Abstract
Rumi O-glucosylates the EGF repeats of a growing list of proteins essential in metazoan development including Notch. Rumi is essential for Notch signaling, and Rumi dysregulation is linked to several human diseases. Despite Rumi’s critical roles, it is unknown how Rumi glucosylates a serine of many but not all EGF repeats. Here we report crystal structures of Drosophila Rumi as binary or ternary complexes with a folded EGF repeat and/or donor substrates. These structures provide insights into the catalytic mechanism, and show that Rumi recognizes structural signatures of the EGF motif, the U-shaped consensus sequence, C-X-S-X-(P/A)-C and a conserved hydrophobic region. We found that five Rumi mutations identified in cancers and Dowling-Degos disease are clustered around the enzyme active site and adversely affect its activity. Our study suggests that loss of Rumi activity may underlie these diseases, and the mechanistic insights may facilitate the development of modulators of Notch signaling.
Introduction
Notch signaling plays essential roles in development of all metazoans, and dysregulation in the Notch pathway leads to a variety of human diseases including cancers and developmental disorders1,2. Differential O-linked glycosylation of Notch extracellular domain (NECD) can regulate Notch signaling. Conserved O-glucose modification of a subset of NECD EGF repeats by Rumi is essential for Notch signaling3–5 while elongation of O-linked glucose by sequential action of GXYLT1/2 and XXYLT1 negatively regulates Notch activation6. Rumi is required for embryonic development from flies to mammals, in a Notch-dependent manner: Rumi aberration in fly leads to a temperature-sensitive loss of Notch signaling3; Mouse embryos lacking Rumi shows early embryonic lethality and severe cardiovascular defects4. In addition to the NECD as the best characterized substrate of Rumi, several new substrates have recently been identified, including Eys in photoreceptor development7, CRUMBS2 in mammalian gastrulation8 and Jag1 in Notch pathway9, all featuring tandem EGF repeats. Moreover, Rumi dysregulation has been linked to several human diseases including Dowling-Degos disease (DDD)10 and Alagille syndrome9.
Rumi is a member of the GT90 family of glycosyltransferases (GTs)11 and adds an O-linked glucose to the serine in the consensus C-X-S-X-(P/A)-C found in EGF repeats. No structural information is available for any member of the GT90 family. In general, there is a paucity of structures of GTs that target protein substrates, although a few studies reported GTs in complex with short peptides of the acceptor protein substrates12–16. More recently, structures of GTs that glycosylate small cysteine-rich protein domains such as EGF repeats and thrombospondin type 1 repeats (TSRs) have been reported. For example, POFUT1 (a GT65 family member) and POFUT2 (a GT68 family member) add O-fucose to EGF repeats and TSRs, respectively, which contain the appropriate consensus sequences17. Proper folding is required for the EGF repeats and TSRs to become efficient glycosylation substrates17. POFUT1 and POFUT2 have classic GT-B folds18–20. A recent crystal structure of C. elegans POFUT2 complexed with a folded TSR and GDP revealed the importance of not only the 3D fold of the TSR acceptor but also the interfacial water molecules in the enzyme substrate recognition20. How POFUT1 recognizes the folded EGF acceptor has not been determined experimentally19. We recently solved the crystal structure of the ER-anchored XXYLT1, a GT-A fold enzyme that adds a xylose to the existing O-glucose disaccharide on an EGF repeat. We found limited contacts between XXYLT1 and the EGF in the enzyme–donor–acceptor ternary complex structure21. Therefore, the molecular basis of how a folded EGF repeat is recognized and glycosylated - either fucosylated by POFUT1 or glucosylated by Rumi - remains unknown.
Here we describe the structures of Drosophila Rumi (dRumi) complexed with its substrates, a folded EGF module and donor ligand (UDP or UDP-Glc). These structures show that Rumi recognizes conserved signatures of EGF module, including the O-glucosylation consensus sequence, C-X-S-X-(P/A)-C, and a previously unknown conserved hydrophobic region (P55 and Y69). Two different ternary complex structures and biochemical data reveal the catalytic mechanism of Rumi where dRumi D151 functions as the catalytic base. We structurally and biochemically characterized Rumi alterations that were identified previously in DDD and demonstrated that they are either activity-abolishing missense mutation or likely non-functional nonsense/frameshift mutations. Moreover, we assessed the functional consequences of Rumi mutations identified in human cancers and found four missense mutations that exhibit severely reduced O-glucosylation activity, suggesting the deleterious impact of Rumi dysfunction in these cancers. Our studies add new knowledge on Notch O-glucosylation and provide a framework for understanding Rumi aberrations in diseases and for the development of modulators of Notch signaling.
Results
Overall structure of Rumi complexed with an EGF repeat
Drosophila Rumi (dRumi) shares high sequence identities with its human (~52%) and mouse (~51%) counterparts (Supplementary Results, Supplementary Fig. 1a). We produced an N-terminally truncated dRumi protein (residues 21–407) in HEK293T cells, purified the protein to homogeneity, and further removed the recombinant purification tag (Supplementary Fig. 1b, Online Methods). Rumi O-glucosylates various EGF repeats of its regulated substrates, all of which share a similar fold of the cysteine knot that is constrained by three conserved disulfide bonds, despite their wide sequence variation22. Therefore, we used the EGF repeat (residues 46–84) of human factor IX (hFA9) as a surrogate in our structural study, which was previously shown to be an authentic Rumi substrate in vitro23. We determined the co-crystal structure of dRumi in complex with hFA9 EGF repeat at 1.9 Å resolution (Supplementary Table 1), using the anomalous signals of native sulfurs24. In this complex, dRumi residues 42–406 and hFA9 EGF repeat residues 48–84 were resolved. Furthermore, we soaked donor ligands UDP or UDP-Glc into the binary complex crystals, and solved the structures of the ternary complexes of dRumi–EGF–UDP at 2.15 Å and dRumi–(Glc–EGF)–UDP at 2.5 Å. We also crystallized and solved the structure of dRumi complexed with UDP at 3.2 Å resolution.
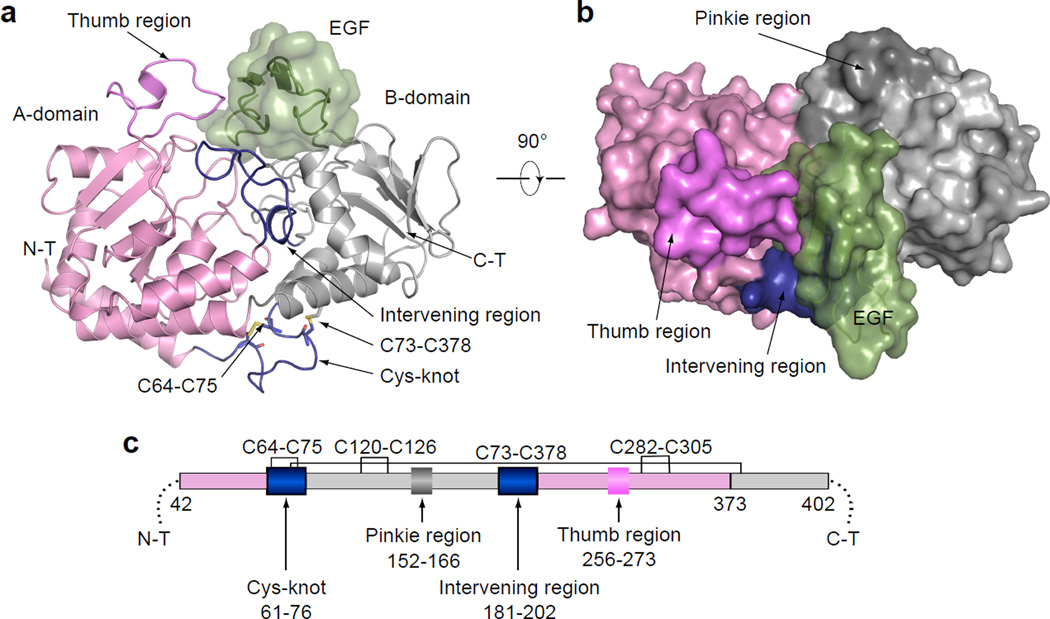
The overall structure of dRumi–EGF binary complex is shown in Fig. 1a–b. dRumi contained two domains: the A-domain and the smaller B-domain, each with a Rossmann fold typical of GT-B superfamily members (Fig. 1c and Supplementary Fig. 1c). The two-domain architecture of dRumi was stabilized by two tandem helixes; one helix at the N-terminal region (residues 42–88) and the other helix was kinked and was located at the C-terminal region (residues 352–390). The two helices were stabilized by two disulfide bonds (C64-C75 and C73-C378). A DALI search25 identified the DNA alpha-glucosyltransferase (AGT, pdb id: 1ya6) as the closest structure of dRumi with a Z score of 12.1 and RMSD of 4.5 Å over 216 aligned residues. Despite the similar fold, these two enzymes had very different functions26.
Figure 1. Overall structure of dRumi complexed with hFA9 EGF repeat.
a, Ribbon diagram of dRumi–EGF binary complex. Rumi A-domain is shown in pink, B-domain in grey, the Intervening linkage region and cys-knot connecting A- and B-domain in blue, the Thumb region in violet, and the Pinkie region in dark grey. The EGF is in dark green. b, Surface representation of the binary complex viewed from the top. c, Schematic of the domain organization of dRumi.
In the dRumi–EGF binary complex structure, the EGF bound to a cleft between the A- and B-domains of Rumi, with an interface area of 827 Å2. Therefore, approximately one third of the 2922 Å2 surface of the EGF was buried by Rumi. Surrounding the cleft were two loops that make additional contacts with the EGF: a “Thumb” region (residues 256–273) of A-domain and an Intervening region (residues 181–202) that bridges A- and B-domains (Fig. 1a).
Rumi recognizes structural features of the EGF repeats
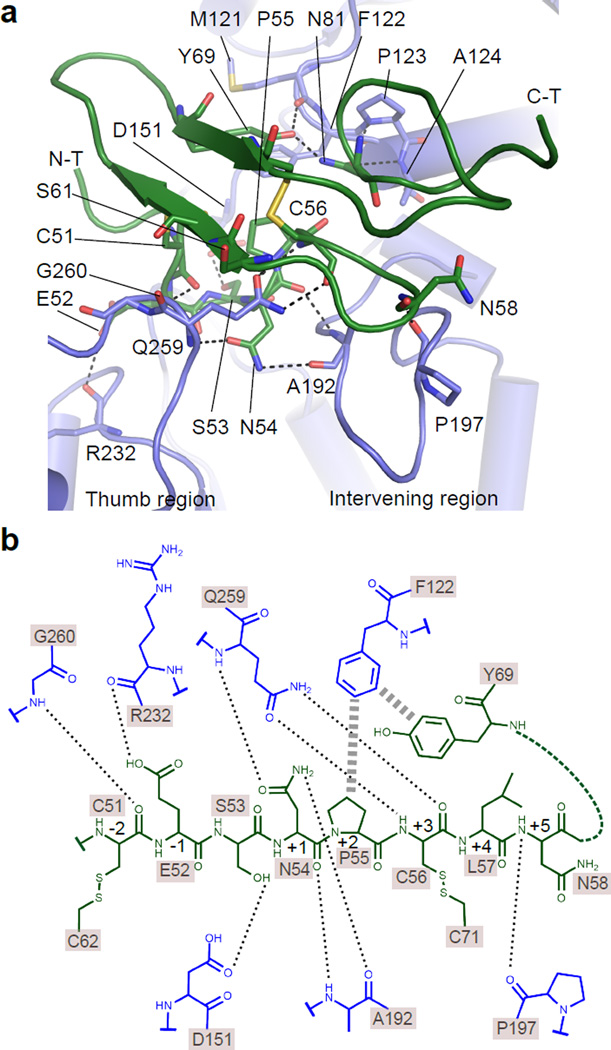
Rumi O-glucosylates folded EGF repeats with the C-X-S-X-(P/A)-C consensus sequence1, but elucidating the underlying molecular basis for this required solution of the binary complex as presented below. We found a remarkable surface complementarity between dRumi and the EGF repeat in the crystal structure (Supplementary Fig. 2). This is mainly imposed by the O-glucosylation consensus sequence C-E-S-N-P-C of hFA9 EGF which is located in the N-terminal region (residues 51–56) of the motif and forms a U-shaped loop structure. The structure of this loop is not affected by the crystal packing (Supplementary Fig. 3). The loop inserts into the substrate-binding cleft of Rumi, and the backbone atoms of EGF C51, N54, C56, and N58 interact extensively with Rumi residues Q259 and G260 in the Thumb region and with residues A192 and P197 in the Intervening region (Fig. 2a, b). Additionally, the side chain of EGF N54 at sub-site +1 is involved in two H-bonds in a narrow space, consistent with the known preference of Rumi for Asn/Ser over Arg at this sub-site23 (Supplementary Fig. 4a–c). Following this region, the backbones of several EGF residues stacked above the Thumb region with the O-fucosylation site S61 facing upward, away from Rumi (Fig. 2a), explaining why Rumi is able to tolerate a fucosylated EGF27. On the other side of the cleft, dRumi F122 stacked against Y69 and P55 of hFA9 EGF repeat (Fig. 3a). These two hydrophobic interactions are likely important because the residue at position 69 is conserved to either a Y or F, while P55 is absolutely conserved among all EGF repeats of Drosophila Notch with O-glucose sites (Fig. 3b and Supplementary Fig. 4a, d). Around this region, the side chain of EGF Y69 forms a H-bond with dRumi M121 main chain, while EGF N81 side chain H-bonds with backbone atoms of dRumi P123 and A124 (Fig. 2a).
Figure 2. Interactions between dRumi and EGF repeat.
a, A cartoon view of the interface between dRumi (blue) and EGF (dark green). Dashed lines show H-bonds. b, Schematic of the interactions between dRumi and the C-X-S-X-(P/A)-C consensus motif and the hydrophobic region of hFA9 EGF repeat. Residues are colored as in (a) with H-bonds indicated by dashed lines and hydrophobic stacking by thicker dash lines. The consensus sequence motif (51-CESNPC-56) of hFA9 EGF is referred to as −2 to +3 sub-sites with O-glucosylation site S53 being sub-site 0.
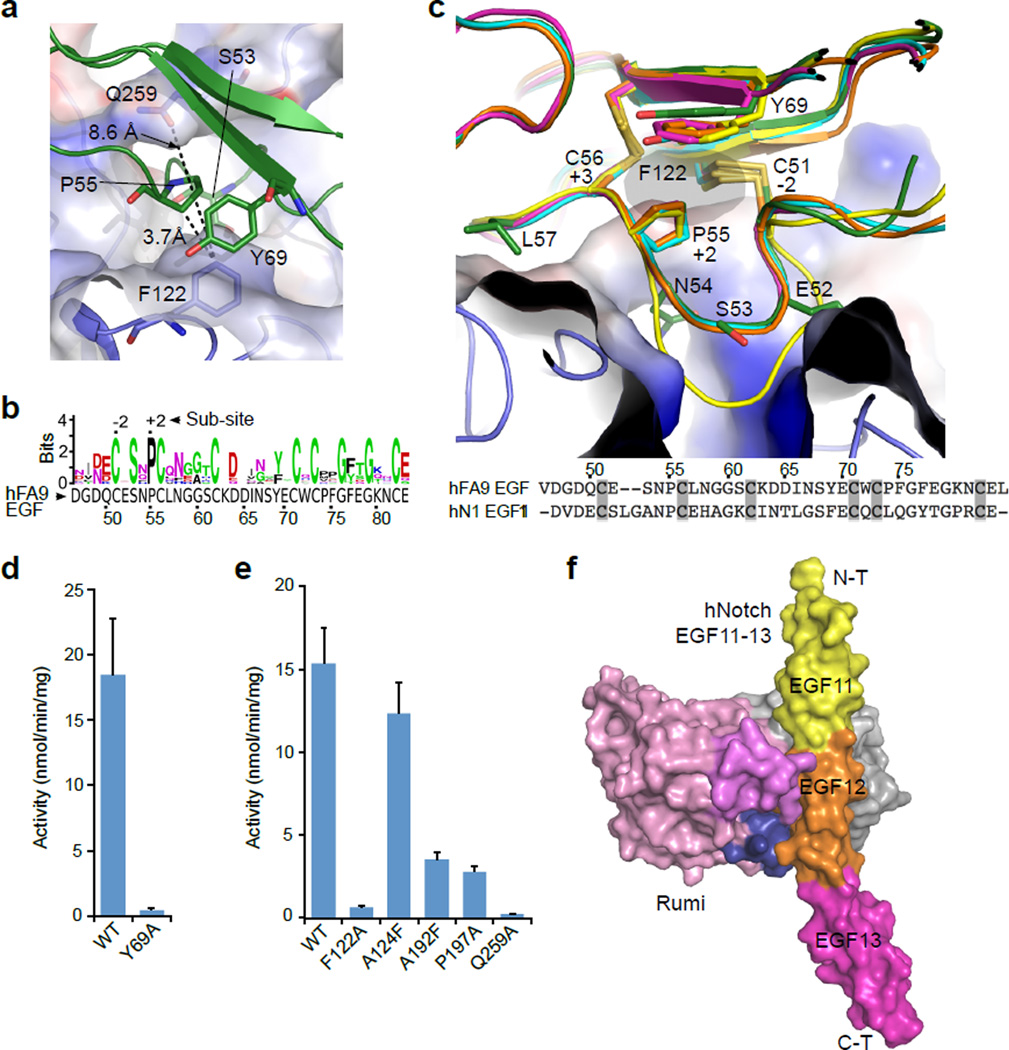
Figure 3. Rumi recognizes conserved 3-D features of EGF repeats with diverse primary sequences.
a, Top view of dRumi’s EGF-binding cleft shown in semi-transparent electrostatic surface representation. Note the hydrophobic region (P55 and Y69) stacks against dRumi F122. b, Sequence conservation of the 16 Drosophila Notch EGFs with confirmed Rumi O-glucosylation (generated by WebLogo). The hydrophobic region identified above is conserved. The logo size is proportional to the level of conservation. c, Side view of dRumi’s EGF-binding cleft. EGF11 (yellow), EGF12 (orange) and EGF13 (magenta) of human Notch1 EGF11–13 crystal structure (pdb id: 2VJ3) are superposed individually onto the hFA9 EGF (dark green) in the binary complex. Human Notch1 EGF11 (yellow cartoon, sequence shown in the bottom with cysteines highlighted) has a shifted serine and elongated loop in the consensus motif and cannot be glucosylated. d, In vitro dRumi activity toward hFA9 EGF WT and Y69A mutant. e, In vitro activity of dRumi mutants located on the interacting interface. For (d) and (e), data were from four and three independent assays, respectively; the values shown indicate mean ± S.E.M. f, A model for Rumi binding to Notch1 EGF12 in the context of tandem EGFs. The model was generated by overlaying EGF12 of human Notch1 EGF11–13 structure (PDB ID 2VJ3) onto the EGF moiety of the dRumi–EGF complex. Rumi is in surface view and colored as in Fig. 1b.
The interface provided detailed insights into how Rumi recognizes its folded EGF repeat substrates. dRumi F122 and Q259 define the narrowest point of the cleft (8.6 Å), which is accessible only to a loop but excludes any other secondary structure elements (Fig. 3a). EGF P55 at sub-site +2 is located right in the middle of the cleft, only 3.7 Å away from dRumi F122. Therefore, residues bulkier than Pro at +2 sub-site are not tolerated as shown previously23. More importantly, P55 and the two disulfide bonds at sub-sites −2 and +3 fashion the C-X-S-X-(P/A)-C motif into the U-like configuration. This configuration is essential for insertion of the glucose-accepting residue Ser deeply into the Rumi active site (Fig. 3c). This observation rationalizes the requirement of the consensus sequence, and perhaps explains why Rumi does not modify EGFs with serine residue either shifted even by one position or with a consensus sequence with even one extra residue (Supplementary Fig. 4b). As for the identified hydrophobic region (hFA9 EGF P55 and Y69, Fig. 3a), we previously showed that the P55A mutant of hFA9 EGF is a very poor Rumi substrate23. We prepared the properly folded Y69A mutant (Supplementary Fig. 4e and f) and found that it was also a poor substrate (Fig. 3d). Taken together, Rumi recognizes the U-shaped loop structure of the consensus motif as well as the conserved hydrophobic region formed by P55 and Y69 of the acceptor EGF (Fig. 2 and Fig. 3a–d). The Rumi recognition surface is largely defined by the conserved F122, A124, and A192, and Q259. Indeed, F122A and Q259A mutations markedly reduced the enzymatic activity, and A124F or A192F mutation disrupted surface complementarity, leading to a reduction of Rumi activity by 20% and 80%, respectively. The P197A mutation in the intervening region also markedly reduced the enzymatic activity (Fig. 3e).
Because the binary Rumi–EGF structure revealed exquisite interface complementarity and a recognition interface that is largely made up of loop-loop interactions (Figs. 1a and 2a, Supplementary Fig. 2a), we wondered if conformational changes were involved in this recognition process. To address this question, we solved the structure of dRumi complexed with UDP but in the absence of the EGF (Supplementary Table 1). We found that dRumi structure in the absence of EGF repeat was essentially the same as the structure in complex with EGF, with an RMSD of 0.24 Å. Thus, the EGF-contacting surface of Rumi apparently did not change upon binding to EGF (Supplementary Fig. 2b, c). On the other hand, hFA9 EGF repeat structure in the presence or absence of Rumi had a slightly higher RMSD of 0.81 Å, but changes were limited to regions distant from the interacting interface (Supplementary Fig. 2d, e). Therefore, neither dRumi nor the EGF repeat undergoes conformational change in the binary complex structure. This exemplifies the classic “lock-and-key” type recognition mechanism, with minimal binding-induced conformational changes.
Our binary complex structure contained only a single EGF repeat, yet all currently known Rumi-regulated targets such as Notch, Eys, Jag1, Factor IX, and Crumbs2, contain multiple EGF repeats3,7–9. We therefore considered how Rumi might recognize these more complicated substrates, of which Notch is the most characterized. As noted earlier, because all EGF repeats have a similar fold, we were able to superimpose the crystal structure of Notch EGF11–13 with the hFA9 EGF in our binary structure (Fig. 3f, Supplementary Fig. 5). We found that, in addition to the primary interface discussed above, a “Pinkie” region (residues 152–166) from the B-domain of dRumi that is on the opposite side of the Thumb region, may make contacts with a second EGF that is upstream of the primary EGF, and the secondary EGF binding may be dependent on the linking angle between the two adjacent EGFs (Supplementary Fig. 5b–e). Notch NECD is known for its flexible or rigid linkage between neighboring EGF repeats28. However, the EGF repeats in these different molecular contexts can still be modified with O-glucose at high stoichiometry6,29. In the crystal structure, the B-factor in this Pinkie region is substantially higher than the rest of the structure (Supplementary Fig. 5f), implying a level of flexibility in this region. We speculate that the flexible Pinkie region may accommodate a certain level of linking angle variation between the tandem EGF repeats of Rumi substrates.
Catalytic mechanism revealed by two ternary complexes
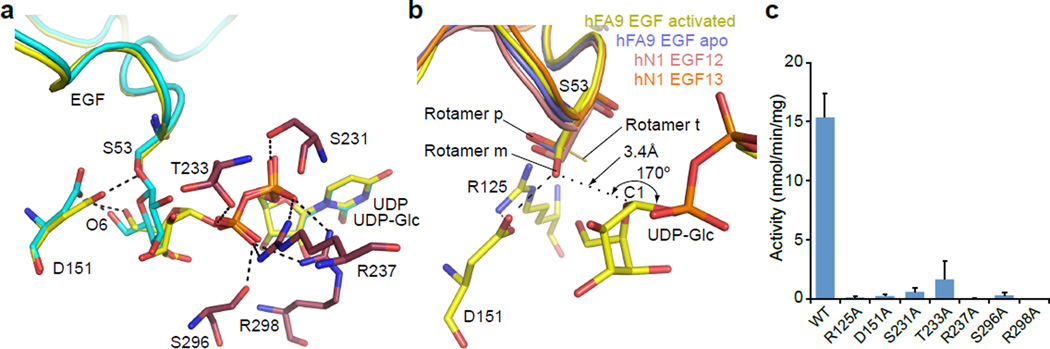
Rumi was previously identified as an inverting GT27. To determine the transfer mechanism employed by Rumi, we soaked donor ligands UDP-glucose (UDP-Glc) or UDP into the dRumi–EGF complex crystals and solved the structures of two ternary complexes: dRumi–(Glc–EGF)–UDP, a product complex in which glucose transfer reaction had occurred in the crystals, and dRumi–EGF–UDP (Supplementary Fig. 6). In the product complex, both the transferred glucose (Supplementary Fig. 6f) and the cleaved UDP (Fig. 4a) were held in place by a network of H-bonds. The UDP diphosphate was sandwiched between and formed salt bridges with Rumi R237 and R298, and further stabilized by side chains of S231, T233, and S296.
Figure 4. Reaction mechanism revealed by two ternary complexes of dRumi.
a, Overlay of the dRumi product complex dRumi–(Glc-EGF)–UDP (cyan) with the modeled Michaelis complex dRumi–EGF–(UDP-Glc) (yellow). Dashed black lines indicate H-bonds. Enzyme residues involved in diphosphate binding are shown in purple sticks. During transfer reaction, the upper half of the glucose ring (C1, C2, C5 and O5) tilts leftward such that the anomeric carbon shifts by 2.3 Å to form the new bond with the acceptor oxygen atom of Ser53. b, A close-up view of the active site of the modeled ternary Michaelis complex (yellow). Three isolated EGF structures are superimposed onto the bound EGF: hFA9 EGF (blue, PDB ID 1EDM), and EGF12 (salmon) and EGF13 (orange) of human Notch1 (hN1) EGF11–13 (PDB ID 2VJ3). Among the three serine rotamers (p: 48%, m: 29%, t: 22%), unbound EGFs are all in the p configuration, only the bound EGF serine is in m configuration, stabilized by the catalytic residue D151. The t rotamer shown as a dark yellow line is not observed in any of the four EGF structures. c, In vitro activity of dRumi active site variants. Data were from three independent assays. The values indicate mean ± S.E.M.
Interestingly, between UDP and acceptor EGF S53 in the Rumi–EGF–UDP ternary complex, we found electron densities of two water molecules and one solvent glycerol. The spatial arrangement of the five hydroxyl groups, 2 from the two water molecules and 3 from glycerol, closely resembled the arrangement of oxygen atoms of a glucose molecule in the transitional boat configuration. Therefore, we modeled a UDP-Glc into the experimental densities in the Rumi active site and obtained a putative Michaelis complex structure of dRumi–EGF–(UDP-Glc) (Fig. 4b and Supplementary Fig. 6b, c). In the model structure, R125 appeared to be crucial, as it formed a salt bridge with the active residue D151, bringing it close to the acceptor S53 (2.4 Å). Consequently, D151 oriented EGF S53 side chain into a less favored rotamer conformation with the oxygen pointed directly towards the anomeric carbon of the donor UDP-Glc. In this arrangement, the acceptor oxygen of EGF S53 is almost linear with the anomeric C–O bond (170°) of the donor, consistent with an SN2 inverting mechanism30,31. Therefore, we propose that dRumi D151 functions as the general base to activate the nucleophile (EGF S53 OH), and R237 and R298 weaken the phosphoester bond by forming salt bridges with the donor β;-phosphate (Supplementary Fig. 6h). Similar mechanisms have been reported in structural studies of other GT-B family members such as POFUT218,20, human OGT12,16, T4 bacteriophage β-glucosyltransferase (BGT)32 where aspartate, glutamate, histidine or donor α-phosphate has been identified as catalytic base while positively charged residues such as arginine or lysine facilitates the departure of leaving group. Indeed, substituting dRumi active site residues individually to alanine all markedly reduced enzyme activity (Fig. 4c). In particular, the four catalytic residue mutants R125A, D151A, R237A, and R298A all completely abrogated enzymatic activity (Supplementary Fig. 7d).
Disease-associated mutations in Rumi
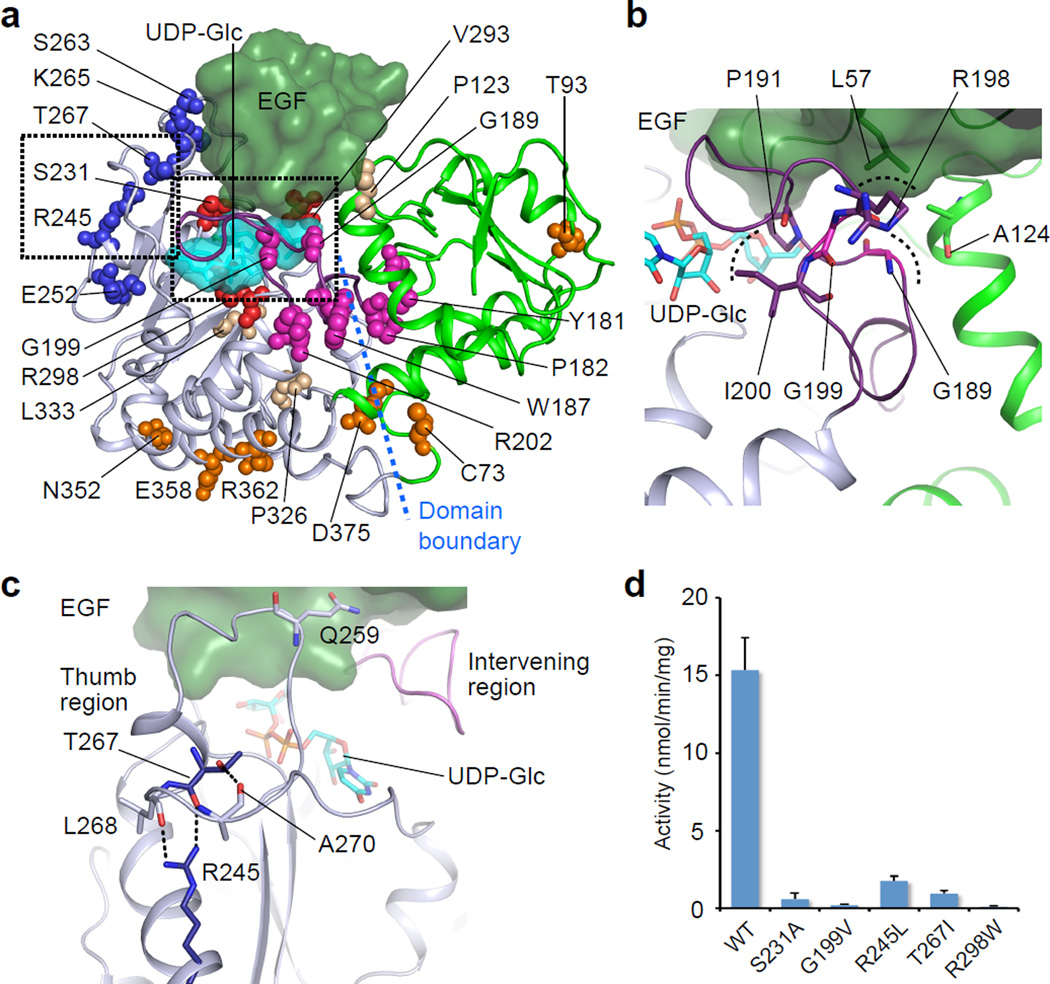
Notch pathway components are frequently implicated in cancer21,33–35. Given the essential roles of Rumi in Notch activation3–5, we hypothesized that Rumi mutations might also be related to cancer. We therefore searched through a cancer genomics database36, and found 26 Rumi mutations (Supplementary Table 2, referred to in dRumi sequence). We subsequently analyzed the mutations in the context of the dRumi–EGF–(UDP-Glc) ternary complex model. We found that the Rumi truncations, resulting from either frame-shift or nonsense mutations, all lacked key structural components, making the enzyme almost certainly inactive (Supplementary Fig. 8a, b). The missense mutations mostly cluster in four regions in A-domain but rarely in B-domain (Fig. 5a–c). Among the identified point mutations, we chose a few representatives from these regions for further characterization. Rumi S231 is at the donor-binding interface (Fig. 4a). The S231A mutant, milder than the S231L found in cancer, already markedly reduced the in vitro activity (Fig. 4c). G189R and G199V are located within the Intervening region that is flexible and likely affect substrate binding (Fig. 5b). G189E was previously reported to abolish the enzyme activity3. Consistently, we found in this study that G199V retained only a trace amount of Rumi activity (Fig. 5d, Supplementary Fig. 7e). R245 and T267 stabilize the flexible Thumb region that anchors the EGF (Fig. 5d). Indeed, destabilizing mutations R245L and T267I markedly reduced the enzyme activity (Fig. 5d). Interestingly, the deleterious Rumi mutations we have characterized appear in cancers where Notch functions as tumor suppressor34,37–39: e.g., S231L, R237* and R386* in endometrial cancer; R245L and S307* in bladder urothelial cancer; G189R in lung squamous cell cancer; and G199V in head and neck squamous cell cancer (Supplementary Table 2). Since Rumi is essential for Notch signaling3–5, these deleterious mutations likely promote tumorigenesis by compromising Notch function. We therefore suggest that Rumi may play a tumor suppressor role.
Figure 5. Disease-related Rumi mutants adversely affect enzyme activity.
a, Rumi mutations from human diseases (numbering based on dRumi sequence) are mapped onto the dRumi–EGF–(UDP-Glc) Michealis complex. These mutants are either DDD-related or identified from cancer genomics database cBioportal. EGF and UDP-Glc are in dark green and cyan surface, respectively. Rumi A-domain is in grey, B-domain in green, the intervening region in purple. The mutated residues are shown in spheres: those involved in donor ligand binding in red; in the Thumb region in blue; in the Intervening region in magenta; in the A- and B-domain linker region in orange; and in other regions in wheat. Mutants chosen for in vitro characterization are enclosed in the dashed boxes and their close-up views are shown in (b, c). b, Mutants at G189 and G199 locate to the sensitive Intervening region with R198 supporting EGF L57 on the top, G189 supporting the helix of B-domain to the right, and I200 and P191 being close to bound UDP-Glc. c, dRumi R245 and T267 function to stabilize the Thumb region for EGF anchoring. d, Effects of Rumi disease-related mutations on enzymatic in vitro activity. Data were from three independent assays. The values indicate mean ± S.E.M.
Dowling-Degos disease (DDD) is a rare autosomal dominant disease characterized by reticulate skin hyperpigmentation. Nine causative mutations of Rumi have been reported in DDD (Supplementary Table 2)10,40,41. It is unknown how these mutations affect Rumi enzymatic activity. By homology modeling and mapping the human mutations onto the Drosophila Rumi crystal structure, we found that the eight truncated Rumi variants resulting from either frame-shift or nonsense mutations all lack key portions of the structure, almost certainly rendering them enzymatically inactive (Supplementary Fig. 8c). The only missense mutation substituted the catalytic residue arginine (R298) for a tryptophan (W) (Fig. 4a). As expected, we found that R298W completely abolished the Rumi activity in vitro (Fig. 5d, Supplementary Fig. 7e). Therefore, all reported Rumi mutations in DDD are deleterious, supporting the role of Rumi loss-of-function in DDD pathogenesis.
Discussion
Here we have described a set of Drosophila Rumi structures, in the presence or absence of a folded EGF acceptor as well as the donor substrates. To our knowledge, Rumi presents the first structure of the GT90 family with over 500 members, and our work is the first demonstration of how a properly folded EGF repeat is recognized and glycosylated by a glycosyltransferase. Importantly, we have elucidated the structural basis for the requirement of the consensus sequence C-X-S-X-(P/A)-C for EGF O-glucosylation. We discovered a conserved hydrophobic region of EGF repeats (corresponding to hFA9 EGF repeat P55 and Y69) as an additional structural feature being recognized by Rumi. Furthermore, by solving the crystal structures of two enzyme–acceptor–donor ternary complexes, we have identified D151 as the catalytic residue and elucidated the SN2 inverting reaction mechanism for Rumi.
In the SN2 mechanism, a properly positioned nucleophile is paramount for the reaction to proceed. We previously observed that Rumi strongly prefers serine over threonine as the nucleophile in the acceptor EGF29. To understand the structural basis of the preference, we computationally substituted S53 with a threonine in the EGF, and found that T53 was too close to the donor glucose ring (Supplementary Fig. 6g). The close contact may prevent the glucose ring from assuming an optimal binding pose at the enzyme active site. This observation may explain why Rumi adds glucose to a serine rather than to a threonine.
Our structural and biochemical studies provide a framework for understanding the functional consequences of Rumi alterations in diseases. Our analyses reveal that the Rumi mutations reported in DDD abolish the enzyme activity. This observation strongly supports a pathogenic role of these Rumi mutations. More importantly, we found that cancer-associated mutations in Rumi alter key structural components and inactivate the enzyme. This result suggests that Rumi may function as a tumor suppressor in contexts where Notch activity inhibits tumor growth. We note that contribution of these Rumi mutations to tumorigenic activity may be more complex than it appears since the Rumi-mutations found in DDD apparently do not cause tumors. Furthermore, the cancer genomics database does not specify the homo- or hetero-zygosity of the mutations. We could assume that at least some of the mutations are from heterozygotes. Because cancer is a complicated disease, and most cancers, unlike DDD, have multiple mutations that in combination lead to a cancer phenotype, there are several possibilities that may reconcile these apparent discrepancies. These include loss of heterozygosity (LOH), which is very common in cancers, or possibly other mutations in the Notch pathway that decrease Notch activity - in combination with mutations in Rumi/POGLUT1 - to the point where they have an effect. In other cases where mutations of several Notch pathway genes (NCSTN, APH1, MAML1 and NOTCH2) are known to be from heterozygotes, these mutations have been associated with a tumor-suppressor role35. While our work points to a tumor-suppressive role of Rumi in cancer, it requires further studies.
Atomic structure of an important enzyme such as Rumi is invaluable for therapeutic development. Inhibitors of γ-secretase, a Notch-processing enzyme, have garnered considerable clinical interest in cancer therapeutics and have also been extensively used in functional studies of Notch42. However, it is widely recognized that down regulating Notch by inhibiting γ-secretase is problematic, because γ-secretase has multiple substrates leading to severe off-target effects42,43. In this regard, Rumi as an essential Notch regulator may provide a novel molecular target for the development of small molecule inhibitors that down-regulate Notch signaling. However, Rumi does not exclusively target Notch either, so it remains to be investigated if Rumi will become a druggable target. Our Rumi structure and mechanistic insights will facilitate future research in this direction.
ONLINE METHODS
Preparation of human factor IX EGF
The procedure was performed as previously described23. Briefly, human factor IX (hFA9) EGF repeat was expressed in BL21 (DE3) E coli and purified by Ni-NTA agarose (QIAGEN) affinity chromatography and subsequent reverse phase HPLC. The final product was lyophilized using Vacuum-centrifuge. Product mass was analyzed by LC-MS/MS. For introducing the Y69A mutation, site-directed mutagenesis was performed by a conventional PCR-based method with the pET20b vector encoding wild type hFA9 EGF repeat as template and the primers (Forward, 5’-CATTAATTCCGCTGAATGTTGGTGTCCCTTTGGATTTGAAGGA-3’, Reverse, 5’-CCAACATTCAAAGGAATTAATGTCATCCTTGCAACTGCC-3’). The introduced mutation was confirmed by DNA sequencing.
Cloning, expression and purification of Drosophila Rumi
Cloning of the Drosophila Rumi (dRumi) of cDNA was previously described3. The cDNA encoding Drosophila melanogaster Rumi without its signal peptide and C-terminal KDEL ER-retention motif was cloned into pSecTag2c vector (Invitrogen) with C-terminal thrombin-cleavable Myc/His6-tag. The whole primary sequence of the expressed dRumi is shown in Supplementary Fig. 1a and b.
Protein expression and purification of dRumi were done as previously described23. Because of the strict quality control system in the endoplasmic reticulum (ER) of mammalian cells, only properly folded and stable proteins are secreted into the medium. By taking advantage of this robust ER quality control system, we successfully solved the structure of XXYLT1 and generated, purified and tested multiple XXYLT1 mutants21. Here we took advantage of this same quality control method by purifying recombinant dRumi protein from the culture medium of transiently transfected HEK293T cells. The expression plasmid (2 µg) was transfected in adherent HEK293T cells using polyethylenimine transfection reagent, and the transfected cells were cultured in a 10-cm plate with 6 ml of DMEM (Invitrogen) containing 10% bovine calf serum (HyClone, GE Healthcare) overnight. The medium was changed to 6 ml of OPTI-MEM I (Invitrogen) and the cells were cultured for another 3 days. The secreted dRumi protein was purified from the culture media using Ni-NTA agarose (QIAGEN) affinity chromatography with gravity flow. The culture media (approximately 240 ml) from forty 10-cm plates were supplemented with 0.5 M NaCl and 10 mM imidazole, and applied to Ni-NTA agarose (column volume, 300 µl). After the column was washed with 20 ml of Tris-buffered saline, pH 7.4 (TBS) containing 0.5 M NaCl and 10 mM imidazole, the dRumi protein was eluted with 2 ml of TBS containing 250 mM imidazole, dialyzed against TBS containing 20% glycerol at 4 °C overnight, and stored at −80 °C until use. The yield of dRumi from one standard transfection was approximately 0.4 mg/L of culture. Protein expression was confirmed by Western blot analysis with anti-Myc antibody (Stony Brook University, Cell culture/Hybridoma Facility) and protein purity and concentration were estimated by 10% SDS-PAGE followed by Coomassie stain with BSA as standard.
For crystallization, the C-terminal tag was removed through thrombin cleavage and then the tag-free Rumi was further purified by size-exclusion chromatography (Superdex 200, GE Healthcare) in 20 mM HEPES, pH 7.5, 150 mM NaCl. The sample was concentrated to 5 mg/ml and stored at −80 °C.
Crystallization, ligand soaking, and heavy atom soaking
For Rumi–UDP binary complex crystallization, Rumi at the concentration of 5 mg/ml was mixed with 3 mM UDP and incubated at 4 °C for 2 hr. The hanging-drop diffusion method was used to produce initial micro-crystals in mother liquor containing 20 mM HEPES, pH 7.4, sodium citrate tribasic and glycerol at 20 °C. The rare crystals with maximum dimensions reaching ~30 µm × 30 µm × 200 µm were only obtained by seeding optimization. Well solution with increasing glycerol concentration (30%) was used as the cryo-protectant for crystal flash freezing in liquid nitrogen.
For crystallization of the binary complex of Rumi and hFA9 EGF acceptor ligand (Rumi–EGF), the purified Rumi at the concentration of 5 mg/ml was mixed with 2 fold molar excess of hFA9 EGF, incubated at 4 °C for 2 hr, then the mixture was used for crystal screen or crystal reproduction. Crystals of the Rumi–EGF binary complex were obtained several days after setting up the hanging-drop vapor diffusion plates at 20 °C using a reservoir solution containing (NH4)2SO4. For ligand soaking in the Rumi–EGF crystals, UDP, or UDP-Glc at the final concentration of 20 mM were added to crystal-containing drops for 1 hr before crystals were picked up and flash-frozen in liquid nitrogen. The Rumi–EGF binary complex or ligand soaked crystals were frozen in the cryo-protectant consisting of the well solution supplemented with 25% glycerol.
For heavy atom soaking of the Rumi–EGF binary complex crystals, we screened several heavy atom compounds and found that the following conditions in the original well solution as soaking solution gave derivatized crystals that diffracted to >4 Å resolution with useful anomalous signals: (1) 1 min soaking in 1 M KI; (2) 1 hr soaking in 20 mM ErCl3.
X-ray diffraction data collection, structural determination and refinement
The datasets were collected at the NSLS beamlines X25 and X29 in Brookhaven National laboratory at 1.1000 Å wavelength, at the APS LRL-CAT at 0.9793 Å wavelength, except for the heavy atom derivative datasets that were collected at the wavelength of 1.7000 Å and 1.4832 Å for KI and ErCl3 derivatized crystals, respectively. The native sulfur SAD datasets were collected at NSLS X4A at 2.0703 Å. Diffraction images were processed and scaled in XDS44, Mosflm45 or HKL200046. The Rumi–EGF binary complex crystals had a space group of H32 with one complex in the asymmetrical unit (AU). The Rumi–UDP binary complex crystals were in space group P31 with six molecules in AU.
Substructure determination with KI or ErCl3 derivative datasets both failed, likely due to the low occupancy of heavy atoms. To solve the phase problem, three datasets of Rumi–EGF binary complex crystals were collected at a wavelength of 2.0703 Å to measure the sulfur anomalous signal. These datasets were processed using XDS44, combined with Pointless, and merged with Scala of the CCP4 suite47 as previously described24. Ten sulfur atoms were found by SHELXD48. The initial SAD phases were obtained using the PHENIX49 AutoSol and a crude model was built automatically with the Phenix AutoBuild. The crude model was then used to generate phases for the 1.9 Å Rumi–EGF binary complex dataset by molecular replacement with the program MOLREP50. With improved electron density map, the Rumi–EGF binary complex model was built by PHENIX AutoBuild, and further corrected and completed in several iterations by manual building in COOT51 followed by refinement with REFMAC in the CCP4 program52. For all ligand-soaked datasets, structures were determined following the similar strategy: molecular replacement by MOLREP using the side chain hydroxyl group of EGF S53 and carboxyl group of Rumi D151 deleted version of the Rumi–EGF binary complex structure as the search model, followed by one round of automatic refinement in REFMAC without building the soaked ligand. Then the Fo-Fc difference maps and the 2Fo-Fc electron density maps were carefully analyzed before building the ligand(s) into the density map. Donor UDP was fit into the map first, and side chain hydroxyl group of EGF S53 and carboxyl group of Rumi D151 were fit next, followed by the building of glycerol (UDP soaking) or transferred glucose (UDP-Glc soaking). After ligand building, several additional rounds of refinement were carried out in REFMAC. Water molecules were added at last.
The Rumi–UDP binary complex structure was solved to 3.2 Å by molecular replacement with program MOLREP using the Rumi moiety of the Rumi–EGF binary complex structure as the search model. There are six molecules in the ASU and the diffraction dataset was highly twinned (twin fractions of ~0.5). By using REFMAC, one round of rigid body refinement and one round of restrained refinement (with twin refinement option enabled) were performed with local NCS restraints. Clear density of intact UDP was identified at this stage, based on which six UDP in ASU were modeled in COOT and refined by one round of restrained refinement with REFMAC.
The crystallographic statistics for data collection and refinement are presented in Supplementary Table 1.
Mutagenesis and enzyme activity measurement and mass spectrometry
Site-directed mutagenesis was performed by a conventional PCR-based method with the pSecTag vector encoding wild type dRumi as template. Primers were listed in Supplementary Table 3. Introduced mutations were confirmed by direct DNA sequencing.
The enzymatic assay with radiolabeled UDP-[6-3H]glucose (Glc) (American Radiolabeled Chemicals, >97%) was performed as previously described23. Briefly, the standard 10-µl reaction mixtures contained 50 mM HEPES pH 6.8, 10 mM MnCl2, 10 µM hFA9 EGF repeat, 10 µM UDP-[6-3H]Glc (7.14 GBq/mmol), 10 ng dRumi enzyme, and 0.1% Nonidet P-40. The reaction was performed at 37 °C for 20 min and stopped by adding 900 µl of 100 mM EDTA pH 8.0. The sample was loaded onto a C18 cartridge (100 mg, Agilent Technologies). After the cartridge was washed with 5 ml of H2O, the EGF repeat was eluted with 1 ml of 80% methanol. Incorporation of [6-3H]Glc into the EGF repeats was determined by scintillation counting of the eluate. Reactions without enzymes were used as background control. Data were from three independent assays. The values indicate mean ± S.E.M.
For overnight reactions with dRumi or its mutants, hFA9 EGF repeat (10 µM) was incubated in the presence of 100 ng of dRumi or its mutants and UDP-Glc at the concentration of 200 µM. The reaction was carried out in 30 µl of 50 mM HEPES pH 6.8, 10 mM MnCl2 at 37 °C overnight. An aliquot of the products was analyzed by LC-MS using an Agilent 6340 ion-trap mass spectrometer with a nano-HPLC CHIP-Cube interface. Extracted ion chromatograms for the most abundant charge state of the unmodified form or O-glucosylated form of EGF repeats were generated.
Supplementary Material
Acknowledgments
We thank members of the Li and Haltiwanger labs for critical comments on this work and Sumreet Singh Johar for technical assistance. The work is supported by NIH grants GM061126 (to R.S.H) and AG029979 (to H.L.) and SBU-BNL Seed Grant (to R.S.H. and H.L.). We acknowledge access to beamlines X25, X29, X4A at NSLS, Brookhaven National Laboratory and LRL-CAT at APS, Argonne National Laboratory and thank the staff at these beamlines. NSLS and APS were supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886 and DE-AC02-06CH11357, respectively. Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly Company, which operates the facility. The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Footnotes
Accession codes
The coordinates have been deposited at the PDB with accession codes 5F84 (Rumi–(Glc–EGF)–UDP), 5F85 (Rumi–EGF–UDP), 5F86 (Rumi–EGF), and 5F87 (Rumi–UDP).
Author contributions
H.Y., H.T., R.S.H. and H.L. designed the research. H.Y. prepared the protein complexes, performed crystallization and solved the structures. H.Y. and Q.L. collected and processed the sulfur-SAD data. H.Y. and H.L. analyzed the structures. H.T., M.T., and J.K. expressed and purified proteins and mutants and performed the enzymatic assays and mass spectrometry. H.Y., H.T., R.S.H. and H.L. designed the mutants, analyzed the mutant data, and performed the cancer-related analysis. H.Y. prepared the initial draft of the manuscript, and H.Y, H.T., R.S.H. and H.L. wrote the paper with input from all authors.
Additional information
Supplementary information is available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
REFERENCES
- 1.Rana NA, Haltiwanger RS. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 2011;21:583–589. doi: 10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acar M, et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez-Valdivia R, et al. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138:1925–1934. doi: 10.1242/dev.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonardi J, Fernandez-Valdivia R, Li YD, Simcox AA, Jafar-Nejad H. Multiple O-glucosylation sites on Notch function as a buffer against temperature-dependent loss of signaling. Development. 2011;138:3569–3578. doi: 10.1242/dev.068361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee TV, et al. Negative regulation of notch signaling by xylose. PLoS Genet. 2013;9:e1003547. doi: 10.1371/journal.pgen.1003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haltom AR, et al. The protein O-glucosyltransferase Rumi modifies eyes shut to promote rhabdomere separation in Drosophila. PLoS Genet. 2014;10:e1004795. doi: 10.1371/journal.pgen.1004795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramkumar N, et al. Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2. PLoS Genet. 2015;11:e1005551. doi: 10.1371/journal.pgen.1005551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakurdas SM, et al. Jagged1 heterozygosity in mice results in a congenital cholangiopathy which is reversed by concomitant deletion of one copy of Poglut1 (Rumi) Hepatology. 2016;63:550–565. doi: 10.1002/hep.28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basmanav FB, et al. Mutations in POGLUT1, encoding protein O-glucosyltransferase 1, cause autosomal-dominant Dowling-Degos disease. Am. J. Hum. Genet. 2014;94:135–143. doi: 10.1016/j.ajhg.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazarus MB, et al. Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol. 2012;8:966–968. doi: 10.1038/nchembio.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lizak C, Gerber S, Numao S, Aebi M, Locher KP. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474:350–355. doi: 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- 15.Lazarus MB, et al. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science. 2013;342:1235–1239. doi: 10.1126/science.1243990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schimpl M, et al. O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nat. Chem. Biol. 2012;8:969–974. doi: 10.1038/nchembio.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasudevan D, Haltiwanger RS. Novel roles for O-linked glycans in protein folding. Glycoconj. J. 2014;31:417–426. doi: 10.1007/s10719-014-9556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CI, et al. Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012;31:3183–3197. doi: 10.1038/emboj.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lira-Navarrete E, et al. Structural insights into the mechanism of protein O-fucosylation. PLoS One. 2011;6:e25365. doi: 10.1371/journal.pone.0025365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valero-Gonzalez J, et al. A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat. Chem. Biol. 2016;12:240–246. doi: 10.1038/nchembio.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, et al. Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat. Chem. Biol. 2015;11:847–854. doi: 10.1038/nchembio.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wouters MA, et al. Evolution of distinct EGF domains with specific functions. Protein Sci. 2005;14:1091–1103. doi: 10.1110/ps.041207005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi H, Kantharia J, Sethi MK, Bakker H, Haltiwanger RS. Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats. J. Biol. Chem. 2012;287:33934–33944. doi: 10.1074/jbc.M112.401315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, et al. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lariviere L, Sommer N, Morera S. Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase. J. Mol. Biol. 2005;352:139–150. doi: 10.1016/j.jmb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi H, et al. Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc. Natl. Acad. Sci. USA. 2011;108:16600–16605. doi: 10.1073/pnas.1109696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu A, Lei L, Irvine KD. Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. J. Biol. Chem. 2005;280:30158–30165. doi: 10.1074/jbc.M505569200. [DOI] [PubMed] [Google Scholar]
- 29.Rana NA, et al. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J. Biol. Chem. 2011;286:31623–31637. doi: 10.1074/jbc.M111.268243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 31.Breton C, Fournel-Gigleux S, Palcic MM. Recent structures, evolution and mechanisms of glycosyltransferases. Curr. Opin. Struct. Biol. 2012;22:540–549. doi: 10.1016/j.sbi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Lariviere L, Gueguen-Chaignon V, Morera S. Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism. J. Mol. Biol. 2003;330:1077–1086. doi: 10.1016/s0022-2836(03)00635-1. [DOI] [PubMed] [Google Scholar]
- 33.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer cell. 2014;25:318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rampias T, et al. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat. Med. 2014;20:1199–1205. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- 35.Klinakis A, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 2011;108:17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jonusiene V, et al. Down-regulated expression of Notch signaling molecules in human endometrial cancer. Med. Oncol. 2013;30:438. doi: 10.1007/s12032-012-0438-y. [DOI] [PubMed] [Google Scholar]
- 40.Hanneken S, et al. [Galli-Galli disease. Clinical and histopathological investigation using a case series of 18 patients] Hautarzt. 2011;62:842–851. doi: 10.1007/s00105-011-2222-x. [DOI] [PubMed] [Google Scholar]
- 41.Mauerer A, Betz RC, Pasternack SM, Landthaler M, Hafner C. Generalized solar lentigines in a patient with a history of radon exposure. Dermatology. 2010;221:206–210. doi: 10.1159/000316091. [DOI] [PubMed] [Google Scholar]
- 42.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling--are we there yet? Nat. Rev. Drug Discov. 2014;13:357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 43.Rizzo P, et al. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
Methods-only References
- 44.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pannu NS, Murshudov GN, Dodson EJ, Read RJ. Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.