Abstract
As sessile organisms, plants must be able to adapt to the environment. Plants respond to the environment by adjusting their growth and development, which is mediated by sophisticated signaling networks that integrate multiple environmental and endogenous signals. Recently, increasing evidence has shown that a bHLH transcription factor PIF4 plays a major role in the multiple signal integration for plant growth regulation. PIF4 is a positive regulator in cell elongation and its activity is regulated by various environmental signals, including light and temperature, and hormonal signals, including auxin, gibberellic acid and brassinosteroid, both transcriptionally and post-translationally. Moreover, recent studies have shown that the circadian clock and metabolic status regulate endogenous PIF4 level. The PIF4 transcription factor cooperatively regulates the target genes involved in cell elongation with hormone-regulated transcription factors. Therefore, PIF4 is a key integrator of multiple signaling pathways, which optimizes growth in the environment. This review will discuss our current understanding of the PIF4-mediated signaling networks that control plant growth.
Keywords: brassinosteroid, high temperature, light, PIF4, signaling integration
INTRODUCTION
Since plants are sessile, they must adapt to their environment. One of major adaptation mechanisms is plasticity in their growth and development. Plants have the ability to adjust their growth and development in response to different environmental conditions. The plasticity of growth and development is achieved by intimate interactions between various environmental and endogenous hormonal signaling pathways (Chaiwanon et al., 2016). How these multiple signaling pathways are integrated and coordinately regulate signaling outputs (e.g., development) has been studied at the molecular level in Arabidopsis model system for the last decade.
Growing evidence indicates that PIF4 acts as a molecular hub that integrates the environmental and hormonal signaling pathways (Lucyshyn and Wigge, 2009; Leivar and Quail, 2011; Quint et al., 2016). PIF4 is a member of the family of PHYTOCHROME INTERACTING FACTORs (PIFs) that directly interact with light-activated phytochromes and regulate various light responses. PIFs redundantly inhibit skotomorphogenesis and individually regulate other light-mediated processes such as shade avoidance responses, chloroplast differentiation and seed germination (Jeong and Choi, 2013). In addition to the light responses, some PIF members including PIF4 are involved in the hormonal responses (de Lucas and Prat, 2014). Studies have shown that the PIF4 activity is regulated by various environment factors (light and temperature), hormones (gibberellic acid (GA), brassinosteroid (BR), and auxin), and the circadian clock. The integrated information by PIF4 is then transduced to downstream output pathways to optimize plant growth in a given environmental conditions. Here, we review the current knowledge related to the PIF4-mediated multiple signal integration mechanism.
PIF4 FUNCTIONS IN THE LIGHT SIGNALING PATHWAYS
PIF4 was initially identified by a genetic screen with T-DNA-mutagenized population to identify new components of the phytochrome signaling pathway (Huq and Quail, 2002). The isolated new mutants are specifically defective in responsiveness to red light. Later, one of the mutants was found to have a T-DNA insertion in a gene encoding basic helix-loop-helix (bHLH) transcription factor that is sequentially similar to PHYTOCHROME INTERACTING FACTOR3 (PIF3), thus named PHYTOCHROME INTERACTING FACTOR4 (PIF4) (Huq and Quail, 2002). PIF3 is the first identified phytochrome-interacting bHLH transcription factor that interacts with the biologically active Pfr form of both phytochrome A (phyA) and phytochrome B (phyB) (Kim et al., 2003; Ni et al., 1998). PIF4 also directly interacts with the red light-activated Pfr phyB through Active Phytochrome Binding (APB) motif (Huq and Quail, 2002; Khanna et al., 2004). Although, unlike PIF3, PIF4 does not have an APA (Active phyA binding) motif that is necessary for interaction with phyA (Al-Sady et al., 2006), it interacts with phyA (Huq and Quail, 2002). The hypocotyl elongation of pif4 null mutant is hypersensitive to red light, but not to far-red light. However, the double mutant of PIF4 and its close homolog PIF5 (pif4;pif5) show hypersensitivity to far-red light, suggesting that they redundantly control the far-red light responses (Lorrain et al., 2009). While the red light hypersensitivity of the pif4 mutant requires phyB (Huq and Quail, 2002), the far-red light hypersensitivity of the pif4;pif5 double mutant requires phyA (Lorrain et al., 2009).
PIF4 protein stability is regulated by light conditions (Fig. 1A). In the dark, PIF4 protein is dephosphorylated and stable (Lorrain et al., 2008). However, upon red light irradiation, PIF4 protein is rapidly phosphorylated by phyB-mediated signaling and subsequently targeted for 26S proteasome-mediated degradation (Lorrain et al., 2008). Deletion of APB motif of PIF4 increases the stability of PIF4 under red light, indicating that the interaction with phyB is required for the red light-induced PIF4 degradation (Lorrain et al., 2008; Shen et al., 2007). In contrast to red light, PIF4 protein is quite stable under far-red and blue light (Lorrain et al., 2008; Ma et al., 2016; Pedmale et al., 2016). Although it has been known for a long time that PIF factors, including PIF4, are rapidly phosphorylated by red light irradiation (Al-Sady et al., 2006), the kinases that mediate the red light-dependent phosphorylation of PIF factors are yet to be identified. In addition to PIF factors, the phyB-PIF interactions reciprocally induce degradation of phyB under red light, forming a mutually-negative feedback-loop (Christians et al., 2012; Ni et al., 2014). It was shown that Bric-a-Brack/Tramtrack/Broad (BTB)-Cullin3-type E3 ubiquitin ligase LIGHT RESPONSE BTB (LRB) proteins directly interact with the red light-activated phyB-PIF3 complex, subsequently polyubiquitinate the complex, and target both phyB and PIF3 for degradation (Ni et al., 2014). The mutually assured degradation of both phyB (light receptor) and PIF3 (its immediate signaling partner) attenuates light signaling, thereby preventing unnecessary excessive light responses. However, the E3 ligases responsible for PIF4 degradation have not been identified to date. It will be interesting to determine if the LRBs also mediate the red light-induced degradation of other PIFs, including PIF4 (Fig. 1A).
Fig. 1.
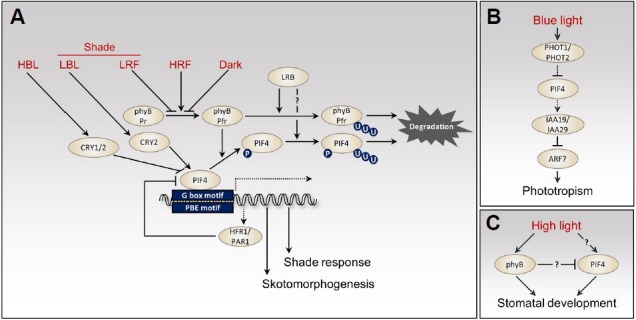
PIF4-mediated light signaling pathways. (A) PIF4 positively regulates skotomorphogenesis and shade response. HBL, high blue light; LBL, low blue light; LRF, low red:far-red ratio; HRF, high red:far-red ratio; PBE, PIF-binding E-box. “P” in a circle indicates phosphorylated status. A group of “U” in a circle indicates polyubiquitination. (B) PIF4 is involved in the blue light-dependent phototropism by regulating auxin signaling. (C) High light promotes stomatal development, which is mediated by phyB and PIF4. Solid lines indicate post-translational modifications or regulation at protein levels. Dashed lines show transcriptional regulation. Question marks indicate unknown or unconfirmed mechanism.
In the dark, a pif4 single mutant does not show any obvious phenotypes, but a pif1;pif3;pif4;pif5 quadruple (pifq) mutant shows strong constitutive photomorphogenic morphology, including short hypocotyls and open cotyledons, indicating that PIF4 together with other PIF members inhibits photomorphogenesis in the dark (Leivar et al., 2009; Shin et al., 2009) (Fig. 1A). Consistently, integrated RNA-Seq and ChIP-Seq analyses showed that four PIFs regulate the expression of a large number of shared target genes (Pfeiffer et al., 2014). However, individual PIF factors also regulate many differential target genes suggesting that each PIF factor has a distinct function in regulating morphogenic responses (Pfeiffer et al., 2014). In the ChIP-Seq analyses of PIF4, the G-box motif (CACGTG) was highly enriched in the PIF4 target promoters, consistent with a result of electrophoretic mobility shift assay (EMSA) showing PIF4 binding to the G-box motif in vitro (Huq and Quail, 2002; Oh et al., 2012; Zhang et al., 2013). However, one of the E-box motifs (CACATG, PIF-binding E-box) was also significantly over-represented in the PIF4 target gene promoters, especially in the PIF4-activated genes, suggesting that PIF4 is able to bind to the E-box motif in vivo (Oh et al., 2012; Zhang et al., 2013) (Fig. 1A).
In addition to skotomorphogenesis (morphogenesis in the dark), PIF4 and PIF5 mediate shade avoidance responses including stem/hypocotyl elongation (Lorrain et al., 2008). Under shade conditions, in which the red light to far-red light ratio is reduced and active phyB is decreased, PIF4 and PIF5 proteins are dephosphorylated and stabilized (Lorrain et al., 2008). The increased PIF4 and PIF5 by shade induce the expression of shade genes and promote hypocotyl elongation (Fig. 1A). Among the genes that are rapidly induced by shade in a PIF4-and PIF5-dependent manner, LONG HYPOCOTYL IN FAR-RED1 (HFR1) and PHYTOCHROME RAPIDLY REGULATED1 (PAR1) negatively regulate PIF4 and PIF5 activities (Hao et al., 2012; Hornitschek et al., 2009). Both HFR1 and PAR1 are atypical bHLH transcription factors that directly interact with PIF4/PIF5, and form a non-DNA-binding heterodimer with them, thereby preventing PIF4/PIF5 from binding to target gene promoters (Hao et al., 2012; Hornitschek et al., 2009). The negative feedback regulation of PIF4/PIF5 by HFR1 and PAR1 provides a regulatory mechanism that prevents exaggerated plant response to shade (Fig. 1A).
PIF4 mediates blue light responses, as well as the red/far-red light responses. In addition to reduction in the red light to far-red light ratio, blue light is also reduced under canopy shade. Similar to the low red to far-red ratio light, reduced or low blue light (LBL) also induces a shade avoidance response, including stem elongation. PIF4 and PIF5 directly interact with blue light receptor cryptochromes CRY1 and CRY2 and mediate the LBL-induced hypocotyl elongation (Pedmale et al., 2016). Unlike red light, blue light does not affect the PIF4 protein stability (Ma et al., 2016; Pedmale et al., 2016). Instead, blue light regulates PIF4 transcriptional activity through CRYs. Under LBL, CRY2 associated with PIF4 is recruited to PIF4 target genes and regulates their expression by modulating PIF4 transcriptional activity (Pedmale et al., 2016). In contrast, under high blue light, blue light-activated CRY1 directly interacts with PIF4 and represses PIF4 transcriptional activity, without affecting PIF4 DNA-binding ability (Ma et al., 2016; Pedmale et al., 2016). Therefore, PIF4-mediated hypocotyl elongation is suppressed under high blue light (Fig. 1A). In addition to the LBL-mediated hypocotyl elongation, PIF4 is also involved in blue light-mediated phototropism downstream of another blue light receptors PHOTOTROPIN1 (PHOT1) and PHOT2 (Sun et al., 2013). PIF4 directly activates the expression of auxin signaling repressors IAA19 and IAA29, which repress ARF7 that mediates the hypocotyl phototropic response (Fig. 1B). PHOT1 and PHOT2 transcriptionally reduce PIF4 to induce phototropic growth in response to blue light.
Stomatal development is highly dependent on light conditions. In Arabidopsis mature leaves, the number of stomata is increased at high intensity of light, compared to low light. Both phyB and PIF4 are required for the high light-induced stomatal development (Casson et al., 2009). Interestingly, while phyB and PIF4 antagonistically regulate photomorphogenesis and shade avoidance responses, they cooperatively regulate the stomatal development in response to high light (Casson et al., 2009) (Fig. 1C). The identification of the molecular mechanisms underlying the context dependent relationship between phyB and PIF4 is an interesting topic for future study.
PIF4 FUNCTIONS IN HORMONE SIGNALING PATHWAYS
Plant development and growth are regulated by several plant hormones. Auxin, gibberellic acid (GA), and brassinosteroid (BR) are major hormones that promote plant growth. Interestingly, these hormone’s biosynthetic and signaling mutants display short hypocotyls and opened cotyledons, even in the dark, indicating that these growth promoting hormones are required for skotomorphogenesis (Chaiwanon et al., 2016). The constitutive photomorphogenic phenotypes of auxin, GA, and BR mutants suggest that these hormone pathways intimately interact with the phytochrome pathway. Growing evidence indicates that PIF4 functions as a molecular hub that links these hormones and phytochrome signaling pathways.
BR-mediated skotomorphogenesis is mainly mediated by BR-regulated transcription factor, BRASSINAZOLE RESISTANT1 (BZR1) (Wang et al., 2012). In the absence of BR, BZR1 is phosphorylated and inactivated by GSK3-like kinase BRA-SSINOSTEROID INSENSITIVE2 (BIN2). BR inactivates BIN2, which leads to dephosphorylation of BZR1. The dephosphorylated BZR1 regulates gene expression and promotes hypocotyl elongation. Surprisingly, genome-wide identification of BZR1 and PIF4 binding sites revealed that two transcription factors share thousands of target genes (Oh et al., 2012). BZR1 and PIF4 cooperatively regulate the expression of co-target genes, including genes involved in cell elongation (Oh et al., 2012). Consistent with their interdependency on the regulation of target gene expression, both BZR1 and PIF4 are required for hypocotyl elongation in the dark (Oh et al., 2012). Thus, BR and phytochrome signaling pathways are integrated by a direct interaction between PIF4 and BZR1 (Fig. 2A). In addition to BZR1, the BR-regulated kinase, BIN2, also interacts with and inactivates PIF4 (Bernardo-Garcia et al., 2014). BIN2 phosphorylates PIF4 as well as BZR1, and this phosphorylation marks PIF4 for 26S proteasome-mediated degradation, thereby reducing cellular PIF4 level. BR signaling increases PIF4 protein stability through the inactivation of BIN2 (Fig. 2A). BZR1 interacts with other PIF factors, including PIF1 and PIF3 (Oh et al., 2012; Zhang et al., 2014). However, it has not been fully determined whether BIN2 phosphorylates and inactivates other PIF factors.
Fig. 2.
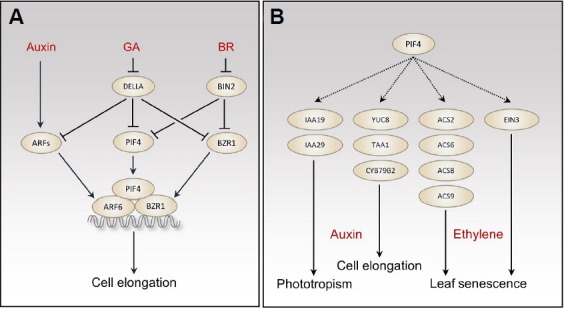
PIF4-mediated hormone signaling pathways. (A) Integration of auxin, gibber-ellic acid (GA), and brassinosteroid (BR) signaling pathways in the regulation of hypocotyl elongation. (B) PIF4 transcriptionally regulates auxin and ethylene signaling and biosynthetic genes, which mediates phototropism, cell elongation, and leaf senescence.
Synergistic actions of auxin and BR in the regulation of cell elongation have been reported in several studies (Hardtke, 2007). At the molecular level, auxin and BR regulate the expression of common target genes, which is mediated by a direct interaction between AUXIN RESPONSE FACTORs (ARF6 and ARF8) and BZR1 (Oh et al., 2014). ARF6/ARF8 redundantly regulates hypocotyl elongation in the dark. ARF6/ARF8 also interacts with PIF4, as well as BZR1. The interactions with BZR1 and PIF4 increase ARF6 activity by enhancing its binding to target promoters (Oh et al., 2014). Combined analyses of ChIP-Seq and RNA-Seq assays found that BZR1, ARF6, and PIF4 share a large number of target genes and cooperatively regulate the expression of genes, including cell elongation-related genes (SAURs and PREs) (Oh et al., 2014). However, it is unclear whether these three transcription factors act as a trimeric complex or form only a dimeric complex. PIF4 and PIF5 also regulate the expression of auxin repressor IAA19 and IAA29 that are involved in auxin-mediated hypocotyl photo-tropic growth (Sun et al., 2013) (Fig. 2B). In addition to auxin signaling, PIF4 increases auxin levels by directly activating the expression of key auxin biosynthesis genes, including YUC8, TAA1, and CYB79B2 (Franklin et al., 2011; Sun et al., 2012). The up-regulation of auxin biosynthesis is required for the PIF4-mediated hypocotyl elongation in response to warm temperature and shade (Fig. 2B).
GA promotes hypocotyl elongation through the destabilization of negative regulators, DELLAs. DELLA proteins directly interact with several PIF factors, including PIF4, and prevent PIFs from binding to target promoters, thereby inactivating PIFs (de Lucas et al., 2008; Feng et al., 2008). In addition, DELLAs also interact with and inactivate BZR1 and ARF6 (Bai et al., 2012; Oh et al., 2014). Consistently, GA-mediated hypocotyl elongation requires PIFs, BZR1, and ARF6 activities. Therefore, GA, BR, auxin, and light signaling pathways are integrated by multiple interactions among their key signaling transcription factors (DELLAs, BZR1, ARFs, and PIFs) (Oh et al., 2014) (Fig. 2A).
In addition to early seedling morphogenesis, PIF4, together with PIF5, regulates dark-induced leaf senescence (Sakuraba et al., 2014; Song et al., 2014), which is mainly mediated by antagonistic interaction of two plant hormones, cytokinin and ethylene. PIF4 positively regulates both ethylene biosynthesis and signaling to promote leaf senescence. PIF4 promotes ethylene biosynthesis by activating the expression of several ethylene biosynthesis genes (ACS2, 6, 8, and 9) and enhances ethylene signaling by directly activating expression of ethylene key signaling transcription factor, ETHYLENE INSENSITIVE3 (Sakuraba et al., 2014; Song et al., 2014) (Fig. 2B). Ethylene is also required for skotomorphogenesis, including apical hook formation and closed cotyledons. In the dark, plants overexpressing PIF5 (PIF5-OX) are morphologically similar to plants overproducing ethylene (Khanna et al., 2007). Consistently, ethylene production is significantly increased in PIF5-OX, compared to wild type (Khanna et al., 2007). In contrast, ethylene production is not significantly altered in pif5 mutants. Considering the reduced ethylene production in pif4 mutants in senescencing leaves (Song et al., 2014), it is highly likely that PIF4, as well as other PIF factors, redundantly regulates ethylene biosynthesis in the etiolated seedlings. However, the defects of ethylene production in pif4;pif5 or pifq etiolated seedlings have not been reported to date.
PIF4 FUNCTIONS IN THE CIRCADIAN CLOCK OUTPUT PATHWAYS
The expression of PIF4 and PIF5 is regulated by the circadian clock (Yamashino et al., 2003). Under diurnal conditions, both PIF4 and PIF5 expression is repressed at the evening by the Evening Complex consisting of ELF3, ELF4, and LUX (Nusinow et al., 2011). The Evening Complex directly binds to the promoters of PIF4 and PIF5 and represses their expression (Fig. 3). As the cellular level of Evening Complex decreases, PIF4 and PIF5 expression gradually increases after the evening, and peaks in the middle of day when the Evening Complex is at its minimum level (Nusinow et al., 2011). Since PIF4 and PIF5 proteins are unstable under light, it was expected that endogenous PIF4 and PIF5 protein levels would be at a maximum at the end of day when the expression of PIF4 and PIF5 is high and their proteins are stable (Nozue et al., 2007). The coincidence mechanism was proposed to underlie observed rhythmic hypocotyl growth under short-day conditions (Nozue et al., 2007). However, in contrast to initial expectations, endogenous PIF4 protein levels are still very high in the daytime under short-day conditions, which is not consistent with a low growth rate of hypocotyl (Yamashino et al., 2013). It was thus suggested that PIF4 protein activity as well as stability is suppressed by light. It seems likely that PIF4 DNA-binding ability is inhibited by phyB under light, because the DNA-binding ability of another PIF factor, PIF3, was shown to be inhibited by the light-activated phyB (Park et al., 2012). Furthermore, endogenous GA levels are low under light, which may lead to the accumulation of DELLA proteins that directly interfere with PIF4 DNA-binding ability (Sun, 2011). In addition to protein stability, PIF4 transcriptional activity is also likely to be repressed under light through the blue light-induced CRY1-PIF4 interaction (Ma et al., 2016). Thus, the combined actions of phyB/DELLA/CRY1 may ensure low PIF4 activity in the daytime to suppress hypocotyl growth. However, this could not explain why the hypocotyl growth rate is higher in the day than in the night under long-day conditions (Nozue et al., 2007).
Fig. 3.
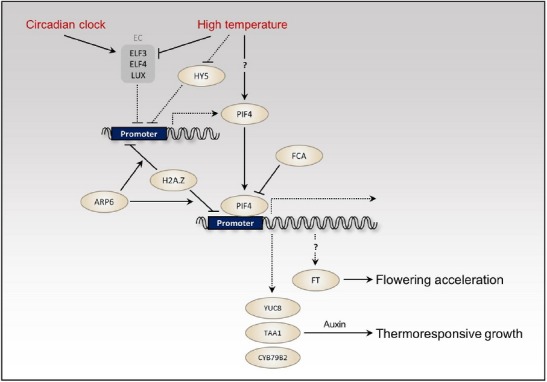
PIF4-mediated high temperature signaling pathways EC indicates the Evening Complex consisting of ELF3, ELF4 and LUX. Solid lines indicate post-translational modification or regulation at protein levels. Dashed lines show transcriptional regulation. A question mark indicates unconfirmed mechanism.
PIF4 FUNCTIONS IN THE TEMPERATURE SIGNALING PATHWAYS
Arabidopsis responds to elevated temperature (20–22°C to 27–29°C) by a suite of morphological changes, including elongation of stems and leaf hyponasty (Gray et al., 1998). This thermoresponsive growth has been observed in various species of plants, implicating its evolutionary benefit in adapting to higher temperatures (Quint et al., 2016). It was indeed shown that the thermoresponsive growth increases leaf transpiration rate, thereby enhancing leaf cooling capacity in well-watered environments (Crawford et al., 2012).
PIF4 is a key regulator of thermoresponsive growth as the hypocotyl growth of pif4 mutants is insensitive to high temperatures (29°C) (Koini et al., 2009). It was suggested that high temperature activates PIF4 RNA expression by inactivating ELF3 that directly represses PIF4 expression (Fig. 3). ELF3 binding to the PIF4 promoter is decreased at high temperatures, compared to normal temperatures (22°C) (Box et al., 2015). Consistently, elf3 mutant displays a constitutive thermomorphogenic phenotype (elongated hypocotyls) at 22°C under short-day conditions, supporting the fact that high temperature inactivation of ELF3 is a major mechanism to induce thermoresponsive growth (Box et al., 2015; Mizuno et al., 2014). However, the high temperature effects on PIF4 expression are restricted to the end of night when ELF3 levels are already low and PIF4 expression is normally repressed in the evening, even at high temperatures (Box et al., 2015), raising questions about the exact role of ELF3 in high temperature regulation of PIF4 expression. Moreover, the hypocotyl growth of elf3 mutants is significantly promoted by high temperature under long-day conditions (Raschke et al., 2015), suggesting that high temperature activates PIF4 post-transcriptionally as well as transcriptionally. Indeed, a study showed that PIF4 protein is dramatically stabilized at high temperature under red light (Foreman et al., 2011). However, the PIF4 protein stability was minimally affected by high temperature under white light conditions in other studies (Kumar et al., 2012; Stavang et al., 2009). Thus, more comprehensive analyses of PIF4 protein stability in different combinations of light and temperature conditions is required to determine how temperature changes affect PIF4 protein stability.
High temperature-activated PIF4 directly up-regulates the expression of auxin biosynthesis genes, including YUC8, TAA1, and CYB79B2 (Franklin et al., 2011; Sun et al., 2012) (Fig. 3). As a result, the accumulated auxin induces hypocotyl/stem elongation and leaf hyponasty. Consistently, auxin biosynthetic and signaling mutants have defects in the thermoresponsive growth (Gray et al., 1998). Since PIF4 activity is dependent on BR (activation of BZR1) and GA (inactivation of DELLAs) hormones, both hormone signals are required for the hypocotyl growth response to high temperature (Bai et al., 2012; Oh et al., 2012; Stavang et al., 2009). Light also profoundly affects the thermoresponsive growth because PIF4 RNA expression (through HY5) as well as PIF4 protein stability and transcriptional activity (through phytochromes and cryptochromes) are reduced by light (Delker et al., 2014; Lorrain et al., 2008; Ma et al., 2016). Therefore, high temperature-induced hypocotyl elongation is suppressed under high intensity of light (Ma et al., 2016).
Chromatin structure also affects PIF4 level and its activity. ACTIN RELATED PROTEIN6 (ARP6) is involved in the deposition of alternative histone H2A.Z into the nucleosome in place of H2A. Surprisingly, arp6 mutants show a constitutive thermoresponsive growth phenotype and the expression levels of many heat-induced genes, including PIF4, are elevated in the arp6 mutant (Kumar and Wigge, 2010; Kumar et al., 2012). Thus, H2A.Z nucleosome deposition at the promoter of PIF4 is likely to determine PIF4 expression. Generally, H2A.Z occupancy in the promoters of heat-regulated genes are decreased as temperature increases (Kumar and Wigge, 2010). Whether the reduction of H2A.Z nucleosomes is indeed required for the high temperature induction of PIF4 expression should be addressed in future studies. A previous study reported that RNA-binding protein FCA suppresses PIF4 activity through chromatin modification (Lee et al., 2014) (Fig. 3). FCA inhibits PIF4 binding to its target YUC8 promoter by removing H3K4me2 from the YUC8 promoter, thereby attenuating the thermoresponsive growth.
In addition to seedling thermomorphogenic growth, PIF4 was reported to mediate thermal acceleration of flowering by directly activating FLOWERING LOCUS T (FT) in a temperature dependent manner (Kumar et al., 2012) (Fig. 3). In short-day conditions, flowering of wild type is dramatically accelerated by high temperature, but the pif4 mutant was shown to be almost insensitive to high temperature-mediated flowering (Kumar et al., 2012). However, a recent study showed that pif4, and also higher-order pif mutants (pif4;pif5, pif3;pif4;pif5), flowered significantly earlier in response to high temperature under short-day conditions, almost indistinguishable from wild type (Galvao et al., 2015). Moreover, the high temperature induction of FT expression was not much affected in the pif4 mutant (Galvao et al., 2015; Seaton et al., 2015). Therefore, it is controversial as to whether the activation of FT by PIF4 constitutes the major mechanism to induce flowering at elevated ambient temperature.
CONCLUSION
Since plant morphological adaptation to the environment is an irreversible process that requires huge energy, it must be elaborately regulated, considering both environmental factors and endogenous conditions. Plants have evolved comprehensive signaling networks that integrate the external and internal information and determine plant growth. PIF4 plays a key role in signal integration for the seedling morphological adaption in Arabidopsis. Cellular PIF4 level is regulated transcriptionally by the circadian clock, light, and temperatures, and post-translationally by light. In addition, PIF4 transcriptional activity is dependent on the light conditions and the transcription factors of auxin, gibberellic acid and brassinosteroid hormone signaling pathways. The morphological adaption is also affected by the other hormones (cytokinin, abscisic acid, salicylic acid, and jasmonic acid) and biotic stresses. Thus, it is likely that these signals are also integrated by PIF4 or other PIF factors to coordinately regulate plant growth with other environmental/hormonal signals.
Acknowledgments
We apologize to the authors whose works could not be cited here due to space limitation. Research was supported by a grant from the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2016R1C1 B2008821).
REFERENCES
- Al-Sady B., Ni W., Kircher S., Schafer E., Quail P.H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-Garcia S., de Lucas M., Martinez C., Espinosa-Ruiz A., Daviere J.M., Prat S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014;28:1681–1694. doi: 10.1101/gad.243675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box M.S., Huang B.E., Domijan M., Jaeger K.E., Khattak A.K., Yoo S.J., Sedivy E.L., Jones D.M., Hearn T.J., Webb A.A., et al. ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Biol. 2015;25:194–199. doi: 10.1016/j.cub.2014.10.076. [DOI] [PubMed] [Google Scholar]
- Casson S.A., Franklin K.A., Gray J.E., Grierson C.S., Whitelam G.C., Hetherington A.M. phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr. Biol. 2009;19:229–234. doi: 10.1016/j.cub.2008.12.046. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J., Wang W., Zhu J.Y., Oh E., Wang Z.Y. Information integration and communication in plant growth regulation. Cell. 2016;164:1257–1268. doi: 10.1016/j.cell.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians M.J., Gingerich D.J., Hua Z., Lauer T.D., Vierstra R.D. The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol. 2012;160:118–134. doi: 10.1104/pp.112.199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford A.J., McLachlan D.H., Hetherington A.M., Franklin K.A. High temperature exposure increases plant cooling capacity. Curr. Biol. 2012;22:R396–397. doi: 10.1016/j.cub.2012.03.044. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Prat S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014;202:1126–1141. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Daviere J.M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blazquez M.A., Titarenko E., Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Delker C., Sonntag L., James G.V., Janitza P., Ibanez C., Ziermann H., Peterson T., Denk K., Mull S., Ziegler J., et al. The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 2014;9:1983–1989. doi: 10.1016/j.celrep.2014.11.043. [DOI] [PubMed] [Google Scholar]
- Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., Chen L., Yu L., Iglesias-Pedraz J.M., Kircher S., et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J., Johansson H., Hornitschek P., Josse E.M., Fankhauser C., Halliday K.J. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 2011;65:441–452. doi: 10.1111/j.1365-313X.2010.04434.x. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao V.C., Collani S., Horrer D., Schmid M. Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J. 2015;84:949–962. doi: 10.1111/tpj.13051. [DOI] [PubMed] [Google Scholar]
- Gray W.M., Ostin A., Sandberg G., Romano C.P., Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant. 2012;5:688–697. doi: 10.1093/mp/sss011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S. Transcriptional auxin-brassinosteroid crosstalk: who’s talking? Bioessays. 2007;29:1115–1123. doi: 10.1002/bies.20653. [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Quail P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Choi G. Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells. 2013;35:371–380. doi: 10.1007/s10059-013-0135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Marion C.M., Tsuchisaka A., Theologis A., Schafer E., Quail P.H. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell. 2007;19:3915–3929. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell. 2003;15:2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Lucyshyn D., Jaeger K.E., Alos E., Alvey E., Harberd N.P., Wigge P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Jung J.H., Cortes Llorca L., Kim S.G., Lee S., Baldwin I.T., Park C.M. FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat. Commun. 2014;5:5473. doi: 10.1038/ncomms6473. [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Lorrain S., Trevisan M., Pradervand S., Fankhauser C. Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 2009;60:449–461. doi: 10.1111/j.1365-313X.2009.03971.x. [DOI] [PubMed] [Google Scholar]
- Lucyshyn D., Wigge P.A. Plant development: PIF4 integrates diverse environmental signals. Curr. Biol. 2009;19:R265–266. doi: 10.1016/j.cub.2009.01.051. [DOI] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., Noel J.P., Liu H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA. 2016;113:224–229. doi: 10.1073/pnas.1511437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Nomoto Y., Oka H., Kitayama M., Takeuchi A., Tsubouchi M., Yamashino T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:958–976. doi: 10.1093/pcp/pcu030. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S.L., Tepperman J.M., Stanley D.J., Maltby D.A., Gross J.D., Burlingame A.L., Wang Z.Y., Quail P.H. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farre E.M., Kay S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Bai M.Y., Arenhart R.A., Sun Y., Wang Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012;72:537–546. doi: 10.1111/j.1365-313X.2012.05114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale U.V., Huang S.S., Zander M., Cole B.J., Hetzel J., Ljung K., Reis P.A., Sridevi P., Nito K., Nery J.R., et al. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell. 2016;164:233–245. doi: 10.1016/j.cell.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H. Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant. 2014;7:1598–1618. doi: 10.1093/mp/ssu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M., Delker C., Franklin K.A., Wigge P.A., Halliday K.J., Zanten M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants. 2016;2:15190. doi: 10.1038/nplants.2015.190. [DOI] [PubMed] [Google Scholar]
- Raschke A., Ibanez C., Ullrich K.K., Anwer M.U., Becker S., Glockner A., Trenner J., Denk K., Saal B., Sun X., et al. Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 2015;15:197. doi: 10.1186/s12870-015-0566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M.Y., Kim J., Paek N.C., Choi G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014;5:4636. doi: 10.1038/ncomms5636. [DOI] [PubMed] [Google Scholar]
- Seaton D.D., Smith R.W., Song Y.H., MacGregor D.R., Stewart K., Steel G., Foreman J., Penfield S., Imaizumi T., Millar A.J., et al. Linked circadian outputs control elongation growth and flowering in response to photoperiod and temperature. Mol. Syst. Biol. 2015;11:776. doi: 10.15252/msb.20145766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Khanna R., Carle C.M., Quail P.H. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Yang C., Gao S., Zhang W., Li L., Kuai B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant. 2014;7:1776–1787. doi: 10.1093/mp/ssu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang J.A., Gallego-Bartolome J., Gomez M.D., Yoshida S., Asami T., Olsen J.E., Garcia-Martinez J.L., Alabadi D., Blazquez M.A. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009;60:589–601. doi: 10.1111/j.1365-313X.2009.03983.x. [DOI] [PubMed] [Google Scholar]
- Sun T.P. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Zhai Q., Li C. PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell. 2013;25:2102–2114. doi: 10.1105/tpc.113.112417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Yamashino T., Matsushika A., Fujimori T., Sato S., Kato T., Tabata S., Mizuno T. A Link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:619–629. doi: 10.1093/pcp/pcg078. [DOI] [PubMed] [Google Scholar]
- Yamashino T., Nomoto Y., Lorrain S., Miyachi M., Ito S., Nakamichi N., Fankhauser C., Mizuno T. Verification at the protein level of the PIF4-mediated external coincidence model for the temperature-adaptive photoperiodic control of plant growth in Arabidopsis thaliana. Plant Signal. Behav. 2013;8:e23390. doi: 10.4161/psb.23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9:e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Jing Y., Jiang Z., Lin R. The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell. 2014;26:2472–2485. doi: 10.1105/tpc.113.121848. [DOI] [PMC free article] [PubMed] [Google Scholar]


