Abstract
AIM
To investigate the effect of microRNA on insulin-like growth factor binding protein-3 (IGFBP-3) and hence on insulin-like growth factor-II (IGF-II) bioavailability in hepatocellular carcinoma (HCC).
METHODS
Bioinformatic analysis was performed using microrna.org, DIANA lab and Segal lab softwares. Total RNA was extracted from 23 HCC and 10 healthy liver tissues using mirVana miRNA Isolation Kit. microRNA-17-5p (miR-17-5p) expression was mimicked and antagonized in HuH-7 cell lines using HiPerFect Transfection Reagent, then total RNA was extracted using Biozol reagent then reverse transcribed into cDNA followed by quantification of miR-17-5p and IGFBP-3 expression using TaqMan real-time quantitative PCR. Luciferase reporter assay was performed to validate the binding of miR-17-5p to the 3’UTR of IGFBP-3. Free IGF-II protein was measured in transfected HuH-7 cells using IGF-II ELISA kit.
RESULTS
Bioinformatic analysis revealed IGFBP-3 as a potential target for miR-17-5p. Screening of miR-17-5p and IGFBP-3 revealed a moderate negative correlation in HCC patients, where miR-17-5p was extensively underexpressed in HCC tissues (P = 0.0012), while IGFBP-3 showed significant upregulation in the same set of patients (P = 0.0041) compared to healthy donors. Forcing miR-17-5p expression in HuH-7 cell lines showed a significant downregulation of IGFBP-3 mRNA expression (P = 0.0267) and a significant increase in free IGF-II protein (P = 0.0339) compared to mock untransfected cells using unpaired t-test. Luciferase assay validated IGFBP-3 as a direct target of miR-17-5p; luciferase activity was inhibited by 27.5% in cells co-transfected with miR-17-5p mimics and the construct harboring the wild-type binding region 2 of IGFBP-3 compared to cells transfected with this construct alone (P = 0.0474).
CONCLUSION
These data suggest that regulating IGF-II bioavailability and hence HCC progression can be achieved through targeting IGFBP-3 via manipulating the expression of miRNAs.
Keywords: Insulin-like growth factor binding protein-3, Insulin-like growth factor signaling pathway, MicroRNA, Insulin-like growth factor-II, Hepatocellular carcinoma
Core tip: microRNA-17-5p (miR-17-5p) was extensively underexpressed in hepatocellular carcinoma tissues, while insulin-like growth factor binding protein-3 (IGFBP-3) mRNA showed significant upregulation in the same set of patients. In HuH-7 cell line, miR-17-5p directly targets and downregulates IGFBP-3, consequently elevating the level of free insulin-like growth factor-II (IGF-II). Thus, manipulation of microRNAs can potentially control the activation of the oncogenic IGF axis.
INTRODUCTION
The insulin-like growth factor (IGF) signaling pathway is composed of IGF ligands, IGF receptors and insulin-like growth factor binding proteins (IGFBPs) which work in unison to regulate cell growth, differentiation, proliferation, and apoptosis. This axis is activated when the IGFs, IGF-I and IGF-II, bind to the insulin-like growth factor-1 receptor (IGF-1R) and activate a series of downstream signaling pathways controlling the cell cycle[1,2]. IGFBPs are transport proteins which bind to IGF-II with high affinity thereby prolonging their half-life and circulation turnover, and negatively regulate the activity of IGFs by controlling their binding to IGF receptors[3]. The levels of IGFBPs are modulated by various IGFBP proteases, such as matrix metalloproteinases (MMPs), which regulate the bioavailability and activity of IGFBPs, by mediating their proteolytic cleavage[4].
Multiple IGF axis members were found to play an important role in hepatocellular carcinoma (HCC) pathogenesis. IGF-II was found to be overexpressed in HCC and to promote tumor cell migration, proliferation and extra-hepatic metastasis[5-8]. Moreover, our research group has shown IGF-II to be overexpressed in peripheral blood monocytes of HCC patients, and this aberrant expression was directly correlated with elevated serum levels of alfa-fetoprotein and poor prognosis[9]. IGF-1R was reported to be upregulated in 59% of HCC tissues in which it was associated with poor prognosis and tumors exceeding the Milan criteria[10]. The tumorigenic effect of IGF-1R was reversed through its efficient blockage by combination of two IGF-1R antibodies which dramatically reduced liver tumor growth[11]. On the other hand, IGFBP-3 expression was found to be inversely correlated to HCC metastasis and proliferation[12,13].
The potential regulation of IGF axis members by microRNAs is an appealing subject of investigation. We have previously shown that miR-615-5p downregulates IGF-II expression and forcing its expression reduces tumorigenesis in HCC[14]. miR-122 was found to suppress IGF-1R expression thus inhibiting HCC progression[15,16]. Conversely, we have demonstrated that forcing the expression of the oncomiR miR-96 leads to the upregulation of IGF-1R and IGFBP-3 expression, while forcing the expression of the oncomiR-182 leads to the downregulation of IGF-1R and the upregulation of IGFBP-3 expression[17]. On the other hand, our research group reported that miR-155 induces the expression of IGF-II and IGF-1R and downregulates IGFBP-3 expression[18]. Nevertheless, the regulation of IGF-axis members by microRNAs still needs further investigation, particularly for the IGFBP-3. In silico analysis revealed IGFBP-3 as a potential downstream target for several microRNAs, one of which is microRNA-17-5p (miR-17-5p). This microRNA is an oncomiR that belongs to miR-17-92 cluster[19]. We have previously shown miR-17-5p to be significantly downregulated in non-metastatic HCC tissues compared to healthy tissues, where forcing its expression in HuH-7 cells resulted in enhancement of tumor cell growth, proliferation, migration, and colony-formation[20]. Therefore, this study aimed at identifying the impact of this important microRNA on IGFBP-3 expression, and consequently on the IGF-II bioavailability, and hence on HCC tumorigenesis.
MATERIALS AND METHODS
Bioinformatics
Bioinformatics algorithms microrna.org, DIANA Lab, and Segal lab were used to predict microRNAs that may target IGFBP-3.
Study subjects
This study included 23 HCC patients who underwent liver transplantation surgery in the Kasr Al Aini Hospital, Cairo University, Egypt. Ten healthy liver tissues were obtained from the healthy liver donors. Healthy donors were non-diabetic, non-hypertensive and negative for hepatitis B and C viruses (Table 1). The study was approved by the ethical review committees of the German University in Cairo and Cairo University, and is in accordance with the standards set by the Declaration of Helsinki. All participants gave their written informed consent. All patients were non-metastatic with no extrahepatic manifestations and no vascular invasion. Most of the patients (65.5%) had more than one focal lesion as indicated in the pathology report and were subjected to clinical assessment as shown in (Table 2).
Table 1.
Characteristic features of non-metastatic hepatocellular carcinoma patients and healthy controls
| Average ± SD | |
| HCC and cirrhotic patient parameters | |
| Mean age | 49 ± 13.5 |
| Sex: Male/female | 22/1 |
| Ethanol abuse | None |
| AST (U/L) | 100.5 ± 65.8 |
| ALT (U/L) | 85.6 ± 95.6 |
| Alkaline phosphatase (U/L) | 110.2 ± 60.7 |
| Serum albumin (g/dL) | 4.6 ± 1.5 |
| Serum AFP (ng/mL) | 155.7 ± 22.3 |
| HCV Ab | 100% (23/23 HCC patients) |
| HBV Ab | 17.3% (4/23 HCC patients) |
| Healthy control (liver donor) parameters | |
| Mean age | 31 ± 10.5 |
| Sex: Male/female | 7/3 |
| Ethanol abuse | None |
| HCV Ab | None |
| HBV Ab | None |
HCC: Hepatocellular carcinoma; HCV: Hepatitis C virus; AFP: Alpha fetal protein; HBV: Hepatitis B virus; AST: Aspartate aminotransferase; ALT: Alanine aminotransferase; SD: Standard deviation.
Table 2.
Number/sizes of focal lesions according to Milan criteria
| Patients | No. of focal lesions | Size of focal lesions (cm) |
| Patient 1 | 3 focal lesions | 1.5, 1 and 1 |
| Patient 2 | Unifocal | 2.5 |
| Patient 3 | 3 focal lesions | 2, 2.5 and 3 |
| Patient 4 | 3 focal lesions | 2, 2 and 3.5 |
| Patient 5 | Unifocal | 1.5-2 |
| Patient 6 | 3 focal lesions | 3-4, 1 and 1 |
| Patient 7 | Unifocal | 4 |
| Patient 8 | 3 focal lesions | 4, 1 and 1 |
| Patient 9 | 3 focal lesions | 1, 1 and 1.5 |
| Patient 10 | Unifocal | 2.5 |
| Patient 11 | 2 focal lesions | 1 and 1.7 |
| Patient 12 | 3 focal lesions | 1, 1 and 1 |
| Patient 13 | Unifocal | 3 |
| Patient 14 | 3 focal lesions | 3, 1.5 and 2 |
| Patient 15 | 3 focal lesions | 1, 1 and 4 |
| Patient 16 | 2 focal lesions | 3 and 1.5 |
| Patient 17 | 2 focal lesions | 1.5 and 3 |
| Patient 18 | 3 focal lesions | 2.5, 2.5 and 1.5 |
| Patient 19 | 3 focal lesions | 1.5, 1 and 1 |
| Patient 20 | Unifocal | 2 |
| Patient 21 | Unifocal | 1.5 |
| Patient 22 | 3 focal lesion | 3, 2.5 and 1 |
| Patient 23 | Unifocal | 3 |
Cell cultures and transfection of microRNA oligonucleotides
HuH-7 cells were maintained in Dulbecco’s modified Eagle’s medium (Lonza, Switzerland) supplemented with 4.5 g/L glucose, 4 mmol/L L-glutamine, 10% fetal bovine serum and Mycozap (1:500, Lonza, Switzerland) at 37 °C in 5% CO2 atmosphere. HuH-7 cells were transfected with mimics and inhibitors of miR-17-5p (Qiagen, Germany) (Qiagen ID: MSY0000070 and MIN0000070, respectively). All transfection experiments were carried out in triplicates using HiPerFect Transfection Reagent (Qiagen, Germany), according to the manufacturer’s protocol; the experiments were repeated three times. Cells that were only exposed to transfection reagent are designated as mock. Cells transfected with miR-17-5p mimics are designated as miR-17-5p, whereas cells transfected with miR-17-5p inhibitor are designated as anti-miR-17-5p.
mRNA and microRNA isolation from liver tissues and HCC cell lines
mRNAs and microRNAs were extracted from liver tissues and HCC cell lines. Fresh liver samples (HCC and healthy tissues) were collected during surgery and were immediately snapfrozen in liquid nitrogen. The specimens were manually pulverized in liquid nitrogen, and about 100 mg of tissues powder were used for large and small RNA extraction using mirVana miRNA Isolation Kit (Ambion, United States), according to the manufacturer’s protocol. HCC cell lines were harvested 48 h after transfection according to HiPerFect Transfection Reagent protocol and total RNA was extracted using Biozol Reagent (Bioer Technology, China).
miRNA and mRNA quantification
The extracted microRNAs were reverse transcribed into single stranded complementary DNA (cDNA) using TaqMan MicroRNA Reverse Transcription Kit (ABI, United States) and specific primers for has-miR-17-5p and RNU6B. mRNA was reverse transcribed into cDNA using the high-capacity cDNA reverse transcription kit (ABI, United States) according to the manufacturer’s instructions. Relative expression of miR-17-5p and RNU6B (for normalization) as well as IGFBP-3 and beta-2 microglobulin (B2M; as housekeeping gene for normalization) was quantified using TaqMan Real-Time quantitative PCR (ABI Assay IDs: 002308, 001093, Hs00365742_g1 and Hs00984230_m1, respectively) using StepOne™ Systems (ABI, United States). Relative expression was calculated using the 2–ΔΔCT method. All PCR reactions including controls were run in duplicate reactions.
IGFBP-3 3’UTR construct and luciferase assay
The two predicted target sites for miR-17-5p on IGFBP-3 3’UTR were each designed as sticky ended oligonucleotides flanked by Sac I and Xba I restriction sites, and ligated into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Germany) to form the two wild-type (WT) constructs. Also, two mutant constructs (MUT) were designed where 3 nucleotides from the binding region had been deleted from each site. The first target site is denoted as WT1 and its mutant form as MUT1; the second target site is WT2 and its mutant form is MUT2. The forward and reverse primer sequences for each construct are as shown in (Table 3). HuH-7 cells were seeded in 24-well plates and either WT or MUT constructs were transfected by lipofection technique using SuperFect Transfection Reagent (Qiagen, Germany). The following day, the cells were co-transfected with miR-17-5p mimics using HiPerFect according to the protocol (Qiagen). After 48 h, luciferase assay was performed using Steady-GLO Luciferase Reporter System (Promega, Germany) according to the manufacturer’s protocol. After 5 min, luminescence was measured at 545 nm. Luciferase experiments were done in triplicates.
Table 3.
The forward and reverse primer sequences used in the wild type 1 and 2, and the mutant type 1 and 2 insulin-like growth factor binding protein-3 3’UTR constructs
| Primer name | Primer sequence |
| WT1 forward | 5’-CAATGGTAAACTTGAGCATCTTTTCACTTTCCAGTAGT-3’ |
| WT1 reverse | 5’-CTAGACTACTGGAAAGTGAAAAGATGCTCAAGTTTACCATTGAGCT-3’ |
| WT2 forward | 5’-CGTCGAAGCGGCCGACCACTGACTTTGTGACTTT-3’ |
| WT2 reverse | 5’-CTAGAAAGTCACAAAGTCAGTGGTCGGCCGCTTCGACGAGCT-3’ |
| MUT1 forward | 5’-CAATGGTAAACTTGAGCATCTTTTCATCCAGTAGT3’ |
| MUT1 reverse | 5’-CTAGACTACTGGATGAAAAGATGCTCAAGTTTACCATTGAGCT-3’ |
| MUT2 forward | 5’-CGTCGAAGCGGCCGACCACTGACGTGACTTT-3’ |
| MUT2 reverse | 5’-CTAGAAAGTCACGTCAGTGGTCGGCCGCTTCGACGAGCT-3’ |
WT: Wild type; MUT: Mutant type.
Quantitative detection of free IGFII protein
Free IGFII protein was measured in the cell culture supernatant from miR-17-5p mimicked, miR-17-5p antagonized, and mock untransfected HuH-7 cells, using the human IGFII ELISA kit (CUSABIO, China), according to the manufacturer’s instructions. Absorbance was measured at 450 nm in a microplate reader.
Statistical analysis
miRNA and gene expression data analysis was performed according to the 2-ΔΔCT method. An assessment of the normality of data was done as a prerequisite for all the statistical tests to identify the correct statistical methods to analyze our data with. We used Shapiro Wilks test since the size of the sample is less than 50. The normality test for miR-17-5p and IGFBP-3 screening experiments of “Healthy controls” and “HCC patients” showed that the dependent variable, “RQ”, isn’t normally distributed since the significant value of the Shapiro Wilks test is less than 0.05, so the data significantly deviate from a normal distribution, with an exception in the data obtained from IGFBP-3 expression in the healthy controls were found to be normally distributed. In view of this fact the statistical significance of the data was analyzed by performing the non-parametric Mann-Whitney test. The degree of the relationship between linear related variables was measured by the Pearson r correlation test. The normality test for the transfection and binding confirmation experiments showed that the data are normally distributed; therefore the parametric unpaired t-test was used. The specific types of tests, when applicable, are indicated in the figure legends. All data are presented as mean ± standard error of the mean (SEM). All tests were 2-tailed and a two-tailed P value < 0.05 was required for statistical significance. All the data were statistically analyzed using GraphPad Prism 5 software.
The statistical methods of this study were reviewed by Dr. Nihal Aly Etman, Department of Statistics, Mathematics and Insurance, Faculty of Commerce, Ain Shams University.
RESULTS
Bioinformatics
miR-17-5p accession number and mature sequence were retrieved using miRBase database (http://www.mirbase.org/). In silico predictions were carried out using three different softwares, and results showed IGFBP-3 to be a potential downstream target to miR-17-5p, where the microRNA was predicted to bind to the 3’UTR of IGFBP-3 at two different regions. The interactions between miR-17-5p seed sequence and its target sequence on the 3’UTR of IGFBP-3 are as shown in (Table 4). Where, the seed sequence of miR-17-5p is shown in bold and italic, while the target sequence of the 3’UTR of IGFBP-3 is underlined. The lines indicate complementarity between the binding region of the mRNA and the seed sequence of the microRNA, while the dots indicate mismatches or GU wobbles.
Table 4.
Predicted target region-seed sequence binding for miR-17-5p on the 3’UTR of insulin-like growth factor binding protein-3
| Target region | hsa-miR-17-5p (seed sequence) binding to IGFBP-3 (target sequence) | Target sequence position on 3’UTR of IGFBP-3 | 6mer/7mer/8mer |
| Region 1 | miR-17-5p 3’gaUGGACGUG-ACAUUCGUGAAAc 5’ | | : | | : | | | | | | | | | IGFBP-3 5’aaACUUGAGCAUCUUUUCACUUUc 3’ | 196-204 | 6mer |
| Region 2 | miR-17-5p 3’GAUGGAC- GUGACAUUCGUGAAAC 5’ | ||__ | | || |___ | | | | || IGFBP-3 5’ CGGCCGACCACUG-----------ACUUUG 3’ | 335-343 | 6mer |
IGFBP-3: Insulin-like growth factor binding protein-3; miR-17-5p: MicroRNA-17-5p.
Expression profile of miR-17-5p and IGFBP-3 in non-metastatic HCC liver tissues
Expression of miR-17-5p in non-metastatic HCC tissues (n = 23) (0.318 ± 0.109) was significantly lower compared to healthy tissues (n = 10) (3.488 ± 1.267, P = 0.0012; Figure 1A). On the other hand, the expression of IGFBP-3 in the same non-metastatic HCC tissues (5.913 ± 1.294) was significantly higher compared to healthy tissues (1.352 ± 0.272, P = 0.0041; Figure 1B).
Figure 1.
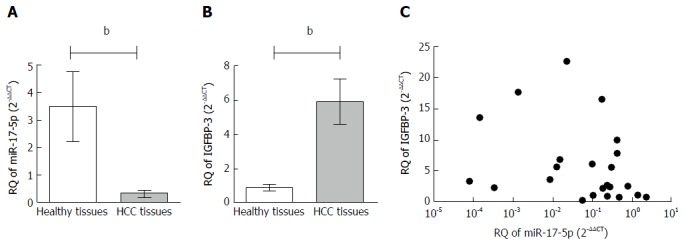
Expression profile of microRNA-17-5p and insulin-like growth factor binding protein-3 and their correlation in liver tissues. The expression of miR-17-5p and IGFBP-3 were investigated in 10 healthy and 23 HCC liver tissues using TaqMan qRT-PCR and normalized in each sample to RNU6B endogenous control for miR-17-5p and B2M for IGFBP-3. A: miR-17-5p expression was down-regulated in non-metastatic HCC patients compared to healthy liver tissues (P = 0.0012); B: Regarding IGFBP-3, its mRNA expression showed a significant higher expression in HCC tissues compared to healthy tissues (P = 0.0041). Statistical analysis was performed using the Mann-Whitney test; C: Relative quantitation (RQ) values of miR-17-5p and IGFBP-3 mRNA in HCC tissues were analyzed using Pearson’s method of correlation. A non-significant inverse correlation was found with Pearson’s r = -0.3244 (P = 0.1310). bP < 0.01. HCC: Hepatocellular carcinoma; IGFBP-3: Insulin-like growth factor binding protein-3; miR-17-5p: MicroRNA-17-5p; qRT-PCR: Real-time quantitative PCR.
Correlation analysis between miR-17-5p and IGFBP-3 mRNA expression in HCC tissues
IGFBP-3 mRNA was quantified in all HCC tissues and correlated to miR-17-5p expression in the same patients. Using Pearson’s statistical method of correlation, miR-17-5p expression was found to be moderately inversely correlated but not statistically significant with IGFBP-3 transcript levels in all HCC tissues studied (r = -0.3244, P = 0.1310; Figure 1C).
Impact of miR-17-5p on IGFBP-3 mRNA in HuH-7 cells
HuH-7 cells were transfected with miR-17-5p mimics and transfection efficiency was achieved with an observed 250 fold increase (P = 0.0470) in miR-17-5p levels in transfected cells (266.6 ± 113.2) compared to their respective untransfected mock cells (1.069 ± 0.1927) (Figure 2A). Mimicking of miR-17-5p in HuH-7 resulted in a significant downregulation of IGFBP-3 mRNA levels (0.6527 ± 0.1021) compared to mock untransfected cells (1.069 ± 0.1502, P = 0.0267). Conversely, inhibitors of miR-17-5p in HuH-7 cells showed a tendency of increase compared to mock untransfected HuH-7 cell lines (Figure 2B).
Figure 2.
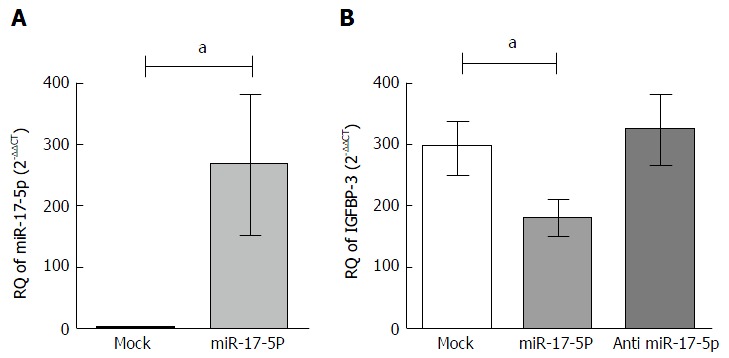
Impact of microRNA-17-5p on insulin-like growth factor binding protein-3 mRNA expression in HuH-7 cell line. A: The expression of miR-17-5p was determined by TaqMan qRT-PCR in HuH-7 cells transfected with oligonucleotide mimics of miR-17-5p, 48 h post-transfection, relative to their expression in mock untransfected HuH-7 cells. The expression of miR-17-5p was normalized to RNU6B endogenous control. A: Transfection of miR-17-5p mimics increased miR-17-5p levels in HuH-7 by 250 fold compared to mock cells (P = 0.0470). Unpaired t-test was performed; B: HuH-7 cells were transfected with miR-17-5p mimics or inhibitors, and the relative expression of IGFBP-3 was determined using TaqMan qRT-PCR, relative to mock untransfected cells, and gene expression was normalized to endogenous control B2M. IGFBP-3 mRNA expression was dramatically suppressed upon mimicking of miR-17-5p compared to mock cells (P = 0.0267), while inhibitors of miR-17-5p showed a tendency of increase compared to mock cells. Unpaired t-test was performed. aP < 0.05. IGFBP-3: Insulin-like growth factor binding protein-3; miR-17-5p: MicroRNA-17-5p; qRT-PCR: Real-time quantitative PCR.
Impact of miR-17-5p on free IGF-II protein in HuH-7 cells
In miR-17-5p mimicked HuH-7 cells, there was a significant upregulation in the amount of the free IGF-II protein (1.045 ± 0.05255) compared to mock untransfected HuH-7 cells (0.8344 ± 0.06783, P = 0.0339). Antagonizing the expression of miR-17-5p had no effect on the amount of the free IGF-II protein compared to the mock HuH-7 cells (Figure 3).
Figure 3.
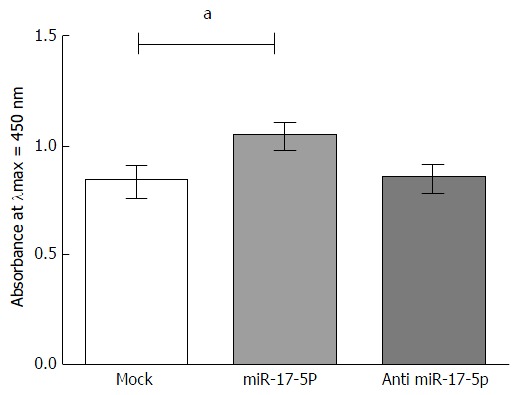
Impact of microRNA-17-5p on free insulin-like growth factor-II protein in HuH-7 cells. HuH-7 cells were transfected with miR-17-5p mimics or inhibitors. The free IGF-II protein was measured in media of mimicked and antagonized HuH-7 cells using an IGF-II ELISA Kit. Free IGF-II protein, measured at λmax = 450, was found to be significantly increased upon mimicking of miR-17-5p expression compared to mock untransfected cells (P = 0.0339), while inhibitors of miR-17-5p showed no effect on the levels of free IGF-II protein levels compared to mock cells. Unpaired t-test was performed. aP < 0.05. IGF-II: Insulin-like growth factor-II; miR-17-5p: MicroRNA-17-5p.
Confirming IGFBP-3 as a direct target of miR-17-5p
To confirm that miR-17-5p directly targets the 3’UTR of IGFBP-3, wild-type constructs (WT1 and WT2) were designed where each of the two predicted 3’UTR target regions were inserted downstream to a luciferase reporter gene in pmiRGLO vector. To assess that the effects were due to specific binding to these binding regions, a mutant construct for each binding site was also prepared in which 3 base pairs were deleted from the predicted binding sequence in the 3’UTR of IGFBP-3, to form mutant constructs MUT1 and MUT2, respectively. Also, in a set of cells, empty pmiRGLO vector was transfected as a control to ensure that miR-17-5p mimics have no effect on the vector itself. For each binding region, experiments were performed by transfecting HuH-7 cells with either the construct containing the WT 3’UTR binding region of IGFBP-3, or the construct containing the MUT 3’UTR binding region. Then miR-17-5p mimics were co-transfected with the vectors or constructs and luciferase reporter activity was assessed. In cells transfected with WT1 construct, luciferase activity was not affected upon co-transfection with miR-17-5p mimics (Figure 4A). On the other hand, luciferase activity was inhibited by 27.5% in cells co-transfected with miR-17-5p mimics and WT2 construct (72.48 ± 2.383) compared to cells transfected with the WT2 construct alone (100.0 ± 9.432, P = 0.0474) (Figure 4B). In contrast, in cells transfected with either MUT1 or MUT2, no change in luciferase activity was observed upon mimicking with miR-17-5p (Figure 4). The inhibition in the luciferase activity observed only in the WT2 construct indicates direct targeting and transcriptional inhibition of IGFBP-3 by miR-17-5p mimics through only one of the two predicted target regions.
Figure 4.
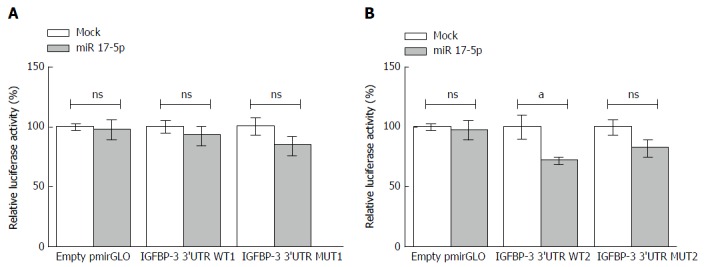
Insulin-like growth factor binding protein-3 is a direct target of miR-17-5p. For each target sequence, experiments were performed by transfecting HuH-7 cells with either empty pmiRGLO vector, or the construct with the wild-type (WT) miR-17-5p target region insert, or the construct with the mutant (MUT) miR-17-5p target region insert. Then miR-17-5p mimics were co-transfected with the vectors or constructs. A: Luciferase activity was not affected in cells co-transfected with miR-17-5p mimics and WT1 construct compared to cells transfected with the WT1 construct alone; B: On the other hand, luciferase activity was inhibited by 27.5%, in cells co-transfected with miR-17-5p mimics and WT2 construct compared to cells transfected with the WT2 construct alone (P = 0.0474). The cells transfected with either of the mutant constructs (MUT1 or MUT2) show no change in the luciferase activity upon mimicking with miR-17-5p. Unpaired t-test was performed. aP < 0.05. NS: Not significant; IGFBP-3: Insulin-like growth factor binding protein-3; miR 17-5p: MicroRNA-17-5p.
DISCUSSION
The regulation of IGFBP-3 by microRNAs has not been extensively studied but recently our research group showed that the oncomiR miR-155 represses IGFBP-3 expression in HCC cell lines[18]. In addition, we showed an increased expression of IGFBP-3 upon forcing the expression of miR-96 and miR-182[17]. To the best of our knowledge the IGF-II bioavailability has never been investigated after targeting IGFBPs with microRNAs, therefore in this study, we aimed at identifying a new microRNA which could regulate the IGFBP-3 and consequently the IGF-II bioavailability, and hence influence HCC tumorigenesis. In silico analysis revealed IGFBP-3 as a potential downstream target for miR-17-5p (Table 4), a microRNA which we have previously shown to have oncogenic properties in HCC[20].
No correlation analysis was previously done between miR-17-5p and IGFBP-3 expression in HCC patients, therefore non-metastatic liver tissues of HCC patients were screened for that purpose. miR-17-5p was markedly downregulated (Figure 1A) while IGFBP-3 was significantly upregulated (Figure 1B) in the non-metastatic liver tissues of HCC patients compared to healthy controls. This goes in line with previous studies showing IGFBP-3 to be highly expressed in breast and esophageal cancer[21,22]. But on the other hand, it contradicts other studies that reported reduced IGFBP-3 mRNA expression and protein levels in metastatic HCC patients[12,13]. Moreover, the repression of miR-17-5p in HCC tissues (Figure 1A) corroborates our previous results that showed a significant downregulation of miR-17-5p expression in non-metasatic HCC patients[20], but nonetheless it contradicts other studies in metastatic HCC tissues[23]. These disparities can, however, be attributed to differences in the cohorts of patients included in the various studies, with regards to stage and etiology of the disease as well as other factors such as ethnicity, gender and age. Of note, the results of the correlation analysis revealed a moderate negative correlation between miR-17-5p and IGFBP-3 expression in HCC patients (Figure 1C), suggesting that IGFBP-3, as predicted by in silico analysis, may in fact be under the posttranscriptional regulation of miR-17-5p.
In order to investigate the effect of miR-17-5p on IGFBP-3, transfection experiments were performed by forcing miR-17-5p expression in HuH-7 cell lines and the expression of IGFBP-3 mRNA was assessed, where it was found that upon forcing miR-17-5p expression in HuH-7 cells, there was a significant downregulation in IGFBP-3 expression (Figure 2B). This finding further implies that miR-17-5p may target and regulate IGFBP-3 expression. As revealed by in silico analysis, the 3’UTR of the IGFBP-3 transcript contains two exclusive putative binding sites for miR-17-5p. In order to validate IGFBP-3 as a direct downstream target of miR-17-5p, a WT and a MUT luciferase reporter gene construct was prepared for each binding region on the 3’UTR of IGFBP-3. Using these microRNA-target expression constructs, it was demonstrated that forcing the expression of miR-17-5p significantly decreased luciferase activity only in the construct harboring the WT2 binding region of the 3’UTR of IGFBP-3 target gene (Figure 4). This interesting finding indicates that only one of the two putative binding sites is in fact functionally active and that miR-17-5p effectively targets and inhibits the transcription of IGFBP-3 by directly associating with this specific target region. This unusual observation has also been found in colon cancer where bioinformatic tools predicted two target sites on the oncogene Friend leukemia virus integration 1 (Fli-1) for the tumor suppressor miR-145; however, upon measuring the luciferase activity only the construct harboring one of these two predicted target sites of Fli-1 showed a decrease in luciferase activity by more than 50% upon miR-145 mimicking, while the other construct harboring the second target site did not respond to miR-145[24].
Since IGFBP-3 is a crucial negative regulator of the bioavailability of IGF-II, therefore the levels of free IGF-II protein were quantified in the media of miR-17-5p mimicked and mock untransfected HuH-7 cells. The results showed a significant increase in unbound IGF-II in miR-17-5p mimicked HuH-7 cells compared to mock untransfected cells (Figure 3). This in turn confirms that miR-17-5p regulates IGF-II bioavailability through direct targeting of IGFBP-3. In this regard, the biological function of miR-17-5p appears to simulate the effect of another regulator of the IGF pathway, the MMPs, whose overexpression leads to the decrease in IGFBP-3 and subsequent increase in IGF-II bioavailability[25].
In conclusion, the findings of this study shed light on the important role of the oncogenic miR-17-5p in hepatocarcinogenesis, where this microRNA was found to increase IGF-II bioavailability by directly targeting and repressing IGFBP-3 expression. Hence, manipulating microRNA expression might be a compelling potential therapeutic approach in preventing HCC progression.
ACKNOWLEDGMENTS
The authors would like to thank the English instructor, Ms. Gilan Hamdi, and the English Department at the German University in Cairo for revising the manuscript. We would like to thank Ms. Nihal Etman, from the Department of Statistics, Mathematics and Insurance at Ain Shams University, Cairo, Egypt for reviewing the statistics of the manuscript.
COMMENTS
Background
Insulin-like growth factor-II (IGF-II) is a major activator of the oncogenic IGF axis, often overexpressed in hepatocellular carcinoma leading to the promotion of tumor cell migration, proliferation and metastasis. IGF-II protein bioavailability is controlled by a class of insulin-like growth factor binding proteins (IGFBPs) 1-6 which regulate the binding of IGF-II to its receptor, IGF-1 receptor. Very few studies have investigated the regulation of IGFBPs by microRNAs.
Research frontiers
Recently, microRNAs have entered the first clinical trials investigating their therapeutic potential in primary liver cancer.
Innovations and breakthroughs
This is the first study to investigate the effect of a microRNA on an IGFBP and consequently on the IGF-II bioavailability.
Applications
microRNA-17-5p affected IGFBP-3 and consequently the level of free IGF-II which could allow for the activation of the oncogenic IGF axis. This suggests that microRNAs can be manipulated to regulate the activation of this axis.
Terminology
microRNAs are approximately 22 nucleotide long single stranded, small, non-coding RNA sequences that post-transcriptionally regulate gene expression by binding to the 3’UTR of their target mRNA, suppressing its translation or causing its degradation.
Peer-review
The study is well planned involves proving of a concept by bioinformatics tools and then confirming in vitro and patient’s tissues.
Footnotes
Institutional review board statement: The study was approved by the ethical review committees of the German University in Cairo and Cairo University.
Informed consent statement: All liver biopsy specimens from patients and healthy donors were taken after informed consent and ethical permission was obtained for participation in the study.
Conflict-of-interest statement: To the best of our knowledge, no conflict of interest exists.
Data sharing statement: All liver biopsy specimens from patients and healthy donors were taken after informed consent was obtained for participation in the study. Any clinical data stated in the manuscript is anonymous.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Egypt
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Peer-review started: March 7, 2016
First decision: May 17, 2016
Article in press: July 13, 2016
P- Reviewer: Patial V S- Editor: Gong ZM L- Editor: A E- Editor: Li D
References
- 1.Zha J, Lackner MR. Targeting the insulin-like growth factor receptor-1R pathway for cancer therapy. Clin Cancer Res. 2010;16:2512–2517. doi: 10.1158/1078-0432.CCR-09-2232. [DOI] [PubMed] [Google Scholar]
- 2.Blundell TL, Bedarkar S, Rinderknecht E, Humbel RE. Insulin-like growth factor: a model for tertiary structure accounting for immunoreactivity and receptor binding. Proc Natl Acad Sci USA. 1978;75:180–184. doi: 10.1073/pnas.75.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 4.Rajah R, Katz L, Nunn S, Solberg P, Beers T, Cohen P. Insulin-like growth factor binding protein (IGFBP) proteases: functional regulators of cell growth. Prog Growth Factor Res. 1995;6:273–284. doi: 10.1016/0955-2235(95)00012-7. [DOI] [PubMed] [Google Scholar]
- 5.Nussbaum T, Samarin J, Ehemann V, Bissinger M, Ryschich E, Khamidjanov A, Yu X, Gretz N, Schirmacher P, Breuhahn K. Autocrine insulin-like growth factor-II stimulation of tumor cell migration is a progression step in human hepatocarcinogenesis. Hepatology. 2008;48:146–156. doi: 10.1002/hep.22297. [DOI] [PubMed] [Google Scholar]
- 6.Qian J, Yao D, Dong Z, Wu W, Qiu L, Yao N, Li S, Bian Y, Wang Z, Shi G. Characteristics of hepatic igf-ii expression and monitored levels of circulating igf-ii mRNA in metastasis of hepatocellular carcinoma. Am J Clin Pathol. 2010;134:799–806. doi: 10.1309/AJCPTFDSE2V3LCZP. [DOI] [PubMed] [Google Scholar]
- 7.Breuhahn K, Longerich T, Schirmacher P. Dysregulation of growth factor signaling in human hepatocellular carcinoma. Oncogene. 2006;25:3787–3800. doi: 10.1038/sj.onc.1209556. [DOI] [PubMed] [Google Scholar]
- 8.Couvert P, Carrié A, Pariès J, Vaysse J, Miroglio A, Kerjean A, Nahon P, Chelly J, Trinchet JC, Beaugrand M, et al. Liver insulin-like growth factor 2 methylation in hepatitis C virus cirrhosis and further occurrence of hepatocellular carcinoma. World J Gastroenterol. 2008;14:5419–5427. doi: 10.3748/wjg.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Tayebi HM, Salah W, El Sayed IH, Salam EM, Zekri AR, Zayed N, Salem ES, Esmat G, Abdelaziz AI. Expression of insulin-like growth factor-II, matrix metalloproteinases, and their tissue inhibitors as predictive markers in the peripheral blood of HCC patients. Biomarkers. 2011;16:346–354. doi: 10.3109/1354750X.2011.573095. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Jiang W, Aucejo F, Kim R, Miller C, Byrne M, Lopez R, Yerian L. Insulin-like growth factor I receptor β expression in hepatocellular carcinoma. Hum Pathol. 2011;42:882–891. doi: 10.1016/j.humpath.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Dong J, Demarest SJ, Sereno A, Tamraz S, Langley E, Doern A, Snipas T, Perron K, Joseph I, Glaser SM, et al. Combination of two insulin-like growth factor-I receptor inhibitory antibodies targeting distinct epitopes leads to an enhanced antitumor response. Mol Cancer Ther. 2010;9:2593–2604. doi: 10.1158/1535-7163.MCT-09-1018. [DOI] [PubMed] [Google Scholar]
- 12.Luo SM, Tan WM, Deng WX, Zhuang SM, Luo JW. Expression of albumin, IGF-1, IGFBP-3 in tumor tissues and adjacent non-tumor tissues of hepatocellular carcinoma patients with cirrhosis. World J Gastroenterol. 2005;11:4272–4276. doi: 10.3748/wjg.v11.i27.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huynh H, Chow PK, Ooi LL, Soo KC. A possible role for insulin-like growth factor-binding protein-3 autocrine/paracrine loops in controlling hepatocellular carcinoma cell proliferation. Cell Growth Differ. 2002;13:115–122. [PubMed] [Google Scholar]
- 14.El Tayebi HM, Hosny KA, Esmat G, Breuhahn K, Abdelaziz AI. miR-615-5p is restrictedly expressed in cirrhotic and cancerous liver tissues and its overexpression alleviates the tumorigenic effects in hepatocellular carcinoma. FEBS Lett. 2012;586:3309–3316. doi: 10.1016/j.febslet.2012.06.054. [DOI] [PubMed] [Google Scholar]
- 15.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Zeng C, Wang R, Li D, Lin XJ, Wei QK, Yuan Y, Wang Q, Chen W, Zhuang SM. A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology. 2010;52:1702–1712. doi: 10.1002/hep.23875. [DOI] [PubMed] [Google Scholar]
- 17.Assal RA, El Tayebi HM, Hosny KA, Esmat G, Abdelaziz AI. A pleiotropic effect of the single clustered hepatic metastamiRs miR-96-5p and miR-182-5p on insulin-like growth factor II, insulin-like growth factor-1 receptor and insulin-like growth factor-binding protein-3 in hepatocellular carcinoma. Mol Med Rep. 2015;12:645–650. doi: 10.3892/mmr.2015.3382. [DOI] [PubMed] [Google Scholar]
- 18.El Tayebi HM, Waly AA, Assal RA, Hosny KA, Esmat G, Abdelaziz AI. Transcriptional activation of the IGF-II/IGF-1R axis and inhibition of IGFBP-3 by miR-155 in hepatocellular carcinoma. Oncol Lett. 2015;10:3206–3212. doi: 10.3892/ol.2015.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Tayebi HM, Omar K, Hegy S, El Maghrabi M, El Brolosy M, Hosny KA, Esmat G, Abdelaziz AI. Repression of miR-17-5p with elevated expression of E2F-1 and c-MYC in non-metastatic hepatocellular carcinoma and enhancement of cell growth upon reversing this expression pattern. Biochem Biophys Res Commun. 2013;434:421–427. doi: 10.1016/j.bbrc.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Figueroa JA, Jackson JG, McGuire WL, Krywicki RF, Yee D. Expression of insulin-like growth factor binding proteins in human breast cancer correlates with estrogen receptor status. J Cell Biochem. 1993;52:196–205. doi: 10.1002/jcb.240520211. [DOI] [PubMed] [Google Scholar]
- 22.Takaoka M, Harada H, Andl CD, Oyama K, Naomoto Y, Dempsey KL, Klein-Szanto AJ, El-Deiry WS, Grimberg A, Nakagawa H. Epidermal growth factor receptor regulates aberrant expression of insulin-like growth factor-binding protein 3. Cancer Res. 2004;64:7711–7723. doi: 10.1158/0008-5472.CAN-04-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Jiang M, Yuan W, Tang H. miR-17-5p as a novel prognostic marker for hepatocellular carcinoma. J Invest Surg. 2012;25:156–161. doi: 10.3109/08941939.2011.618523. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Guo H, Zhang H, Wang H, Qian G, Fan X, Hoffman AR, Hu JF, Ge S. Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene Friend leukemia virus integration 1 gene. Cancer. 2011;117:86–95. doi: 10.1002/cncr.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YM, Bae MH, Lee OH, Moon EJ, Moon CK, Kim WH, Kim KW. Synergistic induction of in vivo angiogenesis by the combination of insulin-like growth factor-II and epidermal growth factor. Oncol Rep. 2004;12:843–848. [PubMed] [Google Scholar]


