The incidence of invasive candidiasis, which includes candidemia and deep tissue infections, continues to rise and is associated with considerable mortality rates. Candida albicans remains the most common cause of invasive candidiasis, although the prevalence of non-albicans species has increased over recent years. Since its first description in 2009, Candida auris has emerged as a serious nosocomial health risk, with widespread outbreaks in numerous hospitals worldwide. However, despite receiving considerable attention, little is known concerning the pathogenicity of this emerging fungal pathogen. Here, using the Galleria mellonella insect systemic infection model, we show strain-specific differences in the virulence of C. auris, with the most virulent isolates exhibiting pathogenicity comparable to that of C. albicans, which is currently accepted as the most pathogenic member of the genus.
KEYWORDS: pathogenicity, Candida auris, pathogenic yeasts, emerging pathogen
ABSTRACT
Candida auris, first described in 2009, has since emerged as an important, multidrug-resistant, nosocomial agent of candidemia, with large outbreaks reported worldwide and high mortality rates associated with therapeutic failure. The current study employed C. auris isolates from a variety of centers in the United Kingdom to evaluate the pathogenicity of this emerging pathogen compared to that of other common pathogenic yeast species in the invertebrate Galleria mellonella infection model. We showed that C. auris isolates differ in their growth characteristics in vitro, with a proportion of isolates failing to release daughter cells after budding, resulting in the formation of large aggregates of cells that cannot be physically disrupted. Our results also demonstrate strain-specific differences in the behavior of C. auris in G. mellonella, with the aggregate-forming isolates exhibiting significantly less pathogenicity than their nonaggregating counterparts. Importantly, the nonaggregating isolates exhibited pathogenicity comparable to that of C. albicans, which is currently accepted as the most pathogenic member of the genus, despite the fact that C. auris isolates do not produce hyphae and produce only rudimentary pseudohyphae either in vitro or in G. mellonella.
IMPORTANCE The incidence of invasive candidiasis, which includes candidemia and deep tissue infections, continues to rise and is associated with considerable mortality rates. Candida albicans remains the most common cause of invasive candidiasis, although the prevalence of non-albicans species has increased over recent years. Since its first description in 2009, Candida auris has emerged as a serious nosocomial health risk, with widespread outbreaks in numerous hospitals worldwide. However, despite receiving considerable attention, little is known concerning the pathogenicity of this emerging fungal pathogen. Here, using the Galleria mellonella insect systemic infection model, we show strain-specific differences in the virulence of C. auris, with the most virulent isolates exhibiting pathogenicity comparable to that of C. albicans, which is currently accepted as the most pathogenic member of the genus.
INTRODUCTION
The incidence of invasive fungal infections caused by unusual Candida spp. continues to rise, driven in part by increased populations of immunocompromised patients and those undergoing invasive procedures (1 – 8). However, to date, Candida albicans remains the most frequently isolated Candida species in the clinical setting, is the principal agent of nosocomial yeast infections (1, 4 – 6), and is widely accepted as being the most pathogenic Candida species (reviewed in references 9 and 10).
In 2009, a novel Candida species in the Candida haemulonii complex (Metchnikowiaceae), Candida auris, was described after isolation from a discharge from a human external ear canal in Japan (11). Subsequent studies confirmed an association with chronic otitis media in 15 patients from South Korea (12), with evidence of clonal transmission and resistance to certain triazole antifungal agents. C. auris has since been reported from a wide spectrum of clinical manifestations, ranging from colonization through deep-seated infections and candidemia (13 – 17). Today, it is evident that C. auris has emerged as an important nosocomial pathogen with clonal inter- and intrahospital transmission, and it has become widespread across several Asian countries and South Africa (13 – 18). C. auris fungemia is associated with a high mortality rate, therapeutic failure (13 – 15), and widespread resistance to several classes of antifungal agents (13, 15 – 21). Furthermore, correct identification of C. auris isolates is complicated by the fact that many commercially available biochemical-based tests can misidentify C. auris as the phylogenetically related species Candida haemulonii (11, 12, 19 – 23), which presents an additional challenge for appropriate patient management.
The first 2 United Kingdom isolates of C. auris were received at the UK National Mycology Reference Laboratory (MRL) in 2013, from blood cultures from 2 unrelated patients in distant geographical localities (MRL unpublished data). Since 2013, we have received a further 19 isolates from at least 6 different hospitals, including 14 isolates suspected of being part of an outbreak. Here we have compared the pathogenicities of 12 United Kingdom isolates of C. auris from 6 different referring National Health Service (NHS) hospitals with the pathogenicities of equivalent isolates of other common pathogenic Candida species, using the Galleria mellonella insect systemic infection model.
RESULTS AND DISCUSSION
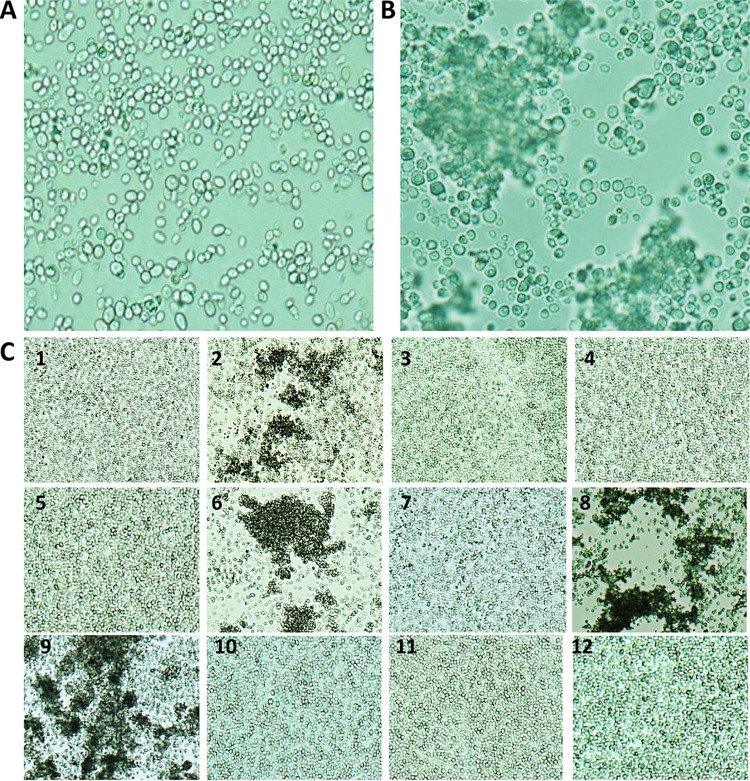
The characteristics of the 12 isolates of C. auris employed in the current study are detailed in Table 1, with antifungal MIC values determined at the MRL. Initial attempts to generate suspensions of C. auris isolates in phosphate-buffered saline (PBS) for larval inoculation revealed striking strain-specific differences in phenotypic behavior. While most isolates readily formed homogeneous suspensions upon thorough vortex mixing, the resulting suspensions seen with 4 independent isolates from 3 different referring hospitals were grossly particulate and contained individual yeast cells mixed with large aggregations (“aggregate” strains) (Table 1 and Fig. 1). For these 4 isolates, aggregates could not be physically disrupted by vigorous vortex mixing or by detergent treatments (data not shown). Since the aggregates were too large to permit larval inoculation and since cell numbers within the aggregates could not be accurately quantified, homogeneous suspensions were instead achieved by allowing initial suspensions to settle for 10 min, followed by removal of the supernatant containing individual yeast cells that had remained in suspension and adjustment of these individual cells to the appropriate concentration for injection into larvae.
TABLE 1 .
Origin of the Candida auris strains employed in this studya
| Isolate | Yr | Site | Hospital | Morphology | MIC value (mg/liter) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMB | FLC | VRC | PSC | ANID | 5FC | |||||
| 1 | 2015 | Sputum | A | Single cells | 0.5 | 8 | 0.06 | 0.25 | ||
| 2 | 2015 | CSF | B | Aggregates | 0.5 | >64 | 0.5 | <0.03 | <0.125 | |
| 3 | 2015 | Not stated | A | Single cells | 1.0 | 8 | 0.06 | 0.25 | ||
| 4 | 2015 | Line | A | Single cells | 0.5 | 8 | 0.06 | 0.03 | ||
| 5 | 2015 | Arterial line | A | Single cells | 0.5 | 16 | 0.125 | 0.5 | ||
| 6 | 2014 | Pleural fluid | C | Aggregates | 0.5 | >64 | 0.25 | 0.125 | ||
| 7 | 2016 | Not stated | D | Single cells | 1 | 16 | 0.125 | 0.25 | ||
| 8 | 2015 | Pustule swab | B | Aggregates | >64 | |||||
| 9 | 2016 | Blood culture | E | Aggregates | 0.5 | 64 | 2 | 0.125 | ||
| 10 | 2016 | Wound swab | Fb | Single cells | 1 | >64 | 16 | 0.25 | ||
| 11 | 2015 | Femoral line | A | Single cells | 0.5 | 8 | 0.06 | 0.06 | ||
| 12 | 2016 | Not stated | E | Single cells | 0.5 | 0.5 | 1.0 | 0.25 | ||
The antifungal susceptibility results expressed as MICs (in milligrams per liter) are given for those antifungal agents requested by referring centers; the susceptibility tests were performed at the MRL. MICs were obtained using CLSI broth microdilution methodologies (26). Abbreviations: AMB, amphotericin B; FLC, fluconazole; VRC, voriconazole; PSC, posaconazole; ANID, anidualfungin; 5FC, flucytosine; CSF, cerebrospinal fluid.
The patient was transferred from hospital A.
FIG 1 .
Microscopic appearance of non-aggregate-forming isolates (A) and aggregate-forming isolates (B) of C. auris in PBS suspensions. Suspensions were subjected to vortex mixing for 1 min prior to examination at ×1,000 magnification. (C) The 12 isolates of C. auris employed in the current study (×100 magnification).
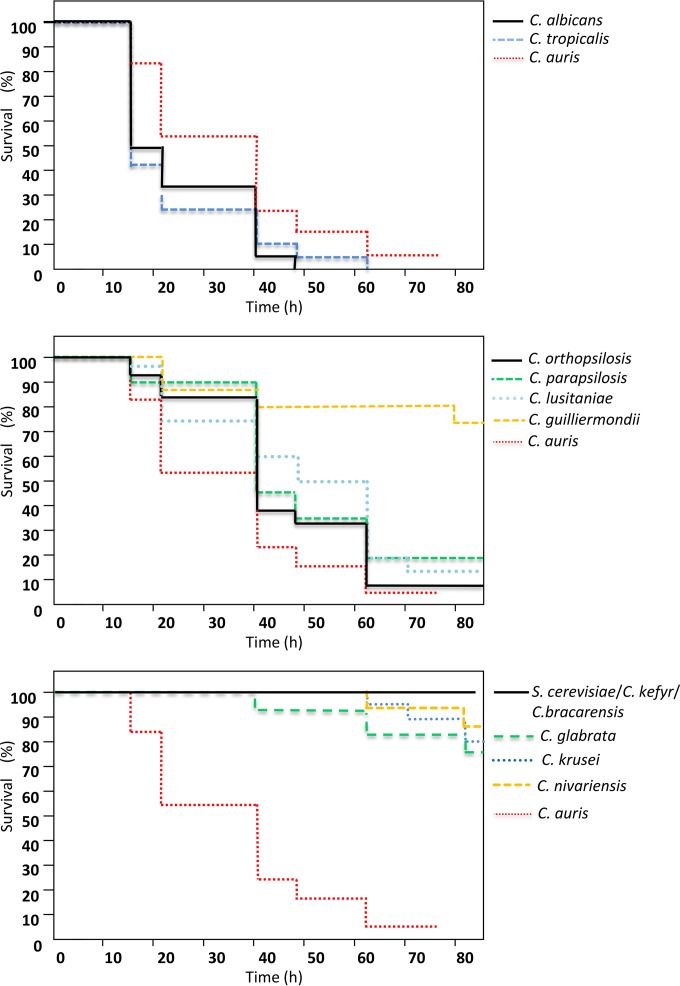
In agreement with previous reports (10, 24), the pathogenicity of the common Candida species at 37°C in G. mellonella was directly related to the ability of individual species to produce hyphal filaments or pseudohyphae (Fig. 2; see also Fig. S1 in the supplemental material), with very little strain-to-strain variation in virulence within each species (see Fig. S1). Thus, C. albicans and C. tropicalis exhibited greater virulence than C. lusitaniae, C. guilliermondii, and members of the C. parapsilosis species complex, and virtually no larval killing was induced by those organisms that form only rudimentary pseudohyphae or no pseudohyphae (C. glabrata, C. nivariensis, C. krusei, C. kefyr, C. bracarensis, and Saccharomyces cerevisiae) (Fig. 2; see Table 2 for full statistical analyses).
FIG 2 .
The virulence of Candida species in Galleria mellonella larvae at 37°C is species specific. Kaplan-Meier plots of G. mellonella survival after injection with 106 CFU/larva of the indicated Candida species, organized as those that produce true hyphae (top panel), pseudohyphae (middle panel), or no hyphae/pseudohyphae (bottom panel), are shown. Equivalent plots obtained with C. auris isolates are included in all three panels for comparison. Four strains were tested per species, with 15 larvae per strain (60 larvae per species), except for C. auris, where 12 strains were included, with 10 larvae per strain. Experiments were performed in duplicate; plots represent the combined (additive) data from all strains and all experiments. No larval killing was observed in control larvae injected with an equivalent volume of PBS.
TABLE 2 .
Statistical analyses of species-specific differences in pathogenicitya
| Species | Pathogenicity difference P value for species: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 1. C. albicans | |||||||||||||||
| 2. C. tropicalis | ns | ||||||||||||||
| 3. C. auris (all) | ns | ns | |||||||||||||
| 4. C. auris (single) | ns | ns | ns | ||||||||||||
| 5. C. auris (aggregative) | 0.008 | 0.007 | ns | 0.02 | |||||||||||
| 6. C. parapsilosis | 0.04 | 0.01 | ns | 0.04 | ns | ||||||||||
| 7. C. orthopsilosis | 0.04 | 0.03 | ns | 0.05 | ns | ns | |||||||||
| 8. C. lusitaniae | 0.01 | 0.01 | ns | 0.04 | ns | ns | ns | ||||||||
| 9. C. guilliermondii | 0.001 | 0.001 | 0.01 | 0.001 | ns | ns | ns | 0.02 | |||||||
| 10. C. glabrata | 0.001 | 0.001 | 0.001 | 0.001 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | ||||||
| 11. C. krusei | 0.001 | 0.001 | 0.001 | 0.001 | 0.002 | 0.003 | 0.002 | 0.003 | 0.009 | ns | |||||
| 12. C. nivariensis | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.003 | 0.004 | 0.009 | ns | ns | ||||
| 13. C. bracarensis | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.007 | 0.03 | ns | ns | |||
| 14. C. kefyr | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.007 | 0.03 | ns | ns | ns | ||
| 15. S. cerevisiae | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.007 | 0.03 | ns | ns | ns | ns | |
P values of <0.05 as determined using the Mann-Whitney (two-sample Wilcoxon) test are given for all species combinations where a given species (horizontal axis) was more pathogenic than another (vertical axis). ns, not statistically significant (P > 0.05). 1, C. albicans; 2, C. tropicalis; 3, C. auris (all); 4, C. auris (single); 5, C. auris (aggregative); 6, C. parapsilosis; 7, C. orthopsilosis; 8, C. lusitaniae; 9, C. guilliermondii; 10, C. glabrata; 11, C. krusei; 12, C. nivariensis; 13, C. bracarensis; 14, C. kefyr; 15, S. cerevisiae.
Individual killing curves for the common Candida species depicted in Fig. 2, with error bars representing the maximum and minimum larval killing rates observed with different isolates of that species at each time point. Download Figure S1, TIF file, 0.05 MB (50.5KB, tif) .
Copyright © 2016 Borman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
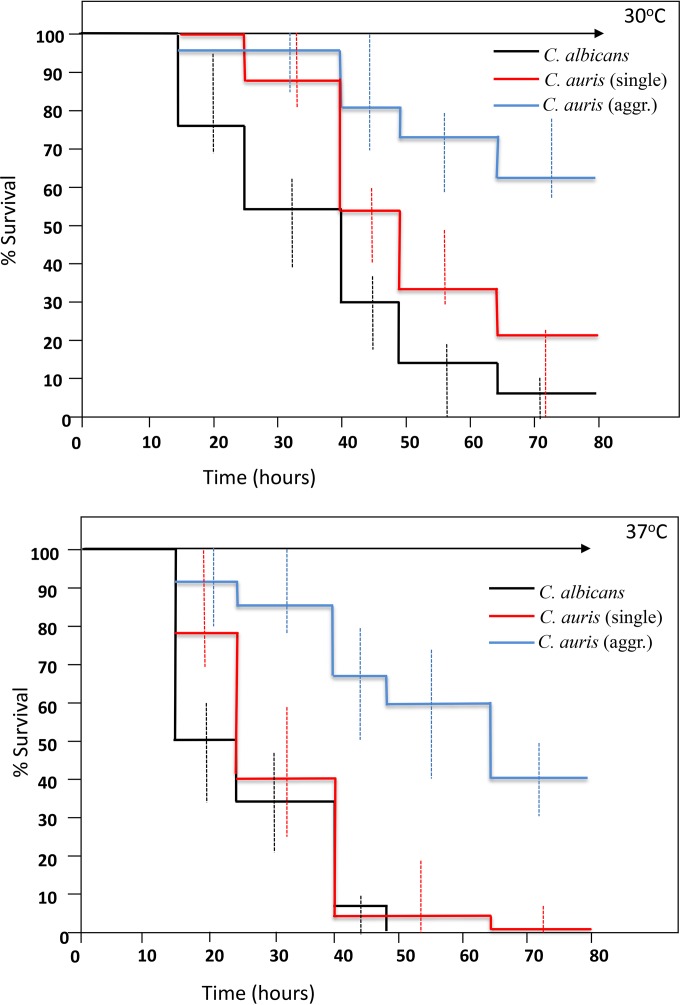
Strikingly, despite most reports suggesting that C. auris does not form significant pseudohyphae in vitro (14, 15, 21), C. auris strains exhibited virulence in G. mellonella that was significantly higher (in terms of the kinetics of larval death and the number of larvae killed) than that seen with most other common pathogenic yeast species, with overall pathogenicity approaching that observed with C. albicans and C. tropicalis isolates (Fig. 2 and Table 2). Dissection of representative larvae that had been inoculated with the various strains and incubated at 37°C for 18 h revealed significant hyphal proliferation in hemolymph form larvae inoculated with C. albicans (Fig. 3A). However, no hyphal or pseudohyphal formation was observed in larvae infected with any C. auris strains at 18 h or any time postinfection (Fig. 3B to D). Interestingly, in larvae that had received nonaggregating strains of C. auris, larval dissection revealed large numbers of individual budding yeast cells, including in phagocytic cells (Fig. 3B and E). However, in larvae inoculated with individual yeast cells prepared from aggregate-forming strains of C. auris, hemolymph contained large aggregates of C. auris cells, with few individual yeast cells, indicating that the ability to produce large aggregates had been maintained in vivo (Fig. 3C and E). In the light of this differential behavior of C. auris isolates in G. mellonella, further experiments compared larval killing with aggregate-forming versus non-aggregate-forming strains, with larvae incubated at both 30°C and 37°C. Strikingly, nonaggregate strains exhibited significantly greater virulence than aggregate-forming strains at both temperatures (Fig. 4 and Table 2) (P = 0.02), with nonaggregate isolates showing virulence that was indistinguishable from that of C. albicans strains at 37°C (Fig. 4).
FIG 3 .
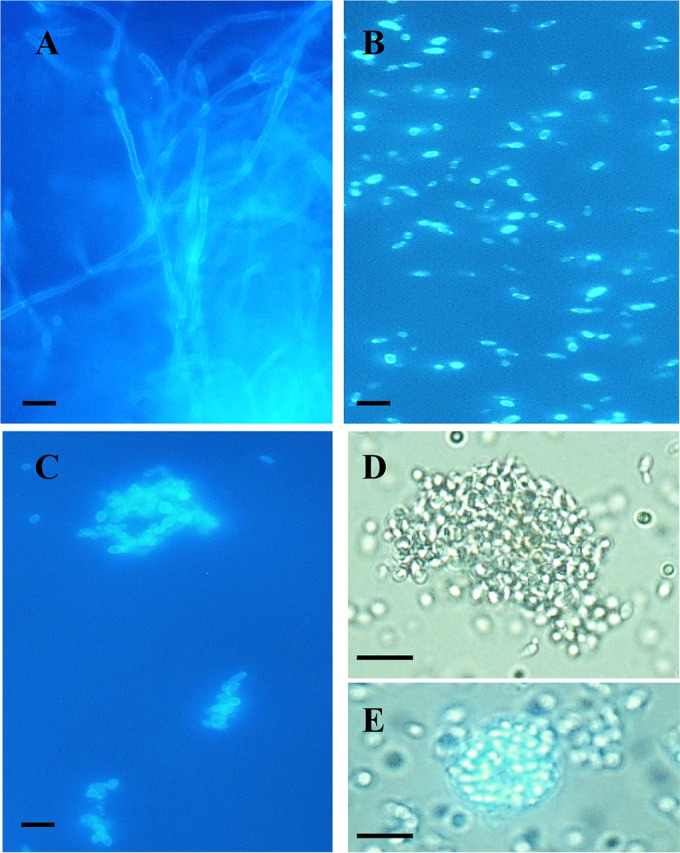
Microscopic appearance of hemolymph from infected larvae. (A to D) Hemolymph recovered after 18 h at 37°C in larvae inoculated with C. albicans (A), a nonaggregating strain of C. auris (strain 1) (B), and single cells prepared from an aggregate-forming isolate of C. auris (strain 2) (C and D). The hemolymph was stained with Calcofluor fluorescent enhancer after KOH treatment and examined under UV illumination (A to C) or was examined directly by light microscopy (D and E). Panel E shows a single phagocytic cell containing many individual budding C. auris cells. Magnification in all panels was ×400. Scale bar = 10 µm.
FIG 4 .
Virulence of aggregate-forming and nonaggregate strains of Candida auris compared to C. albicans in Galleria mellonella larvae at 30°C (upper panel) and 37°C (lower panel). Kaplan-Meier plots of G. mellonella survival after injection with 106 CFU/larva of Candida albicans (black line), nonaggregating C. auris strains (red line), and aggregate-forming C. auris strains (blue line) are shown. Four strains were tested for C. albicans, with 15 larvae per strain, and 8 and 4 strains were tested for nonaggregate and aggregate-forming C. auris, respectively (with 10 larvae per strain). Experiments were performed in duplicate; plots represent the combined (additive) data from all strains and all experiments. Error bars represent the maximum and minimum larval killing observed with different isolates of each species at each time point. No larval killing was observed in control larvae injected with an equivalent volume of PBS (arrowed lines).
In the current report, we present for the first time a comparative study of the pathogenicities of isolates of Candida auris and those of other common pathogenic Candida species and the somewhat surprising finding that C. auris virulence is comparable to that seen with C. albicans in the invertebrate G. mellonella model, despite the fact that C. auris isolates do not undergo significant filamentation in this model organism. This finding is all the more striking since C. auris yeast cells are more comparable in size and growth rate to C. glabrata than to C. albicans (Fig. 2 and data not shown). Moreover, we have demonstrated the novel finding that certain C. auris isolates form large aggregates of cells both in vitro and in vivo in inoculated larvae, even when larvae were inoculated with individual cells prepared from aggregating isolates. Microscopic examination of these aggregates suggests that they form due to reduced daughter cell liberation after budding (see, for example, Fig. 3C), rather than due to flocculation of individual budding cells. This contention would certainly be supported by our inability to disrupt the aggregates with intense vortex mixing and detergent treatments. In G. mellonella, aggregate-forming strains exhibit less virulence than those strains that exist as single budding cells. Further studies will be required to determine if aggregate-forming strains produce less dissemination during infections in humans or, conversely, whether the ability to form large aggregates protects those strains against phagocytic attack or the effects of antifungal agents or detergents used to clean hospital environments.
MATERIALS AND METHODS
Fungal strains.
All C. auris isolates were identified by ribosomal DNA (rDNA) gene sequencing targeting the 28S rRNA or by internal transcribed spacer 1 (ITS1) regions and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis or by a combination of the two methods exactly as described previously (25). For the other Candida species included for comparison, where possible, clinical isolates were from deep-seated infections. Identity to the species level was confirmed by sequencing or MALDI-TOF analysis in all cases.
Killing assays in G. mellonella.
Killing assays were performed in Galleria mellonella exactly as described previously (10), using final (sixth) instar larvae (Livefood UK Ltd., Rooks bridge, Somerset, United Kingdom) weighing approximately 300 mg each that were free of gray markings and that had been maintained at room temperature in the dark and inoculated within 48 h of receipt. Suspensions of individual Candida isolates that had been grown on Sabouraud’s agar for 24 h at 37°C were harvested by gentle scraping of colony surfaces with sterile plastic loops, washed twice in sterile PBS, counted in hemocytometers, and adjusted to 105 cells/μl in sterile PBS. Individual larvae were inoculated in the left rear proleg with 1 × 106 yeast cells–PBS (final inoculum volume, 10 µl) using a 10-µl Hamilton syringe fitted with a 26-gauge blunt needle. At least 10 larvae were inoculated per isolate per experiment (experiments employed 4 independent isolates of each Candida test species [12 isolates in the case of C. auris]). Control groups of larvae received 10 µl of sterile PBS in exactly the same manner. Inoculated larvae were incubated at 30°C or 37°C and scored for viability at 8-h intervals as described previously (10). Differences in resulting Kaplan-Meier survival plots were evaluated using the Mann-Whitney (two-sample Wilcoxon) test. In some experiments, fungal cell filamentation postinfection was assessed by sacrificing representative larvae from each inoculum group at 24 h postinfection and aseptic collection of the fat body/solid internal structures and hemolymph followed by microscopic examination (10).
Antifungal susceptibility testing of C. auris isolates.
Broth microdilution determination of yeast MICs was performed according to CLSI method M27-A3 (26) in round-bottomed 96-well plates with yeast blastospore suspensions prepared in saline solution and then diluted into RPMI 1640 and adjusted to a final concentration of 2.5 × 103 CFU/ml. Inoculated plates were incubated for 24 to 48 h at 35°C. MICs were read at 24 and 48 h as the concentration of drug that elicited 100% inhibition of growth (amphotericin B) or significant (approximately 50%) inhibition of growth compared with that of a drug-free control (fluconazole, voriconazole, posaconazole, anidulafungin, and flucytosine).
ACKNOWLEDGMENT
We are grateful to the other members of the United Kingdom MRL for their assistance with data collation and phenotypic and molecular analyses of isolates.
REFERENCES
- 1.Marr KA. 2004. Invasive Candida infections: the changing epidemiology. Oncology 18:9–14. [PubMed] [Google Scholar]
- 2.Nucci M, Marr KA. 2005. Emerging fungal diseases. Clin Infect Dis 41:521–526. doi: 10.1086/432060. [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Jones RN, Doern GV, Fluit AC, Verhoef J, Sader HS, Messer SA, Houston A, Coffman S, Hollis RJ. 1999. International surveillance of bloodstream infections due to Candida species in the European SENTRY program: species distribution and antifungal susceptibility including the investigational triazole and echinocandin agents. SENTRY Participant Group (Europe). Diagn Microbiol Infect Dis 35:19–25. doi: 10.1016/S0732-8893(99)00046-2. [DOI] [PubMed] [Google Scholar]
- 4.Pfaller MA, Jones RN, Messer SA, Edmond MB, Wenzel RP. 1998. National surveillance of nosocomial bloodstream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn Microbiol Infect Dis 31:327–332. doi: 10.1016/S0732-8893(97)00240-X. [DOI] [PubMed] [Google Scholar]
- 5.Ruhnke M. 2006. Epidemiology of Candida albicans infections and role of non-Candida albicans yeasts. Curr Drug Targets 7:495–504. doi: 10.2174/138945006776359421. [DOI] [PubMed] [Google Scholar]
- 6.Snydman DR. 2003. Shifting patterns in the epidemiology of nosocomial Candida infections. Chest 123:500S–503S. doi: 10.1378/chest.123.5_suppl.500S. [DOI] [PubMed] [Google Scholar]
- 7.Wingard JR. 1994. Infections due to resistant Candida species in patients with cancer who are receiving chemotherapy. Clin Infect Dis 19(Suppl 1):S49–S53. doi: 10.1093/clinids/19.Supplement_1.S49. [DOI] [PubMed] [Google Scholar]
- 8.Wright WL, Wenzel RP. 1997. Nosocomial Candida. Epidemiology, transmission, and prevention. Infect Dis Clin North Am 11:411–425. doi: 10.1016/S0891-5520(05)70363-9. [DOI] [PubMed] [Google Scholar]
- 9.Moran GP, Coleman DC, Sullivan DJ. 2011. Comparative genomics and evolution of pathogenicity in human pathogenic fungi. Eukaryot Cell 10:34–42. doi: 10.1128/EC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borman AM, Szekely A, Linton CJ, Palmer MD, Brown P, Johnson EM. 2013. Epidemiology, antifungal susceptibility, and pathogenicity of Candida africana isolates from the United Kingdom. J Clin Microbiol 51:967–972. doi: 10.1128/JCM.02816-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, Lee JS, Jung SI, Park KH, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW. 2009. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48:e57–e61. doi: 10.1086/597108. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 14.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarma S, Kumar N, Sharma S, Govil D, Ali T, Mehta Y, Rattan A. 2013. Candidemia caused by amphotericin B and fluconazole resistant Candida auris. Indian J Med Microbiol 31:90–91. doi: 10.4103/0255-0857.108746. [DOI] [PubMed] [Google Scholar]
- 17.Kumar D, Banerjee T, Pratap CB, Tilak R. 2015. Itraconazole-resistant Candida auris with phospholipase, proteinase and hemolysin activity from a case of vulvovaginitis. J Infect Dev Ctries 9:435–437. [DOI] [PubMed] [Google Scholar]
- 18.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodero L, Cuenca-Estrella M, Córdoba S, Cahn P, Davel G, Kaufman S, Guelfand L, Rodríguez-Tudela JL. 2002. Transient fungemia caused by an amphotericin B-resistant isolate of Candida haemulonii. J Clin Microbiol 40:2266–2269. doi: 10.1128/JCM.40.6.2266-2269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muro MD, Motta Fde A, Burger M, Melo AS, Dalla-Costa LM. 2012. Echinocandin resistance in two Candida haemulonii isolates from pediatric patients. J Clin Microbiol 50:3783–3785. doi: 10.1128/JCM.01136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 23.Oh BJ, Shin JH, Kim MN, Sung H, Lee K, Joo MY, Shin MG, Suh SP, Ryang DW. 2011. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med Mycol 49:98–102. doi: 10.3109/13693786.2010.493563. [DOI] [PubMed] [Google Scholar]
- 24.Cotter G, Doyle S, Kavanagh K. 2000. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol 27:163–169. doi: 10.1111/j.1574-695X.2000.tb01427.x. [DOI] [PubMed] [Google Scholar]
- 25.Fraser M, Brown Z, Houldsworth M, Borman AM, Johnson EM. 2016. Rapid identification of 6328 isolates of pathogenic yeasts using MALDI-ToF MS and a simplified, rapid extraction procedure that is compatible with the Bruker Biotyper platform and database. Med Mycol 54:80–88. doi: 10.1093/mmy/myv085. [DOI] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution susceptibility testing of yeasts: approved standard, 3rd ed. Document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Individual killing curves for the common Candida species depicted in Fig. 2, with error bars representing the maximum and minimum larval killing rates observed with different isolates of that species at each time point. Download Figure S1, TIF file, 0.05 MB (50.5KB, tif) .
Copyright © 2016 Borman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.