Abstract
Mutations in FGD1 cause Aarskog–Scott syndrome (AAS), an X-linked condition characterized by abnormal facial, skeletal, and genital development due to abnormal embryonic morphogenesis and skeletal formation. Here we report a novel FGD1 mutation in a family with atypical features of AAS, specifically bilateral upper and lower limb congenital joint contractures and cardiac abnormalities. The male proband and his affected maternal uncle are hemizygous for the novel FGD1 mutation p.Arg921X. This variant is the most carboxy-terminal FGD1 mutation identified in a family with AAS and is predicted to truncate the FGD1 protein at the second to last amino acid of the carboxy-terminal pleckstrin homology (PH) domain. Our study emphasizes the importance of the 3′ peptide sequence in the structure and/or function of the FGD1 protein and further demonstrates the need to screen patients with X-linked congenital joint contractures for FGD1 mutations.
Keywords: ankyloglossia; bilateral cryptorchidism; clinodactyly of the fifth finger; congenital foot contractures; decreased palmar creases; deep philtrum; downslanted palpebral fissures; flexion contracture; low-set, posteriorly rotated ears; metatarsus adductus; nonrestrictive ventricular septal defect; shawl scrotum; short nose; unilateral ptosis
INTRODUCTION
Aarskog–Scott syndrome (AAS or faciogenital dysplasia; MIM# 305400) is an X-linked recessive condition typically characterized by facial dysmorphology and skeletal and genital anomalies that result from mutations in FGD1 (Pasteris et al. 1994). Males affected with AAS typically present with dysmorphic facial features, genital hypoplasia, short stature, and brachydactyly (Orrico et al. 2010); however, a broad spectrum of clinical heterogeneity has been reported in patients with a molecular diagnosis of AAS, including intellectual disability, unilateral focal polymicrogyria, and myopathy and distal arthropathy (Lebel et al. 2002; Bottani et al. 2007; Al-Semari et al. 2013). These atypical features may prohibit clinical recognition of AAS, leading to a failure to screen for FGD1 mutations and a delayed diagnosis for patients and families (Orrico et al. 2010).
To date, 63 mutations in FGD1 have been reported in AAS (Orrico et al. 2014; Pérez-Coria et al. 2015). The FGD1 gene contains 18 exons that code for a specific guanine nucleotide exchange factor (GEF) for the Rho protein CDC42 that is critical for embryogenesis (Gorski et al. 2000). The 961-amino-acid FGD1 protein has five discrete domains, including a proline-rich domain, an SH3-binding domain, a Rho GEF homology domain, two PH domains, and a cysteine-rich Zn2+-finger domain. Missense, frameshift, nonsense, splice site, branch point, and large and small in-frame deletion mutations have been reported to cause AAS (Orrico et al. 2014). Interestingly, these mutations are randomly distributed throughout the protein's functional domains, indicating a loss-of-function mechanism of pathogenesis in AAS through the disruption of GEF catalytic activity and/or protein localization of FGD1 (Orrico et al. 2000, 2004, 2010; Schwartz et al. 2000; Bedoyan et al. 2009).
Here, we report the identification of a novel truncating mutation in FGD1 via exome sequencing of genomic DNA from a family that presented with X-linked distal joint contractures, cardiac defects, and mild craniofacial and genital manifestations typical of AAS. This report illustrates the importance of screening boys with distal skeletal abnormalities and a family history suggesting X-linked inheritance for FGD1 mutations.
RESULTS
Clinical Presentation and Family History
The proband (Individual IV-1) (see Fig. 2A) was observed to have bilateral clubfeet at 20 wk gestation via ultrasound. In early infancy, the Ponseti method was used to correct the clubfeet with serial casting, followed by bilateral Achilles tenotomy, and corrective shoes with a foot abduction orthosis (Ponseti et al. 2006). Additionally, it was noted that he had difficulty opening his hands because of flexion contractures of the fingers and was treated with hand splints and occupational therapy that improved his range of motion.
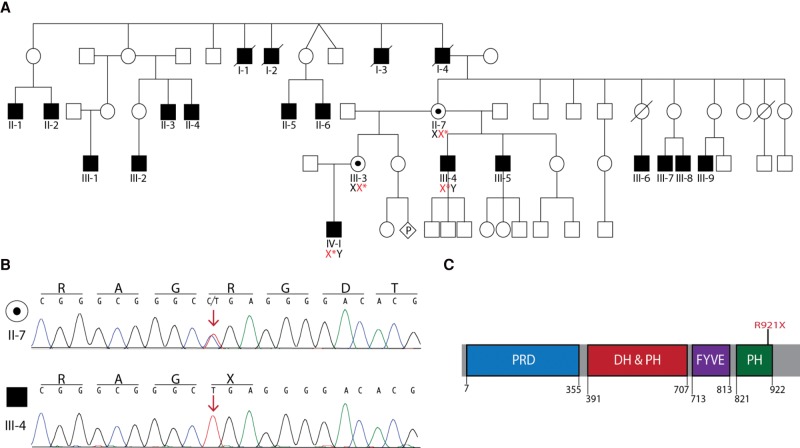
Figure 2.
Identification of a FGD1 mutation that segregates with phenotypic characteristics of Aarskog–Scott syndrome (AAS). (A) Pedigree of the affected family. DNA samples from the proband (IV-1) and his maternal uncle (III-4) were used for whole-exome sequencing. Sanger sequencing of Individuals IV-1 and III-4 confirmed the presence of the FGD1 mutation c.2761 C>T (red X*) in affected individuals and determined carrier status for Individuals II-7 and III-3. Empty symbols indicate unaffected individuals, filled symbols indicate affected individuals, and black dots indicate unaffected mutation carriers. Diamond-shaped symbol with a “P” represents unknown sex of a fetus in utero and slashes through symbols indicate deceased individuals. (B) Representative traces from Sanger sequencing confirming the presence of the c.2761 C>T mutation in an unaffected carrier female (II-7) and an affected male (III-4). Red arrow marks the affected nucleotide. The single-letter abbreviations for the nucleotides and corresponding amino acid codes are provided along the top of each trace. (C) Protein cartoon illustrates the known FGD1 functional domains in blue (proline-rich domain; PRD), red (DBL homology domain and pleckstrin homology domain; DH and PH), purple (FYVE zinc finger domain; FYVE), and green (pleckstrin homology domain; PH). The numbers indicate the amino acid positions for each domain. The position of the FGD1 R921X affected residue is indicated in red.
Upon physical exam, the proband was observed to have some mild craniofacial dysmorphic features including a space between the upper teeth (diastema), ankyloglossia, a round face, down-slanted palpebral fissures with ptosis of the left eyelid, a short, upturned nose with a well-formed philtrum, and small, low-set ears with overfolded helices (Fig. 1A). Additionally, an ophthalmologic exam revealed farsightedness and astigmatism bilaterally, a genital exam revealed a mild shawl scrotum and bilateral undescended testes, and a Still's murmur was appreciated on physical exam (data not shown).
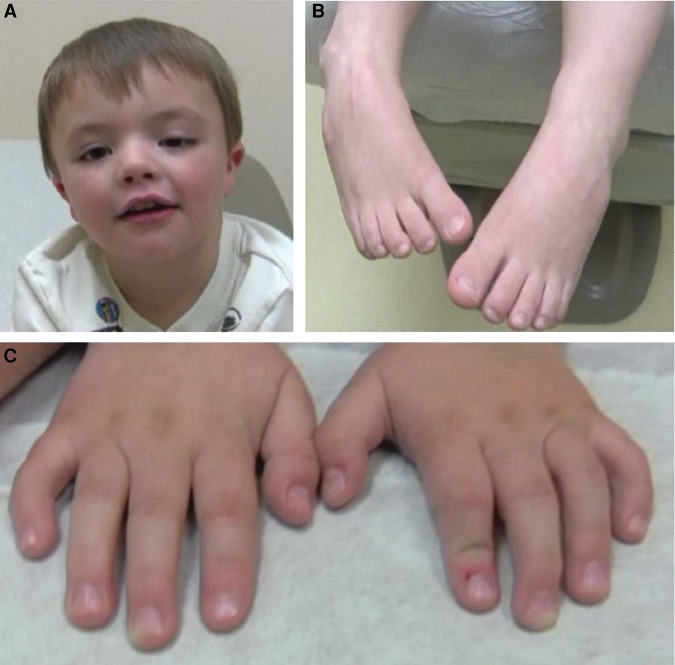
Figure 1.
Phenotypic characteristics of the proband include (A) mild craniofacial dysmorphisms including round face, down-slanted palpebral fissures, ptosis of the left eyelid, short, upturned nose, and small, low-set ears, (B) bilateral metatarsus adductus (partially corrected clubfeet), and (C) mild flexion contractures of the fingers, brachydactyly, and clinodactyly.
His feet at most recent exam have reduced creases on the plantar surfaces and residual metatarsus adductus (Fig. 1B). The individual's hands showed a single palmar crease on the right and a transitional crease on the left with mild flexion contractures of the fingers, brachydactyly, and bilateral clinodactyly (Fig. 1C).
There was a strong family history of craniofacial abnormalities, hand contractures, and clubfeet that segregated in an X-linked pattern (Fig. 2A). The proband has two maternal half uncles with similar features, including craniofacial dysmorphisms and hand contractures (Individuals III-4 and III-5 in Fig. 2A). One uncle also had bilateral clubfeet (III-4 in Fig. 2A), whereas the other uncle has a ventricular septal defect (III-5) (Fig. 2A). In addition, multiple affected male relatives were reported in at least two earlier generations, and two individuals were diagnosed with supraventricular tachycardia in addition to the observed skeletal abnormalities (Table 1). Taken together, this family represented a multigenerational pedigree with a prominent X-linked skeletal syndrome characterized by distal joint contractures and craniofacial abnormalities with the occurrence of cardiac abnormalities in multiple affected individuals.
Table 1.
Phenotypes reported for affected individuals
| Individual | Facial gestalta | Hand contractures | Clubfeet | Cardiac abnormalities | Adult height |
|---|---|---|---|---|---|
| I-1 | Yes | Yes | NA | NA | NA |
| I-2 | Yes | Yes | NA | NA | NA |
| I-3 | Yes | Yes | NA | NA | NA |
| I-4 | Yes | Yes | NA | NA | NA |
| II-1 | Yes | Yes | NA | NA | NA |
| II-2 | Yes | Yes | NA | NA | NA |
| II-3 | Yes | Yes | NA | SVT | NA |
| II-4 | Yes | Yes | NA | NA | NA |
| II-5 | Yes | Yes | NA | NA | NA |
| II-6 | Yes | Yes | NA | NA | NA |
| III-1 | Yes | Yes | NA | NA | NA |
| III-2 | Yes | Yes | NA | SVT | NA |
| III-4 | Yes | Yes | Yes | No | 5′8′′ |
| III-5 | Yes | Yes | No | VSD | 5′9′′ |
| III-6 | Yes | Yes | NA | NA | 5′8′′ |
| III-7 | Yes | Yes | NA | NA | 5′8′′ |
| III-8 | Yes | Yes | NA | NA | 5′8′′ |
| III-9 | Yes | Yes | NA | NA | 5′8′′ |
| IV-1 | Yes | Yes | Yes | Murmur | - |
NA, information not available; SVT, supraventricular tachycardia; VSD, ventricular septal defect.
aIncludes widow's peak, round face, ptosis, down-slanting palpebral fissures, hypertelorism, and anteverted nares. Specific features are not available for each individual except as noted in the text.
Genomic Analysis
Because of the X-linked inheritance pattern, our analysis focused on variants identified on the X Chromosome. To identify variants of interest, whole-exome sequencing was performed on DNA from Individuals III-4 and IV-1. Initial bioinformatic analysis identified three putative nonsynonymous single-nucleotide variants common between the two DNA samples evaluated. Sanger sequencing determined that only one variant was indeed present in both affected individuals and ruled out the other two variants as sequencing errors. Both affected individuals were found to be hemizygous for a nonsense mutation in exon 18 of FGD1 (c.2761 C>T; Table 2; Fig. 2B) (forward 5′-CTGAAGACAGAGCATATGC-3′, reverse 5′-CAAGTATTGACTGAGCTGG-3′). This mutation results in a substitution of a stop codon for an arginine at amino acid residue 921 (p.Arg921X) and is predicted to result in a 40-amino-acid truncation of the gene product (Fig. 2C). Targeted Sanger sequencing revealed that the mutation is carried by both the proband's mother and maternal grandmother (Fig. 2A; Individual II-7 and III-3, respectively). No additional family members were available for segregation studies. Identification of the FGD1 p.Arg921X mutation provided a molecular diagnosis of AAS in this family.
Table 2.
FGD1 (NC_000023.11)
| Chr: position GRCh37 (hg19) | HGVS cDNA | HGVS protein reference | HGVS protein | Variant type | Predicted effect | Genotype |
|---|---|---|---|---|---|---|
| X: 54472667 | c.2761C>T | NP_004454.2 | p.Arg921X | Nonsense | Aarskog–Scott syndrome | Hemizygous |
HGVS, Human Genome Variation Society.
DISCUSSION
Here we describe a novel FGD1 mutation in a large pedigree with X-linked distal joint contractures, cardiac abnormalities, and craniofacial and genital abnormalities consistent with a molecular and clinical diagnosis of AAS. AAS is typically diagnosed based on clinical features, specifically the presence of craniofacial and genital abnormalities in conjunction with short stature and brachydactyly. However, in cases where the presenting phenotypic characteristics are atypical of AAS, such as the congenital joint contractures and clubfeet described in this report, a diagnosis of AAS should be considered and molecular techniques should be used for proper diagnosis.
Previously, Bottani et al. (2007) and Al-Semari et al. (2013) described three individuals from two families with a molecular diagnosis of AAS who presented with atypical arthropathic features (Bottani et al. 2007; Al-Semari et al. 2013). The case presented here reinforces the need to strongly consider screening for FGD1 mutations and additional phenotypic characteristics of AAS in male patients presenting with either X-linked or de novo distal joint contractures or clubfeet. Careful evaluations for more subtle features of AAS and targeted molecular testing in these cases would prevent unnecessary clinical and diagnostic testing for families, and identification of novel FGD1 mutations may improve the understanding of the clinical and molecular spectrum of AAS. Similar to what was observed in members of the family reported here, previous reports have shown the occurrence of cardiac abnormalities in patients with FGD1 mutations, specifically ventricular septal defect (Fernandez et al. 1994; Nouraei et al. 2005; Orrico et al. 2007), which was identified in Individual III-4 (Fig. 2A) in this study. Additionally, supraventricular tachycardia (SVT) was observed in two individuals, which to our knowledge, has not been previously associated with AAS; however, we cannot confirm that the SVT is a component of AAS in this family and not an unrelated abnormality. Still, this further illustrates the need for careful cardiac evaluations in patients with FGD1 mutations.
Identification of novel FGD1 mutations has led to important insights into FGD1 function and the mechanism of AAS pathogenesis. Mutations in FGD1 have been identified in all known functional domains of the FGD1 protein, indicating a loss-of-function mechanism. A complete gene deletion as well as multiple nucleotide insertions that are predicted to result in premature translation termination have been identified in the FGD1 gene in individuals with AAS (Orrico et al. 2004; Bedoyan et al. 2009), indicating that complete loss of FGD1 in hemizygous individuals leads to the AAS phenotype. The mutation described here (p.Arg921X) resides 181 nucleotides into the final exon of FGD1. Therefore, the transcript is not predicted to undergo nonsense-mediated decay (NMD) and thus would be expected to produce a truncated protein (Amrani et al. 2006); however, it is important to note that NMD has been reported when a nonsense mutation occurred in the last exon of the Type X collagen gene COL10A1 (Chan et al. 1998). Thus protein studies would be needed to ensure the corresponding protein product of the p.Arg921X FGD1 mutation is indeed present. FGD1 contains five domains: a proline-rich, SH3-binding domain (residues 7–330); a Rho GEF (DH or DBL) homology domain; a PH domain; a cysteine-rich Zn2+ finger (FYVE) domain; and a second PH domain (Fig. 2C). Functional studies of the FGD1 protein indicated that the amino terminus of FGD1 is responsible for localization to the subcortical actin cytoskeleton and Golgi complex, whereas the carboxy-terminal PH domain interacts with phosphorylated phosphatidylinositol (PtdIns) molecules in the plasma membrane (Estrada et al. 2001). Although the carboxy-terminal 40 amino acids of FGD1 are not part of a known functional motif, their exclusion from the protein likely results in a detrimental effect on FGD1 function, as evidenced by the phenotype seen in this family. Thus, it will be important to evaluate how the carboxy-terminal 40-amino-acid residues affect protein folding and/or the function of FGD1. It should be noted that there is one nonsense mutation (p.Glu938X) listed in the ExAC database for FGD1 that would affect the carboxy-terminal domain of FGD1. This variant was identified as a single allele and is noted in the ExAC database to be a low-confidence variant because of coverage in <80% of sequenced individuals; thus, it is difficult to exclude sequencing errors for the p.Glu938X variant. Additionally, there is no phenotypic data available for the individual carrying p.Glu938X FGD1, including gender or evaluation for subtle AAS characteristics. Thus, the pathogenicity of this allele cannot be determined at this time. However, if p.Glu938X is a nonpathogenic allele, it would serve as an excellent control for studies on the carboxy-terminal region of FGD1.
In conclusion, we report a novel FGD1 mutation, p.Arg921X, in a family with clinical manifestations of AAS and predominant congenital contractures. This study provides a strong rationale for screening male patients with distal skeletal abnormalities for FGD1 mutations.
METHODS
Sample Collection and Whole-Exome Sequencing
DNA was extracted from whole blood using the Wizard Genomic DNA Purification Kit per the manufacturer's instructions (Promega). DNA samples from Individuals III-4 and IV-1 (Fig. 2A) were subjected to whole-exome sequencing to identify variants in common between these two individuals. Genomic DNA was sheared using a Covaris ultrasonicator to 400 bp. The libraries were prepared using the NEBNext DNA Library Prep Master Mix kit and gel size selected for 450-bp fragments. A 250-ng library for each sample was pooled and exome captured with NimbleGen SeqCap EZ System version 3. Paired-end 100-nt Illumina sequencing was performed (Illumina HiSeq 2000). FastQC was used to assess the quality of the data. Pair end reads were aligned to the human genome build GRCh37 (hg19) using Burrows–Wheeler alignment (BWA). To remove duplicates, the resultant mapped reads were processed by Picard. Variants were called using the following GATK (https://www.broadinstitute.org/gatk/) modules: local realignment by “RealignerTargetCreator” and “IndelRealigner”; base quality recalibration by “BaseRecalibrator” and “PrintReads”; variant calling by “UnifiedGenotyper”; and variant quality recalibration by “VariantRecalibrator” and “ApplyRecalibration.”
Variants that passed GATK variant quality score recalibration (VQSR) assessment were output as the final set of variants (Table 3). The detected variants were matched to detected variants in dbSNP database and further annotated by SnpEff.
Table 3.
Sequencing results
| Patient | 10× Coverage | Mean coverage | Yield (Gb) | >Q30 (%) | Mean Q | Filtered variants | FGD1 mean exon coverage | Variant coverage |
|---|---|---|---|---|---|---|---|---|
| III-4 | 97.8 | 82 | 11.3 | 83.9 | 33.64 | 52,632 | 40.11 | 35 |
| IV-I | 97.8 | 89 | 11.2 | 83.8 | 33.69 | 52,813 | 46.49 | 46 |
ADDITIONAL INFORMATION
Data Deposition and Access
Whole-exome sequencing data is not publicly available because patient consent could not be obtained. The variant has been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) under accession number SCV000265836.
Ethics Statement
Written informed consent was obtained from all study participants in accordance with approved protocols from the Institutional Review Board of the University of Michigan.
Acknowledgments
We are indebted to the family described in the manuscript for their participation in the study. We thank the University of Michigan Bioinformatics Core for their help with the bioinformatics analysis.
Author Contributions
L.B.G., A.A., and C.E.K. conceived and designed the experiments. L.B.G. performed DNA preparation, mutation validation, and data analysis. L.B.G. drafted the manuscript. C.E.K. and F.A.F. provided the clinical data. C.E.K., A.A., and F.A.F. critically reviewed the manuscript.
Funding
L.B.G. was supported by a National Institutes of Health (NIH) F30 National Research Science Award (NRSA) from the National Institute of Neurological Disorders and Stroke (NINDS) (NS092238), the National Institutes of Health Cellular and Molecular Biology Training Grant (T32 GM007315), and the National Institutes of Health Medical Scientist Training Grant (T32 GM007863). A.A. was supported by the NINDS (NS073748).
Competing Interest Statement
The authors have declared no competing interest.
Referees
Christopher Cassa
Anonymous
REFERENCES
- Al-Semari A, Wakil SM, Al-Muhaizea MA, Dababo M, Al-Amr R, Alkuraya F, Meyer BF. 2013. Novel FGD1 mutation underlying Aarskog–Scott syndrome with myopathy and distal arthropathy. Clin Dysmorphol 22: 13–17. [DOI] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, Jacobson A. 2006. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7: 415–425. [DOI] [PubMed] [Google Scholar]
- Bedoyan JK, Friez MJ, DuPont B, Ahmad A. 2009. First case of deletion of the faciogenital dysplasia 1 (FGD1) gene in a patient with Aarskog–Scott syndrome. Eur J Med Genet 52: 262–264. [DOI] [PubMed] [Google Scholar]
- Bottani A, Orrico A, Galli L, Karam O, Haenggeli C-A, Ferey S, Conrad B. 2007. Unilateral focal polymicrogyria in a patient with classical Aarskog–Scott syndrome due to a novel missense mutation in an evolutionary conserved RhoGEF domain of the faciogenital dysplasia gene FGD1. Am J Med Genet A 143A: 2334–2338. [DOI] [PubMed] [Google Scholar]
- Chan D, Weng YM, Graham HK, Sillence DO, Bateman JF. 1998. A nonsense mutation in the carboxyl-terminal domain of type X collagen causes haploinsufficiency in Schmid metaphyseal chondrodysplasia. J Clin Invest 101: 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada L, Caron E, Gorski JL. 2001. Fgd1, the Cdc42 guanine nucleotide exchange factor responsible for faciogenital dysplasia, is localized to the subcortical actin cytoskeleton and Golgi membrane. Hum Mol Genet 10: 485–495. [DOI] [PubMed] [Google Scholar]
- Fernandez I, Tsukahara M, Mito H, Yoshii H, Uchida M, Matsuo K, Kajii T. 1994. Congenital heart defects in Aarskog syndrome. Am J Med Genet 50: 318–322. [DOI] [PubMed] [Google Scholar]
- Gorski JL, Estrada L, Hu C, Liu Z. 2000. Skeletal-specific expression of Fgd1 during bone formation and skeletal defects in faciogenital dysplasia (FGDY; Aarskog syndrome). Dev Dyn 218: 573–586. [DOI] [PubMed] [Google Scholar]
- Lebel RR, May M, Pouls S, Lubs HA, Stevenson RE, Schwartz CE. 2002. Non-syndromic X-linked mental retardation associated with a missense mutation (P312L) in the FGD1 gene. Clin Genet 61: 139–145. [DOI] [PubMed] [Google Scholar]
- Nouraei SM, Hasan A, Chaudhari MP, Dunning J. 2005. Aarskog syndrome with aortic root dilatation and sub-valvular aortic stenosis: surgical management. Interact Cardiovasc Thorac Surg 4: 47–48. [DOI] [PubMed] [Google Scholar]
- Orrico A, Galli L, Falciani M, Bracci M, Cavaliere ML, Rinaldi MM, Musacchio A, Sorrentino V. 2000. A mutation in the pleckstrin homology (PH) domain of the FGD1 gene in an Italian family with faciogenital dysplasia (Aarskog–Scott syndrome). FEBS Lett 478: 216–220. [DOI] [PubMed] [Google Scholar]
- Orrico A, Galli L, Cavaliere ML, Garavelli L, Fryns J-P, Crushell E, Rinaldi MM, Medeira A, Sorrentino V. 2004. Phenotypic and molecular characterisation of the Aarskog–Scott syndrome: a survey of the clinical variability in light of FGD1 mutation analysis in 46 patients. Eur J Hum Genet 12: 16–23. [DOI] [PubMed] [Google Scholar]
- Orrico A, Galli L, Obregon MG, de Castro Perez MF, Falciani M, Sorrentino V. 2007. Unusually severe expression of craniofacial features in Aarskog–Scott syndrome due to a novel truncating mutation of the FGD1 gene. Am J Med Genet A 143: 58–63. [DOI] [PubMed] [Google Scholar]
- Orrico A, Galli L, Faivre L, Clayton-Smith J, Azzarello-Burri SM, Hertz JM, Jacquemont S, Taurisano R, Arroyo Carrera I, Tarantino E, et al. 2010. Aarskog–Scott syndrome: clinical update and report of nine novel mutations of the FGD1 gene. Am J Med Genet A 152A: 313–318. [DOI] [PubMed] [Google Scholar]
- Orrico A, Galli L, Clayton-Smith J, Fryns J-P. 2014. Clinical utility gene card for: Aarskog–Scott syndrome (faciogenital dysplasia)—Update 2015. Eur J Hum Genet 19: 10.1038/ejhg.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteris NG, Cadle A, Logie LJ, Porteous ME, Schwartz CE, Stevenson RE, Glover TW, Wilroy RS, Gorski JL. 1994. Isolation and characterization of the faciogenital dysplasia (Aarskog–Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell 79: 669–678. [DOI] [PubMed] [Google Scholar]
- Pérez-Coria M, Lugo-Trampe JJ, Zamudio-Osuna M, Rodríguez-Sánchez IP, Lugo-Trampe A, de la Fuente-Cortez B, Campos-Acevedo LD, Martínez-de-Villarreal LE. 2015. Identification of novel mutations in Mexican patients with Aarskog–Scott syndrome. Mol Genet Genomic Med 3: 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponseti IV, Zhivkov M, Davis N, Sinclair M, Dobbs MB, Morcuende JA. 2006. Treatment of the complex idiopathic clubfoot. Clin Orthop Relat Res 451: 171–176. [DOI] [PubMed] [Google Scholar]
- Schwartz CE, Gillessen-Kaesbach G, May M, Cappa M, Gorski J, Steindl K, Neri G. 2000. Two novel mutations confirm FGD1 is responsible for the Aarskog syndrome. Eur J Hum Genet 8: 869–874. [DOI] [PubMed] [Google Scholar]