast cell identity.
Figure 6. Wnt/β-catenin dependent gene expression of embryonic dermal fibroblast signature genes.
Relative FPKM values for each gene from the whole genome RNA-seq of E13.5 control (white column) and β-catenin mutant (black column) dorsal upper dermal fibroblasts. (A) Five genes in the dermal fibroblast gene expression signature were significantly dependent on Wnt/β-catenin activity in vivo by RNA-seq. (B,C) Twist2 and (D,E) Wnt5a expression in dorsal dermis at E13.5 in control and conditional mesenchyme β-catenin deleted mutants.
Dermal fibroblast gene expression signature is enriched for disease-associated risk loci
Finally, we queried the Genome-wide association studies (GWAS) catalog and calculated the enrichment of traits near the genes in dermal fibroblast gene expression signature genes to identify disease associations. GWAS studies for character traits including congenital skin and dermal defects are currently lacking. Recently, GWAS studies have identified genomic loci which confer risk for human androgenic alopecia. At the 2q37 risk locus, the lead single nucleotide polymorphism (SNP) rs9287638 is within 500kb from the Twist2 (R. Li et al., 2012). In embryonic dermal fibroblasts, Twist2 is one of the earliest expressing transcription factors and a Wnt/β-catenin signaling-responsive gene. We also performed an unbiased search for any diseases or traits that were enriched near the genes that comprise our signature. Recent studies have demonstrated that the majority of GWAS risk loci lie within noncoding regulatory regions, rather than coding regions (Ernst et al., 2011; Trynka et al., 2013). Thus, we evaluated all disease risk loci within 250-kb of the TSS of the dermal fibroblast gene expression signature compared to 1000 random gene sets (n-matched) and summarized our findings for lead SNPs (Table S7). We found significant enrichment for SNPs associated with lung disease (P=0.018). Seventeen lead SNPs associated with pulmonary disease, pulmonary function or lung cancer were found within 250-kb from the TSS of Acta2, Crabp1, Dpysl4, Mfap2, Rasl11b, SSt, Tril, or Twist2. We also found significant enrichment of SNPs associated with electrical heart function (P=0.0004). Twenty-one lead SNPS found within 250-kb from the TSS of GWAS were associated with atrial fibrillation, electrocardiographic traits or QT interval trait. Thus, we have identified two categories of traits that are most enriched near our gene set and our approach demonstrates that these results are statically unlikely to occur for any gene list. Whether these results are reflective of the impact of risk loci on early (mesoderm) development, which contributes to altered function in adults, or are reflective of additional role of dermal fibroblast genes in adult tissues, remains to be determined. Futures GWAS studies will reveal if the dermal fibroblast signature genes are associated with congenital skin, dermal, hair follicle defects.
In summary, we identified a statistically enriched fibroblast gene expression signature of 63 genes in mouse dermal fibroblast progenitors. Expression of a subset of these genes was validated by different techniques and only a small subset of these genes were dependent on Wnt/β-catenin signaling that is required for instructing dermal fibroblast cell identity (Table S8). Promoter sequence analysis of genes in the expression signature revealed new candidate transcription factors, but did not include Wnt/β-catenin signaling transcriptional cofactors, TCF4/Lef1. Transcriptional access to the genes in the expression signature in other cell types confirmed that the expected somitic and lateral plate mesoderm shared the most similarity. Finally, a subset of genes in the expression signature was repressed in distantly related or adult cell types, suggesting they are important for dermal fibroblast identity. Given the limited available data for genes in skin diseases, we identified only a handful of genes that were associated with skin and SNPs in the GWAS catalog. Thus, despite the inherent limitations of each technique and availability of relevant data sets, we were able to identify candidate mediators of dermal identity and obtain insights in the regulation and disease association of the fibroblast gene expression signature.
DISCUSSION
In this study, we sought to define an embryonic dermal fibroblast gene expression signature to identify genes that may govern identity and function. Based on the emerging literature about dermal fibroblast development and epigenetic regulation of cell identity, we tested the hypothesis that the gene expression signature was actively regulated and dependent on Wnt/β-catenin signaling for dermal identity (Atit et al., 2006, Ohtola et al., 2008, Song et al, 2011). Our statistical and comparative analyses of original and preexisting datasets revealed a actively regulated gene expression signature for dermal fibroblast progenitors that was validated with various techniques and partially dependent on Wnt/β-catenin signaling.
We used several techniques to validate the gene expression signature. Due to inherent noise, sensitivity, and technical limitations of each technique to assay gene expression, we were only able to validate a subset of the gene expression signature obtained from E12.5 dermal fibroblast progenitors. Most notably, the genes in the myosin complex and hemoglobin complex previously identified in the gene expression signature obtained by Shannon Entropy analysis were not expressed in E11.5 LPM (DNase hypersensitivity score=0) and in E13.5 or E14.5 dermal fibroblasts progenitors (FPKM ≤ 1) (Table S4) (Sennett et al., 2015). However, we cannot rule out embryonic stage specific differences in gene expression and less sensitivity in the Shannon entropy analysis done with a limited number of available microarray data sets.
We used several techniques on the dermal fibroblast gene expression signature to identify genes that may govern identity. Mouse trunk dermal fibroblasts originate from LPM and somite-derived mesoderm (Ohtola et al., 2008; Atit et al., 2006a; Candille et al., 2004). As expected, our results show that our statistically enriched dermal fibroblast gene expression signature is most similar to its recent precursors, namely the E11.5 somitic mesoderm and LPM. In contrast, the signature has the highest degree of difference from adult dermal fibroblasts and from distantly related cell-types such as brain. Genes in the expression signature that had a robust DNase hypersensitivity score (≥10) only in the promoters of E11.5 LPM include the following: Apccd1, Crapb1, Cdkn1c, Collagen genes, F2r, Gpc3, Grb10, Igf2, Mest, Mid1, Sema5a, Tril, Tmem132c, Tead2, Twist2, Wnt5a. These genes may govern embryonic dermal fibroblast identity but their functional role in development remains to be tested. Other genes in our fibroblast gene expression signature such as Nfatc4, PDGFRa, Tbx3, and Twist1 had promoters associated with DNase hypersensitivity in E11.5 mesoderm, NIH 3T3 cells, 8wk adult fibroblasts, signifying a shared fibroblast signature. In the absence of accompanying gene expression data for all the cell types, we can only infer epigenetically poised for active gene transcription from DNase hypersensitivity scores alone. We also intersected the gene expression signature with the differentially expressed genes in the mesenchyme Wnt/β-catenin signaling mutant embryonic dermal fibroblasts to find genes that govern cell identity. Only Apccd1, Crabp1, Twist2, Tmem132c, and Wnt5a had significantly lower expression in the absence of Wnt/β-catenin signaling in E13.5 dermis and had promoters associated with DNase hypersensitivity in the E11.5 LPM. Since we have established that Wnt/β-catenin signaling is an instructive cue for embryonic dermal fibroblast identity, we would have predicted a larger subset of the signature would be transcriptionally regulated and be dependent on Wnt/β-catenin signaling. It is likely we may not be able to validate all the candidates in our RNA-seq results. For instance, we previously showed the expression of Twist1 mRNA and protein are clearly dependent on ectodermal Wnts and mesenchymal Wnt/β-catenin signaling in vivo (Goodnough et al., 2014; 2012). However, differential expression of Twist1 did not reach statistical significance by RNA-seq of E13.5 β-catenin mutant dermis. One of the inherent limitations of single-end RNA-seq includes differences in genome wide coverage between samples. Alternatively, our analysis and approach revealed that the expression signature may be regulated by different set of transcription factors and may direct other functional roles. Despite the limitations in the various techniques and approaches, our studies predict that Apccd1, Crabp1, Twist2, Tmem132c, and Wnt5a are most likely to be involved in acquisition of dermal identity, but their function remains to be examined.
We used available data sets for histone modification and DNase hypersensitivity to understand the regulation of fibroblast gene expression signature. DNase hypersensitivity at the promoter of the fibroblast gene expression signature in other cell types allowed us to draw conclusions regarding lineage relationships between E12.5 dermal fibroblasts and other embryonic and adult cell types. As expected, the closest relationship was with E11.5 lateral plate mesoderm, and distantly with immortalized fibroblast cell line and adult fibroblast. Assessing the predictive epigenetic histone modifications near human Pdgfrα and Nfatc4 revealed that the same regulatory elements were epigenetically repressed by PRC2 in HepG2 liver cell line. These elements were transcriptionally active in adult human dermal fibroblasts and may regulate gene expression of Pdgfrα and Nfatc4 in dermal fibroblasts. Both Pdgfrα and Nfatc4 displayed regions upstream of the 5′ TSS that serve as potential fibroblast lineage-specific enhancer regions and warrant further investigation in transgenic reporter mice. Comparative histone modification analysis in the mouse dermal fibroblast progenitors and distantly related cell types, will provide a more comprehensive view of fibroblast gene regulation. In addition, enhancer elements for the transcription factor binding profiles may provide additional insights into the molecular mechanisms controlling dermal fibroblast lineage.
A small subset of the fibroblast expression signature genes (Apcdd1, Crabp1, Twist2, Tmem 132c, and Wnt5a) were positively dependent on Wnt/β-catenin signaling pathway and were also associated with skin defects. Twist2/Dermo1 gene is a basic helix loop helix transcription factor that is expressed in upper dermal fibroblast progenitors starting at E11.5–15.5 (Atit et al., 2006a; Franco et al., 2011; L. Li et al., 1995). Non-sense mutation in human TWIST2 is associated with Setleis syndrome in humans and mice with characteristic bi-temporal dysplasia lacking hair (Tukel et al., 2010). Apcdd1 and Wnt5a are known signature genes of the dermal papilla, the specialized embryonic dermal fibroblasts that associate with hair follicles (Hu et al., 2010; Rendl et al., 2008; van Amerongen et al., 2012). Mutations in human APCDD1 is associated with hair loss in hereditary hypotrichosis (Jukkola et al., 2004; Shimomura et al., 2010). Dermal Wnt5a is involved in hair follicle cell fate and overexpression of Wnt5a in mouse whole skin leads to decrease in secondary hair follicle formation (Hu et al., 2010; van Amerongen et al., 2012). Wnt5a is a non-canonical extracellular Wnt ligand and is uniformly expressed in dermal progenitors between E12.5–E14.5 and then in the developing dermal condensate (Andl et al., 2002; Cadau et al., 2013; Goodnough et al., 2014). Wnt5a signaling controls distal limb elongation by creating planar cell polarity in chondrocytes and embryonic patterning (Andre et al., 2015; Gao et al., 2011). Its role in embryonic dermis migration and formation remains to be established with conditional genetic mutants and live cell imaging analysis. Crapb1, a retinoic acid binding protein, and Tmem132c have restricted expression in embryonic dermis from E13.5 onwards, but their function in skin development is unknown (Collins and Watt, 2008; Diez-Roux et al., 2011). We have identified new candidate downstream effectors of β-catenin, and future functional analysis will elucidate if these genes singularly or in concert determine the embryonic dermal fibroblasts progenitor identity or promote hair follicle initiation.
Taken together, we have identified a gene expression signature for embryonic dermal fibroblast. Our results extend our knowledge regarding the putative mediators of dermal identity in development and disease, and is informative in the context of tissue engineering.
MATERIALS AND METHODS
Mice
Control (En1Cre/+; R26R) or (En1Cre/+; RRYFP/+; β-catenin flox/+), conditional β-catenin dermal fibroblast mutant (En1Cre/+; RRYFP/+; β-catenin flox/deleted) were generated as previously described (Atit et al., 2006b; Brault et al., 2001; Kimmel et al., 2000; Soriano, 1999; Srinivas et al., 2001). For each experiment, a minimum of three mutants with litter-matched controls were studied. Case Western Reserve Institutional Animal Care and Use Committee approved all animal procedures. Mice and embryos were genotyped as described previously. Mice were time-mated, and the vaginal plug day was assigned as E0.5. At desired time points, embryos were harvested and processed for frozen or paraffin sections as previously described (Atit et al., 2006a).
DNA Microarray
The population of E12.5 ventral dermal fibroblast progenitor cells was isolated by Laser Capture Microdissection (LCM) from the ventral body wall from five En1Cre/+; R26R/+ embryos. LCM was performed as previously described (Simone et al., 2000). Total RNA was extracted and pooled before amplification (Ambion) and labeling. Three technical replicates were analyzed on Illumina MouseRef-8 v2.0 expression bead chip platform and normalized using rank invariant method (GSE75913).
Shannon Entropy Analysis
Shannon Entropy Analysis was completed using the datasets acquired from our DNA Microarray against seven different datasets acquired from Gene Expression Omnibus (GEO) (Edgar et al., 2002) (Table S2). The datasets were normalized in LumiR (Illumina) to account for background signal and condense the data into single column format. Then, the Shannon Entropy was performed as previously described in Schug et al (2005) (Supplemental Figure 2). Shannon Entropy calculates both an H value, that measures the degree of overall tissue specificity of a gene, and a Q value that incorporates overall tissue specificity and relative expression level (Schug et al., 2005). Relative expression, H, and Q formulae are shown below:
Bioinformatics Tools
The genes identified above the fold-change associated Q Score cutoff (9.246775) were clustered into basic ontologies using Mouse Genome Informatic’s (MGI) batch query tool (http://www.informatics.jax.org/batch) (Blake et al., 2014). Transcription factor binding site over-representation analysis within 5Kb of the TSS was performed using using OPOSSUM 3.0 (Ho Sui et al., 2007; Kwon et al., 2012). Statistical thresholds were calculated as previously described (Kwon et al., 2012).
UCSC Genome Browser
Using the Human Genome Browser Gateway (http://genome.ucsc.edu/cgi-bin/hgGateway), query of histone modification and binding analysis were performed for NFATC4, PDGFRA, and COL3A1 on HepG2 liver carcinoma and normal human adult dermal fibroblast cell lines (Bernstein et al., 2005; 2006; Ernst et al., 2011; Guttman et al., 2010; Mikkelsen et al., 2007). Using the Broad Histone ChIP-seq data from the ENCODE Histone Modification Tracks, EZH2, H3K4me1, H3K27Ac, H3K27me3, and H3K36me3 binding profiles were assessed (ENCODE Project Consortium, 2011; Karolchik et al., 2014; Rosenbloom et al., 2013). UCSC genome browser supplies density graphs of signal enrichment for each histone modification, computed by the number of sequenced tags overlapping the particular region of interest, with signal peaks referring to regions of statistically significant signal enrichment. Data was visualized using the UCSC Genome Browser. Putative transcriptionally active regions are associated with H3K27Ac and H3K36Me3 and repressed regions are marked by EZH2 binding and H3K27me3.
Identification of DNase hypersensitivity and H3K27me3 peaks
Alignment files (BAM format) were downloaded from UCSC (http://hgdownload.cse.ucsc.edu/) and HOMER (Hypergeometric Optimization of Motif EnRichment) tools were used to find and annotate DNase hypersensitivity peaks for 6 mouse-derived cell/tissue types: E14.5 brain, E14.5 liver, E11.5 somitic and lateral plate mesoderm (LPM), NIH3T3 immortalized cells, adult fibroblasts, and adult lung (Heinz et al., 2010) (wgEncode EM001726, EM003401, EM001935, EM001936, EM001719, EM001723, EM002725). These tools were also used to find and annotate H3K27me3 ChIP-seq peaks for E14.5 whole brain (wgEncodeEM002725). Peaks were annotated with the nearest transcription start site (TSS) and the distance to that TSS, which allowed us to extract all peaks within 2kb of the TSS for our 63 genes of interest. Default settings and threshold values were used for all HOMER analysis. Maximum peak values for each gene in each tissue of interest were used to generate a DNase hypersensitivity heat map and a “peak score” (the sum of maximum peak values over all genes) for each tissue. We defined all transcriptionally active promoters with DNase hypersensitivity scores ≥10 as 1 and <10 as “0” (Song et al., 2011). We created a matrix for all 6 cell types and performed hierarchical clustering using Matlab (average score settings).
Tissue Extraction and qRT-PCR
For the qRT-PCR validation and quantification work was performed on E13.5 β-catenin mutants (En1Cre/+; RRYFP/+; β-catenin flox/deleted) and control (En1Cre/+; RRYFP/+; β-catenin flox/+) dermal fibroblast progenitors. Dorsal skin was microdissected from E13.5 embryos and treated with dispase treatment to isolate the dermal progenitors from the surface ectoderm. E13.5 dorsal dermal progenitors and whole liver were flash frozen in liquid nitrogen and stored in −80 ° C. Primers were chosen from the list of known primers at Primer Depot (mouseprimerdepot.nci.nih.gov) and shown in Table S3. qPCR quantifies relative amounts of mRNA between samples by the ΔΔCT method (Livak and Schmittgen, 2001).
RNA-Seq and Statistical Analysis
Poly-A-selected RNA was isolated from the dorsal dermal fibroblasts that were obtained by FACS from two E13.5 En1Cre/+; RRYFP/+ control and three En1Cre/+; RRYFP/+; β-cateninlox/deleted mutant embryos (GSE75944). The libraries were prepared with Illumina Truseq kit and sequenced using the Illumina Genome Analyzer IIX platform with single-end 50bp reads. Raw reads were aligned to the mouse reference sequences NCBI mm10 downloaded from UCSC genome databases using GSNAP program. Only reads that were mapped to a unique location in mm10 genome were retained for further analysis. The numbers of raw reads that were mapped to mouse genes annotated in RefSeq database were counted using HTSeq program (http://wwwhuber.embl.de/users/anders/HTSeq/doc/count.html#count) and subsequently used for differential gene expression analysis after normalizing the values to the total of mapped reads in each condition (Anders et al., 2014). Genes with P values less than 0.05 after adjusting for false discovery rate with Benjamini and Hochberg method in a negative binomial test and 1.5 fold changes between compared conditions were considered statistically significant (Benjamini and Hochberg, 1995). Expression levels of RefSeq annotated genes were calculated in unit of fragments per kilobase of transcript per million mapped reads (FPKM) values. Genes with low FPKM (≤1) were filtered out as not expressed (Benjamini and Hochberg, 1995; Wu and Nacu, 2010).
Immunohistochemistry and Immunofluorescence
Histology, and immunofluorescence on dorsal mouse trunk skin were performed as described previously (Ohtola et al., 2008; Atit et al., 2006a). The antibodies used were anti-Col3〈1 (Rockland, cat#600-401-105, 1:100) and anti-Igf2 (SantaCruz, cat#sc-7435, 1:200).
GWAS analysis
SNPs associated with disease were obtained from the NHGRI-EBI GWAS catalog. We retrieved all SNPs in linkage disequilibrium (LOD >2 and D’>.6) with GWAS SNPs using publically available data on the CEPH ancestry from HapMap. We identified all GWAS loci for which the GWAS SNP or its LD partners were within 250-kb of the TSS of the genes of the dermal fibroblast expression signature. For all disease-associations within 250-kb of the TSS, we determined which proportion were contributed by each individual trait. We compared this to expected proportion, i.e. the proportion of all disease-associations in the GWAS catalog that were attributed to this trait. The traits with the highest odds ratios fell within two categories, lung disease and electrical heart function. To investigate whether these enrichments were significant, we generated 1000 random (n-matched) gene sets comprised of genes that are not a part of dermal fibroblast expression signature. We then determined the number of GWAS loci for which the GWAS SNP (or its LD partners) was within 250-kb of the TSS of the genes of each random set. This generated a distribution of the expected results for both lung disease and electrical heart function. We then compared the expected distribution to the number of GWAS loci within 250-kb of the genes of the dermal fibroblast gene signature to obtain p-values.
Supplementary Material
Table S7: Summary of GWAS trait enrichment within 250kb and 100kb of the TSS for the dermal fibroblast genes compared to 1000 random gene sets.
Table S1: Tissue microarrays used for comparison in Shannon Entropy Analysis. Stage and type of tissue, list of GEO Accession Numbers, and references for the microarray datasets.
Table S2: List of 63 genes identified by Shannon entropy in order of ascending Q-scores in three biological replicates.
Table S3: Mouse qRT-PCR primer sequences.
Table S4: FPKM values of gene expression signature in E13.5 mouse embryonic dermal fibroblast progenitors. Up or down regulated expression in β-catenin mutant compared to control fibroblasts in vivo.
Table S5: Peak values of all DNaseI hypersensitivity sites within 2kb of the TSS for the dermal fibroblast gene expression signature.
Table S6: Peak scores assigned to H3K27me3 ChIP-seq peaks for E14.5 brain.
Table S8: Summary of the signature genes’ expression in each test which agrees (green) or disagrees (orange) with our hypothesis. Not tested genes are indicated in grey.
Acknowledgments
Funding: This work was supported by NIH R01DE01870 (RPA), Case Startup Funds from Case Western Reserve University (RPA), Scleroderma Research Foundation (RPA), F30AG045009 (EH-S) Case Western Reserve University SPUR and SOURCE Programs (IB).
We profusely thank Drs. Howard Y. Chang, Rajnish Gupta, and Stanford Sequencing core for generating the microarray and RNA-seq data sets. We thank ENCODE Project Consortium, the NIH and Howard Hughes Medical Institute for funding the cited project (ENCODE Project Consortium, 2011). We thank past and present members of the lab, specially Henry L. Goodnough, Venkata Sai Kumar Thulabandu, and Gregg DiNuoscio, for their invaluable assistance.
References
- Anders S, Pyl PT, Huber W. HTSeq A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Andre P, Song H, Kim W, Kispert A, Yang Y. Wnt5a and Wnt11 regulate mammalian anterior-posterior axis elongation. Development. 2015;142:1516–1527. doi: 10.1242/dev.119065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Developmental Biology. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Barrott JJ, Cash GM, Smith AP, Barrow JR, Murtaugh LC. Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proceedings of the National Academy of Sciences. 2011;108:12752–12757. doi: 10.1073/pnas.1006437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE Mouse Genome Database Group. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Research. 2014;42:D810–7. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, Mcmahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Byrne C, Hardman M, Nield K. 23 Converting ectoderm to functional skin - differentiation pathways and formation of barrier. J Anat. 2002;201:423. [PMC free article] [PubMed] [Google Scholar]
- Cadau S, Rosignoli C, Rhetore S, Voegel J, Parenteau-Bareil R, Berthod F. Early stages of hair follicle development: a step by step microarray identity. Eur J Dermatol. 2013 doi: 10.1684/ejd.2013.1972. [DOI] [PubMed] [Google Scholar]
- Caiazzo M, Colucci-D’Amato L, Esposito MT, Parisi S, Stifani S, Ramirez F, di Porzio U. Transcription factor KLF7 regulates differentiation of neuroectodermal and mesodermal cell lineages. Exp Cell Res. 2010;316:2365–2376. doi: 10.1016/j.yexcr.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Candille SI, Van Raamsdonk CD, Chen C, Kuijper S, Chen-Tsai Y, Russ A, Meijlink F, Barsh GS. Dorsoventral patterning of the mouse coat by Tbx15. Plos Biol. 2004;2:E3. doi: 10.1371/journal.pbio.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Jarrell A, Guo C, Lang R, Atit R. Dermal beta-catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development. 2012;139:1522–1533. doi: 10.1242/dev.076463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Kretzschmar K, Watt FM. Reprogramming adult dermis to a neonatal state through epidermal activation of β-catenin. Development. 2011;138:5189–5199. doi: 10.1242/dev.064592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Watt FM. Dynamic regulation of retinoic acid-binding proteins in developing, adult and neoplastic skin reveals roles for beta-catenin and Notch signalling. Developmental Biology. 2008;324:55–67. doi: 10.1016/j.ydbio.2008.08.034. [DOI] [PubMed] [Google Scholar]
- Dhouailly D, Olivera-Martinez I, Fliniaux I, Missier S, Viallet JP, Thelu J. Skin field formation: morphogenetic events. Int J Dev Biol. 2004;48:85–91. doi: 10.1387/ijdb.15272373. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nürnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dollé P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. Plos Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Clavel C, Rendl M, Watt FM. Hair follicle dermal papilla cells at a glance. Journal of Cell Science. 2011;124:1179–1182. doi: 10.1242/jcs.082446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons Ben D, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, Watt FM. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Watt FM. Understanding fibroblast heterogeneity in the skin. Trends Cell Biol. 2015;25:92–9. doi: 10.1016/j.tcb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Eames BF, Schneider RA. Quail-duck chimeras reveal spatiotemporal plasticity in molecular and histogenic programs of cranial feather development. Development. 2005;132:1499–1509. doi: 10.1242/dev.01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) Plos Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Shekar PC, Ang YS, Fujiwara Y, Orkin SH, Wang J. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. STEM CELLS. 2011;29:1705–1716. doi: 10.1002/stem.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Casasnovas JJ, Leon RG, Friesel R, Ge Y, Desnick RJ, Cadilla CL. Nonsense mutations of the bHLH transcription factor TWIST2 found in Setleis Syndrome patients cause dysregulation of periostin. Int J Biochem Cell Biol. 2011;43:1523–1531. doi: 10.1016/j.biocel.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Hsu W. Epidermal Wnt Controls Hair Follicle Induction by Orchestrating Dynamic Signaling Crosstalk between the Epidermis and Dermis. Journal of Investigative Dermatology. 2012 doi: 10.1038/jid.2012.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, Andre P, Robinson J, Sood R, Minami Y, Economides AN, Yang Y. Wnt Signaling Gradients Establish Planar Cell Polarity by Inducing Vangl2 Phosphorylation through Ror2. Dev Cell. 2011;20:163–176. doi: 10.1016/j.devcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnough LH, Chang AT, Treloar C, Yang J, Scacheri PC, Atit RP. Twist1 mediates repression of chondrogenesis by beta-catenin to promote cranial bone progenitor specification. Development. 2012;139:4428–38. doi: 10.1242/dev.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnough LH, DiNuoscio GJ, Ferguson JW, Williams T, Lang RA, Atit RP. Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors. PLoS Genet. 2014;10:e1004152. doi: 10.1371/journal.pgen.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik KH, Bornholdt D, Oeffner F, Knig A, del Carmen Boente MA, Enders H, Fritz B, Hertl M, Grasshoff U, Hfling K. Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nat Genet. 2007;39:833–835. doi: 10.1038/ng2052. [DOI] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Research. 2007;35:W245–52. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Lefort K, Qiu W, Nguyen BC, Rajaram RD, Castillo E, He F, Chen Y, Angel P, Brisken C, Dotto GP. Control of hair follicle cell fate by underlying mesenchyme through a CSL-Wnt5a-FoxN1 regulatory axis. Genes Dev. 2010;24:1519–1532. doi: 10.1101/gad.1886910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukkola T, Sinjushina N, Partanen J. Drapc1 expression during mouse embryonic development. Gene Expr Patterns. 2004;4:755–762. doi: 10.1016/j.modgep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hinrichs AS, Learned K, Lee BT, Li CH, Raney BJ, Rhead B, Rosenbloom KR, Sloan CA, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. The UCSC Genome Browser database: 2014 update. Nucleic Acids Research. 2014;42:D764–70. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes & Development. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2012;2:987–1002. doi: 10.1534/g3.112.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cserjesi P, Olson EN. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Developmental Biology. 1995;172:280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- Li R, Brockschmidt FF, Kiefer AK, Stefansson H, Nyholt DR, Song K, Vermeulen SH, Kanoni S, Glass D, Medland SE, Dimitriou M, Waterworth D, Tung JY, Geller F, Heilmann S, Hillmer AM, Bataille V, Eigelshoven S, Hanneken S, Moebus S, Herold C, den Heijer M, Montgomery GW, Deloukas P, Eriksson N, Heath AC, Becker T, Sulem P, Mangino M, Vollenweider P, Spector TD, Dedoussis G, Martin NG, Kiemeney LA, Mooser V, Stefansson K, Hinds DA, Nöthen MM, Richards JB. Six novel susceptibility Loci for early-onset androgenetic alopecia and their unexpected association with common diseases. PLoS Genet. 2012;8:e1002746. doi: 10.1371/journal.pgen.1002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 Translates Epigenetic Signatures into Enhancer-Driven Lineage-Specific Transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackool RJ, Gittes GK, Longaker MT. Scarless healing. The fetal wound. Clin Plast Surg. 1998;25:357–365. [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar SE. An Ideal Society? Neighbors of Diverse Origins Interact to Create and Maintain Complex Mini-Organs in the Skin. Plos Biol. 2005;3:e372. doi: 10.1371/journal.pbio.0030372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A, Yardimci GG, Sheffield NC, Crawford GE, Ohler U. Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res. 2012;22:1711–1722. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtola J, Myers J, Akhtar-Zaidi B, Zuzindlak D, Sandesara P, Yeh K, Mackem S, Atit R. Beta-Catenin has sequential roles in the survival and specification of ventral dermis. Development. 2008;135:2321–2329. doi: 10.1242/dev.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, Nguyen N, Paten B, Zweig AS, Karolchik D, Kent WJ. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30:1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Polak L, Fuchs E. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes & Development. 2008;22:543–557. doi: 10.1101/gad.1614408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, Januszyk M, Krampitz GW, Gurtner GC, Lorenz HP, Weissman IL, Longaker MT. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science. 2015;348:aaa2151–aaa2151. doi: 10.1126/science.aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, Liu H, Ridky TW, Stadler HS, Nusse R, Helms JA, Chang HY. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes & Development. 2008;22:303–307. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Research. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Edelkraut U, Daniel G, Hoffmann A, Spengler D. Zac1 regulates cell cycle arrest in neuronal progenitors via Tcf4. Mol Cell Biol. 2014;34:1020–1030. doi: 10.1128/MCB.01195-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ. Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengel P, Mauger A. Peridermal cell patterning in the feather-forming skin of the chick embryo. Developmental Biology. 1976;51:166–171. doi: 10.1016/0012-1606(76)90132-9. [DOI] [PubMed] [Google Scholar]
- Sennett R, Wang Z, Rezza A, Grisanti L, Roitershtein N, Sicchio C, Mok KW, Heitman NJ, Clavel C, Ma’ayan A, Rendl M. An Integrated Transcriptome Atlas of Embryonic Hair Follicle Progenitors, Their Niche, and the Developing Skin. Dev Cell. 2015a:1–16. doi: 10.1016/j.devcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo KW, Roh KH, Bhandari DR, Park SB, Lee SK, Kang KS. ZNF281 knockdown induced osteogenic differentiation of human multipotent stem cells in vivo and in vitro. Cell Transplant. 2013;22:29–40. doi: 10.3727/096368912X654948. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Agalliu D, Vonica A, Luria V, Wajid M, Baumer A, Belli S, Petukhova L, Schinzel A, Brivanlou AH, Barres BA, Christiano AM. APCDD1 is a novel Wnt inhibitor mutated in hereditary hypotrichosis simplex. Nature. 2010;464:1043–1047. doi: 10.1038/nature08875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone NL, Paweletz CP, Charboneau L, Petricoin EF, Liotta LA. Laser capture microdissection: beyond functional genomics to proteomics. Mol Diagn. 2000;5:301–307. doi: 10.1007/BF03262091. [DOI] [PubMed] [Google Scholar]
- Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Berghout J, Campbell J, Corbani LE, Forthofer KL, Frost PJ, Miers D, Shaw DR, Stone KR, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2014 update. Nucleic Acids Research. 2014;42:D818–24. doi: 10.1093/nar/gkt954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee B-K, Sheffield NC, Gräf S, Huss M, Keefe D, Liu Z, London D, McDaniell RM, Shibata Y, Showers KA, Simon JM, Vales T, Wang T, Winter D, Zhang Z, Clarke ND, Birney E, Iyer VR, Crawford GE, Lieb JD, Furey TS. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sorrell JM, Caplan AI. Fibroblasts-a diverse population at the center of it all. Int Rev Cell Mol Biol. 2009;276:161–214. doi: 10.1016/S1937-6448(09)76004-6. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriwiriyanont P, Lynch KA, Maier EA, Hahn JM, Supp DM, Boyce ST. Morphogenesis of chimeric hair follicles in engineered skin substitutes with human keratinocytes and murine dermal papilla cells. Exp Dermatol. 2012;21:783–85. doi: 10.1111/exd.12003. [DOI] [PubMed] [Google Scholar]
- Supp D, Boyce S. Engineered skin substitutes: practices and potentials. Clinics in Dermatology. 2005;23:403–412. doi: 10.1016/j.clindermatol.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Tran TH, Jarrell A, Zentner GE, Welsh A, Brownell I, Scacheri PC, Atit R. Role of canonical Wnt signaling/beta-catenin via Dermo1 in cranial dermal cell development. Development. 2010a;137:3973–3984. doi: 10.1242/dev.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Jarrell A, Zentner GE, Welsh A, Brownell I, Scacheri PC, Atit R. Role of canonical Wnt signaling/β-catenin via Dermo1 in cranial dermal cell development. Development. 2010b;137:3973–3984. doi: 10.1242/dev.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda T, Markova D, Wang H, Evangelisti L, Pan TC, Chu ML. Zinc finger protein Zac1 is expressed in chondrogenic sites of the mouse. Dev Dyn. 2004;229:340–348. doi: 10.1002/dvdy.10439. [DOI] [PubMed] [Google Scholar]
- Tukel T, Sosic D, Al-Gazali LI, Erazo M, Casasnovas J, Franco HL, Richardson JA, Olson EN, Cadilla CL, Desnick RJ. Homozygous nonsense mutations in TWIST2 cause Setleis syndrome. Am J Hum Genet. 2010;87:289–296. doi: 10.1016/j.ajhg.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente T, Junyent F, Auladell C. Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: differential phenotype of the Zac1-expressing cells during development. Dev Dyn. 2005;233:667–679. doi: 10.1002/dvdy.20373. [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Fuerer C, Mizutani M, Nusse R. Wnt5a can both activate and repress Wnt/β-catenin signaling during mouse embryonic development. Developmental Biology. 2012;369:101–114. doi: 10.1016/j.ydbio.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Reid Sutton V, Omar Peraza-Llanes J, Yu Z, Rosetta R, Kou YC, Eble TN, Patel A, Thaller C, Fang P, Van den Veyver IB. Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat Genet. 2007;39:836–838. doi: 10.1038/ng2057. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Teh CHL, Chan CMY, Chu C, Rossbach M, Kunarso G, Allapitchay TB, Wong KY, Stanton LW. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. STEM CELLS. 2008;26:2791–2799. doi: 10.1634/stemcells.2008-0443. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa S, Onizuka T, Shimoji K, Ohno Y, Kageyama T, Yoon SH, Egashira T, Seki T, Hashimoto H, Nishiyama T, Kaneda R, Murata M, Hattori F, Makino S, Sano M, Ogawa S, Prall OWJ, Harvey RP, Fukuda K. Zac1 is an essential transcription factor for cardiac morphogenesis. Circ Res. 2010;106:1083–1091. doi: 10.1161/CIRCRESAHA.109.214130. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Afink GB, Svensson K, Jacobs JJ, Günther T, Forsberg-Nilsson K, van Zoelen EJ, Westermark B, Nistér M. Specific expression in mouse mesoderm- and neural crest-derived tissues of a human PDGFRA promoter/lacZ transgene. Mechanisms of Development. 1998;70:167–180. doi: 10.1016/s0925-4773(97)00190-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, Morrisey EE, Overbeek PA, Scheidereit C, Millar SE, Schmidt-Ullrich R. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev Cell. 2009;17:49–61. doi: 10.1016/j.devcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S7: Summary of GWAS trait enrichment within 250kb and 100kb of the TSS for the dermal fibroblast genes compared to 1000 random gene sets.
Table S1: Tissue microarrays used for comparison in Shannon Entropy Analysis. Stage and type of tissue, list of GEO Accession Numbers, and references for the microarray datasets.
Table S2: List of 63 genes identified by Shannon entropy in order of ascending Q-scores in three biological replicates.
Table S3: Mouse qRT-PCR primer sequences.
Table S4: FPKM values of gene expression signature in E13.5 mouse embryonic dermal fibroblast progenitors. Up or down regulated expression in β-catenin mutant compared to control fibroblasts in vivo.
Table S5: Peak values of all DNaseI hypersensitivity sites within 2kb of the TSS for the dermal fibroblast gene expression signature.
Table S6: Peak scores assigned to H3K27me3 ChIP-seq peaks for E14.5 brain.
Table S8: Summary of the signature genes’ expression in each test which agrees (green) or disagrees (orange) with our hypothesis. Not tested genes are indicated in grey.