Abstract
We describe here the main characteristics of “Prevotellamassilia timonensis” gen. nov., sp. nov., strain Marseille-P2831T (CSUR P2831), a new member of the Prevotellaceae family that was isolated from stool samples from a 45-year-old patient.
Keywords: Culturomics, Prevotellamassilia timonensis, taxonomy, taxonogenomics, human gut
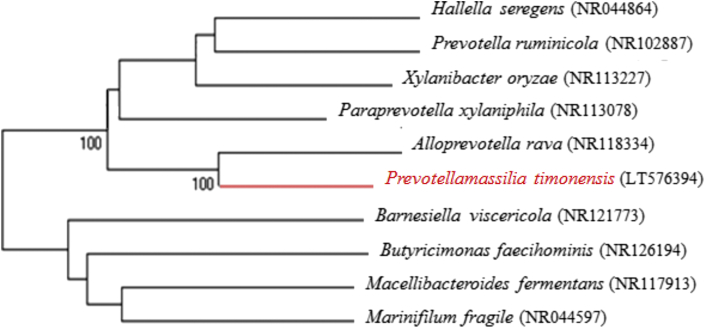
By culturomics study [1], we isolated a new bacterial strain from the stool specimen of a 45-year-old patient hospitalized in March 2016 for the treatment of a melanoma in Marseille, France. We obtained the patient's consent, and the study was approved by the Institut Fédératif de Recherche 48 (Faculty of Medicine, Marseille, France) under agreement number 09-022. Growth of the strain Marseille-P2831 was obtained on 5% sheep’s blood–enriched Columbia agar (bioMérieux, Marcy l'Etoile, France) after 48 hours' incubation at 37°C under an anaerobic atmosphere generated by AnaeroGen (bioMérieux). Strain Marseille-P2831 was a strictly anaerobic, Gram-negative cocci, non–spore forming, motile, with no catalase and no oxidase activities. The colonies were irregular and beige, with a diameter of 0.8 to 1.5 mm on blood-enriched Colombia agar (bioMérieux) after 72 hours' incubation. Individual cells exhibited a diameter of 0.7 to 1.2 μm and a length of 1.5 to 2.5 μm measured by electron microscopy using a DM1000 photonic microscope (Leica, Wetzlar, Germany). Our systematic matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) screening using a MicroFlex spectrometer (Bruker Daltonics, Bremen, Germany) [2] was not be able to identify this strain. The complete 16S rRNA gene was sequenced using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France) using the universal primers FD1 and RP2 (Eurogentec, Angers, France) as previously described [3]. The sequence of strain Marseille-P2831 (GenBank accession number LT576394) showed a similarity of 90% with Alloprevotella rava strain F0323T (GenBank accession number GU470887), the phylogenetically closest species with standing in nomenclature (Fig. 1), which classifies it as a new genus within the Prevotellaceae family in the Bacteroidetes phylum [4]. Isolated from the human oral cavity and described in 2013, Alloprevotella rava is an obligatory anaerobic, Gram-negative nonmotile bacilli.
Fig. 1.
Phylogenetic tree highlighting position of “Prevotellamassilia timonensis” strain Marseille-P2831T relative to other Bacteroidetes. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Scale bar represents 2% nucleotide sequence divergence.
Strain Marseille-P2831 exhibited a 16S rRNA sequence divergence of >5% with the other phylogenetically closest species within the Alloprevotella genus with a validly published name with standing in nomenclature [5]. From these results, we propose the creation of a new genus “Prevotellamassilia” (pre.vo.tel′la.ma.si.lia, related to pre.vo.tel′la; N.L. fem. n. Prevotella, a bacterial generic name, and ma.si.lia, “of Massilia,” the Latin name of Marseille, where the strain was isolated; N.L. fem. n. “Prevotellamassilia,” organism different from, but related to, the genus Prevotella and Massilia, where our hospital is situated). “Prevotellamassilia timonensis” gen. nov., sp. nov. (ti.mo.nen′sis, L. masc. adj. timonensis, related to Timone, the name of the main university hospital in Marseille, France, from where the strain was isolated) is a new species within this new genus.
MALDI-TOF MS spectrum accession number
The MALDI-TOF MS spectrum of “P. timonensis” is available online (http://www.mediterraneeinfection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in GenBank under accession number LT576394.
Deposit in a culture collection
Strain Marseille-P2831T was deposited in the Collection de Souches de l'Unité des Rickettsies (CSUR) under number P2831.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Downes J., Dewhirst F.E., Tanner A.C.R., Wade W.G. Description of Alloprevotella rava gen. nov., sp. nov., isolated from the human oral cavity, and reclassification of Prevotella tannerae Moore et al. 1994 as Alloprevotella tannerae gen. nov., comb. nov. Int J Syst Evol Microbiol. 2013;63:1214–1218. doi: 10.1099/ijs.0.041376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]