Abstract
Alkyl hydroperoxide reductase subunit C from Pseudomonas aeruginosa PAO1 (PaAhpC) is a member of the 2-Cys peroxiredoxin family. Here, we examined the peroxidase and molecular chaperone functions of PaAhpC using a site-directed mutagenesis approach by substitution of Ser and Thr residues with Cys at positions 78 and 105 located between two catalytic cysteines. Substitution of Ser with Cys at position 78 enhanced the chaperone activity of the mutant (S78C-PaAhpC) by approximately 9-fold compared with that of the wild-type protein (WT-PaAhpC). This increased activity may have been associated with the proportionate increase in the high-molecular-weight (HMW) fraction and enhanced hydrophobicity of S78C-PaAhpC. Homology modeling revealed that mutation of Ser78 to Cys78 resulted in a more compact decameric structure than that observed in WT-PaAhpC and decreased the atomic distance between the two neighboring sulfur atoms of Cys78 in the dimer-dimer interface of S78C-PaAhpC, which could be responsible for the enhanced hydrophobic interaction at the dimer-dimer interface. Furthermore, complementation assays showed that S78C-PaAhpC exhibited greatly improved the heat tolerance, resulting in enhanced survival under thermal stress. Thus, addition of Cys at position 78 in PaAhpC modulated the functional shifting of this protein from a peroxidase to a chaperone.
Keywords: 2-Cys peroxiredoxin, alkyl hydroperoxide reductase subunit C, chaperone, heat tolerance, Pseudomonas aeruginosa
INTRODUCTION
Pseudomonas aeruginosa possesses efficient machinery equipped with several components to counterbalance the potential risks of the oxidative stress during aerobic metabolism. Alkyl hydroperoxide reductase C (AhpC) is one of these components known to effectively detoxify H2O2 and maintain an optimal concentration of H2O2 to facilitate metabolic processes (Ochsner et al., 2000). Due to strong sequence homology, AhpC protein from P. aeruginosa PAO1 (PaAhpC) belongs to the typical 2-Cys peroxiredoxin (2-Cys Prx) family, and, in addition to H2O2, PaAhpC can also detoxify organic peroxides and peroxynitrite in cells (Parsonage et al., 2008). Like typical 2-Cys Prx, AhpC contains two conserved cysteines, the peroxidatic and resolving cysteines, which are essential for catalysis and regeneration, respectively. The peroxidatic cysteine (CysP) reacts with peroxide (ROOH) to yield the corresponding alcohol (ROH), or water in the case of H2O2, and cysteine sulfenic acid (CysP-SOH). The peroxidatic CysP-SOH is then reduced by the free thiol of the resolving cysteine (CysR) residue to form a disulfide bond between the two residues. After the catalytic cycle, AhpD or AhpF, in coordination with the cellular reductants NADPH or NADH, regenerate the reduced form of AhpC for further enzymatic activity (Bryk et al., 2002; Poole, 1996; Wood et al., 2003).
The active form of AhpC exists as a homodimer of two identical subunits arranged in a head to tail configuration, such that the CysP of one subunit is parallel to the CysR of the other subunit. Wood et al. (2003) reported that AhpC protein crystallizes as a dimer or decamer composed of five homodimers organized into a doughnut-shaped structure. In physiological solution, AhpC exists as a mixture of both dimers and decamers. The extent of decamer formation is influenced by the oxidation-reduction state of the protein. Reduction or overoxidation favors decamer formation, whereas a moderate level of oxidation induces conformational changes favoring dissociation to dimers (Parsonage et al., 2005; Wood et al., 2002).
The functional significance of 2-Cys Prx in all types of organisms is very well established (Hall et al., 2009; Tripathi et al., 2009). The capability of these proteins to perform multiple functions, e.g., peroxidase, chaperone, thiol oxidase, and signaling modulator, depending on the circumstances, has attracted a great deal of interest during recent years (Jang et al., 2004; König et al., 2013). As a typical 2-Cys Prx, AhpC is also known to perform peroxidase and molecular chaperone functions (Chuang et al., 2006). To the best of our knowledge, the mechanisms mediating the functional shifting of AhpC from a peroxidase to a chaperone, and vice versa, are unclear. While some studies have demonstrated the dual functionality and functional switching of AhpC from a peroxidase to a chaperone function in a gastrointestinal bacterium, Helicobacter pylori (Chuang et al., 2006; Huang et al., 2010), H. pylori AhpC (HpAhpC) is highly homologous to human peroxiredoxins and significantly different from bacterial AhpC (Huang et al., 2010).
As described above, PaAhpC possess two active cysteines, CysP and CysR, located at positions 47 and 166 in the N- and C-terminal regions, respectively. The cysteine-sulfenic acid intermediate formed after interaction with peroxides is susceptible to further oxidation to sulfinic acid and subsequently switches the function of the protein from a peroxidase to a chaperone due to conformational changes, altering the protein structure from a low-molecular-weight (LMW) structure to a high-molecular-weight (HMW) structure (König et al., 2013). The oligomerization of Prx monomers increases surface hydrophobicity, thereby enhancing the chaperone function of the protein. Oxidative stress or heat shock can trigger changes in the conformation of 2-Cys Prx protein, leading to functional switching from a peroxidase to a chaperone (Chuang et al., 2006; Jang et al., 2004; König et al., 2013; Lee et al., 2015). However, the molecular basis of this functional switching is still not completely understood. We recently determined the importance of an additional cysteine residue, distinct from the two conserved active cysteines in a typical 2-Cys Prx, involved in the functional switching of 2-Cys Prx proteins (An et al., 2015). Moreover, during alignment of amino acid sequences of PaAhpC with other AhpC in different bacterial species, we noticed the presence of two additional cysteines (Cys78 and Cys105) in Azospirillum brasilense located between the two active Cys residues of AhpC. In contrast, in PaAhpC, positions 78 and 105 are occupied by Ser and Thr, respectively.
Therefore, in the current study, we aimed to determine the roles of these two additional cysteines in modulating the dual functions of PaAhpC. For this purpose, two mutant PaAhpC proteins were generated by substituting Ser and Thr at positions 78 and 105 with Cys through site-directed mutagenesis and then evaluated the ability of the mutated PaAhpC to confer resistance to oxidative and heat stress in complemented E. coli. The structure and function of 2-Cys Prx of Pseudomonas species have already been well characterized and standardized in our laboratory (An et al., 2010; 2011; 2015), therefore, we have used PaAhpC to manipulate based on two additional cysteines in AhpC of A. brasilense. Our results provided important insights into the functional shifting of PaAhpC between its peroxidase and chaperone functions and the capacity for the mutant proteins to deal with abiotic stresses.
MATERIALS AND METHODS
Bacterial strains and growth
P. aeruginosa PAO1 and E. coli strains BL21 and KRX were cultured aerobically in Luria broth (LB) (0.5% sodium chloride, 0.5% yeast extract, and 1% tryptone; DB, Franklin Lakes, USA). The cultures were incubated at 30°C for P. aeruginosa PAO1 or 37°C for E. coli with continuous shaking. For cloning and overexpression studies, the transformed E. coli was grown in LB medium containing 50 μg ml−1 ampicillin.
Cloning of AhpC genes and protein expression in E. coli
The AhpC gene was cloned from P. aeruginosa PAO1 genomic DNA by polymerase chain reaction (PCR). Two mutants, S78C-PaAhpC and T105C-PaAhpC, were generated by substitution of Ser at position 78 and Thr at position 105 of WT-PaAhpC with Cys through PCR-mediated site-directed mutagenesis. Various AhpC DNA constructs were subcloned into the pRSET-A expression vector and transformed into E. coli strain BL21(DE3). The His6-fused PaAhpC proteins were purified using a Ni-NTA column (Peptron, Korea) and eluted with a linear gradient of 40 to 500 mM imidazole in phosphate-buffered saline (PBS) as described previously with slight modifications (Woo et al., 2014). The purified proteins were dialyzed into 50 mM HEPES buffer (pH 8.0) and used for biochemical analyses.
Assays for peroxidase and chaperone activities
The peroxidase activity of PaAhpC and its mutants was measured with 0.05 μM PaAhpF (already purified in our laboratory) and 150 μM of NADH as a reductant using methods described earlier (Nelson and Parsonage, 2011). The molecular chaperone activity was assessed by measuring the ability of PaAhpC proteins to inhibit the thermal aggregation of the substrate protein MDH (Jang et al., 2004). Briefly, 1 μM of MDH was mixed with various concentrations of PaAhpC proteins in a degassed 50 mM HEPES (pH 8.0) solution. The reaction mixture was incubated at 43°C for 15 min, and the increase in light scattering as a result of the thermal aggregation of substrate proteins was monitored at 340 nm with a DU 800 spectrophotometer equipped with a thermostatic cell holder (Beckman, USA).
CD spectroscopy
WT and mutant PaAhpC proteins in 10 mM sodium phosphate buffer (pH 7.4) were used for far UV-CD spectral analysis with a Jasco J-715 spectrophotometer (Jasco, UK), and the spectra were accumulated five times from independent experiments (Ito et al., 2001).
Fluorescence measurement
Exposure of the hydrophobic domains of PaAhpC proteins was determined by monitoring the binding of 10 μM bis-ANS with 100 μg protein in 50 mM HEPES buffer (pH 8.0) using an SFM25 spectrofluorometer (Kontron, Germany). The excitation wavelength was set at 380 nm, and the emission spectra were monitored from 400 to 600 nm (Sharma et al., 1998).
SEC and PAGE analyses
The recombinant proteins isolated from WT-PaAhpC and its mutants were further purified based on size using fast protein liquid chromatography (FPLC; AKTA, Amersham Biosciences, Sweden) with a Superdex 200 10/300 GL column (Amersham Biosciences Inc., USA) at 4°C, as described earlier (An et al., 2011). The column was equilibrated and run at a flow rate of 0.5 ml min−1 with 50 mM HEPES buffer (pH 8.0) containing 100 mM NaCl. Reducing SDS-PAGE, nonreducing SDS-PAGE, and native PAGE were performed as described previously (Moon et al., 2005).
Homology modeling
The protein sequence of PaAhpC was set as the query in the protein data bank, and PSI-BLAST was selected as the algorithm for the BLAST search. The sequence alignment was performed using Clustal W2 web-based software in EMBL-EBI (www.ebi.ac.uk). The Generate Homology Modeling protocol in Discovery Studio (DS) 3.5 (Accelrys Software Inc.; Accelrys, USA) was used for homology modeling of PaAhpC. After homology modeling, the stereochemical qualities of the homology models were validated using PROCHECK (Laskowski et al., 1993) and the ProSA-web server (Wiederstein and Sippl, 2007). Decameric structures of the two mutated proteins (S78C-PaAhpC and T105C-PaAhpC) were built from the decameric structure of WT-PaAhpC. The two mutant structures were generated by substituting each residue using the Build and Edit Protein protocol in DS 3.5. All structures (WT-PaAhpC, S78C-PaAhpC, and T105C-PaAhpC) were refined using the Standard Dynamics Cascade protocol in DS 3.5. For energy minimization, the steepest descent algorithm was used initially at 10 kcal (mol Å)−1 root mean square (RMS) energy gradient followed by the conjugate gradient algorithm until the RMS gradient converged to 0.01 kcal (mol Å)−1. The system was then heated from 50 to 300 K and equilibrated at 300 K for 1000 steps. To conduct distance analyses, 100-step conformations were selected from 1000 steps of production run with NVT Ensemble. The distance between the oxygen atoms in the side-chain of Ser78 was measured in WT-PaAhpC, whereas the distance between sulfur atoms in the side-chain of Cys78 was measured in S78C-PaAhpC. The average distances of residues at position 78 for five different interface regions and differences in distances between WT-PaAhpC and S78C-PaAhpC were calculated.
Complementation assay for oxidative stress and heat tolerance in WT-PaAhpC and mutant PaAhpC proteins
WT-PaAhpC and the two mutant PaAhpC constructs, inserted into the pRSET-A plasmid, were transformed individually into ΔAhpC E. coli, which is transposon insertion disruptant (JD20825) obtained from NBRP (National BioResource Project; www.nbrp.jp) of National Institute of Genetics. To test the expression levels of PaAhpC protein, we examined the immunoblot assay probed with a polyclonal PaAhpC antibodies using the transformed E. coli or P. aerusinosa with optical density (OD) of 0.9–1.0 at 600 nm.
For complementation assay, the transformed bacteria were grown aerobically in 5 ml LB medium supplemented with ampicillin (50 μg ml−1) and then transferred to 50 ml of fresh LB medium in a shaking incubator at 37°C. Bacterial cells from the optimally grown culture (having an OD of 0.5 at 600 nm) were harvested by centrifugation and concentrated until the OD reached 1.0 at 600 nm in fresh medium. The concentrated cells were serially diluted from 10−1 to 10−5. The diluted cells were poured on LB agar plates and incubated at 37°C for normal conditions or 50°C for thermal stress. In another set of experiments, the diluted cells were poured onto LB plates and subsequently treated with 0.4, 0.6, 0.8, or 1.0 mM H2O2 for the induction of oxidative stress, with incubation at 37°C (Gnanasekar et al., 2009).
RESULTS
Sequence comparison of PaAhpC with other homologous AhpC proteins
A comparison of the sequences of PaAhpC with homologous AhpC proteins of other microbes through multiple sequences alignment confirmed the presence of two conserved cysteine residues at positions 47 and 166 (Fig. 1). Further sequence alignment also showed the presence of additional cysteines at positions 78 and 105 in AbAhpC (Azosprillium brasiliense AhpC) and at position 78 in AsAhpC (Azosprilium sp. B510 AhpC). However, in PaAhpC, positions 78 and 105, which were located between the two active cysteines, were occupied by Ser and Thr, respectively (Fig. 1).
Fig. 1.
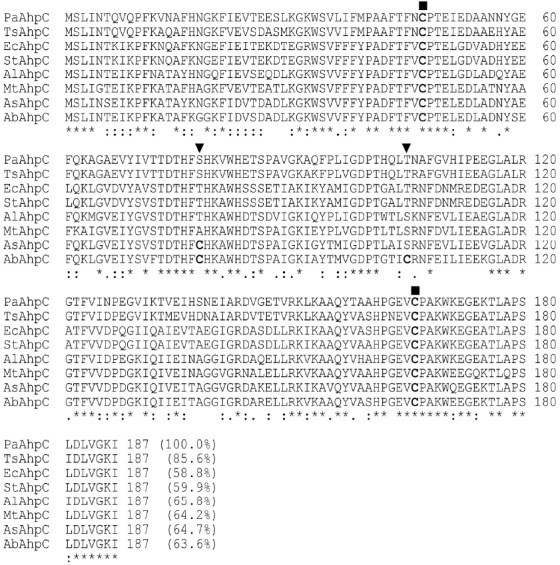
Alignment and sequence comparison of PaAhpC with AhpC proteins from other bacteria. The encoded amino acid sequences were aligned according to the two highly conserved active cysteines (Cys47 as the catalytic Cys and Cys166 as the resolving Cys), which are indicated by squares. The positions of the introduction of additional cysteine residues (Cys78 and Cys105) are indicated by a triangle. ‘*’ indicates positions which have a single, fully conserved residue. The percentage of the number of identical amino acids shared with PaAhpC is shown in parentheses at the end of the last lines. The abbreviations for the amino acid sequences of AhpC proteins from various species are as follows: Pseudomonas aeruginosa PAO1 (PaAhpC; NP_248829); Thauera sp. MZ1T (TsPrx; ACR02206); E. coli (EcAhpC; AJE54932); Salmonella enterica subsp. enterica serovar Typhimurium (StAhpC; AIE04547); Acinetobacter lwoffii SH145 Prx (AlPrx; EEY91350); Methylosinus trichosporium (MtPrx; WP_003609555); Azospirillum sp. B510 (AsAhpC; BAI75915); Azospirillum brasilense Sp245 (AbAhpC; AAY88877).
Dual functions of PaAhpC as a peroxidase and a molecular chaperone
PaAhpC showed the capacity for performing dual functions, e.g., peroxidase and molecular chaperone functions, in a protein concentration-dependent manner. Increasing concentrations of PaAhpC significantly reduced H2O2 content, complete degradation of H2O2 by 20 μM PaAhpC within 10 min (Fig. 2A), demonstrating the peroxidase activity of PaAhpC. Moreover, PaAhpC also exhibited chaperone activity, which was measured by monitoring the ability of the protein to suppress thermal aggregation of malate dehydrogenase (MDH) at 43°C. The aggregation of MDH was effectively inhibited at a 1:20 molar ratio of MDH to PaAhpC (Fig. 2B). The chaperone activity of PaAhpC was ∼2-fold lower than compared to that of PaPrx (An et al., 2010).
Fig. 2.
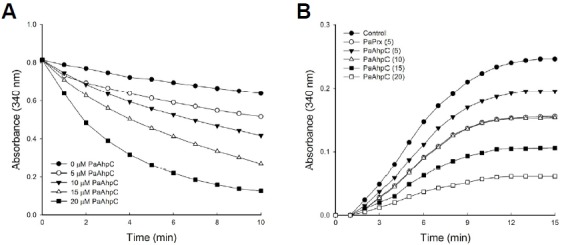
Peroxidase and chaperone activities in vitro. (A) In vitro peroxidase assay of PaAhpC with PaAhpF in the presence of 150 μM of H2O2 according to the methods of Nelson and Parsonage (Lee et al., 2007). (B) The molecular chaperone activity of PaAhpC protein. Chaperone activities of AhpC were measured at different molar ratios of MDH/PaAhpC or PaPrx: control, 1:0 (•); 1:5 AhpC (▾); 1:10 AhpC (▵); 1:15 AhpC (▪); 1:20 AhpC (□); 1:5 PaPrx (○).
Site-directed mutagenesis of PaAhpC enhances molecular chaperone activity but reduces peroxidase activity
Purification of wild-type PaAhpC (WT-PaAhpC) and two mutants (S78C-PaAhpC and T105C-PaAhpC) generated single major band (24 kDa) in reducing sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), indicating the high degree of purity obtained under reducing condition (Fig. 3A). In non-reducing SDS-PAGE, WT-PaAhpC formed a single major band of approximately 48 kDa, supporting the presence of a dimeric structure similar to that observed for other 2-CysPrx proteins (Chuang et al., 2006; Hall et al., 2009; Jang et al., 2004; König et al., 2013; Parsonage et al., 2005; Wood et al., 2002; 2003). In contrast, S78C-PaAhpC produced several high-molecular-weight (HMW) bands corresponding to an HMW structure, whereas T105C-PaAhpC showed a slightly lower band than WT-PaAhpC under non-reducing conditions (Fig. 3B). In native PAGE analysis, WT-PaAhpC and T105C-PaAhpC showed similar sizes, and S78C-PaAhpC generated relatively HMW bands compared with that of other proteins (Fig. 3C). The occurrence of HMW bands of S78C-PaAhpC in native gels was further supported by the results of size-exclusion chromatography (SEC; Fig. 3D). WT-PaAhpC and T105C-PaAhpC generated a single peak corresponding to the decameric form in SEC, whereas S78C-PaAhpC generated three peaks, including a prominent HMW peak, the decameric form, and the dimeric form in SEC (Fig. 3D).
Fig. 3.
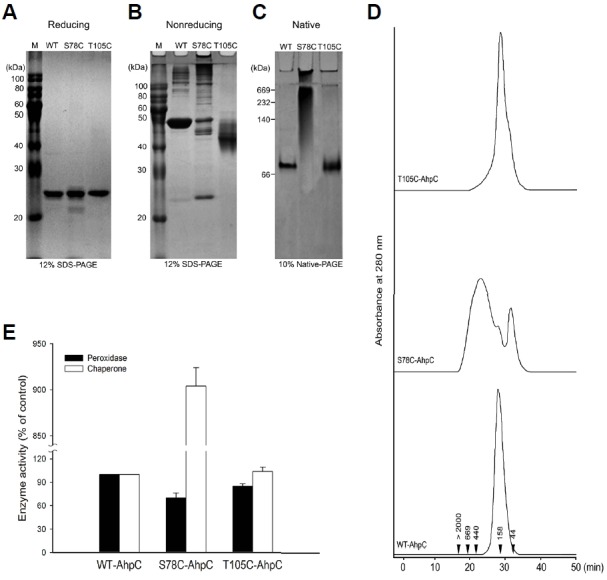
Effect of additional cysteine substitution on the structural and functional change in PaAhpC based on PAGE analysis under reducing (A), non-reducing (B), and native conditions (C) and based on SEC analysis (D). The numbers in the chromatogram represent the molecular weights of the standard proteins: blue dextran (> 2000 kDa), thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), and ovalbumin (44 kDa). (E) Comparison of the enzymatic activities of wild-type and mutant AhpC proteins. The relative activities of the two mutated PaAhpC proteins were compared with the activity of WT-PaAhpC. The peroxidase and chaperone activities of WT-AhpC were set to 100%. Data are the means of at least three independent experiments.
Next, the relative peroxidase and chaperone activities of the two mutants were compared with that of WT-PaAhpC (Fig. 3E). S78C-PaAhpC and T105C-PaAhpC showed approximately 30.0% and 15.0% lower peroxidase activity than WT-PaAhpC. However, S78C-PaAhpC exhibited approximately 9-fold higher chaperone activity than WT-PaAhpC. In contrast, the chaperone activity of T105C-PaAhpC was almost equal to that of WT-PaAhpC (Fig. 3E).
Influence of the S78C mutation on the secondary structure and hydrophobicity of PaAhpC
We evaluated the effects of the S78C and T105C substitutions on the secondary structure of the protein using UV-circular dichroism (CD) spectra analysis. The α-helix content was decreased from 42.0% to 30.0%, β-sheet content was increased from 26.0% to 38.0%, and random coil and turn contents remained almost unchanged in S78C-PaAhpC compared with those in WT-PaAhpC (Fig. 4A). In T105C-PaAhpC, β-sheet content was increased by about 10% compared with that of WT-PaAhpC. Previous report has suggested that chaperones bind to the non-native forms of protein substrates through hydrophobic interactions to protect target substrates from stress-induced aggregation (Mayer and Bukau, 2005). Indeed, the fluorescence intensity of bis-ANS was significantly higher for S78C-PaAhpC than for WT-PaAhpC and T105C-PaAhpC (Fig. 4B), suggesting that the S78C substitution greatly increased the exposure of hydrophobic domains, leading to polymerization of the protein into HMW complexes and providing binding sites for partially denatured substrate proteins during chaperone assays.
Fig. 4.
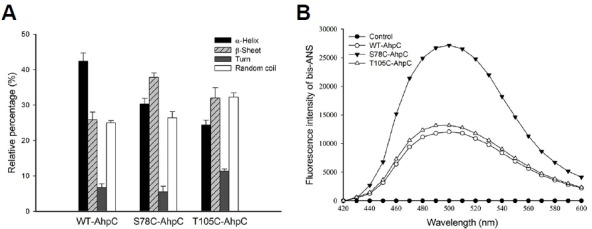
Changes in the secondary structures of WT and mutant PaAhpC proteins in vitro. (A) Comparison of the secondary structure index values based on the far UV-CD spectra results of the proteins. Data are the means of at least three independent experiments. (B) Changes in cysteine-mediated hydrophobicity of the WT and mutated PaAhpC proteins in vitro. The control was measured in the absence of PaAhpC protein.
Protein homology modeling revealed the basis of higher chaperone activity in S78C-PaAhpC
A homology model of PaAhpC was generated from AhpC of Salmonella enterica subsp. enterica serovar Typhimurium (PDB ID: 1N8J) to measure changes in the three-dimensional structures of mutant proteins. PROCHECK results showed that about 94.0% of residues from the generated model were present in the most favored regions, and a proper z-score value of −7.11 was obtained through ProSA-web (Fig. 5). After validation of the homology model, a decameric structure of PaAhpC was constructed using the template protein based on sequence alignment information. From the constructed decameric structure, two mutants (S78C-PaAhpC and T105C-PaAhpC) were generated by mutating each residue on the homology model. Thus, three different decameric structures were constructed and refined by standard dynamics cascade simulation. Based on the analysis of these decameric structures, the structural information of two mutation sites (Ser78 and Thr105) was predicted. Ser78 was located in an α2 helix facing another α2 helix of the neighboring dimer at the dimer-dimer interface. In contrast, the Thr105 residue was located on the inside surface of the ring, limiting its interaction with other residues (Figs. 6A and 6B).
Fig. 5.
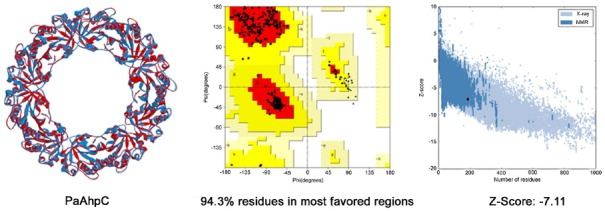
Overall structure and validation results of the homology model. Superimposition between WT-PaAhpC and the template is shown in the left panel (red: template, blue: homology model of PaAhpC). A Ramachandran plot is shown in the middle panel (red: most favored regions, yellow: additional allowed regions, light yellow: generously allowed regions). The z-score plot obtained from ProSA-web is shown in the right panel.
Fig. 6.
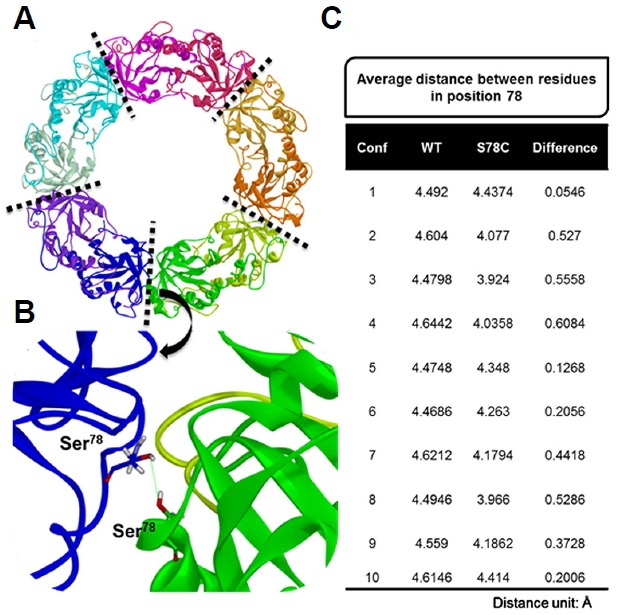
Results of distance analysis for residue 78 in WT-PaAhpC and S78C-PaAhpC. (A) Refined structure of PaAhpC obtained from standard dynamics cascade simulation. The five interface regions between the dimers in the decameric structure of PaAhpC are represented as dotted lines. Each monomer is shown in a different color. (B) Representative model of PaAhpC used to measure the distance for residues at position 78 in each monomer. The distance between oxygen atoms in the side-chain of Ser78 was measured in the WT, whereas the distance between sulfur atoms in the side-chain of Cys78 was measured in the S78C mutant. (C) Average distances of residues at position 78 for five different interface regions and differences in the distances between WT-PaAhpC and S78C-PaAhpC. The conformation (conf.) numbers represent the frame snapshots obtained from standard dynamics simulation for every 100 steps from 100 to 1000 steps.
We then calculated the distances between residues at position 78 located in different monomers for WT and S78C mutant systems. For the 10 different conformations generated from standard dynamics cascade simulation, the average distances of residues at position 78 were measured for five different interface regions, and the differences in distances between WT and S78C systems were then calculated (Fig. 6C). For all conformations, the average distance for the S78C-PaAhpC mutant was closer than that for WT-PaAhpC, suggesting that mutation of Ser78 to Cys created a more compact decameric structure or long filaments of stacked 2Cys-Prx rings. The increased compactness may be associated with the higher chaperone activity observed in the S78C mutant system.
The S78C substitution provided enhanced resistance against high-temperature stress in E. coli
The physiological function of mutant PaAhpC proteins against oxidative and heat stress was examined through the complementation assay in ΔAhpC E. coli mutant, transposon insertion disruptant. Production of the complemented PaAhpC proteins in ΔAhpC E. coli mutant was tested by immunoblot analysis probed with a polyclonal PaAhpC antiserum (Fig. 7A). The AhpC protein was not detected in ΔAhpC E. coli mutant. However, the complemented PaAhpC protein and its mutants (∼24 kDa) were highly expressed in ΔAhpC E. coli mutant and the intact PaAhpC protein (∼22 kDa) was detected in P. aerusinosa. The level of resistance against oxidative stress induced by various concentrations of H2O2 was similar in ΔAhpC E. coli complemented with WT-PaAhpC, S78C-PaAhpC, and T105C-PaAhpC (data not shown). However, S78C-PaAhpC-complemented ΔAhpC E. coli cells showed markedly higher survival rates during heat stress than those transformed with WT-PaAhpC or T105C-PaAhpC (Fig. 7B). S78C-PaAhpC-complemented cells exhibited even better survival than wild-type E. coli cells.
Fig. 7.
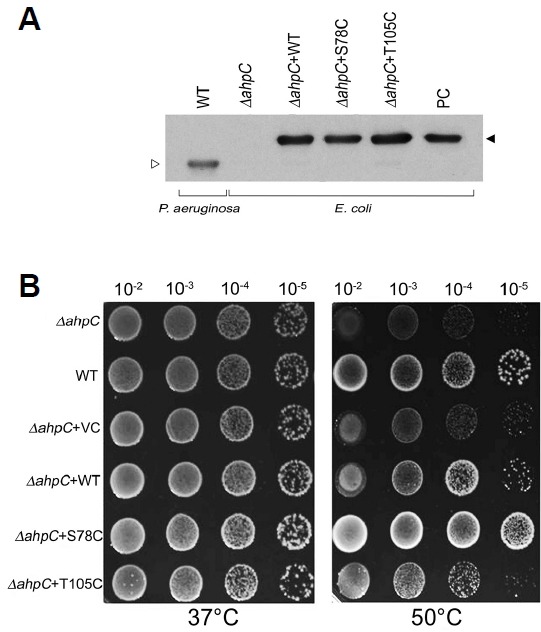
Sensitivity and complementation assay of mutants to heat. (A) Expression of PaAhpC protein in P. aerusinosa or ΔAhpC E. coli mutant. PC is positive control which is His-tagged recombinant PaAhpC protein; open triangle indicates the intact form of PaAphC; closed triangle is His-tagged PaAhpC. (B) Sensitivity of E. coli PaAhpC deletion mutants to heat and complementation by expression of AhpC homologous genes, PaAhpC and its derivatives.
DISCUSSION
In this study, we aimed to determine the role of protein structure and amino acid sequence in mediating the dual functions of PaAhpC. Our results provided important insights into the functional switching of PaAhpC between its peroxidase and chaperone functions and the capacity for the mutant proteins to support the bacterial cell to cope up with abiotic stresses.
Display of dual functions, e.g., peroxidase and chaperone functions, is a common feature of 2-Cys Prx proteins and has been reported in various organisms (Jang et al., 2004; König et al., 2013), including bacteria (An et al., 2010; 2011; Chuang et al., 2006). WT-PaAhpC exhibits AhpF-dependent peroxidase activity that depends directly on the protein concentration. The peroxidase activity of PaAhpC may be a reflection of the well-organized defense system of P. aeruginosa, which allows this organism to successfully colonize diverse habitats by mitigating the continuous threat of oxidants. Furthermore, the chaperone activity of PaAhpC allows P. aeruginosa to adapt to the continuously fluctuating environment. Thus, the functional shifting of this protein from a peroxidase to a chaperone is linked to oxidative and other environmental stresses (König et al., 2013). The mechanism underlying the structural/functional shifting of PaAhpC is thought to be associated with hyperoxidation of the partially oxidized peroxidatic Cys (sulfenic acid intermediate) after reacting with hydroperoxide to form sulfinic acid (An et al., 2011), thereby changing the conformation of the protein from an LMW peroxidase to an HMW chaperone (Jang et al., 2004). After the reaction with peroxide, the partially oxidized peroxidatic Cys is regenerated to its reduced form with the help of various reductants (Bhatt and Tripathi, 2011), such as AhpF in the case of PaAhpC. However, high concentrations of peroxides and lipid hydroperoxides, which occur due to severe oxidative stress, accelerate the hyperoxidation of 2-Cys Prx (Jang et al., 2004) and thereby induce functional switching from a peroxidase to a chaperone. Therefore, it is possible that functional switching of PaAhpC from a peroxidase to a molecular chaperone is a type of physiological adaptation of P. aeruginosa to prevent misfolding/unfolding of proteins or to recover the damaged proteins during severe stress conditions (Chuang et al., 2006). Development of heat resistance in P. aeruginosa due to the improved chaperone activity of PaAhpC, as shown in the present study, also supports this hypothesis.
The molecular chaperone activity of WT-PaAhpC was lower than that of other 2-Cys Prx proteins of other organisms. AhpC protein is originally a peroxidase by nature and it may shift to chaperone depending on the conditions. Therefore, lower molecular chaperone activity of WT-PaAhpC is quite understandable. Further, bacterial cells possess many other chaperones (Chuang et al., 2006), therefore display of chaperone function of a peroxidase protein seems unlikely unless required or triggered by conditions. Therefore, substitution of Ser78 with Cys greatly enhanced the chaperone activity of PaAhpC in the present study. Previous studies have also reported the enhanced chaperone activity of 2-Cys Prx proteins after site-specific mutagenesis (An et al., 2010; König et al., 2013). The improved chaperone activity of S78C-PaAhpC may be coupled with its increased β-sheet content and hydrophobicity, as shown in our current study. Changes in the secondary structure from α-helices to β-sheets and increased hydrophobicity have already been shown to be associated with enhanced chaperone activity in 2-Cys Prx proteins (Angelucci et al., 2013; Saccoccia et al., 2012). Additionally, native PAGE and SEC analyses also showed that the HMW structure was more common in the S78C-PaAhpC mutant than in WT-PaAhpC. The HMW fraction from SEC corresponded to low peroxidase activity and high chaperone activity. Consistent with this, Chuang et al. (2006) demonstrated structural/functional switching of HpAhpC from the LMW structure, with higher peroxidase function, to the HMW structure, with stronger molecular chaperone function, in H. pylori under oxidative stresses.
Homology modeling of WT-PaAhpC and mutated PaAhpC also explained the enhanced molecular chaperone activity of S78C-PaAhpC. The distance (O-O distance in WT-PaAhpC and S-S distance in S78C-PaAhpC) between the two Cys78 residues in neighboring dimers of the decameric structure was decreased in S78C-PaAhpC compared with that between the two Ser78 residues in WT-PaAhpC. The reduced molecular distances between the two neighboring dimers further favored strong interaction at the dimer-dimer interface, thereby leading to conversion of the LMW structure to the HMW structure with higher chaperone activity in S78C-PaAhpC. An earlier study on human PrxI also found that Cys83, which was present between the two conserved reactive cysteines, localized at the dimer-dimer interface and affected the balance between the LMW and HMW structures (Park et al., 2011). Similarly, in the case of Arabidopsis 2-Cys Prx, introduction of the F84R mutation in the dimer-dimer interface disturbs the hydrophobic interaction and yields exclusively the dimeric form of the Prx with a lower KM value for hydrogen peroxide (Huang et al., 2010). Tairum et al. (2012) suggested that the decameric structure of yeast PrxI is stabilized by van der Waals, hydrophobic, and polar interactions, primarily involving the amino acids at the dimeric interface. Furthermore, studies have also established the critical role of the intermediate cysteine of human PrxI at the dimer-dimer interface (Cys83, placed between the two active cysteines [Cys52 and Cys173] of human PrxI) in the regulation of oligomerization and chaperone activity through reversible glutathionylation (Park et al., 2011). Like a S78C-PaAhpC, where an additional Cys residue was inserted by replacing Ser78, human PrxI also possesses an additional cysteine between the two active cysteine residues and shows higher chaperone activity than human PrxII, which lacks this additional cysteine (Lee et al., 2007). Therefore, we hypothesize that the reduced distance between Cys78 residues in the dimer-dimer interface of S78C-PaAhpC enhances the hydrophobic interactions and/or van der Waals forces and facilitates the conversion of PaAhpC from the LMW structure to the HMW structure, which exhibits increased chaperone function.
Heat stress causes inactivation of cellular protein molecules, followed by proteolytic degradation. Thus, augmentation of the molecular chaperone function of AhpC may provide a survival advantage in organisms subjected to heat stress. Furthermore, PaAhpC-complemented ΔAhpC E. coli showed higher heat tolerance than wild-type E. coli and other variants. The higher heat tolerance in S78C-PaAhpC-complemented ΔAhpC E. coli compared with that in WT-PaAhpC- and T105C-PaAhpC-complemented cells could be explained by the enhanced chaperone activity of S78C-PaAhpC achieved after site-directed mutagenesis (S78C) of this protein. Such enhanced chaperone activity may play an instrumental role in preventing misfolding or unfolding of proteins and/or salvage of damaged proteins (Chuang et al., 2006), thereby conferring heat tolerance.
In this study, we demonstrated that PaAhpC exhibited dual functionalities as a peroxidase and a molecular chaperone. However, as a member of the 2-Cys Prx family of proteins and due to its intrinsic antioxidant properties, WT-PaAhpC preferred to act as a peroxidase rather than a molecular chaperone. Condition-dependent conformational changes from a LMW structure to HMW structures triggered the shift in PaAhpC function from a peroxidase to a chaperone. Furthermore, the ability of PaAhpC to perform chaperone activity was greatly improved by substituting Cys for Ser at position 78 (S78C-PaAhpC), located between the two conserved catalytic cysteines, using site-directed mutagenesis. S78C-PaAhpC, which exhibited enhanced chaperone activity, had markedly increased heat tolerance. Thus, our results indicated that the addition of a Cys residue at position 78 in PaAhpC modulated the functional shifting of this protein from a peroxidase to a chaperone. This feature may be exploited in the further development of heat-tolerant bacteria for industrial applications. Additional studies are required to further elucidate the mechanisms modulating the dual functions of PaAhpC and other 2-Cys Prx proteins to improve our understanding of the functional shifting from a peroxidase to a chaperone and enhancement of abiotic stress tolerance.
Acknowledgments
This project was supported by the Nuclear R&D Program of the Ministry of Science, ICT and Future Planning (MSIP), Republic of Korea and in part by the Next-Generation BioGreen 21 program (PJ008038) from Rural Development Administration (RDA) of Republic of Korea.
REFERENCES
- An B.C., Lee S.S., Lee E.M., Lee J.T., Wi S.G., Jung H.S., Park W., Chung B.Y. A new antioxidant with dual functions as a peroxidase and chaperone in Pseudomonas aeruginosa. Mol. Cells. 2010;29:145–151. doi: 10.1007/s10059-010-0023-1. [DOI] [PubMed] [Google Scholar]
- An B.C., Lee S.S., Lee E.M., Lee J.T., Wi S.G., Jung H.S., Park W., Lee S.Y., Chung B.Y. Functional switching of a novel prokaryotic 2-Cys peroxiredoxin (PpPrx) under oxidative stress. Cell Stress Chap. 2011;16:317–328. doi: 10.1007/s12192-010-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B.C., Lee S.S., Jung H.S., Kim J.Y., Lee Y., Lee K.W., Lee S.Y., Tripathi B. N., Chung B.Y. An additional cysteine in a typical 2-Cys peroxiredoxin of Pseudomonas promotes functional switching between peroxidase and molecular chaperone. FEBS Lett. 2015;589:2831–2840. doi: 10.1016/j.febslet.2015.07.046. [DOI] [PubMed] [Google Scholar]
- Angelucci F., Saccoccia F., Ardini M., Boumis G., Brunori M., Di Leandro L., Ippoliti R., Miele A.E., Natoli G., Scotti S., et al. Switching between the alternative structures and functions of a 2-Cys peroxiredoxin, by site-directed mutagenesis. J. Mol. Biol. 2013;425:4556–4568. doi: 10.1016/j.jmb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Bhatt I., Tripathi B.N. Plant peroxiredoxin: catalytic mechanisms, functional significance and future perspectives. Biotechnol. Adv. 2011;29:850–859. doi: 10.1016/j.biotechadv.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Bryk R., Lima C.D., Erdjument-Bromage H., Tempst P., Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- Chuang M.H., Wu M.S., Lo W.L., Lin J.T., Wong C.H., Chiou S.H. The antioxidant protein alkyl hydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc. Natl. Acad. Sci. USA. 2006;103:2552–2557. doi: 10.1073/pnas.0510770103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasekar M., Dakshinamoorthy G., Ramaswamy K. Translationally controlled tumor protein is a novel heat shock protein with chaperone-like activity. Biochem. Biophy. Res. Comm. 2009;386:333–337. doi: 10.1016/j.bbrc.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Karplus P.A., Poole L.B. Typical 2-Cys peroxiredions-structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.H., Chuang M.H., Wu Y.H., Chuang W.C., Jhuang P.J., Chiou S.H. Characterization of site-specific mutants of alkylhydroperxide reductase with dual functionality from Helicobacter pylori. J. Biochem. 2010;147:661–669. doi: 10.1093/jb/mvp209. [DOI] [PubMed] [Google Scholar]
- Ito H., Kamei K., Iwamoto I., Inaguma Y., Nohara D., Kato K. Phosphorylation-induced change of the oligomerization state of alpha B-crystallin. J. Biol. Chem. 2001;276:5346–5352. doi: 10.1074/jbc.M009004200. [DOI] [PubMed] [Google Scholar]
- Jang H.H., Lee K.O., Chi Y.H., Jung B.G., Park S.K., Park J.H., Lee J.R., Lee S.S., Moon J.C., Yun J.W., et al. Two enzymes in one, two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- König J., Galliardt H., Jütte P., Schäper S., Dittmann L., Dietz K.J. The conformational bases for the two functionalities of 2-cysteine peroxiredoxins as peroxidase and chaperone. J. Exp. Bot. 2013;64:3483–3497. doi: 10.1093/jxb/ert184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- Lee W., Choi K.S., Riddell J., Ip C., Ghosh D., Park J.H., Park Y.M. Human peroxiredoxin 1 and 2 are not duplicate proteins: the unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J. Biol. Chem. 2007;282:22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- Lee E.M., Lee S.S., Tripathi B.N., Jung H.S., Cao G.P., Lee Y., Singh S., Hong S.H., Lee K.W., Lee S.Y., et al. Site-directed mutagenesis substituting cysteine for serine in 2-Cys peroxiredoxin (2-Cys Prx A) of Arabidopsis thaliana effectively improves its peroxidase and chaperone functions. Ann. Bot. 2015;116:713–725. doi: 10.1093/aob/mcv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P., Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J.C., Hah Y.S., Kim W.Y., Jung B.G., Jang H.H., Lee J.R., Kim S.Y., Lee Y.M., Jeon M.K., Kim C.W., et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J. Biol. Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- Nelson K.J., Parsonage D. Measurement of peroxiredoxin activity. Curr. Protoc. Toxicol. 2011;49:7.10.1–7.10.28. doi: 10.1002/0471140856.tx0710s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U.A., Vasil M.L., Alsabbagh E., Parvatiyar K., Hassett D. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 2000;182:4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.W., Piszczek G., Rhee S.G., Chock P.B. Glutathionylation of peroxiredoxin induces decamer to dimers dissociation with concomitant loss of chaperone activity. Biochemistry. 2011;50:3204–3210. doi: 10.1021/bi101373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D., Youngblood D.S., Ganapathy N.S., Wood Z.A., Karpus A.P., Poole L.B. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D., Karplus P.A., Poole L.B. Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc. Natl. Acad. Sci. USA. 2008;105:8209–8214. doi: 10.1073/pnas.0708308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole L.B. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium 2. Cystine disulfides involved in catalysis of peroxide reduction. Biochemistry. 1996;35:65–75. doi: 10.1021/bi951888k. [DOI] [PubMed] [Google Scholar]
- Saccoccia F., Di Micco P., Boumis G., Brunori M., Koutris I., Miele A.E., Morea V., Sriratana P., Williams D.L., Bellelli A., et al. Moonlighting by different stressors: crystal structure of the chaperone species of a 2-Cys peroxiredoxin. Structure. 2012;20:429–439. doi: 10.1016/j.str.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K.K., Kaur H., Kumar G.S., Kester K. Interaction of 1,1′-bi(4-anilino) naphthalene-5,5′-disulfonic acid with alpha-crystallin. J. Biol. Chem. 1998;273:8965–8970. doi: 10.1074/jbc.273.15.8965. [DOI] [PubMed] [Google Scholar]
- Tairum C.A., de Oliveira M.A., Horta B.B., Zara F.J., Netto L.E.S. Disulfide biochemistry in 2-Cys peroxiredoxin: requirement of Glu50 and Arg146 for the reduction of yeast Tsa1 by thioredoxin. J. Mol. Biol. 2012;424:28–41. doi: 10.1016/j.jmb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Tripathi B.N., Bhatt I., Dietz K.J. Peroxiredoxins: a less studied component of hydrogen peroxide detoxification in photosynthetic organ’isms. Protoplasma. 2009;235:3–15. doi: 10.1007/s00709-009-0032-0. [DOI] [PubMed] [Google Scholar]
- Wiederstein M., Sippl M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:407–410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M.-H., Kim M.S., Chung N., Kim J.-S. Expression and characterization of a novel 2-deoxyribose-5-phosphate aldolase from Haemophilus influenzae Rd KW20. J. Korean Soc. Appl. Biol. Chem. 2014;57:655–660. [Google Scholar]
- Wood Z.A., Poole L.B., Hantgan R.R., Karpus A.P. Dimers to doughnut: redox sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5505. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- Wood Z.A., Schroder E., Robin H.J., Poole L.B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]


